Abstract
The availability of complete genome sequences for a number of biologically important fungi has become an important resource for fungal research communities. However, the functions of many open reading frames (ORFs) identified through annotation of whole genome sequences have yet to be determined. The disruption of ORFs is a practical method for loss-of-function gene analyses in fungi that are amenable to transformation. Unfortunately, the construction of knockout cassettes using traditional digestion and ligation techniques can be difficult to implement in a high-throughput fashion. Knockout cassettes for all annotated ORFs in Neurospora crassa were constructed using yeast recombinational cloning. Here, we describe a high-throughput knockout cassette construction method that can be used with any fungal transformation system.
Keywords: Gene targeting, Filamentous fungi, ku70, ku80, mus-51, mus-52
1. Introduction
Many ORFs identified through whole-genome sequencing of numerous fungi cannot be functionally characterized based on sequence alone. Gene disruption serves as a fundamental approach for understanding the role of many ORFs that do not have an assigned function. We have undertaken the task of disrupting all ∼10,000 ORFs in the annotated genome of Neurospora crassa as part of a functional genomics consortium (1). This project aims to provide fungal researchers with a set of individual gene knockouts that covers the whole N. crassa genome similar to the set of knockouts that encompasses the entire genome of the yeast Saccharomyces cerevisiae (2–4). While the scale of a whole-genome knockout project may not be feasible for all fungal researchers, systematic disruption of groups of functionally relevant ORFs can be quite informative. For example, the disruption of 103 putative transcription factors in N. crassa (5) demonstrated the usefulness of a high-throughput knockout procedure for fungi with completed genome sequences on a scale that is realistic for many suitably equipped labs.
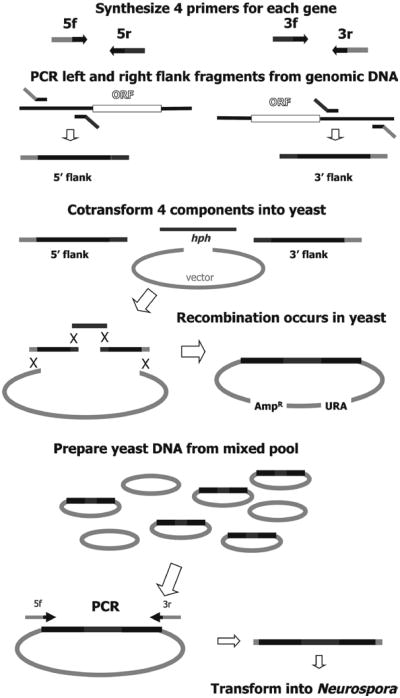
One of the first hurdles to overcome with a large-scale knockout project like this is the construction of the knockout cassettes. The use of traditional restriction digestion and ligation, for example, is too cumbersome to be a favorable option for high-throughput knockout cassette assembly. For this reason, we employed a method that utilizes S. cerevisiae to assemble cassette components that possess short complementary overlapping sequence ends (6, 7). Components are synthesized in separate PCR reactions and cotransformed into S. cerevisiae for assembly by the host cell's own recombination machinery. An overview schematic for the construction of knockout cassettes using the yeast recombinational system is presented in Fig. 1.
Fig. 1.
Schematic of KO cassette construction using yeast recombinational cloning techniques. Primers were designed to amplify flanks both upstream and downstream of the ORF. Each primer also contained 29 nucleotides of “common” sequence to overlap with the yeast cloning vector (light grey) or the hygromycin B phosphotransferase (hph) resistance cassette (dark grey).
Crucial to the success of this project was the work from Ninomiya et al. (8) demonstrating that mutation of single genes (mus-51 and mus-52) involved in nonhomologous end-joining (NHEJ) leads to a high rate of homologous recombination in Neurospora. Prior to this work, high-throughput gene disruption in N. crassa was extremely difficult due to the low frequency of homologous recombination relative to ectopic insertions. We created two disruption strains, Δmus-51 and Δmus-52, using the selectable marker bar, which confers resistance to phosphinothricin (9, 10). Knockout cassettes are transformed into one of the mus mutants to produce primary transformants, which are then crossed to wild type to generate knockout mutants in a genetic background lacking the mus mutation. Subsequent to our work, mutants deficient in the NHEJ-pathway have been shown to display high homologous recombination frequencies in a number of filamentous fungi (11). Therefore, assessment of targeting efficiency and generation of recipient strains that are deficient in the NHEJ-pathway are important areas of consideration prior to undertaking a large-scale gene deletion effort.
The basic protocol for the high-throughput construction of knockout cassettes in N. crassa genes is described here with some aspects of the procedure elaborated upon in greater detail. Additional details of the entire N. crassa process (including N. crassa electroporation and verification of targeted gene disruptions) can be found in the methods and Supplementary Methods of Colot et al. (5). Many of the protocols that were developed for N. crassa are easily transferred to other flamentous fungi. However, depending on the fungal organism of interest, Agrobacterium-mediated, biolistic or protoplast transformation methods may be more suitable than electroporation using this high-throughput knockout cassette construction protocol.
2. Materials
2.1. PCR of Knockout Cassette Components
-
Gene-specific PCR products to be cloned. Typically, these consist of three fragments: the 5′ and 3′ flanks of the gene(s) of interest and a resistance cassette. There should be approximately 30 bp of overlapping complementary sequence between fragments to be assembled. Four gene-specific primers are needed for each knockout cassette flank: 5′ flank forward (5f), 5′ flank reverse (5r), 3′ flank forward (3f), and 3′ flank reverse (3r). The following 5′ common sequences were added to the respective primers for each gene to allow cassette assembly in the yeast recombinational system:
5f, GTAACGCCAGGGTTTTCCCAGTCACGACG…
5r, ATCCACTTAACGTTACTGAAATCTCCAAC…
3f, CTCCTTCAATATCATCTTCTGTCTCCGAC…
3r, GCGGATAACAATTTCACACAGGAAACAGC…
Plasmid pCSN44 (12, 13). This plasmid contains the dominant drug-resistance marker gene hygromycin B phosphotransferase (hph) from E. coli, flanked by the promoter and terminator of the trpC gene from Aspergillus nidulans.
Primers for the amplification of the hph resistance cassette: hphF (GTCGGAGACAGAAGATGATATTGAAGGAGC) hphR (GTTGGAGATTTCAGTAACGTTAAGTGGAT)
A high-fidelity, high-yield Taq polymerase to be used for the generation of gene flanks and resistance cassettes that minimizes errors in the final knockout cassettes. We use LA Taq by TaKaRa (Fisher Scientifc, Pittsburgh, PA).
2.2. Yeast Transformation and Cassette Assembly
Cloning vector pRS426 (14). This is a 2 μm vector that contains a URA3 marker for selection in S. cerevisiae.
S. cerevisiae strain FY834 (MATα his3 Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63) (15).
YPD medium. Dissolve 10 g yeast extract, 20 g peptone and 20 g dextrose in dH2O to a final volume of 1 L. Add 15 g agar and autoclave.
96PEG solution. Dissolve 45.6 g PEG (MW 3350) in dH2O to a final volume of 88 mL. Add 10 mL 1 M lithium acetate and 2 mL 50× TE buffer. Autoclave or flter-sterilize.
Sheared salmon sperm DNA, 10 mg/mL (Fisher Scientifc, Pittsburgh, PA). Dilute to 2 mg/mL in ddH2O, boil for 5 min and store at −20°C. This does not need to be boiled each time it is thawed for use.
SC-Ura medium. To prepare 1 L, mix 26.7 g Drop-out Base with Glucose (US Biologicals, Swampscott, MA) and 2 g Drop-out Mix Synthetic Minus Uracil w/o Yeast Nitrogen Base (US Biologicals, Swampscott, MA) in H2O. Autoclave.
Airpore tape sheets (Qiagen, Valencia, CA).
2.3. Yeast DNA Purification
Gentra Puregene Tissue Kit (Qiagen, Valencia, CA).
Zymolyase T-100 (Seikagaku, Tokyo, Japan).
CleanSeq beads (Agencourt, Beverly, MA).
Agencourt SPRIPlate® 96R – Ring Magnet Plate (Agencourt, Beverly, MA).
3. Methods
Although many of the cassette construction steps were performed using a Biomek NX robot, these methods may also be easily scaled-down for small groups of genes.
3.1. PCR of Knockout Cassette Components
Gene-specific knockout cassettes should be designed to have 1–1.3 kb of gene-specific 5′ and 3′ flanks and a selectable marker (the hygromycin B phosphotransferase gene, hph, in our case). To ensure appropriate expression of the selectable marker, we use the A. nidulans promoter trpC (13).
For the primer design of 5′ and 3′ flanks, software written for us by John Jones (John Jones Consulting, Moreno Valley, CA) was used to retrieve regions adjacent to each ORF in the annotated genome of N. crassa. This information was then passed to PRIMER3 (http://primer3.sourceforge.net/), which would automatically select a list of candidate primers based on defined conditions (5).
Primers for the Neurospora knockout project are ordered from Illumina (San Diego, CA) in 96-well plates at 50 μM. These primers are mixed in three pairs (5f+5r, 3f+3r and 5f+3r) using a Beckman Biomek NX robot to a final concentration of 10 μM each. (The 5f+3r primer combination is required for the amplification of the final knockout cassette to be used for transformation.)
Perform separate 25 μL PCR reactions for the 5′ and the 3′ flanks using primer pair mixtures 5f+5r or 3f+3r. PCR reactions consist of: 2.5 μL 10× buffer (LA Taq), 4 μL dNTP's (LA Taq), 1 μL 10 μM primer mixture, 0.25 μL genomic N. crassa DNA (prepared using the Puregene DNA kit; ∼150 ng/μL), 17 μL water, and 0.25 μL LA Taq (5 units/μL).
Generate the hph cassette fragment in a 50 μL PCR reaction containing: 5 μL 10× buffer (LA Taq), 8 μL dNTP's (LA Taq), 1 μL 10 μM primer hphF, 1 μL 10 μM primer hphR, 0.5 μL pCSN44 DNA (from 1:100 dilution of plasmid miniprep), 34 μL water, and 0.5 μL LA Taq (5 units/μL).
PCR cycling parameters for both flanks and the hph resistance marker are: 94°C for 1:00 min followed by 35 cycles of: {94°C for 30 s, 60°C for 30 s, 72°C for 2:00 min}, and a final extension of 72°C for 10:00 min.
Digest several μg of pRS426 vector with EcoRI and XhoI and dilute to a final concentration of 100 ng/μL.
3.2. Yeast Transformation and Cassette Assembly
The PCR products and digested vector do not need to be cleaned up prior to yeast transformation. Yeast transformation was adapted from a 96-well protocol described in http://depts.washington.edu/sfelds/protocols/cloning_protocol.html
Inoculate 50 mL of YPD with 0.3 mL of a FY834 saturated culture. Grow overnight at 30°C with shaking (up to 300 rpm).
Pellet cells in a 50 mL conical tube (2,530 × g for 3 min). Discard supernatant, resuspend cells in 2 mL 100 mM lithium acetate and transfer to two microcentrifuge tubes.
Spin at top speed in a microcentrifuge for 30 s and discard the supernatant. Resuspend cell pellets in 100 mM lithium acetate to a total final volume of 1.8 mL (approximately 700 μL to each microcentrifuge tube). At this point, it is safe to keep cells on bench at room temperature until use.
Prepare fresh CT110 mixture (16). For 100 transformations mix 20.7 mL 96PEG, 580 μL boiled salmon sperm (2 mg/mL), 210 μL hph cassette (directly from PCR reaction of Section 3.1, step 5), 105 μL cut vector pRS426 (100 ng/μL) and 2.62 mL DMSO. Mix thoroughly for 30 s, then add 1.8 mL yeast cells and mix thoroughly for an additional 60 s.
Pipette 200 μL of CT110 mixture into each well of a 96-well deep-well plate.
Add 4 μL of 5′ flank and 4 μL of 3′ flank PCR reaction for each gene. Seal plate, vortex 4 min, and incubate at 42°C for 30 min.
Spin plate at 2,000 × g for 7 min and aspirate off supernatant.
Add 200 μL SC-Ura to each well and resuspend by pipetting up and down.
Transfer 80 μL of resuspended transformed cells to 1 mL of SC-Ura liquid in each well of a 96-well deep-well plate. Seal the plate with Airpore tape (Qiagen) and grow 3 days at 30°C with shaking.
3.3. Yeast DNA Purification
The following steps are described as they were performed using the Biomek NX robot, with the exception of centrifugation and vortexing steps. The Gentra Puregene™ Tissue kit (Qiagen) was used on the robot with a few modifications that are described below.
After 3 days, centrifuge the plate at 2,000 × g for 3 min.
Remove supernatant and resuspend pellet in 300 μL of Gentra cell suspension solution (Qiagen) containing 120 μg/mL Zymolyase T-100 (Seikagaku).
Incubate the plate at 37°C for 40 min, then centrifuge at 830 × g for 3 min.
Remove the supernatant and add 300 μL Gentra cell lysis solution (Qiagen) to the pellet and mix.
Add 100 μL Gentra protein precipitation solution (Qiagen) and mix.
Vortex the plate for 20 s and centrifuge at 3,070 × g for 12 min.
Transfer 90 μL of supernatant into a round-bottomed 96-well plate, and add 10 μL CleanSeq beads and 143 μL fresh 85% ethanol. Mix components and incubate statically for 3 min.
Place the plate on an SPRI magnetic plate (Agencourt) for 10 min and aspirate the cleared solution.
With the plate remaining on the magnet, add 200 μL of 85% ethanol to each well. After 30 s, remove the ethanol and repeat this rinse with 200 μL of 85% ethanol.
Allow the plate to air-dry for 10 min and remove from the magnet.
Add 40 μL 1× TE to each well and mix.
Return the plate to the magnet for 5 min and elute DNA to a new 96-well plate.
3.4. PCR of Final Full-Length Knockout Cassette
50 μL PCR reactions are performed to generate each deletion cassettes. PCR reactions consist of: 5 μL 10× buffer (LA Taq), 8 μL dNTP's (LA Taq), 2 μL 10 μM primer 5f+3r primer mixture, 4 μL yeast DNA, 30.5 μL water, and 0.5 μL LA Taq.
PCR cycling parameters for the final full-length knockout cassette is: 94°C for 1:00 min followed by 35 cycles of: {94°C for 30 s, 60°C for 30 s, 72°C for 5:00 min} and a final extension of 72°C for 10:00 min.
Estimate DNA yields by agarose gel electrophoresis.
At this point, the cassettes we use for creating the Neurospora Genome Project knockout strains are cleaned up with the Qiagen QIAquick 96 PCR Purification Kit #28181 according to the manufacturer's protocol prior to N. crassa transformation. 5 μL of purifed PCR product usually yields a sufficient number of N. crassa transformants by electroporation, but this amount will vary depending on the recipient organism and the transformation technique.
Acknowledgments
This work was supported by grant P01 GM068087 from the National Institute of General Medical Sciences. We would like to thank John Jones for software design and LIMS implementation.
References
- 1.Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, Glass N, et al. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet. 2007;57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 3.Martin AC, Drubin DG. Impact of genome-wide functional analyses on cell biology research. Curr Opin Cell Biol. 2003;15:6–13. doi: 10.1016/s0955-0674(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 4.Ooi SL, Pan X, Peyser BD, Ye P, Meluh PB, Yuan DS, et al. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Colot HV, Park G, Turner G, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Nat Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymond CK, Pownder TA, Sexson SL. General method for plasmid construction using homologous recombination. Biotechniques. 1999;26:134–141. doi: 10.2144/99261rr02. [DOI] [PubMed] [Google Scholar]
- 8.Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pall ML. The use of Ignite (basta; glufosinate; phosphinothricin) to select transformants of bar-containing plasmids in Neurospora crassa. Fungal Genet Newsl. 1993;40:58. [Google Scholar]
- 10.Pall ML, Brunelli JP. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet Newsl. 1993;40:59–62. [Google Scholar]
- 11.Meyer V. Genetic engineering of flamentous fungi – Progress, obstacles and future trends. Biotechnol Adv. 2008;26:177–185. doi: 10.1016/j.biotechadv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of the hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 13.Staben C, Jensen B, Singer M, Pollock J, Schechtman M, Kinsey J, et al. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet Newsl. 1989;36:79–81. [Google Scholar]
- 14.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopala SV, Titz B, Uetz P. Array-based yeast two-hybrid screening for protein-protein interactions. Method Microbiol. 2007;36:139–163. [Google Scholar]