Abstract
Bone disease in myeloma occurs as a result of complex interactions between myeloma cells and the bone marrow microenvironment. A custom-built DNA single nucleotide polymorphism (SNP) chip containing 3404 SNPs was used to test genomic DNA from myeloma patients classified by the extent of bone disease. Correlations identified with a Total Therapy 2 (TT2) (Arkansas) data set were validated with Eastern Cooperative Oncology Group (ECOG) and Southwest Oncology Group (SWOG) data sets. Univariate correlates with bone disease included: EPHX1, IGF1R, IL-4 and Gsk3β SNP signatures were linked to the number of bone lesions, log2 DKK-1 myeloma cell expression levels and patient survival. Using stepwise multivariate regression analysis, the following SNPs: EPHX1 (P = 0.0026); log2 DKK-1 expression (P = 0.0046); serum lactic dehydrogenase (LDH) (P = 0.0074); Gsk3β (P = 0.02) and TNFSF8 (P = 0.04) were linked to bone disease. This assessment of genetic polymorphisms identifies SNPs with both potential biological relevance and utility in prognostic models of myeloma bone disease.
Keywords: myeloma, bone disease, SNP, molecular, prognosis
Introduction
Multiple myeloma is a tumor of plasma cells that depends on the bone marrow microenvironment for growth and survival.1,2 Bone disease in myeloma occurs as a result of the complex interactions between myeloma cells and the bone marrow osteoclasts, osteoblasts plus other accessory cells and microenvironmental components.2
Myeloma bone disease is characterized by a unique combination of enhanced osteoclast numbers and function plus reduced osteoblast differentiation and function.1–12 The important elements in osteoclast activation are myeloma cellderived MIP-1α, which activates osteoclast CCRX-5 plus microenvironmental-derived RANK-ligand (RANK-L), which activates osteoclast RANK and competes with stromal-derived osteoprotegrin (OPG).10–12 Recent studies have emphasized the central role of the Wnt (Wingless-type MMTV integration site family (mammalian homologue))-signaling inhibitor DKKK-1 in the pathogenesis of the osteolytic bone lesions in myeloma.6 DKK-1 inhibits both osteoblast differentiation and function and increases osteoclast activity. Attention is focused both on the mechanisms responsible for the upregulation of DKK-1 synthesis in plasma cells and the interactions with the microenvironment. 7–10 Expression of DKK-1 is regulated by a combination of intrinsic genomic factors and interactions with the bone marrow microenvironment.8
To assess the predilection to bone disease, it was elected to study the effect of single nucleotide DNA polymorphisms (SNP) in a well-characterized population of myeloma patients for whom DKK-1 expression and gene expression profile (GEP) gene signature data were also available.13 We focused on several pathways involved in the pathogenesis of myeloma bone disease, including the Wnt pathway, in particular GSK3, as well as insulin growth factor, interleukin 4, bradykinin receptors and β3 adrenergic receptors.
Peripheral blood DNA from 282 patients enrolled in the UARK 2003–33 ‘Total Therapy 2’ (TT2) protocol was studied using the previously reported Affymetrix 3k BOAC custom DNA chip to assess the presence or absence of relevant genetic polymorphisms.14–16 Here, we present evidence that several SNPs significantly correlate with both the clinical extent of the bone disease, as well as DKK-1 expression.
Patients, materials and methods
Patients
These analyses included 282 patients with previously untreated multiple myeloma enrolled in the TT2 trial between October 1998 and February 2004. Details of patient characteristics plus treatment and clinical outcomes have been reported.14 All participants had provided written informed consent in keeping with institutional and National Cancer Institute (NIH, Bethesda, MD, USA) guidelines. All details of the protocol had been approved by institutional guidelines and the United States Food and Drug Administration, and were monitored by a data safety and monitoring board as required for Phase III trials. The multiple myeloma baseline evaluation included serum and urine protein electrophoresis, quantitative immunoglobulin measurements, total 24-h urine protein excretion, serum β2-microglobulin (Sβ2M), C-reactive protein, and lactic dehydrogenase (LDH) plus bone marrow aspirate and biopsy evaluations.
Bone studies
Imaging included baseline magnetic resonance imaging (MRI) and complete skeletal survey radiological examination (myeloma bone survey (MBS)) in a prospective manner.14 The MRI included the axial skeleton and pelvis plus any additional areas requiring diagnostic evaluation for pain or other medical issues. MRI studies were carried out with a series of sequences to permit identification of focal or diffuse bone marrow involvement, including spin echo (T2-wt), short T, inversion recovery (STIR) and gadolinium-enhanced spin echo sequences with and without fat suppression. Myeloma bone survey encompassed the long bones and were carried out with digital radiographs incorporating two views of the chest; views of ribs, lateral skull, vertebral column; anteroposterior views of the pelvis, shoulders; and the extremities including hands and feet.
Focal lesions on both MRI and myeloma bone survey were identified as areas with an axial diameter of at least 0.5 cm. The MRIs were reviewed independently by four individuals who recorded the size, number and location of all focal lesions compatible with myeloma. Full details have been previously published.14
Classification of bone disease
X-ray was the primary classification system for bone disease. The exception was 12 patients with extensive focal MRI disease, but no focal changes on X-ray. On the basis of detailed previous analyses,14 this 4% subset was added to the ‘extensive bone disease’ category to give 183/282 (65%) within this extensive bone disease group. The remaining 99 patients (35%) all had negative X-rays and no extensive focal disease on MRI.
Using X-ray results only, validation of the TT2 findings was conducted comparing results in separate Eastern Cooperative Oncology Group (ECOG) and Southwest Oncology Group (SWOG) data sets.15,16 For these analyses, patients with completely negative X-rays were compared with those having > 3 focal lesions on X-ray.
Genotyping
Peripheral blood was collected in heparinized green top tubes and centrifuged to recover mononuclear cell pellets. DNA was extracted from the mononuclear cell pellets and genotyped using the Affymetrix (Santa Clara, CA, USA) Genchip scanner 3000 Targeted Genotyping System (GCS 3000 TG System) using molecular inversion probes to simultaneously identify the 3404 pre-selected SNPs in 983 genes.15,16 All genotyping experiments were carried out in strict adherence to the manufacturer’s protocol.
Custom SNP Chip design and content
A directed, custom SNP chip design was developed with specific criteria from public and commercial databases. Full details are described elsewhere.15,16 In essence, a custom SNP chip was developed, focusing on functionally relevant polymorphisms known to have a role in normal and abnormal cellular functions related to inflammation, immunity and drug responses.
Statistical analysis
Overview
Several methods were used to assess possible correlations between SNPs and the presence or absence of bone disease.
Univariate correlations of individual SNPs were assessed. This was first carried out for the TT2 data set and then for validation with the Eastern Cooperative Oncology Group and Southwest Oncology Group data sets.
Recursive partitioning was used to identify the best combinations of SNPs correlated with bone disease.
The validity of correlations with individual SNPs and combinations of SNPs was assessed using multivariate logistic regression analyses that incorporated known standard prognostic factors, gene expression profile results (risk groups: TT2 only) and Dkk-1expression results (TT2 only).
Correlations between individual SNPs as combinations of SNPs and patient outcomes were assessed including progression- free (PFS) and overall survivals (OS).
The Eastern Cooperative Oncology Group and Southwest Oncology Group data sets were evaluated with respect to SNP signatures identified in the TT2 data.
Statistical analysis details
We used Fisher’s exact test as a univariate screening tool to determine the association of SNPs with bone disease. The top 50 rank-ordered SNPs were selected and a recursive-partitioning algorithm was carried out to determine the combination of SNPs that best distinguished the bone disease subgroups. In recursive partitioning, each genotype was evaluated on its ability to make a correct prediction, creating a decision node.17 Recursive partitioning allowed for interactions of SNPs and also included SNPs further down the rank-ordered list. Univariate association between clinical parameters was assessed using continuous and categorical variables.18 The non-parametric Kruskal–Wallace test was used for continuous variables and the χ2-test was used for categorical variables.19 Multivariate logistic regression was used to test for associations of SNPs and clinical parameters with bone disease.20 Survival curves were constructed according to Kaplan and Meier.21
Results
Classification of bone disease
The 282 patients were divided into 99 patients (35%) with no bone disease (X-rays negative) and 183 patients (65%) with definite/extensive bone disease (X-rays positive and/or extensive focal lesions on MRI (12 patients)). This separation best identified the two sub-populations in detailed analyses of the imaging results for the TT2 data set.14
1. Univariate correlations between bone disease and SNPs in TT2 data set: Fisher’s exact test was used as a univariatescreening tool to determine the association of SNPs with bone disease. Results are shown in Table 1, which displays the top SNPs most highly correlated with bone disease. The top-ranked SNP, EPHX1(P = 0.0003), is rs3766934 (GG), which is an expoxide hydrolase SNP. Several SNPs linked to bone biology were among the top-ranked SNPs, including IGFIR (P=0.003: #6), IL-4 (P=0.009: #16) and Gsk3β (P=0.015: #23).
Table 1.
‘Top 30’ SNPs: Univariate correlation using TT2 model
| RS. Number | Univariate P-value | SNP function | Gene symbol | Rank |
|---|---|---|---|---|
| rs3766934 | 0.000309446 | mRNA-UTR | EPHX1 | 1a |
| rs514658 | 0.00168139 | 3'UTR | TATDN2 | 2 |
| rs2307340 | 0.002064057 | Coding non-synonymous | MCM5 | 3 |
| rs4646227 | 0.002516714 | Coding non-synonymous | SLC15A1 | 4 |
| rs520354 | 0.002945155 | Intron | APOB | 5 |
| rs2684773 | 0.003235843 | Intron | IGF1R | 6 |
| rs2303428 | 0.004614435 | Intron (boundary) | MSH2 | 7 |
| rs934197 | 0.00468514 | Promoter | APOB | 8 |
| rs3176162 | 0.005103038 | Coding non-synonymous | POLG | 9 |
| rs4905475 | 0.005428397 | Promoter | BDKRB1 | 10 |
| rs730566 | 0.005494049 | 3'UTR | TREX1 | 11 |
| rs7102464 | 0.00622144 | Coding non-synonymous | SBF2 | 12 |
| rs698708 | 0.007090614 | Promoter | FVT1 | 13 |
| rs7009367 | 0.007336344 | UTR | ADRB3 | 14 |
| rs693 | 0.009103226 | Coding synonymous | APOB | 15 |
| rs2243289 | 0.009591359 | Intron (boundary) | IL4 | 16 |
| rs2274405 | 0.011215443 | Coding synonymous | ABCC4 | 17 |
| rs1805403 | 0.012224939 | Intron (boundary) | PARP1 | 18 |
| rs2280712 | 0.012409921 | Intron (boundary) | PARP1 | 19 |
| rs2664538 | 0.013514229 | Coding non-synonymous | MMP9 | 20 |
| rs2974938 | 0.014298606 | Coding non-synonymous | PPP1R3A | 21 |
| rs2274750 | 0.01464092 | Coding non-synonymous | TNC | 22 |
| rs3783408 | 0.015425683 | Promoter | Gsk3β | 23a |
| rs7080536 | 0.015452676 | Coding non-synonymous | HABP2 | 24 |
| rs1329568 | 0.015522769 | Promoter | PAX5 | 25 |
| rs1052637 | 0.01560948 | Coding non-synonymous | DDX18 | 26 |
| rs8187710 | 0.015968731 | Coding –non-synonymous | ABCC2 | 27 |
| rs1399291 | 0.015974764 | Intron, TagSNP:DPYD | DPYD | 28 |
| rs3181366 | 0.016310866 | Intron | TNFSF8 | 29a |
| rs12659 | 0.016329684 | Coding synonymous | SLC19A1 | 30 |
Abbreviations: mRNA, messenger RNA; SNP, single nucleotide polymorphism; TT2, total therapy 2 and UTR, untranslated region.
Identified with recursive partitioning and other correlations.
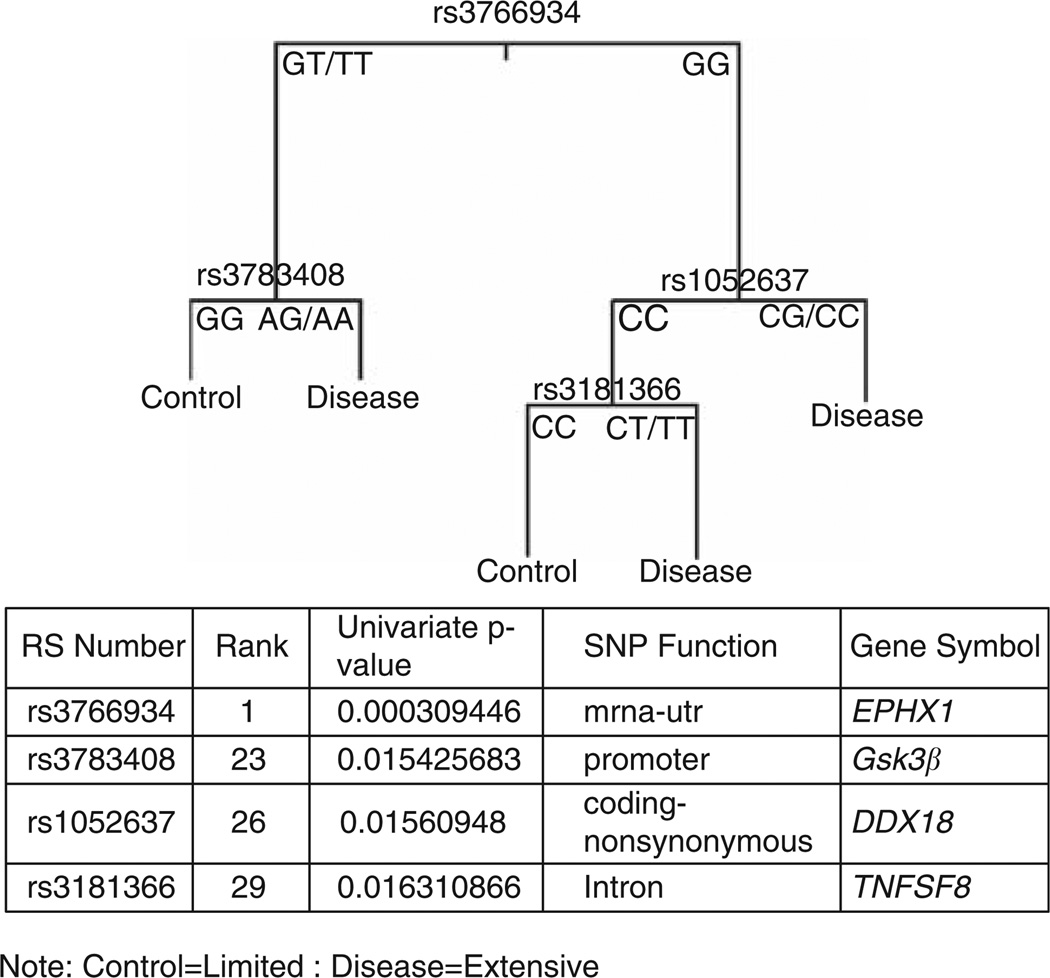
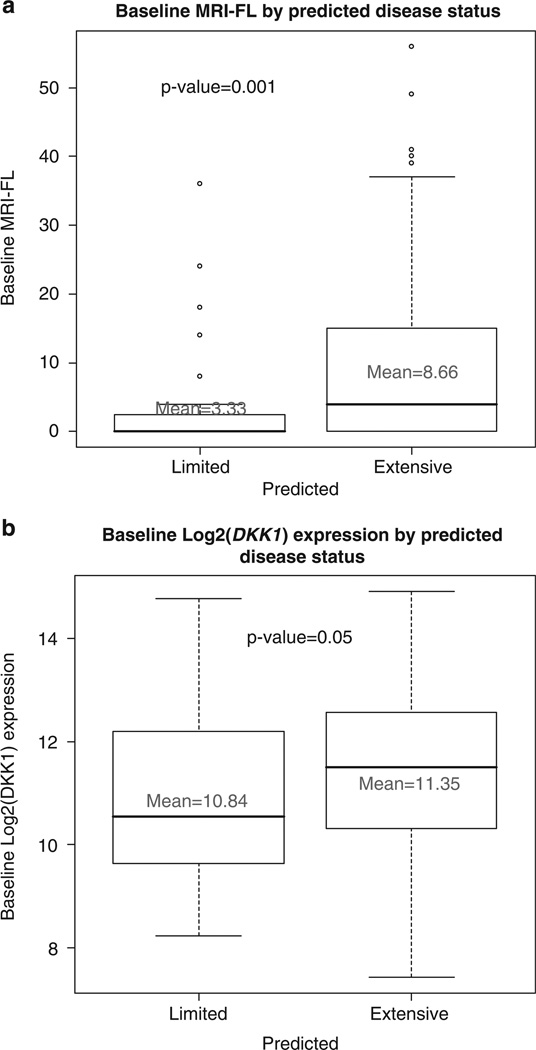
2. Recursive partitioning: The top 50 SNPs with the lowest P-values were selected for recursive partitioning analysis. The results of recursive partitioning analysis are shown in Figure 1. The 4 SNPs providing the best correlation were: rs3766934, EPHX1, RANK #1; rs3783408, Gsk3β, RANK #23; rs1052637, DDX18, RANK #26; and rs3181366, TNFSF8, RANK #29 in the univariate correlations (Table 1). The 4 SNP combination was then used as a search engine to identify further correlations. The results are shown in Figures 2a and b, respectively. There were excellent correlations with both numbers of individual focal bone lesions (P values=0.001) and the directly measured DKK-1 expression levels for individual patients (P=0.05).
Figure 1.
Recursive partitioning using ‘Top SNPs’ with Total Therapy 2 (TT2) model. Recursive partitioning branching tree displaying the four single nucleotide polymorphisms (SNPs) used in the model: rs3766934 (EPHX1); rs3783408 (Gsk3β); rs1052637 (DDX18); and rs3181366 (TNSF8). The SNP genotypes are identified: EPHX1(GT/TT versus GG); Gsk3 β (GG versus AG/AA); DDX18 (CC versus CG/CC); and TNFSF8 (CC versus CT/TT). The appended table shows the univariate P-values for each SNP and SNP function.
Figure 2.
Baseline focal bone lesions and baseline log2 DKK-1 by predicted disease using the recursive-partitioning model. (a) The number of focal bone lesions (per patient) is plotted for patients with limited bone disease and extensive bone disease predicted by the four single nucleotide polymorphism (SNP) model illustrated in Figure 1. The mean values are identified. The P-value for the difference is P = 0.001. (b) The directly measured log2 DKK-1 expression values are plotted for patients with limited bone disease and extensive bone disease predicted by the four SNP model illustrated in Figure 1. The P-value for the difference is P = 0.05.
3. Stepwise multivariate regression analyses: Several logistic regression models were used to further assess the correlations with the identified SNPs. Results are displayed in Table 2. Again, the previously identified SNPs prove to be statistically significantly associated with the bone disease status. The individual SNPs (EPHX1, Gsk3β and TNSF8), DKK-1 and lactic dehydrogenase (serum level) are predictive in the displayed multivariate analysis.
Table 2.
Stepwise multivariate regression analyses for the TT2 dataset†
| Variable | Bone |
|||||
|---|---|---|---|---|---|---|
| N | With factor | Without factor | OR (95% CI) | P-value | SNP/GEP | |
| Univariate | ||||||
| rs3766934 = 0 | 282 | 166/241 (69%) | 17/41 (41%) | 3.12 (1.59,6.16) | 0.0010 | EPHX1 |
| dkk1 | 282 | N/A | N/A | 1.24 (1.07,1.44) | 0.0053 | Dkk1 |
| ldh | 282 | N/A | N/A | 1.01 (1.00,1.01) | 0.0131 | LDH |
| g17high risk | 282 | 32/40 (80%) | 151/242 (62%) | 2.41 (1.06, 5.46) | 0.0348 | G17high |
| rs3181366>0 | 280 | 123/177 (69%) | 59/103 (57%) | 1.70 (1.03,2.81) | 0.0396 | TNSF8 |
| rs1052637>0 | 282 | 159/237 (67%) | 24/45 (53%) | 1.78 (0.94,3.40) | 0.0788 | DDX18 |
| rs3783408<2 | 279 | 82/116 (71%) | 99/163 (61%) | 1.56 (0.94,2.59) | 0.0870 | Gsk3β |
| crp | 279 | N/A | N/A | 1.01 (1.00,1.03) | 0.1404 | CRP |
| Multivariate | ||||||
| rs3766934 = 0 | 275 | 162/234 (69%) | 17/41 (41%) | 3.05 (1.48,6.29) | 0.0026 | EPHX1 |
| ddk1 | 275 | N/A | N/A | 1.27 (1.08,1.50) | 0.0046 | Dkk1 |
| ldh | 275 | N/A | N/A | 1.01 (1.00,1.01) | 0.0074 | LDH |
| rs3783408<2 | 275 | 81/114 (71%) | 98/161 (61%) | 1.93 (1.11,3.37) | 0.0202 | Gsk3β |
| rs3181366>0 | 275 | 120/173 (69%) | 59/102 (58%) | 1.73 (1.01,2.98) | 0.0470 | TNSF8 |
Abbreviations: CI, confidence interval; GEP, gene expression profile; OR, odds ratio and TT2, total therapy 2.
P-value from Wald’s χ2-Test in Logistic Regression.
NS2-Multivariate results not statistically significant at 0.05 level. Univariate P-values reported regardless of significance.
Multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the 0.05 level.
A multivariate P-value greater than 0.05 indicates variable forced into model with significant variables chosen using stepwise selection.
Using the variables already identified as significant in preliminary analyses, we used stepwise logistic regression to find the best prognostic model in all 282 of the TT2 patients. The multivariate results are in order of selection into the model.
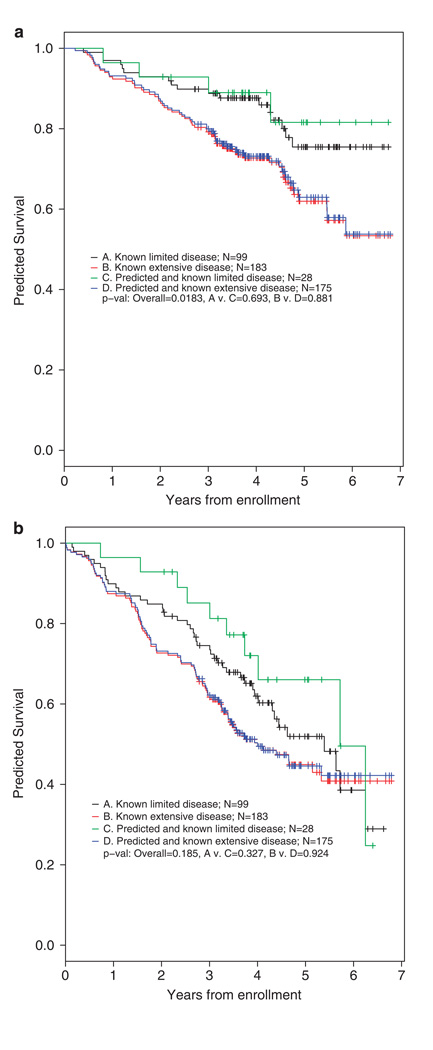
4. Correlations with progression-free and overall survival: Figure 3 shows the correlations between SNP pattern and outcomes. The cross correlations between known and predicted survivals are highly significant.
Figure 3.
Overall survival (OS) and Event-free survival (EFS) for both actual and predicted bone disease (Total Therapy 2 (TT2) model). (a) OS is shown for patients with known limited and extensive bone disease and compared with the survival for patients predicted by the four single nucleotide polymorphism (SNP) model to have limited and extensive disease. The listed P-values indicate that OS is statistically inferior for patients with both actual and predicted extensive versus limited bone disease (P = 0.0183). The actual versus predicted outcomes are not different (P-values 0.693 and 0.881, respectively). (b) EFS is shown for patients with known limited and extensive bone disease and compared with EFS for patients predicted to have limited and extensive bone disease based on the four SNP model (Figure 1). The P-values indicate that EFS is not different for limited versus extensive disease, but this is true for both the actual and predicted patient populations (P-values: overall 0.185; and 0.327 and 0.924 for comparisons).
Cross-validation in additional clinical data sets with bone disease defined by X-ray only. These statistical analyses used 207 patients with zero or more than three X-ray focal lesions from the original TT2 data set plus 62 patients from Southwest Oncology Group (S9321) and 69 patients from Eastern Cooperative Oncology Group (E1A00 and E9486). Collectively, there were 163 patients with no X-ray evidence of bone disease and 175 patients with more then three focal lesions evident on X-ray. A majority of the SNPs from the TT2 only analyses were again highly ranked in combined analyses. For example, EPHX1 (previously ranked #1, now #9); IGFIR (previously ranked #6, now #13) and IL-4 (previously ranked #16, now #19) again showed significant correlations. Conversely, the SNPs for BDKRB1, ADRB3 and DDX18 were not highly ranked.
Stepwise multivariate logistic regression analysis was then repeated incorporating top SNPs identified by cross-validation. The results are displayed in Table 3. This further cross-validation assessment shows that the EPHX1 SNP is still the top SNP in both the univariate and multivariate regressions. The TREX1 SNP, previously ranked number 11, acquires greater significance in these univariate and multivariate regressions. Other significant correlations were with DDK-1, lactic dehydrogenase, the 17-gene gene expression profile high risk, plus again the SNPs for Gsk3β and TNFSF8.
Table 3.
Stepwise multivariate regression analyses incorporating SNPs identified with bone disease classified by X-rays only (0 versus >3 lesions)a
| Variable |
Bone |
|||||
|---|---|---|---|---|---|---|
| N | With factor | Without factor | OR (95% CI) | P-value | SNP/GEP | |
| Univariate | ||||||
| rs3766934 = 0 | 282 | 166/241 (69%) | 17/41 (41%) | 312 (1.59,6.16) | 0.0010 | EPHX1 |
| rs730566<2 | 282 | 172/254 (68%) | 11/28 (39%) | 3.24 (1.45,7.23) | 0.0041 | TREX1 |
| dkk1 | 282 | N/A | N/A | 1.24 (1.07,1.44) | 0.0053 | Dkk1 |
| ldh | 282 | N/A | N/A | 1.01 (1.00,1.01) | 0.0131 | LDH |
| g17high | 282 | 32/40 (80%) | 151/242 (62%) | 2.41 (1.06,5.46) | 0.0348 | G17high |
| rs3181366>0 | 280 | 123/177 (69%) | 59/103 (57%) | 1.70 (1.03,2.81) | 0.0396 | TNSF8 |
| rs7120118 = 0 | 281 | 21/25 (84%) | 161/256 (63%) | 3.10 (1.03,9.30) | 0.0437 | NRIH3 |
| rs1052637>0 | 282 | 159/237 (67%) | 24/45 (53%) | 1.78 (0.94,3.40) | 0.0788 | DDX18 |
| rs3783408<2 | 279 | 82/116 (71%) | 99/163 (61%) | 1.56 (0.94,2.59) | 0.0870 | Gsk3β |
| crp | 279 | N/A | N/A | 1.01 (1.00,1.03) | 0.1404 | CRP |
| Multivariate | ||||||
| rs3766934 = 0 | 274 | 161/233 (69%) | 17/41 (41%) | 2.90 (1.36,6.17) | 0.0057 | EPHX1 |
| rs730566<2 | 274 | 168/247 (68%) | 10/27 (37%) | 3.40 (1.38,8.37) | 0.0077 | TREX1 |
| ldh | 274 | N/A | N/A | 1.01 (1.00,1.01) | 0.0105 | LDH |
| rs3783408<2 | 274 | 80/113 (71%) | 98/161 (61%) | 2.09 (1.17,3.70) | 0.0121 | GSK3β |
| ddk1 | 274 | N/A | N/A | 1.23 (1.04,11.08) | 0.0178 | Dkk1 |
| rs3181366>0 | 274 | 119/172 (69%) | 59/102 (58%) | 1.78 (1.02,3.10) | 0.0423 | TNSF8 |
| rs7120118 = 0 | 274 | 21/25 (84%) | 157/249 (63%) | 3.39 (1.04,11.08) | 0.0430 | NRIH3 |
Abbreviations: CI, confidence interval; GEP, gene expression profile; OR, odds ratio and SNP, single nucleotide polymorphism.
P-value from Wald’s χ2-Test in Logistic Regression.
NS2-Multivariate results not statistically significant at 0.05 level. Univariate P-values reported regardless of significance.
Multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the 0.05 level.
A multivariate P-value greater than 0.05 indicates variable forced into model with significant variables chosen using stepwise selection.
Logistic regression on all 282 Total Therapy 2 (TT2) patients. The variables considered in Table 2 together with top two SNPs from the X-ray only analysis are considered.
Discussion
In this study, several SNPs are correlated with the likelihood of bone disease. The top SNP is EPHX1 (rs3766934: GG genotype versus GT/TT), an epoxide hydrolase. Although EPHX1 has been evaluated in multiple studies of genetic polymorphisms of biotransformation enzymes related to cancer, the functional significance of this specific GG genotype is currently unclear.22 Nonetheless, it is known that epoxide hydrolase is involved in both the inflammatory response linked to the bioactivation of leukotoxins23 and the activation of the dioxin response element by benzo[a] pyrene compounds.24 Further studies are necessary to investigate the potential significance of this EPHX1 SNP genotype in laboratory, clinical and epidemiological studies.
The Gsk3β SNP (Table 1 and Figure 1) was the second SNP selected as part of the recursive partitioning decision tree. This SNP is especially interesting as binding of GSK3βi with axin and APC forms a critical complex involved in Wnt-activated release or stabilization of β-catenin.25–29 This pathway is central to osteoblast function.30 Increased Wnt signaling through Wnt 3A results in an increase in the bone mineral density and a decrease in the osteoclast/osteoblast ratio.31–34 Gsk3 β is the target of upregulation by thalidomide and is central to reactive oxygen species-mediated thalidomide-induced apoptosis.28
Other identified SNPs linked to bone related pathways (see Table 4) included the following: insulin-like growth factor 1 receptor (ranked number 6: Table 1);35–39 bradykinin receptor B1 (ranked number 10: Table 1);40 adrenergic receptor B3 (ranked number 14: Table 1);41,42 and interleukin-4 (ranked number 16: Table 1).43 Several SNPs linked to drug and/or toxin metabolism and/or DNA metabolism and repair were noted and are summarized in Table 4. As dioxins have been linked to the etiology of myeloma,44 it is noteworthy that EPHX145–47 is important in dioxin and polycyclic aromatic hydrocarbon metabolism. In addition, the DPYD SNP (rs1399291) ranked number 28 (Table 1) is involved with pyrimidine metabolism, and has, in addition, been identified in a separate recent largescale screening.48
Table 4.
Biological significance of correlated SNPs
| SNP | Identification | Comments/Discussion |
|---|---|---|
| rs 3783408 | Gsk3β | Binding to GSK3βi in the Wnt pathway stabilizes β-catenin |
| rs 2684773 | IGF1R | Insulin-like growth factor triggers osteoblast functions |
| rs 2243289 | IL-4 | Interleukin-4 modulates the activity of osteoblasts |
| rs 3766934 | EPHX1 | Epoxide hydrolase is a multifunctional protein involved in the metabolism of carcinogenic xenobiotics |
| rs 730566 | TREX-1 | Trex-1 is an exonuclease involved in processing and clearing anomalous DNA structures. Absence is linked to familial lupus with DNA auto antibodies. In this study the SNP is linked to the absence of bone lesions. |
| rs 7120118 | NRIH3 | Key regulator in cholesterol homeostasis: absence results in the rapid accumulation of cholesterol esters and failure to induce CYP7A |
| rs 1052637 | DDX18 | Dead box viral RNA helicase that allows unwinding of double stranded RNA. Linked to C-myc function and oncogenic cell activation |
| rs 4905475 | BDKRB1 | Bradykinin receptor B1 involved in pro inflammatory cytokine and prostaglandin (PGE2) signaling and bone disease |
| rs 7009367 | ADR B3 | β3-adrenergic receptor linked to bone mass index, bone mineral density and fracture risk |
| rs 3760413 | EME1 | Essential meiotic endonuclease 1, which has a key role in DNA repair and maintenance of genome integrity. |
| rs 1399291 | DPYD | DPYD encodes the rate-limiting enzyme in the catabolism of uracil and thymidine. |
| rs 10916 | CYP 1B1 | Cytochrome-P450 enzyme B1: multifunctional enzyme involved in estrogen metabolism and aryl hydrocarbon receptor expression |
| rs 520354 | APOB | Apolipoprotein B the structural protein required for lipoprotein assembly and secretion. Crucial for triglyceride transfer |
Abbreviation: SNP, single nucleotide polymorphism.
Testing with the 3400 SNP custom chip has, thus, revealed several SNPs that are significantly correlated with the likelihood of bone disease in patients with myeloma. Larger studies are currently underway, for example, in collaboration with the National Cancer Institute (NCI) epidemiology branch, to further explore the relationships with identified SNPs.48
Acknowledgements
This investigation was supported in part by an unrestricted grant from the International Myeloma Foundation (Bank on a Cure project), as well as by the following PHS Cooperative Agreement grant numbers awarded by the National Cancer Institute, DHHS: CA32102 and CA38926 (SWOG); and CA21115 (ECOG); plus CA 97513 (JDS and BB).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 2.Giuliani N, Rizzoli V, Roodman GD. Multiple myeloma bone disease: pathophysiology of osteoblast inhibition. Blood. 2006;108:3992–3996. doi: 10.1182/blood-2006-05-026112. [DOI] [PubMed] [Google Scholar]
- 3.Durie BGM, Salmon SE, Mundy GR. Relation of osteoclast activating factor production to extent of bone disease in multiple myeloma. Br J Hematol. 1981;47:21–30. doi: 10.1111/j.1365-2141.1981.tb02758.x. [DOI] [PubMed] [Google Scholar]
- 4.Harada S, Rodan G. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–355. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- 5.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483–2494. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 7.Yaccoby S, Ling W, Zhan F, Walker R, Barlogie B, Shaughnessy JD., Jr Antibody-based inhibition of DKK1 suppresses tumor-induced bone resorption and multiple myeloma growth in vivo. Blood. 2007;109:2106–2111. doi: 10.1182/blood-2006-09-047712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colla S, Zhan F, Xiong W, Wu X, Xu H, Stephens O, et al. The oxidative stress response regulates DKK1 expression through the JNK signaling cascade in multiple myeloma plasma cells. Blood. 2007;109:4470–4477. doi: 10.1182/blood-2006-11-056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian J, Xie J, Hong S, Yang J, Zhang L, Han X, et al. Dickkopf-1 (DKK-1) is a widely expressed and potent tumor-associated antigen in multiple myeloma. Blood. 2007;110:1587–1594. doi: 10.1182/blood-2007-03-082529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi SJ, Oba Y, Gazitt Y, Alsina M, Cruz J, Anderson J, et al. Antisense inhibition of macrophage inflammatory protein 1-alpha blocks bone destruction in a model of myeloma bone disease. J Clin Invest. 2001;108:1833–1841. doi: 10.1172/JCI13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lentzsch S, Gries M, Janz M, Bargou R, Dorken B, Mapara MY. Macrophage inflammatory protein 1-alpha (MIP-1 alpha) triggers migration and signaling cascades mediating survival and proliferation in multiple myeloma (MM) cells. Blood. 2003;101:3568–3573. doi: 10.1182/blood-2002-08-2383. [DOI] [PubMed] [Google Scholar]
- 12.Vallet S, Raje N, Ishitsuka K, Hideshima T, Podar K, Chhetri S, et al. MLN3897, a novel CCR1 inhibitor, impairs osteoclastogenesis and inhibits the interaction of multiple myeloma cells and osteoclasts. Blood. 2007;110:3744–3752. doi: 10.1182/blood-2007-05-093294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker R, Barlogie B, Haessler J, Tricot G, Anaissie E, Shaughnessy JD, Jr, et al. Magnetic resonance imaging in multiple myeloma: diagnostic and clinical implications. J Clin Oncol. 2007;25:1121–1128. doi: 10.1200/JCO.2006.08.5803. [DOI] [PubMed] [Google Scholar]
- 15.Johnson DC, Corthals S, Ramos C, Hoering A, Cocks K, Dickens NJ, et al. Genetic associations with thalidomide mediated venous thrombotic events in myeloma identified using targeted genotyping. Blood. 2008;112:4924–4934. doi: 10.1182/blood-2008-02-140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Ness B, Ramos C, Haznadar M, Hoering A, Haessler J, Crowley J, et al. Genomic variation in myeloma: design, content, and initial application of the bank on a cure SNP panel to analysis of survival. BMC Med. 2008;6:26. doi: 10.1186/1741-7015-6-26. [pages not specified] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terry MT, Elizabeth J, Atkinson R. An introduction to recursive partitioning using the rpart routines. Technical report 61, Mayo Clinic. 1997 2Available at http://mayoresearch.mayo.edu/mayo/research/biostat/techreports.cfm##R package available at http://cran.r-project.org/src/contrib/Descriptions/rpart.html.
- 18.Agresti A. An introduction to categorical data analysis. NJ, USA: Wiley; 1996. [Google Scholar]
- 19.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 20.Efron B, Tibshirani R. An introduction to the Bootstrap. FL, USA: Chapman & Hall/CRC; 1994. [Google Scholar]
- 21.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 22.Hirschhorn JN, Lohmueller K, Byrne E, Hurschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Moghaddam MF, Grant DF, Cheek JM, Green JF, Williamson KC, Hammock BD. Bioactivation of leukotoxins to their toxic diols by epoxide hydrolase. Nature Med. 1997;3:562–566. doi: 10.1038/nm0597-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burchiel SW, Thompson TA, Lauer FT, Oprea TI. Activation of dioxin response element (DRE)-associated genes by benzo (A) pyrene3,6-quinone and benzo (A) pyrene1,6-quinone in MCF-10A human mammary epithelial cells. Toxicol Appl Pharmacol. 2007;221:203–214. doi: 10.1016/j.taap.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi C-S, Huang N-N, Harrison K, Han S-B, Kehrl JH. The mitogen-activated protein kinase kinase kinase kinase GCKR positively regulates canonical and noncanonical Wnt signaling in B lymphocytes. Mol Cell Biol. 2006;26:6511–6521. doi: 10.1128/MCB.00209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JA, Chatterjee-Kishore M, Yaworsky PJ, Cullen DM, ZhaoW LC, et al. Wnt/β signaling is a normal physiological response to mechanical loading in bone. J Biol Chem. 2006;281:31720–31728. doi: 10.1074/jbc.M602308200. [DOI] [PubMed] [Google Scholar]
- 27.Shi C-S, Tuscano JM, Witte ON, Kehrl JH. GCKR links the Bcr-Abl oncogene and Ras to the stress-activated protein kinase pathway. Blood. 1999;93:1338–1345. [PubMed] [Google Scholar]
- 28.Knobloch J, Reimann K, Klotz L-O, Ruther U. Thalidomide resistance is based upon the capacity of the glutathione-dependent antioxidant defense. Mol Pharmaceutics. 2008;5:1138–1144. doi: 10.1021/mp8001232. [DOI] [PubMed] [Google Scholar]
- 29.Edwards CM, Edwards JR, Lwin ST, Mundy GR. Target Wnt signaling in myeloma in vivo; differential effects on tumor burden and myeloma bone disease. Blood. 2008;111:2833–2842. doi: 10.1182/blood-2007-03-077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspi M, Zilberberg A, Eldar-Finkelman H, Rosin-Arbesfeld R. Nuclear GSK-3β inhibits the canonical Wnt signalling pathway in a β-catenin phosphorylation-independent manner. Oncogene. 2008;27:3546–3555. doi: 10.1038/sj.onc.1211026. [DOI] [PubMed] [Google Scholar]
- 31.Staal FJT, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Immunology. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 32.Qiang Y-W, Chen Y, Stephens O, Brown N, Chen B, Epstein J, et al. Myeloma-derived Dixkkopf-1 disrupts Wnt-regulated osteoprotegerin and RANKL production by osteoblasts: a potential mechanism underlying osteolytic bone lesions in multiple myeloma. Blood. 2008;112:196–207. doi: 10.1182/blood-2008-01-132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiang Y-W, Shaughnessy JD, Yaccoby S. Wnt3a signaling within bone inhibits multiple myeloma bone disease and tumor growth. Blood. 2008;112:374–382. doi: 10.1182/blood-2007-10-120253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edward CM. Wnt signaling: bone’s defense against myeloma. Blood. 2008;112:216–218. doi: 10.1182/blood-2008-04-149278. [DOI] [PubMed] [Google Scholar]
- 35.Ferlin M, Noraz N, Hertogh C, Brochier J, Taylor N, Klein B. Insulin-like growth factor induces the survival and proliferation of myeloma cells through an interleukin-6-independent transduction pathway. Br J Hematology. 2000;111:626–634. doi: 10.1046/j.1365-2141.2000.02364.x. [DOI] [PubMed] [Google Scholar]
- 36.Ge N-L, Rudikoff S. Insulin-like growth factor 1 is a dual effector of multiple myeloma cell growth. Blood. 2000;96:2856–2861. [PubMed] [Google Scholar]
- 37.Mitsiades CS, Mitsiades N, Poulaki V, Schlossman R, Akivama M, Chauhan D, et al. Activation of NF-kB and upregulation of intracellular anti-apoptotic proteins via the IGF-1/Akt signaling in human multiple myeloma calls: therapeutic implications. Oncogene. 2002;21:5673–5683. doi: 10.1038/sj.onc.1205664. [DOI] [PubMed] [Google Scholar]
- 38.Podar K, Tai Y-T, Cole CE, Hideshima T, Sattler M, Hamblin A, et al. Essential role of caveolae in interleukin-6 and insulin-like growth factor I-triggered Akt-1-mediated survival of multiple myeloma cells. J Biol Chem. 2003;278:5794–5801. doi: 10.1074/jbc.M208636200. [DOI] [PubMed] [Google Scholar]
- 39.DiGirolamo DJ, Mukherjee A, Fulzele K, Gan Y, Cao X, Frank SJ, et al. Mode of growth hormone action in osteoblasts. J Biol Chem. 2007;282:31666–31674. doi: 10.1074/jbc.M705219200. [DOI] [PubMed] [Google Scholar]
- 40.Brechter AB. Umea Univ Odontological Dissertation. Umea, Sweden: Department of Oral Cell Biology, Umea University; 2006. Kinins-important regulators in inflammation induced bone resorption. (ISBN 91-7264-195-9) [Google Scholar]
- 41.Fujisawa T, Ikegami H, Kawaguchi Y, Ogihata T. Meta-analysis of the association of Trp Arg polymorphism of β3-adrenergic receptor gene with body mass index. J Clin Endocrinol Metab. 1998;83:2441–2444. doi: 10.1210/jcem.83.7.4922. [DOI] [PubMed] [Google Scholar]
- 42.Wang CY, Nguyen ND, Morrison NA, Eisman JA, Center JR, Nguyen TV. β3-adrenergic receptor gene, body mass index, bone mineral density and fracture risk in elderly men and women: the dubbo osteoporosis epidemiology study (DOES) BMC Med Genet. 2006;7:57. doi: 10.1186/1471-2350-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riancho JA, Zarrabeitia MT, Olmos JM, Amado JA, Gonzalez MJ. Effects of interleukin-4 on human osteoblast-like cells. Bone Miner. 1993;21:53–61. doi: 10.1016/s0169-6009(08)80120-1. [DOI] [PubMed] [Google Scholar]
- 44.Durie BGM, Urnovitz HB, Murphy WH. RT-PCR amplicons in the plasma of multiple myeloma patients, clinical relevance and molecular pathology. Acta Oncologica. 2000;39:789–796. doi: 10.1080/028418600750063514. [DOI] [PubMed] [Google Scholar]
- 45.Hassett C, Robinson KB, Beck NB, Omiecinski CJ. The human microsomal expoxide hydrolase gene (EPHX1): complete nucleotide sequence and structural characterization. Genomics. 1994;23:433–442. doi: 10.1006/geno.1994.1520. [DOI] [PubMed] [Google Scholar]
- 46.Fretland AJ, Omiecinski CJ. Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 47.Omiecinski CJ, Hassett C, Hosagrahara V. Epoxide hydrolase-polymorphism and role in toxicology. Toxicol Lett. 2000;112–113:365–370. doi: 10.1016/s0378-4274(99)00235-0. [DOI] [PubMed] [Google Scholar]
- 48.Berndt SI, Johnson D, Crowley J, Durie BGM, Hoover R, Katz M, et al. Large scale evaluation of genetic variation and the risk of multiple myeloma. Blood. 2008;112 Abstract # 1679. [Google Scholar]