Summary
TET enzymes including TET1, 2 and 3 convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC)1 and regulate gene transcription2-5. However, this molecular mechanism by which TET family enzymes regulate gene transcription remains elusive5-6. Here, using protein affinity purification, we searched for functional partners of TET proteins, and found that TET2 and TET3 associate with OGT, an enzyme that by itself catalyzes O-GlcNAcylation in vivo7-8. TET2 directly interacts with OGT, which is important for the chromatin association of OGT in vivo. Although this specific interaction does not regulate the enzymatic activity of TET2, it facilitates OGT-dependent histone O-GlcNAcylation. Moreover, OGT associates with TET2 at transcription starting sites (TSS). Down-regulation of TET2 reduces the amount of H2B S112 GlcNAc marks in vivo, which are associated with gene transcription regulation. Taken together, these results reveal a TET2-dependent O-GlcNAcylation of chromatin. The double epigenetic modifications on both DNA and histones by TET2 and OGT coordinate together for the gene transcription regulation.
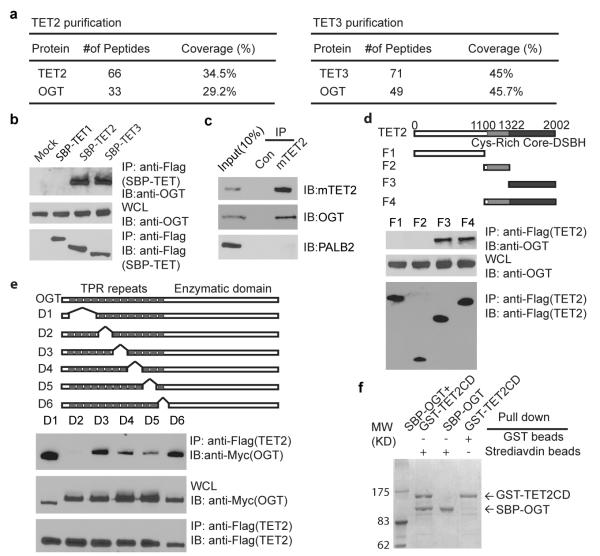
Using protein affinity purification, we sought functional partners to the TET family enzymes. Mass spectrometry analysis showed that both TET2 and TET3 associate with OGT, a enzyme that catalyzes O-GlcNAcylation (Fig. 1a, Supplementary Table 1, and Supplementary Fig. 1a and b) 7-8. To confirm this interaction, we performed a co-immunoprecipitation assay with overexpressed TET1, 2, 3, and OGT. TET2 and TET3, but not TET1, associated with OGT (Fig. 1b, Supplementary Fig. 2a). To study the endogenous interaction, we used mouse ES cells in which TET2, but not TET3, was expressed1,6,9. Again, TET2 associated with OGT endogenously in ES cells (Fig. 1c, Supplementary Fig. 2d). To map the regions of interaction between TET2 and OGT, we generated a series of TET2 deletion mutants, and found that the C-terminal catalytic DSBH (double-strand beta-helix) domain of TET2 interacts with OGT (Fig. 1d, Supplementary Fig. 2b). The tertiary structure of OGT has been examined10-12. There are two major functional domains in OGT. One is the N-terminal regulatory domain that is composed of 13.5 tetratricopeptide repeats (TPR); the other is the C-terminal catalytic domain that transfers GlcNAc from UDP-GlcNAc to Ser/Thr residues of the substrate. Using the internal deletion mutants, we found that TPR5 and 6 of OGT are required for the interaction with TET2 (Fig. 1e and Supplementary Fig. 2c, e, f). We also generated baculoviruses encoding TET2 catalytic domain (TET2CD) and OGT respectively. Co-infecting Sf9 cells with these two types of baculovirus, the recombinant proteins of SBP-tagged TET2CD and GST-tagged OGT could be co-purified at 1:1 stoichiometry from cell lysates, suggesting TET2 tightly binds OGT (Fig. 1f). Moreover, the D4 and D5 mutants of OGT slightly reduced the interaction with TET2, indicating that other TPR repeats may also regulate the interaction between OGT and TET2 (Fig. 1e and Supplementary Fig. 2f). In addition, recombinant TET3 catalytic domain also directly binds OGT (Supplementary Fig. 2g). Taken together, these results demonstrate that TET2 and TET3 form a complex with OGT both in vitro and in vivo.
Figure 1. TET2 forms a complex with OGT.
a, Purification of TET2 and TET3-associated proteins. TET2 and TET3-associated proteins were analyzed by mass spectrometry and are shown in the Supplementary Fig. 1a. b, OGT interacts with TET2 and TET3 but not TET1. SBP tagged TET1-3 were expressed and examined with indicated antibodies. The whole cell lysates (WCL) was used as the input. c, OGT interacts with TET2 endogenously in ES cells. Irrelevant IgG was used as the IP controls (Con). PALB2 was used as the negative control. d, The DSBH domain of TET2 interacts with OGT. Deletion mutants of TET2 mutants were expressed. The F3 mutant with the DSBH domain (catalytic domain) interacts with OGT. e, TPR5 and 6 of OGT interacts with TET2. The D2 mutant of OGT that is deleted TPR5 and 6 abolished the interaction with TET2. f, OGT directly binds TET2. Sf9 cells were infected with baculoviruses encoding SBP-OGT and/or GST-TET2 catalytic domain (TET2CD). The protein complex was purified by streptavidin beads or GST beads and examined by coomassie blue staining.
TET enzymes were originally identified as homologs of JBP1 and JBP2 in Trypanosoma brucei, which are two thymidine hydroxylases involved in β-D-glucosyl-hydroxymethyluracil (aka Base J) synthesis1. Synthesis of J contains two steps in which it is directly converted from thymidine bases incorporated in DNA strands. In the first step JBP1 and JBP2 catalyze thymidine into hydroxymethyluracil (HOMedU). During the second step a yet unknown enzyme transfers a glucose residue onto the hydroxyl group of HOMedU to form J13. Next, we wondered whether OGT could catalyze the glycosylation on hydroxyl groups of 5hmC. To test this hypothesis, 5mC was incubated with recombinant TET2 and OGT in vitro. However, OGT failed to convert 5hmC into any novel base in vitro (Supplementary Fig 3a and b). Co-expression OGT with TET2 in cells did not alter the TET2-dependent 5hmC synthesis (Supplementary Fig. 3c and Supplementary Fig. 4). Finally, we could not detect glycosylated 5hmC in vivo using mass spectrometry (Supplementary Fig. 3d, e and Supplementary Fig. 5). These results suggested that OGT does not affect TET2-dependent 5hmC synthesis.
Next, we asked if TET2 regulates the function of OGT. We fractionated ES cell lysates using different salt concentrations and pH levels. A subset of TET2 and OGT could only be eluted from the chromatin by 300 mM NaCl or 0.2 M HCl, indicating that these TET2 and OGT species tightly associate with the chromatin (Supplementary Fig. 6a). Interestingly, knockdown of TET2 by shRNA in ES cells abolished the chromatin-associated OGT, suggesting that TET2 may target OGT to chromatin (Supplementary Fig. 6a). To verify this phenomenon, we used 293T cells stably expressing TET2. Since the exogenous TET2 level was much higher than the endogenous TET2 level (Supplementary Fig. 7), the chromatin-bound OGT was significantly increased in 293T cells stably expressing TET2 (Supplementary Fig. 6b). Moreover, since the D2 mutant of OGT abolished the interaction with TET2, only wild type OGT but not the D2 mutant existed in the chromatin fraction, suggesting that interaction with TET2 is important for the chromatin localization of OGT (Supplementary Fig. 6c). In addition, knockdown of OGT by shRNA did not significantly affect the chromatin retention of TET2 (Supplementary Fig. 6d). Collectively, these results suggest that TET2 recruits OGT to the chromatin.
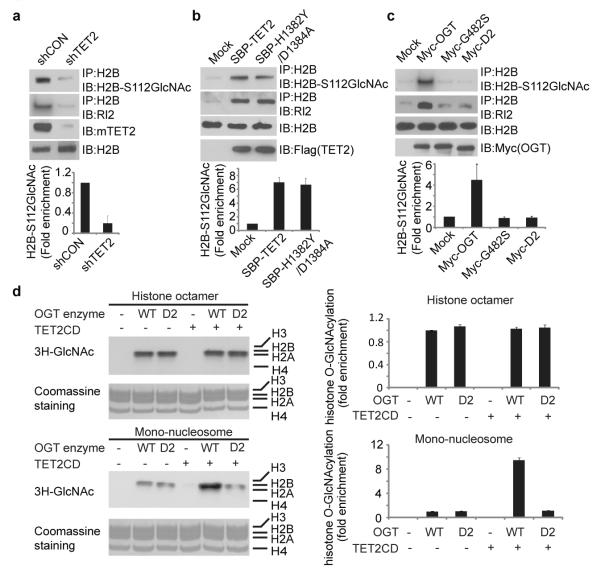
Recently, it has been shown that histones can be modified by OGT at different sites14-17. Particularly, OGT regulates GlcNAcylation of histone H2B at Serine112 in vivo15. Since TET2 targets OGT to chromatin, we wondered whether TET2 regulates OGT-dependent H2B GlcNAcylation. With a specific antibody for H2B S112 GlcNAc, we found that H2B S112 GlcNAc was significantly reduced in TET2-depleted ES cells (Fig. 2a). Moreover, TET2 also regulates other histone GlcNAcylation (Supplementary Fig. 8). Concurrent with TET2 overexpression inducing OGT association with chromatin, H2B S112 GlcNAcylation was increased (Fig. 2b). Interestingly, the TET2 enzymatic dead mutant (H1382Y/D1384A) that still interacted with OGT also increased H2B S112 GlcNAcylation, suggesting that the enzymatic activity of TET2 is not required for the regulation of H2B GlcNAcylation (Fig. 2b and Supplementary Fig. 9). Moreover, only wild type OGT but not the enzymatic dead mutant of OGT (G482S) nor the D2 mutant that lacking the interaction with TET2 could induce H2B S112 GlcNAcylation in vivo (Fig. 2c). Thus, these data suggest that the interaction between TET2 and OGT is important for OGT-dependent histone glycosylation in vivo. To examine the mechanism by which TET2 boosts the enzymatic activity of OGT, we performed an in vitro glycosylation assay using wild type OGT and its D2 mutant. With histone octamers as the substrate, both wild type OGT and the D2 mutant only weakly glycosylate histones. The enzymatic activity of wild type OGT was indistinguishable with that of the D2 mutant as both protein can auto-glycosylate themselves (Supplementary Fig. 10). Moreover, supplementation of TET2 did not affect the enzymatic activity of either wild type OGT or the D2 mutant (Fig. 2d). However, when mono-nucleosomes were used as the substrates, supplementation of recombinant TET2 significantly increased the enzymatic activity of wild type OGT but not the D2 mutant (Fig. 2d, Supplementary Fig. 11), suggesting that the interaction with TET2 facilitates OGT to recognize the substrate. Although H3 and H4 were glycosylated in vivo, only GlcNAcylated H2A and H2B could be clearly detected in this in vitro glycosylation assay (Supplementary Fig. 11). One possibility is that glycosylated H2A and H2B suppresses H3 and H4 glycosylation by OGT. Alternatively, the glycosylation sites on H3 and H4 either in the histone octamer or as mono-nucleosomes are not well exposed to the enzyme. In contrast to many other enzymes, OGT only efficiently glycosylates the substrates that it associates with18-21. Thus, it is likely that TET2 recognizes the chromatin and recruits OGT to the chromatin, and the chromatin-associated OGT glycosylates nucleosomal histones at its vicinity. Consistently, only TET2 but not OGT recognizes double-stranded DNA or mono-nucleosome with 5mC (Supplementary Fig. 12).
Figure 2. TET2 enhanced histone glycosylation.
a, Down-regulation of TET2 impairs H2B GlcNAcylation in ES cells. H2B GlcNAcylation was examined by IP with anti-H2B antibody and Western blot with anti-GlcNAc antibody (RL2) or anti-H2B S112 GlcNAc antibody. Histogram shows the relative level of H2B S112 GlcNAc in TET2 down-regulated cells compared to that in control shRNA treated cells. b, Up-regulation of wild type TET2 or TET2 enzymatic dead mutant (H1382Y/D1384A) induced H2B S112 GlcNAcylation in 293 cells. c, The interaction between OGT and TET2 is important for H2B GlcNAcylation and H2B S112G GlcNAcylation. Wild type OGT, the enzymatic dead mutant of OGT (G482S) and the D2 mutant were expressed in 293T cells. H2B GlcNAcylation and H2B S112 GlcNAcylation were examined. d, TET2 facilitated OGT-dependent histone glycosylation in mono-nucleosome but not in recombinant core histones. Tritium-labeled GlcNAc was incorporated into the histones in the in vitro GlcNAcylation assay. All error bars denote s.d., n=3.
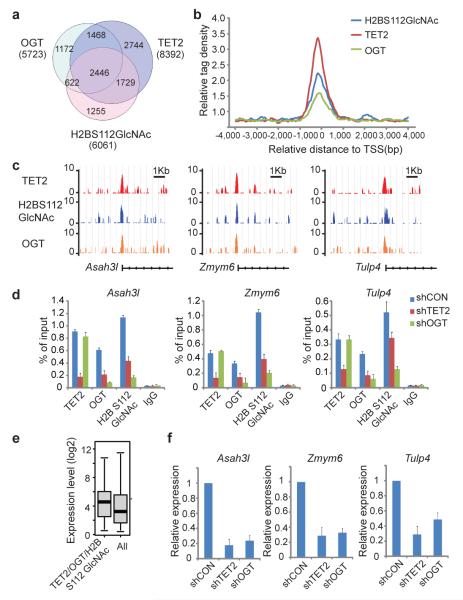
To examine the distribution of OGT and TET2 on the chromatin of ES cells, we performed genome-wide ChIP sequencing analysis (ChIP-seq) using anti-OGT, anti-H2B S112 GlcNAc and anti-mTET2 antibodies. We validated our ChIP-seq results using ChIP-qPCR to examine 45 different loci that represent a broad range of ChIP-seq fragment counts (Supplementary Fig. 13). Next, we compared the TET2 target genes with a published hmeDIP database3, and found that 47 % of hmC positive genes are bound by TET2 (Supplementary Fig. 14a). The majority of the TET2 target genes are associated with high and intermediate density CpG promoters (Supplementary Fig. 14b, c), which are also positive for H3K4me322(Supplementary Fig. 14d). Gene Ontology analysis showed that OGT, H2BS112G and TET2 are involved in a variety of basic cellular processes (Supplementary Fig. 15). The analysis showed that OGT and H2B S112 GlcNAc display a significant overlap of target genes with TET2 (Fig. 3a), and similar binding profiles to TET2 at transcriptional start sites (TSS) (Fig. 3b, c). Moreover, the binding sites of OGT, H2B S112 GlcNAc and TET2 have the highest density around TSS (Fig. 3c). Knockdown of TET2 significantly suppressed the recruitment of OGT to tested target genes and reduced the level of H2B S112 GlcNAc at those loci (Fig. 3d). In contrast, knockdown of OGT had little effect on TET2 at these loci, suggesting that OGT does not contribute to the recruitment of TET2 to these loci (Fig. 3d). Next, we asked if there was a correlation between TET2-mediated OGT targeting and the targeted gene transcription levels. In wild type ES cells, we found that genes that were occupied by OGT and TET2 and enriched with H2B S112 GlcNAc were associated with high levels of transcription in ES cells (Fig. 3e, Supplementary Fig. 16). Moreover, we examined alterations to the gene transcription profile in TET2 knockdown ES cells. The percentage of down-regulated genes that were occupied by OGT, H2B S112 GlcNAc and TET2 was significantly higher than that of genes not occupied by OGT, H2B S112 GlcNAc and TET2 (Supplementary Fig. 17). To validate these results, we performed qPCR to confirm the down-regulation of a number of OGT, H2B S112 GlcNAc and TET2 target genes (Fig. 3f, Supplementary Fig. 18). In addition, we reconstituted TET2-depleted ES cells with the H1382Y/D1384A mutant that abolishes the enzymatic activity of TET2. The H1382Y/D1384A mutant could not restore 5hmC at the TSS of a set of the TET2 and OGT common target genes. However, the H1382Y/D1384A mutant restored H2B S112 GlcNAc and partially rescued the TET2 and OGT target gene expression (Supplementary Fig. 19). These results indicate that OGT-dependent histone GlcNAcylation contributes to TET2-dependent gene transcription.
Figure 3. TET2 regulates H2B S112 GlcNAc and gene transcription.
a, Venn diagram shows a significant overlap between OGT, H2B S112 GlcNAc and TET2 target genes. b, Mean distribution of tags at gene TSS (± 4 kb). c, Examples of OGT, H2B S112 GlcNAc and TET2 ChIP-seq results in mouse ES cells. d, ChIP-qPCR was performed to examine TET2, OGT and H2B S112 GlcNAc in control, TET2 knockdown or OGT knockdown cells (shCon, shTET2 and shOGT). e, The expression of TET2 and OGT common target genes is higher than average gene expression in ES cells. Data analysis was explained in the Material and Method. Boxplots show median, 25th and 75th percentile expression levels in ES cells. p<0.00001. f, Loss of TET2 or OGT is associated with the reduced expression of their common target genes. All error bars denote s.d., n=3.
Taken together, in this study, we have shown that TET2 and OGT form a complex, which might regulate gene transcription (Supplementary Fig. 20). Besides TET2, TET3 also interacts with OGT. Like TET2, TET3 might also target OGT to chromatin for gene transcription regulation. Due to the different tissue distribution9,23-25, it is TET2 that targets OGT to the chromatin and regulates H2B GlcNAcylation in ES cells. Recently, it has been reported that H2B GlcNAcylation is associated with active TSS and positively regulate transcription15. However, 5hmC enriched regions are associated with both transcriptional activation and repression2-4,26. Since TET1 is associated with the Sin3A complex, a transcriptional repression complex, it is likely that TET1-dependent 5hmC is associated with transcriptional repression27. In contrast, TET2 exists in a totally different complex with OGT but not Sin3A. Thus, the TET2/OGT complex might be involved in transcriptional activation in ES cells. Further analysis on each individual 5hmC enriched locus could distinguish the identity of 5hmC in transcription regulation. Moreover, OGT also affects other histone modifications during mitosis28. It is possible that chromatin remodeling regulated by histone GlycNAcylation could be a general phenomenon in other biological processes. In addition to the transcription activation, OGT is also involved in transcription repression19,29-30. Thus, TET2 is only one of the functional partners of OGT. OGT in other complexes may play an important role in gene silencing19,29-30.
METHODS
Plasmids and antibodies
For protein affinity purification and other analyses, full length cDNA of TET1, TET2 and TET3 were cloned into pS-FLAG-SBP (SBP) vector respectively. For generating internal deletion mutants, OGT cDNA fragments were PCR amplified and cloned into pCMV-Myc. For generating truncation mutants, TET2 cDNA was PCR amplified and cloned into SBP vector. TET2 catalytic domain (TET2CD: a.a. 916-1921) was cloned to SBP vector and pFast-Bac vector respectively. Internal deletion mutants of OGT, enzymatic dead mutant (ED: G482S) of OGT and enzymatic dead mutant (H1382Y/D1384A) of TET2 were generated using the QuikChange site-directed mutagenesis kit (Stratagene).
Rabbit anti-mouse TET2 antibody was raised against a.a. 1-200 of mTET2. Rabbit anti-mTET1 antibody was raised against the C-terminus (a.a.1500-2008) of mTET1. Anti-OGT antibody was purchased from Novaus. Anti-FLAG, anti-Myc and anti-β-actin antibodies were purchased from Sigma. Anti-5hmC polyclonal antibody was purchased from Zymo Research. Antibodies against histone H2A, H2B, H3, H4, H3K4me3, H3K27me3 and H3K14Ac were purchased from Millipore. RL2 monoclonal antibody against O-GlcNAc was purchased from Abcam. CTD110.6 monoclonal antibody against O-GlcNAc was purchased from Covance Monoclonal antibody against H2B S112 GlcNAc was generated as described before15.
Purification of TET enzymes associated proteins
To search for binding partners of TET2 and TET3, we harvested 1 liter 293T cells stably expressing SBP-TET2 or SBP-TET3 respectively, and washed the cell pellets with PBS. Cells were lysed with 30 ml ice-cold NETN300 buffer (50 mM Tris-HCl, pH 7.4, 2 mM EDTA, and 300 mM NaCl). The soluble fraction was incubated with 0.5 ml streptavidin-conjugated agarose beads. The beads were washed with NETN100 buffer (50 mM Tris-HCl, pH 7.4, 2 mM EDTA, and 100 mM NaCl) for three times. Associated proteins were eluted with 2 mM Biotin (Sigma) in PBS and further incubated with 50 μl S protein-conjugated beads (Novagen). The bound proteins were eluted with SDS sample buffer (20 % Glycerol, 120 mM Tris-HCl pH 6.8, 4% SDS, 0.02 % bromophenol blue, 2.5 % beta-mercaptoethanol) and analyzed with 10 % SDS-PAGE and mass spectrometry. Cells expressing empty vector were used as purification controls.
Silver Staining
Purified materials were load onto 8 % SDS-PAGE for electrophoresis. The gel was fixed with 50 % methanol, 10 % acetic acid for 15 minutes and followed by additional fixer 10 % ethanol, 5 % acetic acid for 6 minutes. For staining, the gel was agitated in 50 ml 20 mg/l Na2S2O4 for 30 minutes, then stained in 50 ml AgNO3 solution (100 mg AgNO3 in 0.1 % formaldehyde) for 30 minutes. After washed with ddH2O, the gel was treated with 50 ml image developer (3 % Na2CO3, 50 μl 37 % formaldehyde, 250 μl 0.2 % Na-thiosulfate). The reaction was stopped by stop solution (12.5 ml acetic acid, 25 g Tris in 500 ml ddH2O) when clear bands were observed.
Recombinant proteins
Recombinant proteins were purified from Sf9 insect cells. For generating baculovirus, DNA fragments containing full-length OGT, deletion mutants of OGT, TET2CD, and TET3CD were subcloned into pFastBac vector with a GST or SBP tag. Baculoviruses were generated according to manufacturer’s instructions (Invitrogen). After Sf9 cells were infected with baculoviruses for 48 hours, the cells were harvested, washed with PBS and lysed with ice-cold NETN100 buffer. The soluble fraction was incubated with Glutathione-Sepharose beads (for GST-tag proteins) or Streptavidin-conjugated beads (for SBP-tag proteins) and eluted with Glutathione or Biotin. To purify protein complex, Sf9 cells were co-infected with baculoviruses encoding GST-TET2CD and SBP-OGT for 48 hours. The cells were then lysed and the soluble fraction was incubated with Glutathione Sepharose beads or Streptavidin-conjugated beads as indicated.
Cell lysis, Immunoprecipitation and Western blotting
For immunoprecipitation, cells were lysed with ice-cold NETN300 buffer containing 10 mM NaF and 50 mM β–glycerophosphate. Supernatants were incubated with indicated antibodies and protein G-conjugated Sepharose beads (Amersham Pharmacia). Precipitates were washed five times with NETN100, subjected to SDS-PAGE and Western blotting with indicated antibodies.
To examine the localization of OGT (Supplementary Fig. 6), cell pellets were lysed with 400 μl NETN100 buffer. After centrifugation, the supernatants were named as 100 mM NaCl samples. The insoluble pellets were collected, washed with ice-cold PBS, and incubated with 400 μl NETN300 buffer on ice. After centrifugation, the supernatants were named as 300 mM NaCl samples. The remaining pellets were washed with ice-cold PBS twice and then treated with 200 μl 0.2 N HCl. The supernatants were neutralized with 200 μl Tris-HCl, pH8.8 and named as 0.2 N HCl fractions. 20 μl of each sample was loaded onto 7.5 % of SDS-PAGE and immunoblotted by indicated antibodies. The total OGT was calculated by adding OGT from these three different fractions in each sample. The percentage of OGT in each fraction was presented.
Histone fraction
To examine histone glycosylation by immunoprecipitation and Western blot, we followed a protocols from Kaoru Sakabe et al16. Briefly, cells were lysed in 20 mM Tris-HCl (pH8.0), 600 mM NaCl, 0.5 % NP-40, 0.5 % SDS, 0.5 % deoxycholate, 1 mM EDTA and 2 μM PUGNAc (OGA inhibitor). The cells were sonicated and clarified by centrifugation. The lysate was then diluted by NETN100 and histones were immunoprecipitated with antibodies indicated.
To examine histone glycosylation by Western blot, cells were lysed by NETN300 buffer with 2 μM PUGNAc. The pellets were washed with NETN300 twice and treated with 0.2 N HCl. After centrifugation, the supernatants were neutralized by same volume Tris-HCl (pH8.8). Samples were examined by 10%-20% gradient SDS-PAGE or 15 % TAU gel.
Triton-Acetic Acid-Urea (TAU) gel electrophoresis
15 % TAU gel was prepared as reported31. Briefly, the gel was containing 15 % acryl:bisacrylamide 60: 0.4, 6 M urea, 5 % acetic acid and 0.37 % Triton X-100. Acid extracted histones, histone octamers or mono-nucleosomes were dissolved in 2 X loading buffer (8 M Urea, 10 % glycerol, 2.5 % Acetic acid) and loaded onto the gel. The gel was run in 5 % Acetic acid at 200 V for 140 minutes. For Coomassie blue staining, the gel was stained with Coomassie blue 250. For autoradiography, the gel was treated with EN3HANCE (from PerkinElmer) before exposed to X-ray film. For Western blot, the gel was washed twice in 300 ml of 50 mM acetic acid and 0.5 % SDS for 30 minutes of each time, and twice in 300 ml of transfer buffer (25 mM Tris, 192 mM glycine, 20 % (v/v) methanol, and 0.1 % SDS) for 30 minutes of each time. The electrotransfer was carried out at 400 mA constant current at room temperature for 2 hours.
RNA interference
ShRNA sequences targeting human OGT (5′ GCACATAGCAATCTGGCTTCC 3′32 or 5′ CCAAACTTTCTGGATGCTTAT 3′), mouse OGT (5′ GCACACAGCAATCTGGCCTCC 3′ or 5′ AGGGAACTAGATAACATGCTT 3′33) and mouse TET2 (5′ CACTACTAACTCCACCCTAAA 3′34 or 5′ GCTCTGAACAGTATTCAAAGC 3′6) were inserted into the pMSCV-puro-pSilencer vector. Generating retrovirus was carried out as previously described35. pMSCV-puro-shmock, pMSCV-puro-shOGT or pMSCV-puro-shTET2 vectors were used to transfect packaging cell line 293T with 2 other helper packaging plasmids pMD-MLV-OGP (gag-pol) and pVSV-G (env). Tissue culture supernatants from 293T packaging cells transfected with the above plasmids were harvested. Viral particles were concentrated by centrifugation at 50,000 g for 3 hours and then added to the culture media of ES cells. The viral infected cells were selected by puromycin for 72 hours and then used in the following experiments. Similar results were obtained from shRNA treatment with different sequences.
In vitro GlcNAcylation assay
Recombinant SBP-OGT protein (0.5 μg) was incubated with 2 μg substrates (histone octamers, mono-nucleosome, or 210 bp 5hmC-containing oligos with sequence listed in the Supplementary Table 7) and 0.5 mM (0.5 μCi) UDP-[3H]GlcNAc (from ARC) in 25 μl reaction buffer (50 μM Tris-HCl pH7.5, 12.5 mM MgCl2, 1 mM DTT) for 1 hour at 37 degree. The reaction was stopped by 10 mM EDTA. To detect potential DNA glycosylation, DNA was pull down by streptavidin beads. The beads were subjected to scintillation counter measurement. β-GT was purchased from Zymo Research and used as a positive control to produce 5ghmC following the protocol of Zymo Research with [3H] UDP-GlcNAc as the donor. For histone glycosylation analyses, the reaction was resolved with SDS-PAGE, and then subjected to autoradiography after incubation with EN3HANCE (from PerkinElmer). Nucleosome packaging was performed as described below with recombinant core histones purified from bacteria.
Electrophoretic mobility shift assay (EMSA)
157 bp DNA oligo (Supplementary Table 7) containing 5mC was produced by PCR using dATP, dGTP, dmCTP and dTTP. The oligo was labeled at 5′ end with 32P labeled by T4 DNA kinase and purified by G50 spin column (Pharmacia Biotech). The labeled oligo (0.1 pmol) was incubated with GST (0.4 μg), GST-OGT (0.4 μg) and/or GST-TET2CD (0.1, 0.2, 0.4 μg) respectively in the reaction buffer [50 mM HEPES, pH 7.9, 10 mM MgCl2, 50 mM NaCl, 75 μM Fe (NH4)2, 0.1 mg/ml Bovine serum albumin] for 30 minutes at room temperature. The samples were loaded onto 6 % native polyacrylamide gel and run at 150 V in 0.5×TBE (Tris-Borate-EDTA) buffer. The gels were then fixed with 30 % methanol and 10 % acetic acid and exposed to X-ray film.
For EMSA using mono-nucleosomes, we generated oligos containing C or 5mC by PCR. PCR products were end-labeled with 32P and purified by gel electrophoresis. To generate mono-nucleosome, labeled DNA oligos were incubated with core histones at a 1:1 molar ratio in a buffer containing 2 M NaCl and 1 mM DTT with a total volume of 30 μl. Mixtures were then serially dialyzed against 2 M, 0.85 M, 0.65M, and 0.2 M NaCl in 10 mM Tris-HCl pH 7.5, 1 mM EDTA, and 1 mM DTT for 4 to 10 hours at each step. Mono-nucleosomes were separated from free DNA by ultra-centrifugation in 5-30 % linear glycerol gradients. After centrifugation at 35,000 rpm in a SW50.1 rotor for 18 hours at 4°C, gradient fractions were collected. Fractions containing mono-nucleosomes were pooled, and dialyzed against 10 mM Tris-HCl pH 8.0, 1 mM DTT. For nucleosome binding reactions, GST-OGT (0.2 μg) and GST-TET2CD (0.05, 0.1, 0.2 μg) were incubated with 20 nM mono-nucleosomes in 20 μl with final buffer conditions of 50 mM HEPES, pH 7.9, 10 mM MgCl2, 75 μM Fe (NH4)2, 50 mM NaCl, 1 mM DTT, 1 % glycerol, 1 mg/ml Bovine serum albumin. The reactions were incubated at room temperature for 1 hour. Binding reactions were loaded onto 4 % native polyacrylamide gels at 150 V in 0.5×TBE buffer. After electrophoresis, gels were dried and examined by autoradiography.
Chromatin immunoprecipitation assay and DNA immunoprecipitation assay
Chromatin immunoprecipitation assays (ChIP) were preformed according to the protocol described by Upstate. ES cell DNA was sonicated to an averge size between 300 and 600 bp. Solubilized chromatin was immunoprecipitated with antibody against TET2, OGT, H2BS112G, H3K4me3 and H3K27me3. Antibody-chromatin complexes were pulled-down using protein A-sepharose, washed and then eluted. After cross-link reversal and proteinase K treatment, immunoprecipitated DNA was extracted with phenol-chloroform, ethanol precipitated, and treated with RNase. ChIP DNA was quantified using PicoGreen.
For hmC DIP assay, genomic DNA was extracted using Qiagen QIAamp DNa Mini kit. The purified genomic DNA was sonicated using Diagenode Bioruptor 200. The fragment size of DNA was determined by 2 % agarose gel. The fragment size should be 200-800 bp. The samples were heat-denatured and mixed with 1 μg 5hmC antibody. The mixtures were incubated overnight at 4 °C and then pulled down by protein-A sepharose beads. The beads were extensively washed and digested by Proteinase K. DNA was further purified by phenol-chloroform extraction and ethanol precipitation. 5hmC DIP DNA was quantified using PicoGreen.
ChIP sequencing
DNA fragments isolated from ChIP were repaired to blunt ends by T4 DNA polymerase and phosphorylated with T4 polynucleotide kinase using the END-IT kit (Epicentre). A single “A” base was added to 3′ end with Klenow. Double-stranded adaptors (75 bp with a “T” overhang) were ligated to the fragments with DNA ligase. Ligation products between 200 and 600 bp were gel purified to remove unligated adaptors and subjected to 20 PCR cycles. Completed libraries were quantified with PicoGreen. For ChIP-seq analyses, the DNA libraries were analyzed by Solexa/Illumina high-throughput sequencing. The read quality of each sample was determined by FastQC software. After prefiltering the raw data by removing sequence adapters and low quality reads, the tags were mapped to the mouse genome (assembly mm9) by Bowtie software. Parameter settings were listed as follows: -v, 3 (reported alignments with at most 3 mismatches), −5, 3 and −3, 3 (trim3 bases from 5′ and 3′ end to remove low-quality bases). Peak detection was performed using MACS software from Galaxy browser (www.galaxy.psu.edu). Parameters settings were as follows: IgG ChIP-seq aligned reads were used as control file, tag size with 25 bp, band width with 300 bp. Venn diagram analysis was performed with Galaxy browser. The peaks obtained from ChIP sequencing were matched to the annotated reference genome (mouse mm9) using Cisgenome 2.0. To view the peak density and position, Cisgenome 2.0 was used. The relative accumulations of tags around TSS were performed using SEQMINER software. The TSS position of each gene was downloaded from UCSC genome browser. Gene Ontology analysis was performed using Panther. All the statistics and plottings were obtained using the Statistical Program R. To compare TET2 and 5hmC, hmeDIP database was downloaded from NCBI3. A set of 30 PCR primer pairs (Supplementary Table 5) was designed to amplify 100-140 bp fragments from genomic regions showing a wide range of signals for TET2, OGT and H2BS112G by ChIP-seq. ChIP-qPCR values reflect two independent ChIP assays; each was evaluated in duplicate by qPCR.
mRNA analysis
To examine gene expression, total RNA was purified from mouse ES cells using RNeasy (Qiagen). The RNA was reverse transcribed using TaqMan reverse transcription reagents from ABI. Quantitative real-time PCRs (qPCRs) were carried out in 20 μl using SYBR green Master Mix (NEB) containing 10 nM 6-carboxyfluorescein (Sigma) as a reference dye, 50 to 100 ng of cDNA, and 2 μM primers. The reactions were performed on a Bio-Rad iCycler and quantified using the iCycler iQ software. The relative quantitiesy of each mRNA wereas determined for each sample based on the threshold cycle (CT) value normalized to the corresponding values for GAPDH. Oligo sequences of the primers for qPCR assays were listed in the Supplementary Table 6. All reactions were performed in triplicates.
RNA expression data for shCON and shTET2 ES cells were generated from polyA RNA using GeneChip Mouse Genome 430 Arrays (Affymetrix) following Affymetrix procedures and analysis. The results were shown in Supplementary Table S3. The expression value of each transcript was showed in log2 scale. The data analysis was performed using Cluster 3.0 and Java Treeview.
In vitro 5hmC assay
To examine enzymatic activity of TET2 in vitro, 0.4 μg double-stranded DNA oligo (listed in Supplementary Table 7) that contain C, 5mC or 5hmC were incubated with 1 μg TET2CD or 1μg TET2CD together with 1 μg OGT proteins in the presence of 50 mM HEPES (pH 7.9), 100 mM NaCl, 75 μM Fe (NH4)2, 2 mM ascorbate, and 1 mM a-ketoglutate at 37 degree for 40 minutes. The DNA substrates then were purified by ethanol precipitation and digested with TapI. Following the treatment with calf intestinal alkaline phosphatase, the DNA substrates were labeled with [r-32P] ATP by T4 polynucleotide kinase, purified by ethanol precipitation and digested with 10 μg of Dnase I and 10 μg phosphodiesterase I. The samples were separated by thin layer chromatography (TLC) on PERcellulose TLC plates (Merck) using isobutyric acid: NH4OH: H2O=66:2:20. After air dry, the TLC plates were exposed to X-ray film.
Dot blotting assay
Genomic DNA isolated from 293T cells expressing TET2, OGT or both was denatured by 0.2 N NaOH and dotted on Hybond-N+ nitrocellulose membrane (Amersham Pharmacia Biotech). After ultraviolet cross-linking, membranes were blocked overnight with 10 % non-fat milk and 1 % BSA in TBST (150 mM NaCl, 10 mM Tris pH 8.0 + 0.1 % Tween20) at 4 degree followed by 1 hour incubation with either anti-5mC or anti-5hmC antibodies at room temperature. Membranes were washed four times with TBST, incubated with HRP conjugated goat anti-mouse or goat anti-rabbit antibodies (GE healtcare), washed with TBST, and developed using the ECL+ detection system (GE Healthcare).
Mass spectrometry analysis
For 5ghmC standard, β-GT was used to produce 5ghmC following the protocol of Zymo Research using UDP-GlcNAc as the donor. The DNA was digested with DNA Degradase Plus (Zymo Research). The samples were subjected to LC-MS. Liquid chromatography was performed using a 2.1 × 50 mm HSS T3 1.8 um column (from Waters) with gradient elution at flow rate of 500 μl/min using 0.02 % acetic acid in water as mobile phase A and methanol as mobile phase B. The gradient was 96 % A + 4 % B to 70 % A + 30 B in 4.7 minutes. The elutes were directed to the mass spectrometer that was running in the product scan mode (60-500 m/z) selecting 461 as precursor ion (GlcNAc-hmC MW). The collision energy used was 30 V. For detecting GlcNAc-hmC or Glc-hmC, genomic DNA from 293T expressing TET2 and OGT was digested as above and subjected to LC-MS/Ms analysis using the same column and conditions as above. Mass spectrometer was running in multiple reaction monitoring (MRM) mode, monitoring the transition of m/z 242.0 to 126.0 (5mC), m/z 258.0 to 142.0 (5hmC), 461.0 to 345.0 (GlcNAC-hmC) and m/z 420.0 to 304.0 (Glc-hmC) with the collision energy as 15 V.
Supplementary Material
Acknowledgments
We thank Henry Kuang for proof reading the manuscript and Jeanie Hutchins for research support. This work was supported by the National Institute of Health (CA132755 and CA130899 to X.Y.), the University of Michigan Cancer Center and GI Peptide Research Center. X.Y. is a recipient of the Era of Hope Scholar Award from the Department of Defense.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions X.Y. conceived the project and designed the experiments; Q.C performed the experiments with Y.C., C.B., R.F.; Q.C. and X.Y. analyzed the data and wrote the manuscript.
Author Information ChIP-seq and microarray data have been deposited in the Gene Expression Omnibus under accession number GSE41720.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 3.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vosseller K, Sakabe K, Wells L, Hart GW. Diverse regulation of protein function by O-GlcNAc: a nuclear and cytoplasmic carbohydrate post-translational modification. Curr Opin Chem Biol. 2002;6:851–857. doi: 10.1016/s1367-5931(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 8.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 9.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 10.Clarke AJ, et al. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008;27:2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Fleites C, et al. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 12.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annu Rev Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Roche K, Nasheuer HP, Lowndes NF. Modification of histones by sugar beta-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem. 2011;286:37483–37495. doi: 10.1074/jbc.M111.284885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiki R, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong JJ, et al. beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2012;287:12195–12203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capotosti F, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 20.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 21.Iyer SP, Akimoto Y, Hart GW. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J Biol Chem. 2003;278:5399–5409. doi: 10.1074/jbc.M209384200. [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 24.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. J Biol Chem. 2010;285:34460–34468. doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gambetta MC, Oktaba K, Muller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 30.Sinclair DA, et al. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci U S A. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for full methods
- 31.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 32.Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 33.Robinson KA, Ball LE, Buse MG. Reduction of O-GlcNAc protein modification does not prevent insulin resistance in 3T3-L1 adipocytes. Am J Physiol Endocrinol Metab. 2007;292:E884–890. doi: 10.1152/ajpendo.00569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, et al. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011;20:259–267. doi: 10.1089/scd.2010.0072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.