SUMMARY
Survivin is an inhibitor of apoptosis (IAP) family protein implicated in apoptosis and mitosis. In apoptosis, it has been shown to recognize the Smac/DIABLO protein. It is also a component of the chromosomal passenger complex, a key player during mitosis. Recently, Survivin was identified in vitro and in vivo as the direct binding partner for phosphorylated Thr3 on histone 3 (H3T3ph). We have undertaken structural and binding studies to investigate the molecular basis underlying recognition of H3T3ph and Smac/DIABLO N-terminal peptides by Survivin. Our crystallographic studies establish recognition of N-terminal Ala in both complexes, and identify intermolecular hydrogen bonding interactions in the Survivin phosphate-binding pocket that contribute to H3T3ph mark recognition. In addition, our calorimetric data establish that Survivin binds tighter to the H3T3ph-containing peptide relative to the N-terminal Smac/DIABLO peptide, and that this preference can be reversed through structure-guided mutations that increase the hydrophobicity of the phosphate-binding pocket.
INTRODUCTION
Human Survivin was originally identified as an anti-apoptotic protein, which is overexpressed in most human tumors and fetal tissue, but not in terminal differentiated cells (Ambrosini et al., 1997; Reed, 2001; Sah et al., 2006). It is a small protein (142 residues) belonging to the inhibitor of apoptosis (IAP) protein family (Salvesen and Duckett, 2002). The IAP protein family shares a common baculovirus IAP repeat (BIR) domain, which is usually located at the N-terminus of the protein (Srinivasula and Ashwell, 2008). The BIR domain functions as a mediator for protein-protein interactions, through utilization of a deep surface peptide-binding groove to recognize the N-terminal conserved Ala-containing IAP-binding motif (IBM) of the target protein, A well-studied published example involves the recognition of N-terminus Smac/DIABLO (SmacN) IBM by the BIR3 domain of XIAP, in which the N-terminal Ala of Smac inserts into a deep surface pocket of BIR3, with complex formation mediated by extensive intermolecular interactions (Liu et al., 2000; Wu et al., 2000). Although Survivin contains the BIR domain, the role of Survivin in the apoptotic pathway remains to be elucidated (Lens et al., 2006; Yue et al., 2008). One suggested mechanism has proposed that Survivin can bind to Smac, thereby neutralizing the inhibition of XIAP by Smac (Song et al., 2003). Binding of Survivin to Smac has been monitored by NMR with the mapped binding surface overlapping with a region corresponding to the Smac binding site on BIR3 of XIAP (Sun et al., 2005).
It has been established that Survivin plays multiple essential roles during mitosis and meiosis as a subunit of the chromosomal passenger complex (CPC), which is enriched on the centromere in early M phase but is relocated to the spindle midzone in anaphase (Ruchaud et al., 2007). The CPC contains two distinct modules, a kinase subcomplex domain composed of the Aurora B kinase and the C-terminal fragment of the inner centromere protein (INCENP), and a chromosomal localization subcomplex composed of the N-terminal fragment of INCENP, Survivin and Borealin (Kelly et al., 2007). Further studies have established that Survivin was the CPC component responsible for direct binding of histone H3 phosphorylated at Thr3 (H3T3ph), thereby recruiting CPC to mitotic chromatin during mitosis (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). Such recruitment results in the activation of the Aurora B kinase, which can promote spindle assembly and inhibit nuclear reformation (Kelly et al., 2010), and contributes to regulation of the spindle assembly checkpoint and kinetochore-microtubule attachment (Wang et al., 2010; Yamagishi et al., 2010).
Survivin has been shown to be overexpressed in most cancer cells but not normal terminal differentiated tissues (Ambrosini et al., 1997; Reed, 2001; Sah et al., 2006), and downregulation of Survivin expression and/or its functions can sensitize tumor cells to therapeutics (Kanwar et al., 2010a). Thus, Survivin is a promising drug target against cancer, as well as a biomarker for angiogenesis and cancer diagnosis (Kanwar et al., 2010a; Kanwar et al., 2010b; Pennati et al., 2008; Ryan et al., 2009). Crystal structures are available for both human and mouse Survivin in the free state (Chantalat et al., 2000; Muchmore et al., 2000; Verdecia et al., 2000). Structurally, human Survivin is composed of an N-terminal BIR domain and a C-terminal long α-helix, with the latter shown structurally to form an intermolecular three-helix bundle with the N-terminal fragment of INCENP and Borealin (Jeyaprakash et al., 2007). NMR chemical shift mapping studies have shown that the N-terminal BIR domain of Survivin forms a complex with the N-terminal domain of Smac/Diablo (Sun et al., 2005) and with the N-terminal H3T3ph (Kelly et al., 2010) peptides. Nevertheless, these studies have not identified the detailed intermolecular contacts associated with molecular recognition on complex formation. We report below on crystal structures of human Survivin in complex with unmodified H3, H3T3ph-containing and SmacN peptides, as well as isothermal titration calorimetry (ITC)-based studies on the wild-type complex and those containing modified peptides and mutant Survivin. These studies have defined the importance of peptide N-terminal Ala1 recognition in both complexes, and identified details of the intermolecular hydrogen bonding interactions in the Survivin binding pocket that contribute to H3T3ph mark recognition. Notably, our calorimetric data establish that Survivin binds tighter to the H3T3ph-containing peptide relative to the N-terminal Smac/DIABLO peptide, and that this preference can be reversed through structure-guided mutations in the binding pocket. During preparation of our paper for submission, Jeyaprakash et al., (2011) published their structures of human Survivin in complex with H3T3ph peptide and N-terminal peptide of human Shugoshin 1.
RESULTS
Overall Structure of Survivin Bound to H3(1–15)T3ph Peptide
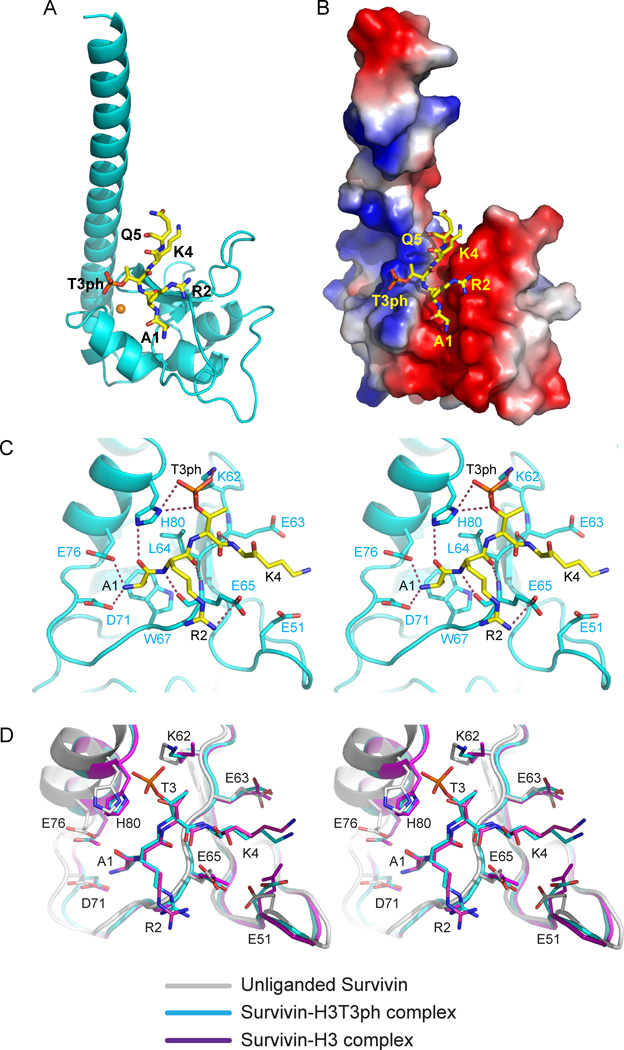
The structure of human Survivin bound to H3(1–15)T3ph was solved by molecular replacement using the structure of an unliganded Survivin (PDB code 1F3H) (Verdecia et al., 2000) as a search model and refined to 2.4 Å resolution, yielding R-work of 21.3% and R-free of 25.0% (Table 1). The overall structure of Survivin in the complex (ribbon representation in Figure 1A and electrostatic surface representation in Figure 1B) resembles its structure in the unliganded state (PDB: 1F3H) (Chantalat et al., 2000; Verdecia et al., 2000), with an r.m.s.d. between structures of 0.45 Å. As anticipated, Survivin in the complex is composed of an N-terminal BIR domain (aa 15–89) and a long C-terminal α-helix (aa 100–140), with the fold stabilized by a Zn2+ ion coordinated by Cys57, Cys60, His77, and Cys84 (Figure 1A). The asymmetric unit in the complex (C2 space group) contains two Survivin molecules that align to form a homodimer (Figure S1, Supplementary Materials), with the dimer interface formed by residues 6–10 and 89–102. We can trace N-terminal residues Ala1 to Gln5 segment of the bound H3(1–15)T3ph peptide in the structure of the complex (Figures 1A,B).
Table 1.
Summary of Diffraction Data and Structure Refinement Statistics of H3(1–10) and H3(1–15)T3ph Peptide Complexes with wild-type Survivin and Survivin(K62Y/H80W) Double Mutant.
| Summary of diffraction data | |||
|---|---|---|---|
| Crystal | H3(1–15)T3ph- Survivin |
H3(1–10)- Survivin |
H3(1–10) Survivin(K62Y/H80W) |
| PDB code | 3UIG | 3UII | 3UIK |
| Beamline | APS-24ID-C | APS-24ID-E | APS-24ID-E |
| Wavelength (Å) | 0.9795 | 0.9792 | 0.9792 |
| Space group | C2 | C2 | C2 |
| Cell parameters | |||
| a (Å) | 115.8 | 115.3 | 114.8 |
| b (Å) | 71.0 | 71.3 | 71.0 |
| c (Å) | 82.3 | 81.6 | 81.6 |
| β (°) | 128.8 | 128.5 | 129.3 |
| Resolution (Å) | 50.0-2.4 (2.49-2.40)a | 50.0-2.6 (2.69-2.60) | 30.0-2.7 (2.80-2.70) |
| Rmerge (%) | 5.2 (52.7) | 8.6 (52.9) | 7.4 (64.0) |
| Observed reflections | 82,484 | 66,350 | 50,310 |
| Unique reflections | 20,244 | 16,412 | 13,470 |
| Redundancy | 4.1 (3.9) | 4.0 (3.9) | 3.7 (3.5) |
| Average I/σ(I) | 17.2 (2.0) | 10.1 (2.0) | 31.4 (1.8) |
| Completeness (%) | 97.9 (86.9) | 98.9 (96.4) | 96.6 (83.6) |
| Refinement and structure model | |||
| R / R free | 21.3 / 25.0 | 22.2 / 27.3 | 21.9 / 28.1 |
| Number of atoms | 2,329 | 2,293 | 2290 |
| Protein / Peptide | 2,215 / 90 | 2,210 / 64 | 2224 / 64 |
| Water | 22 | 17 | - |
| Zn2+ ion | 2 | 2 | 2 |
| Average B factor (Å2) | 96.3 | 89.1 | 119.6 |
| Protien / Peptide | 95.9 / 110.0 | 88.4 / 115.1 | 118.2 / 169.1 |
| Water | 85.5 | 82.3 | - |
| Zn2+ ion | 73.0 | 76.5 | 98.6 |
| RMS deviations | |||
| Bond lengths (Å) | 0.010 | 0.013 | 0.010 |
| Bond angles (°) | 1.224 | 1.430 | 1.408 |
Values in parentheses are for highest-resolution shell.
Figure 1. Interactions between H3(1–15)T3ph Peptide and Survivin in the Complex.
- Overall interaction between residues Ala1 to Gln5 of H3(1–15)T3ph peptide and Survivin in the complex. The bound peptide (yellow) in a stick representation is positioned within the BIR domain (cyan) of Survivin in a ribbon representation.
- An electrostatics surface representation of Survivin with bound residues Ala1 to Gln5 of H3(1–15)T3ph peptide (yellow) in a stick representation. The N-terminus and Ala1 insert into a negatively charged pocket, the Arg2 and Lys4 side chains lie on a flat negatively charge surface, while Thr3ph lies on a positively charged shallow cleft. There is electrostatic complementarity between the bound H3T3ph peptide and Survivin in the complex.
- Stereo view highlighting details of the intermolecular interactions between the A1-R2-T3ph-K4 segment of the bound H3(1–15)T3ph peptide and binding pocket residues of the BIR domain of Survivin. Intermolecular hydrogen-bonding interactions are designated by dashed red lines.
- Stereo view of superpositioned structures of unliganded Survivin (grey), H3(1–15)T3ph peptide-bound Survivin (cyan) and H3(1–10) peptide-bound Survivin (magenta). These views emphasize binding pocket interactions. Note the shift in the α-helix (on left) on complex formation. See also Supplementary Figures S1, S2, S4, S5, S6 and S7.
We have also solved the structure of Survivin bound to unmodified H3(1–10) peptide at 2.6 Å resolution (Figure S2), with refinement yielding an R-work of 22.2% and R-free of 27.3% (Table 1). The overall structure of Survivin in the Survivin-H3(1–10) complex is more similar to the Survivin-H3(1–15)T3ph complex (r.m.s.d. of 0.31 Å for 135 aligned Cα atoms) than to Survivin in the free state (r.m.s.d. of 0.48 Å for 135 aligned Cα atoms). We can trace N-terminal residues Ala1 to Lys4 of the bound H3(1–10) peptide in the structure of the complex.
Recognition of N-terminal H3 A1-R2-T3ph-K4 Segment by Survivin
We can monitor the first five residues from the N-terminus of H3(1–15)T3ph and first four residues of H3(1–10) peptides bound to Survivin in the crystal structures of the complexes (Figures 1A,B;S2A,B). The bound peptides are positioned in a shallow cleft on the surface of the BIR domain of Survivin, with the N-terminal Ala1 inserted into a deep negatively-charged pocket (Figure 1B). The positively-charged Arg2 and Lys4 residues of the bound peptide are oriented towards a negatively charged surface (red color, Figure 1B), while the negatively-charged T3ph is oriented in the opposite direction towards a positively-charged surface (blue color, Figure 1B).
We initially focus on the intermolecular interactions in the complex of H3(1–15)T3ph peptide bound to Survivin (stereo pair in Figure 2A). The N-terminal Ala1 residue is recognized by the BIR domain of Survivin using recognition principles in common with those observed for the complex of Smac/DIABLO peptide bound to the BIR3 domain of XIAP (Liu et al., 2000; Wu et al., 2000). The N-terminal NH3+ group of Ala1 of the bound peptide is anchored through hydrogen bond formation with the negatively-charged side chains of Asp71 and Glu76, while the carbonyl group of Ala1 forms a hydrogen bond with the ring nitrogen of His80 (Figures 1C). The side chain methyl group of Ala1 inserts into a small hydrophobic pocket generated by the side chains of Leu64 and Trp67 (Figure 1C). The side chain of Arg2 of the bound peptide forms a hydrogen bond with the side chain of Glu65, while the backbone of Arg2 forms two main chain hydrogen bonds in an anti-parallel β-sheet-like manner.
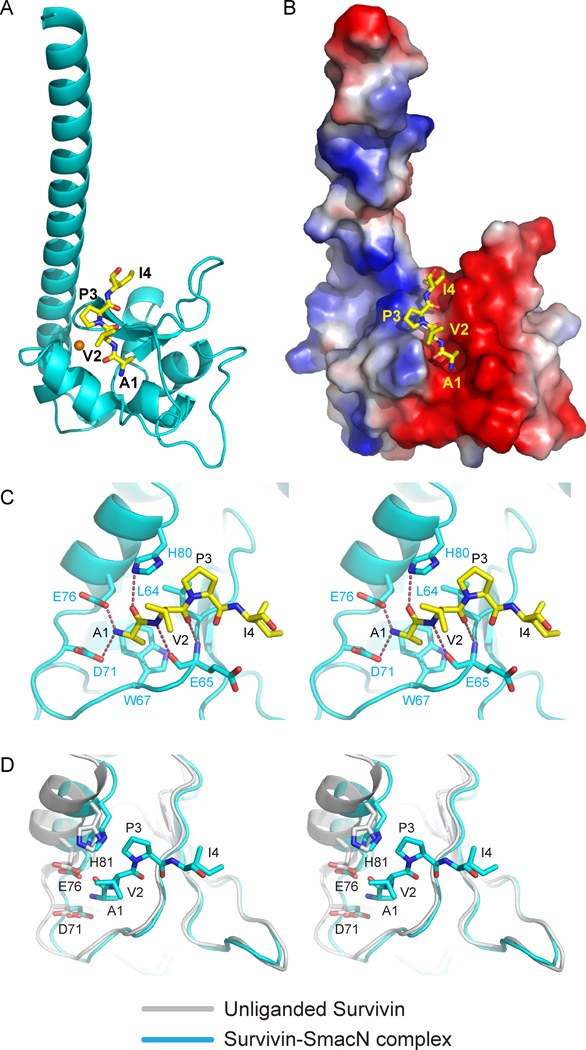
Figure 2. Interactions between SmacN(1–15) Peptide and Survivin in the Complex.
- Overall interaction between residues Ala1 to Ile4 of SmacN(1–15) peptide and Survivin in the complex. The bound peptide (yellow) in a stick representation is positioned within the BIR domain (cyan) of Survivin in a ribbon representation.
- An electrostatics surface representation of Survivin with bound residues Ala1 to Ile4 of SmacN peptide (yellow) in a stick representation.
- Stereo view highlighting details of the intermolecular interactions between the A1-V2-P3-I4 segment of the bound SmacN(1–15) peptide and binding pocket residues of the BIR domain of Survivin. Intermolecular hydrogen-bonding interactions are designated by dashed red lines.
-
Stereo view of superpositioned structures of unliganded Survivin (grey) and SmacN peptide-bound Survivin (cyan). These views emphasize binding pocket interactions. Note the shift in the α-helix (on left) on complex formation.See also Supplementary Figures S2 and S7.
The phosphate group of Thr3ph is positioned in a positively-charged shallow cleft formed by the side chains of Lys62 and His80. Two of the three non-bridging phosphate oxygens are involved in hydrogen-bonding interactions, as is the bridging phosphate oxygen, with the acceptors being the ring nitrogen of His80 and the side chain of Lys62 (Figure 1C). The side chain of Lys4 is positioned between the side chains of Glu51 and Glu63, with its backbone amide forming a main chain-main chain hydrogen bond.
We observe similar intermolecular interactions in the H3(1–10) peptide bound to Survivin, with the only difference being that unmodified Thr3 forms a single hydrogen bond between its side chain and the ring nitrogen of His80 (Figure S2C).
Conformational Transition on Complex Formation
We have superpositioned the structure of Survivin in the free state (PDB: 1F3H) with the corresponding structures in complex with H3(1–10) and H3(1–15)T3ph peptides. We note a conformational transition encompassing a segment spanning a long loop and an α-helix (Pro69 to Gly83), which moves towards the bound peptide on complex formation (stereo pair in Figure 1D). This conformational transition makes the BIR domain more compact on complex formation. In addition, while most residues participating in the interaction with phosphorylated and unmodified H3 peptides have almost identical conformation in the two complex structures, the side chain of Lys62 shows a notable difference. It is directed towards the phosphate of the bound peptide in the H3(1–15)T3ph-Survivin complex, while being directed away from the bound peptide in the H3(1–10)-Survivin complex (Figure 1D), consistent with a contributing role for the side chain of Lys62 in the recognition of the phosphate.
Recognition of N-terminal A1-V2-P3-I4 Segment of SmacN Peptide by Survivin
We have also solved the crystal structure of the N-terminal Smac/DIABLO(1–15) peptide (designated SmacN) bound to Survivin, with the 2.4 Å resolution structure refined to a R-work of 20.9% and a R-free of 24.5% (Table 2). We can trace the N-terminal A1-V2-P3-I4 segment of the bound peptide in the complex (Figures 2A,B).
Table 2.
Summary of Diffraction Data and Structure Refinement Statistics of SmacN(1–15) Peptide Complexes with wild-type Survivin and Survivin(K62Y/H80W) Double Mutant.
| Summary of diffraction data | ||
|---|---|---|
| Crystal | SmacN(1–15)- Survivin |
SmacN(1–15)- Survivin(K62Y/H80W) |
| PDB code | 3UIH | 3UIJ |
| Beamline | NSLS-X29A | APS-24ID-E |
| Wavelength (Å) | 1.2900 | 0.9792 |
| Space group | C2 | C2 |
| Cell parameters | ||
| a (Å) | 114.2 | 114.4 |
| b (Å) | 71.3 | 71.0 |
| c (Å) | 81.1 | 82.3 |
| β (°) | 127.5 | 129.2 |
| Resolution (Å) | 50.0-2.4 (2.49-2.40) | 50.0-2.7 (2.80-2.70) |
| Rmerge (%) | 5.9 (59.5) | 5.5 (51.2) |
| Observed reflections | 152,848 | 43,337 |
| Unique reflections | 20,178 | 13,947 |
| Redundancy | 7.6 (6.5) | 3.1 (3.1) |
| Average I/σ(I) | 22.9 (2.6) | 23.1 (1.4) |
| Completeness (%) | 98.4 (90.0) | 99.4 (97.8) |
| Refinement and structure model | ||
| R / R free | 20.9 / 24.5 | 20.5 / 24.2 |
| Number of atoms | 2,292 | 2297 |
| Protein / Peptide | 2,212 / 55 | 2234 / 46 |
| Water | 23 | 15 |
| Zn2+ ion | 2 | 2 |
| Average B factor (Å2) | 86.8 | 91.8 |
| Protien / Peptide | 86.5 / 102.4 | 91.4 / 121.1 |
| Water | 77.8 | 69.7 |
| Zn2+ ion | 76.2 | 69.5 |
| RMS deviations | ||
| Bond lengths (Å) | 0.005 | 0.004 |
| Bond angles (°) | 0.817 | 0.728 |
Values in parentheses are for highest-resolution shell.
The N-terminal NH3+ and side chain of Ala1 in the SmacN(1–15) peptide-Survivin complex are recognized using the same intermolecular contacts (stereo view in Figure 2C) as reported above for the H3(1–15)T3ph peptide-Survivin complex (Figure 1C). The backbone of Val2 of the bound SmacN peptide forms two intermolecular hydrogen bonds involving main chain-main chain interactions in an anti-parallel β-sheet-like manner (Figure 2C). The hydrophobic ring of Pro3 of bound SmacN peptide forms van der Waals contacts with the side chains of Leu64 and His80 (Figure 2C). The segment spanning a long loop and an α-helix (Pro69 to Gly83) also moves towards the bound peptide on formation of this complex (stereo view in Figure 2D).
ITC-based Binding Affinities for Complex Formation
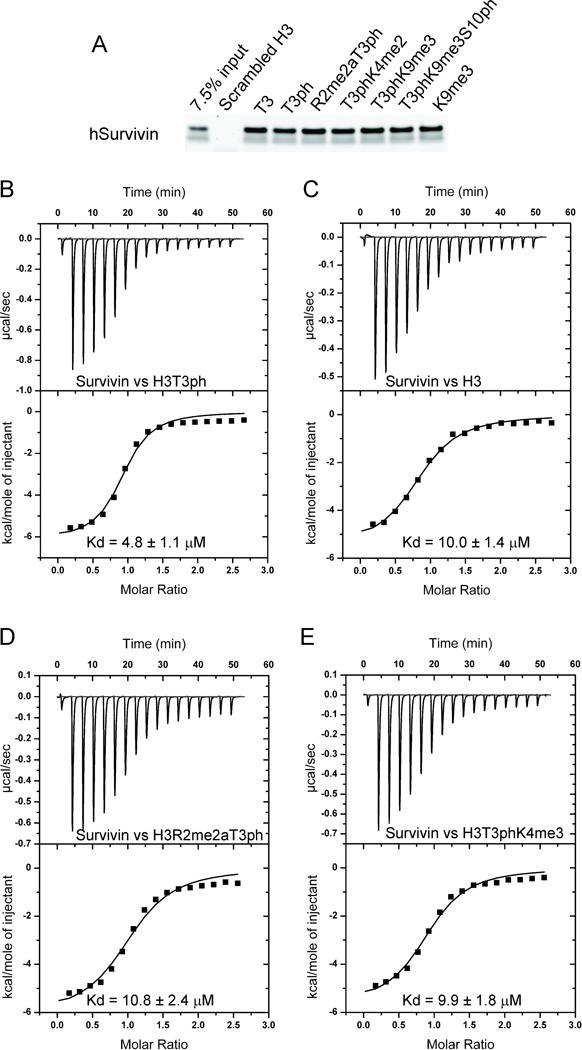
To further characterize the BIR domain of Survivin as a phosho-recognition domain, we performed pull-downs in vitro with purified human Survivin. These studies established that Survivin bound H3 and H3T3ph peptides with similar binding affinities and that the binding was not perturbed by the presence of histone modifications at R2 (dimethylation), K4 (dimethylation), K9 (trimethylation) and S10 (phosphorylation) (Figure 3A).
Figure 3. Pull-down and Isothermal Titration Calorimetry (ITC) Measurements of Binding Affinities between Survivin and H3 Peptides containing Modifications.
- A pull-down assay testing binding of human Survivin to histone H3 containing different modifications. Purified human Survivin (1–142) was incubated with the indicated peptide beads. Coomassie staining of input and bead fractions is shown.
- ITC binding curve for complex formation between H3(1–15)T3ph peptide and Survivin.
- ITC binding curve for complex formation between H3(1–10) peptide and Survivin.
- ITC binding curve for complex formation between H3(1–20)R2me2aT3ph peptide and Survivin.
- ITC binding curve for complex formation between H3(1–20)T3phK4me3 peptide and Survivin.
We have quantitated the binding parameters for the H3T3ph peptide bound to Survivin and its binding pocket mutants by isothermal titration calorimetry (ITC) (Figure 3B-3E and Table 3). We measure a binding affinity of 4.80 µM between Survivin and H3T3ph peptide (Figure 3B). We observe an approximately 2-fold drop in binding affinity for the K62A mutant (10.2 µM), which would disrupt a single hydrogen bond to the T3ph phosphate, while more pronounced drops in binding affinities are observed for the D71A (150 µM) and E76A (99 µµM) mutants, which would disrupt recognition of the N-terminus, and the E65A mutant (93 µM), which would disrupt a single hydrogen bond to the guanidinium group of Arg2 (Table 3). The binding affinities are significantly reduced for L64A, W67A and H80A (all >200 µM, too weak to be measured accurately) residues, given that the H80A mutant would disrupt Ala1 recognition, while the L64A and W67A mutants would decrease the stability of the protein, since they are embedded in the inner part of the protein. Mutation of two other acidic residues, Glu51 and Glu63, which flank the side chain of Lys4, give contrasting results, with a modest drop for the E51A mutant (14.3 µM) and a significant drop for the E63A mutant (>200 µM) (Table 3).
Table 3.
Isothermal Titration Calorimetry Binding Assays for H3T3ph Peptide Bound to Survivin and its Mutants (Buffer: 100 mM NaCl, 10 mM HEPES, pH 7.5, 2 mM β-mercaptoethanol).
| Protein | Peptide | N value | Kd (µM) | ΔH (kcal/mol) |
ΔS (cal/ mol/deg) |
|---|---|---|---|---|---|
| Wild type | H3T3ph | 0.90 ± 0.03 | 4.80 ± 1.13 | −6.13 ± 0.24 | 3.74 |
| E51A | H3T3ph | 0.97 ± 0.03 | 14.3 ± 2.2 | −6.26 ± 0.26 | 1.18 |
| K62A | H3T3ph | 0.84 ± 0.02 | 10.2 ± 1.0 | −5.16 ± 0.13 | 5.52 |
| E63A | H3T3ph | - | NDB | - | - |
| L64A | H3T3ph | - | NDB | - | - |
| E65A | H3T3ph | 0.83 ± 0.05 | 93.4 ± 6.5 | −4.89 ± 0.35 | 2.03 |
| W67A | H3T3ph | - | NDB | - | - |
| D71A | H3T3ph | 0.83 ± 0.09 | 150 ± 13 | −6.21 ± 0.80 | −3.33 |
| E76A | H3T3ph | 0.91 ± 0.06 | 99.0 ± 9.5 | −6.40 ± 0.60 | −3.13 |
| H80A | H3T3ph | - | NDB | - | - |
| Wild type | H3 | 0.86 ± 0.02 | 10.0 ± 1.4 | −5.51 ± 0.20 | 4.38 |
| K62A | H3 | 0.92 ± 0.06 | 10.8 ± 2.9 | −7.71 ± 0.64 | −3.11 |
| Wild type | H3R2me2aT3ph | 1.05 ± 0.03 | 10.8 ± 2.4 | −6.09 ± 0.30 | 2.30 |
| Wild type | H3T3phK4me3 | 0.92 ± 0.03 | 9.90 ± 1.81 | −5.69 ± 0.24 | 3.82 |
| Wild type | SmacN | 0.84 ± 0.09 | 121 ± 15 | −3.48 ± 0.49 | 6.24 |
| K62Y/H80W | H3T3ph | - | NDB | - | - |
| K62Y/H80W | H3 | 0.85 ± 0.01 | 10.6 ± 0.7 | −3.52 ± 0.06 | 10.9 |
| K62Y/H80W | SmacN | 0.82 ± 0.02 | 19.8 ± 2.3 | −4.44 ± 0.19 | 6.64 |
NDB, no detectable binding.
Unexpectedly, only a modest drop in binding affinity was observed for Survivin binding to unmodified H3 peptide (10.0 µM) (Figure 3C). Modest drops in binding affinities were also observed for binding to H3R2me2aT3ph (9.90 µM, Figure 3D) and H3T3phK4me3 (9.90 µM, Figure 3E) peptides, where residues on either side of T3ph were modified (Table 3).
Finally, Survivin bound the SmacN peptide with a binding affinity of 121 µM, which is 25-fold lower than the binding affinity to the H3T3ph peptide (4.8 µM) (Table 3).
Structural Basis for Discrimination between H3T3ph and SmacN peptides
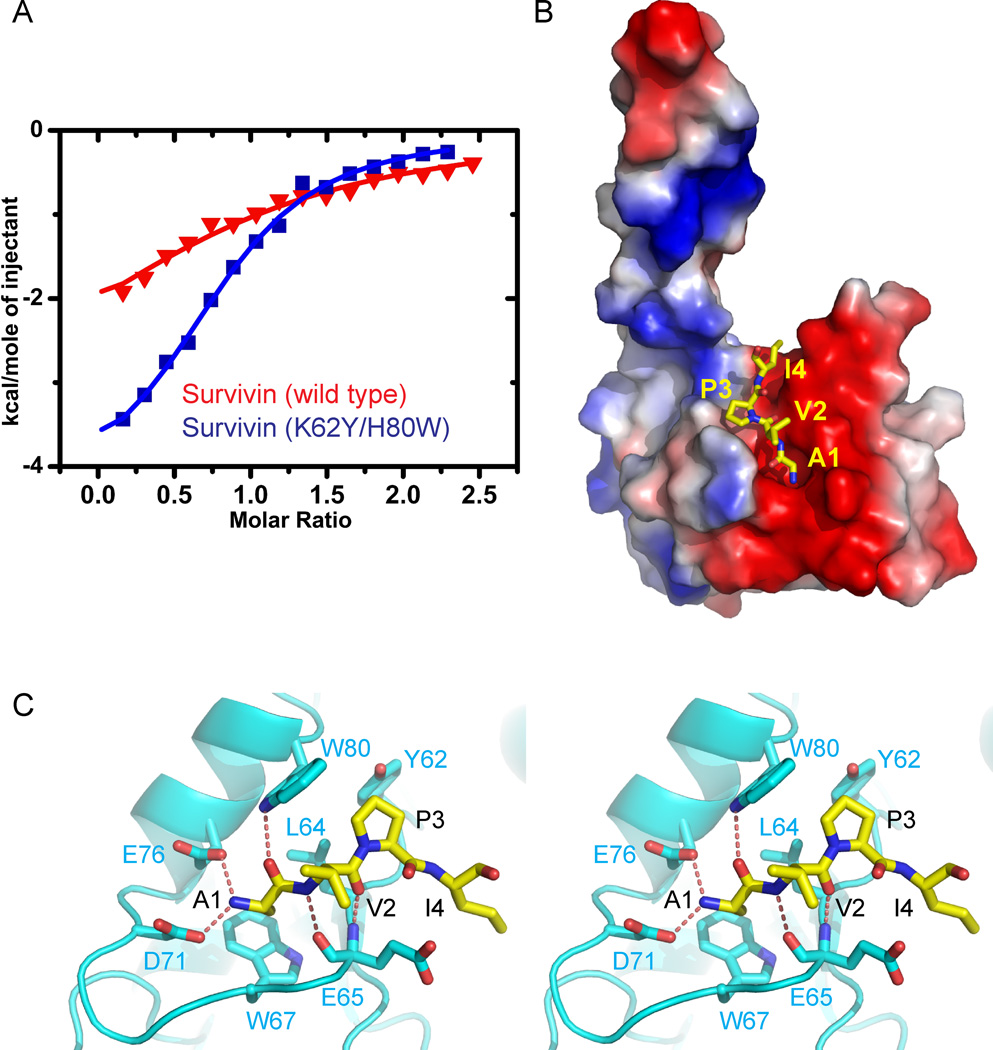
Our binding data indicate that Survivin binds to H3T3ph peptide with higher affinity than the SmacN peptide (Table 3). Although the N-terminal Ala1 of both bound peptides contributes to recognition in both complexes, there are additional intermolecular interactions involving other peptide residues in the H3T3ph peptide complex (Figure 1C) compared with its SmacN counterpart (Figure 2C). In efforts to change the specificity for peptide recognition by Survivin in favor of the SmacN complex, we introduced K62Y/H80W double mutation in Survivin, so as to increase the hydrophobic propensity of the phosphate-binding pocket. Indeed, the H3T3ph peptide no longer binds to Survivin(K62Y/H80W) double mutant, while the unmodified H3 peptide binds with unperturbed binding affinity (10.6 µM, Table 3). By contrast, SmacN peptide binds a factor of six-fold tighter to Survivin(K62Y/H80W) double mutant (19.8 µM, Table 3) when compared to wild-type Survivin (121 µM, Table 3) as plotted in Figure 4A.
Figure 4. Interactions between SmacN(1–15) Peptide and Survivin (K62Y/H80W) Mutants in the Complex.
- ITC binding curves for complex formation between SmacN(1–15) peptide and wild-type Survivin (red curve) and Survivin (K62Y/H80W) mutant (blue curve).
- An electrostatics surface representation of Survivin (K62R/H80W) mutant with bound residues A1 to I4 of SmacN(1–15) peptide (yellow) in a stick representation. The N-terminus and Ala1 insert into a negatively charged pocket, while the Pro3 ring lies on a hydrophobic surface formed by Tyr62 and Trp80.
-
Stereo view highlighting details of the intermolecular interactions between the A1-V2-P3-I4 segment of the bound SmacN(1–15) peptide and binding pocket residues of the BIR domain of Survivin (K62R/H80W) mutant. Intermolecular hydrogen-bonding interactions are designated by dashed red lines.See also Supplementary Figure 3.
The crystal structure of the SmacN(1–15)-Survivin(K62Y/H80W) double mutant complex (x-ray statistics in Table 2) reveals that the K62Y/H80W double mutation converts the positively charged T3ph binding surface from basic to hydrophobic (Figure 4B), with the large side chains of tryptophan and tyrosine form a hydrophobic pocket that readily accommodates the hydrophobic ring of Pro3 of the SmacN peptide on complex formation (stereo view in Figure 4C). The loss in binding affinity for the H3T3ph peptide must reflect both steric clashes and electrostatic incompatibility within the binding pocket of the Survivin(K62Y/H80W) double mutant. By contrast, the H3 peptide lacking a phosphate at the Thr3 position is readily accommodated in the binding pocket of the Survivin(K62Y/H80W) double mutant (x-ray statistics in Table 1) as shown in stereo in Figure S3.
DISCUSSION
Protein phosphorylation, one of the most common post-translation modifications from bacteria to humans, occurs on serine, threonine, histidine and tyrosine residues. Phosphorylation has the potential for causing modification-dependent conformational changes within the target protein, while also changing its local electrostatics environment. This in turn generates a binding site for phosphorylation mark-specific binding proteins, thereby mediating signaling pathways. Several domains are known to bind phosphorylated threonine, including the forkhead-associated (FHA) domain (Pennell et al., 2010), 14-3-3 (Schumacher et al., 2010; Rajagopalan et al., 2008) and polo-box domain (Elia et al., 2003; Yun et al., 2009) proteins. Although the structures of these three proteins exhibit considerable diversity, they use common principles for recognition, whereby protein surface pockets can accommodate the phosphorylated threonine (Figure S4). Individual oxygens of the phosphate group are coordinated by positively-charged residues, with the interactions mediated by a hydrogen-bonding network. For these proteins, the phosphate group provides the fundamental driving force for phosphorylated peptide-protein recognition, with the loss of the phosphate group impacting on complex formation (Elia et al., 2003; Pennell et al., 2010; Rajagopalan et al., 2008; Schumacher et al., 2010; Yun et al., 2009).
Structural Basis for Specificity of H3T3ph Phosphate Group Recognition
The phosphate group of H3T3ph is positioned within a positively-charged patch in Survivin (Figure 1B) and hydrogen-bonded to the side chains of Lys62 and His80 (Figures 1C), with partial conservation observed for these two residues amongst Survivin family members (Figure S5). Thus, in Schizosaccharomyces pombe, which contains two Survivin homologs, one contains the conserved Lys at position 62, while the other does not. In Xenopus laevis Survivin, His80 is replaced by an Arg, with this positively charged replacement residue capable of hydrogen-bonding with the phosphate group, indicative of a conserved function associated with recognition of phosphorylated threonine by Survivin homologs. These two residues (Lys62 and His80) are essential for target recognition in other BIR domains as well, but their unique pairing in Survivin allows for specific recognition of H3T3ph.
Unexpectedly, the binding affinity of Survivin for H3T3ph peptide (Kd = 4.8 µM, Figure 3B) is only 2-fold stronger than for unmodified H3 peptide (Kd = 10.0 µM, Figure 3C), while the K62A mutant of Survivin, that disrupts the hydrogen bond to H3T3ph (Figure 1C), binds the H3T3ph peptide with only a 2-fold reduced binding affinity (Kd = 10.2 µM, Table 3). Since both H3T3ph and H3 peptides differ in their binding affinities for Survivin by only two-fold, phosphorylation of T3 is a contributing factor, but not an absolute requirement, for recognition of N-terminal H3 peptides by Survivin.
Our binding results contrast with the 18-fold preference for H3T3ph relative to unmodified H3 for recombinant Xenopus laevis INCENP(1–58)-Survivin-Borealin subcomplex by fluorescence anisotropy (Wang et al., 2010). This difference may not be due to contributions of INCENP and Borealin to recognition, since the H3T3ph peptide only interacts with the BIR domain of Survivin without any significant contact with the INCENP fragment and/or Borealin moieties, following superposition of our H3T3p-Survivin complex structure with the INCENP-Survivin- Borealin subcomplex structure (PDB: 2QFA) (Jeyaprakash et al., 2007) (Figure S6), while the contribution of the C-terminal region of Borealin, which is not included in the structural study cannot be ruled out.
His80 appears to play a key role in the H3T3ph peptide-Survivin complex. It forms hydrogen bonds to both bridging and non-bridging phosphate oxygens of T3ph, as well as the peptide backbone (Figure 1C). It is therefore not surprising that one observes complete loss of binding affinity for the H80A mutant (Table 3).
Survivin Predominantly Recognizes N-terminal Ala1 of Bound Peptide
Survivin belongs to the IAP protein family that capitalizes on its BIR domain for protein recognition (Srinivasula and Ashwell, 2008), mediated by its IAP-binding motif (IBM) containing tetrapeptide peptide region containing a critical N-terminal Ala (Pop and Salvesen, 2009). The N-terminus of histone H3 and SmacN peptides do not exhibit much consensus in sequence, but do share the same binding mode by Survivin (Figures 1A and 2A). The only residue common to the two peptides is Ala at position 1, with the majority of Survivin residues involved in the recognition of this position strictly conserved from human to yeast, except for Leu64 and His80, which are relatively conserved and occasionally replaced by residues with similar properties in some species (Figure S5). This indicates that other residues of the peptide may be less critical for recognition by Survivin, a conclusion consistent with a previous NMR titration experiment against different SmacN derived peptides (Sun et al., 2005). Nevertheless, Arg2, Thr3ph and Lys4 of the H3 peptide exhibit significant interactions with Survivin (Figure 1C), while only Val2 of the SmacN peptide exhibits main chain-main chain interactions (Figure 2C). This disparity in the number of intermolecular interactions could explain the higher binding affinity between Survivin and H3 peptides (Kd = 4.8 µM) compared to the SmacN peptide (Kd = 121 µM) (Table 3).
The importance of N-terminal recognition of the H3T3ph peptide by Survivin is attested by the pronounced drop in binding affinities on mutating Asp71 and Glu76 to Ala (Table 3), residues involved in intermolecular hydrogen bond and salt bridge formation with the N-terminus (Figure 1C). Similarly, there is complete loss of binding affinities on mutating Leu64, Trp67 and His80 to Ala in Survivin (Table 3), residues that contact the methyl group of Ala1 (Figure 1C), and are also most likely important for formation of the hydrophobic core of the BIR domain.
Impact of Histone Modifications Adjacent to H3T3ph Site
The side chains of Arg2 and Lys4 are directed towards an acidic patch on Survivin and hence we anticipated that H3T3ph recognition by Survivin could be measurably impacted by methylation of adjacent Arg2 and Lys4 within the H3 A1-R2-T3ph-K4 sequence context. Instead, the binding affinities were reduced by only approximately 2-fold for both the dual H3R2me2aT3ph (Kd = 10.8 µM, Table 3 and Figure 3) and the dual H3T3phK4me3 (Kd = 9.9 µM, Table 3 and Figure 3) modifications. There is room in our structure of the complex (Figure 1C) to accommodate methyl groups on both Arg2 and Lys4, thereby explaining the modest decrease in binding affinities. Our results contrast with a pull down assay involving mitotic Hela cell lysate, where trimethylation of Lys4 adjacent to Thr3ph resulted in a strong diminishment of the interaction between CPC complex and histone peptide (Wang et al., 2010).
Peptide Discrimination by Survivin Binding Pocket
Our binding and structural studies on H3 and SmacN peptide binding to Survivin and its binding pocket mutants have provided insights into the factors contributing to molecular recognition. In addition to the contribution of N-terminal Ala1 to recognition, peptide position 3 and the residues that it interacts with in the binding pocket of Survivin, also contribute to recognition. Indeed, our studies establish that the observed 25-fold preference for H3T3ph peptide over SmacN peptide by wild-type Survivin (Table 3), reflecting a basic patch in the pocket that favors the negatively charged phosphate of T3ph, can be reversed in favor of SmacN peptide following incorporation of K62Y/H80W double mutation in the binding pocket of Survivin, which on forming a hydrophobic cage, favors Pro3 over the charged and bulky T3ph (Table 3). By contrast, there is no impact on the binding affinity for Thr at position 3, which binds with unchanged affinity to both wild-type and K62Y/H80W double mutant of Survivin (Table 3).
Our thermodynamic characterization of the interaction of Survivin and Survivin mutants with target peptides has demonstrated that the interaction of H3 peptides with or without phosphorylated Thr3 have comparable binding affinities. This must reflect a complex binding mode, whereby each residue within the ARTK motif of the H3 peptide makes distinct contributions. The repression of Haspin function has quite dramatic cellular effects and leads to a substantial depletion of the CPC from the centromere, an observation that contrasts with the comparable binding affinities of Survivin for H3T3ph and H3 peptides in the present. We conclude that our current structural understanding of this interaction might be insufficient for a full account of the role of Thr3 in CPC recruitment.
Comparison with a Related Contribution on Structures of Survivin Complexes
During preparation of our paper for submission, another group published their structures of human Survivin in complex with H3T3ph peptide and N-terminal peptide of human Shugoshin 1 (Jeyaprakash et al., 2011). The structures of Survivin-H3T3ph peptide complexes reported by Jeyaprakash et al., (2011) and our group are almost identical, with both groups using structural and biochemical data to invetigate the principles underlying binding specificity. In addition, our group also studied the binding between Survivin and unmodified H3 peptide and with its Smac/DIABLO binding partner. These studies highlight that the binding is dependent on Ala1, together with a preference for the third position of the peptide, which is further confirmed by an engineered mutant that reverses the binding specificity. Our studies of Survivin complexes with both H3T3ph and Smac/DIABLO peptides provide a plausible connection between apoptosis and mitosis by Survivin. The studies by Jeyaprakash et al (2011) focused on the structural role of Survivin in mitosis. They identified a potential putative Survivin-binding epitope and showed that Surivin can also bind to human Shugoshin 1 in votro, thereby raising the possibility that Survivin engages in mutually exclusive interactions with other cell cycle machinery proteins in mitosis (Jeyaprakash et al., 2011).
Summary
The research presented here defines the structural relationship between the BIR domain of human Survivin and the N-terminal tails of histone H3 and Smac/DIABLO peptides. Unexpectedly, phosphorylation of Thr3 on the histone H3 tail does not significantly enhance binding to human Survivin in vitro. Our structure-function data indicate that Survivin engages H3 and H3T3ph utilizing the same residues, which led to the identification of Survivin(K62Y/H80W) double mutant that blocks binding to H3T3ph but not H3. The interaction of human Survivin with Smac/DIABLO utilizes the same binding pocket as histone H3, suggesting that Survivin has physiological targets other than histone H3T3ph depending on the cellular context.
EXPERIMENTAL PROCEDURES
Protein Preparation
The gene encoding full length human Survivin was purchased from Open Biosystems and was inserted into a self-modified vector, which fuses an N-terminal hexa-histidine plus yeast Sumo tag to the target gene. The fusion protein was expressed in E. coli strain BL21 (DE3) RIL (Stratagene). The cells were cultured at 37 °C until OD600 reached 1.0, following which the media was cooled to 20 °C and IPTG was added to a final concentration of 0.2 mM to induce protein expression overnight. The cells were harvested by centrifuge at 4 °C and disrupted by sonication in buffer A (500 mM NaCl, 20 mM imidazole, and 50 mM Tris, pH 8.0) supplemented with 1 mM PMSF and 2 mM β-mercaptoethanol. After centrifugation, the supernatant was loaded onto a HisTrap FF column (GE Healthcare). After extensive washing by buffer A, the target protein was eluted with buffer A supplemented with 300 mM imidazole. The hexahistidine-Sumo tag was cleavage by Ulp1 protease and removed by further passing through a HisTrap FF column. The pooled target protein was further purified by a Hiload Superdex G75 26/60 column (GE Healthcare) with buffer B (150 mM NaCl, 20 mM Tris, pH 8.0, and 5 mM DTT). All the mutants were constructed using a QuikChange Mutagenesis Kit (Stratagene) and purified with the same protocol as the wild type protein. The unmodified H3(1–10), unmodified H3(1–15), H3(1–15)T3ph and SmacN (1–15) peptides were synthesized by the Tufts University peptide synthesis facility. The peptides H3(1–21)R2me2aT3ph and H3(1–21)T3phK4me3 were ordered from the Rockefeller University Proteomics Resource Center.
Crystallization and Synchrotron Data Collection
For crystallization of Survivin complexes with different peptides, the Survivin protein was concentrated to 15 mg/ml and mixed with peptides in molar ratios of 1:2 at 4 °C for 1 hour. Crystallization was carried out at 20 °C using the hanging drop vapor diffusion method by mixing 1 µl protein-peptide complex sample with 1 µl reservoir solution and equilibrated against 0.4 ml reservoir. Several conditions yielded crystallization of the complexes within 2 days. The best crystals of H3(1–15)T3ph-Survivin complex grew under conditions containing 0.1 M DL-malic acid, pH 7.0, 12% PEG 3350 while those of H3(1–10)-Survivin complex grew under conditions of 0.1 M ammonium citrate tribasic, pH 7.0, 12% PEG 3350. The best crystals of SmacN(1–15)-Survivin complex grew under conditions of 0.2 M potassium thiocyanate, 10% PEG 3350, while those of of SmacN(1–15)-Survivin K62Y/H80W mutant complex grew under conditions of 0.2 M sodium bromide, 12% PEG 3350. The best crystals of H3(1–10)-Survivin K62Y/H80W mutant complex grew under conditions of 0.2 M succinic acid, pH 7.0, 12% PEG 3350. All crystals were dehydrated by soaking into the corresponding reservoir solution that was supplemented with 20% glycerol for 2 minutes. Then the crystals were directly mounted on a nylon loop for diffraction data collection.
The data for H3(1–15)T3ph-Survivin complex were collected at NE-CAT beamline 24IDC, Advanced Photon Source (APS) at Argonne National Laboratory, Chicago. The data for H3(1–10)-Survivin complex, SmacN(1–15)-Survivin K62Y/H80W mutant complex, and H3(1–10)-Survivin K62Y/H80W mutant complex were collected at APS beamline 24ID-E. The data for SmacN(1–15)-Survivin complex were collected at beamline X29A, National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory, New York. All the data were processed with the program HKL2000 (Otwinowski Z, 1997). The statistics of the diffraction data are summarized in Table 1.
Structure Determination and Refinement
All of the structures of the complexes were solved using the molecular replacement method implemented in the program Phenix (Adams et al., 2010), using the structure of unliganded Survivin (PDB code 1F3H) (Verdecia et al., 2000) as the search model. The model building was carried out using the program Coot (Emsley et al., 2010) and structural refinement carried out using the program Phenix (Adams et al., 2010) with TLS parameters generated by the TLS Motion Determination server (Painter and Merritt, 2006). Free R-factor was calculated using 5% random chosen reflections. For the H3(1–15)T3ph-Survivin complex, the peptide had a well defined electron density and peptide residues from Ala1 to Gln5 were traced without ambiguity (Figure S7A). For the H3(1–10)-Survivin complex, the main chain of peptide from Ala1 to Lys4 could be well traced but some of the side chains exhibited poor density (Figure S7B). For the SmacN-Survivin complex, residues from Ala1 and Ile4 could be traced without ambiguity (Figure S7C). The stereochemistry of the structures were analyzed using the program Procheck (Laskowski, 1993). A summary of diffraction data and structure refinement statistics are listed in Table 1. All molecular graphics were generated with the program Pymol (DeLano Scientific LLC). Sequences were aligned using the program ClustalX (Larkin et al., 2007) and illustrated using the ESPript server (Gouet et al., 1999).
Pull-downs and Isothermal Titration Calorimetry Binding Assays
For pull-downs with purified proteins, High Capacity Streptavidin Agarose (Thermo Scientific) was used. After washing beads three times in Binding/Wash buffer (20 mM Tris-Cl, pH 7.6, 150 mM NaCl, 0.1% Triton X-100, 0.1 mM TCEP), they were incubated with 0.4 nmol of peptide per μl of beads for 2 hours at room temperature. Ten μl of beads (1 nmol peptide) were washed three times and 100 μl of protein mixture (1 uM) in Binding/Wash buffer was incubated with beads for three hours at 4°C, washed four times and eluted with 2X SDS sample buffer and gels stained with Coomassie R-250.
All the binding experiments were performed on a Microcal calorimeter ITC 200 instrument at 25 °C. First, wild-type and mutant Survivin protein samples were dialysis for 3 hours against buffer C (100 mM NaCl, 2 mM β-mercaptoethanol, and 20 mM HEPES, pH 7.5) at room temperature. Then, the protein samples were diluted with buffer C and the lyophilized peptides were dissolved in buffer C. The titration was according to standard protocol and the data were fit using the program Origin 7.0 with a 1:1 binding model.
Supplementary Material
Highlights.
Survivin primarily recognizes N-terminal Ala1 of H3, H3T3ph, and SmacN peptides.
The binding preference is determined by Lys62 and His80 side chains of Survivin.
Survivin exhibits a 25-fold preference for H3T3ph over SamcN peptide.
K62Y/H80W Survivin mutant reverses specificity in favor of SmacN peptide.
Acknowledgments
We are grateful to the staff members at APS and NSLS synchrotrons for support in diffraction data collection. This work was supported by a STARR grant to DJP and by NIH grant (R01GM075249-06A1) to HF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Codes
Coordinates and structure factors have been deposited in the RCSB Protein Data Bank with codes of 3UIG for H3(1–15)T3ph-Survivin complex, 3UII for H3(1–10)-Survivin complex, 3UIH for SmacN(1–15)-Survivin complex, 3UIJ for SmacN(1–15)-Survivin(K62Y/H80W) complex, and 3UIK for H3(1–10)-Survivin(K62Y/H80W) complex, respectively.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Chantalat L, Skoufias DA, Kleman JP, Jung B, Dideberg O, Margolis RL. Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual alpha-helical extensions. Mol. Cell. 2000;6:183–189. [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phospho-dependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash AA, Basquin C, Jayachandran U, Conti E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 2011;19:1625–1634. doi: 10.1016/j.str.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- Kanwar JR, Kamalapuram SK, Kanwar RK. Targeting survivin in cancer: patent review. Expert. Opin. Ther. Pat. 2010a;20:1723–1737. doi: 10.1517/13543776.2010.533657. [DOI] [PubMed] [Google Scholar]
- Kanwar RK, Cheung CH, Chang JY, Kanwar JR. Recent advances in anti-survivin treatments for cancer. Curr. Med. Chem. 2010b;17:1509–1515. doi: 10.2174/092986710790979935. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lens SM, Rodriguez JA, Vader G, Span SW, Giaccone G, Medema RH. Uncoupling the central spindle-associated function of the chromosomal passenger complex from its role at centromeres. Mol. Biol. Cell. 2006;17:1897–1909. doi: 10.1091/mbc.E05-08-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- Muchmore SW, Chen J, Jakob C, Zakula D, Matayoshi ED, Wu W, Zhang H, Li F, Ng SC, Altieri DC. Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell. 2000;6:173–182. [PubMed] [Google Scholar]
- Otwinowski Z, M W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 2006;39:109–111. [Google Scholar]
- Pennati M, Folini M, Zaffaroni N. Targeting survivin in cancer therapy. Expert. Opin. Ther. Targets. 2008;12:463–476. doi: 10.1517/14728222.12.4.463. [DOI] [PubMed] [Google Scholar]
- Pennell S, Westcott S, Ortiz-Lombardia M, Patel D, Li J, Nott TJ, Mohammed D, Buxton RS, Yaffe MB, Verma C, Smerdon SJ. Structural and functional analysis of phosphothreonine-dependent FHA domain interactions. Structure. 2010;18:1587–1595. doi: 10.1016/j.str.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res. 2008;36:5983–5991. doi: 10.1093/nar/gkn598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC. The Survivin saga goes in vivo. J. Clin. Invest. 2001;108:965–969. doi: 10.1172/JCI14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Ryan BM, O'Donovan N, Duffy MJ. Survivin: a new target for anti-cancer therapy. Cancer Treat. Rev. 2009;35:553–562. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244:164–171. doi: 10.1016/j.canlet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Mondry J, Thiel P, Weyand M, Ottmann C. Structure of the p53 C-terminus bound to 14-3-3: implications for stabilization of the p53 tetramer. FEBS Lett. 2010;584:1443–1448. doi: 10.1016/j.febslet.2010.02.065. [DOI] [PubMed] [Google Scholar]
- Song Z, Yao X, Wu M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 2003;278:23130–23140. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ashwell JD. IAPs: what's in a name? Mol. Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Nettesheim D, Liu Z, Olejniczak ET. Solution structure of human survivin and its binding interface with Smac/Diablo. Biochemistry. 2005;44:11–17. doi: 10.1021/bi0485171. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 2000;7:602–608. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- Yue Z, Carvalho A, Xu Z, Yuan X, Cardinale S, Ribeiro S, Lai F, Ogawa H, Gudmundsdottir E, Gassmann R, et al. Deconstructing Survivin: comprehensive genetic analysis of Survivin function by conditional knockout in a vertebrate cell line. J. Cell. Biol. 2008;183:279–296. doi: 10.1083/jcb.200806118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SM, Moulaei T, Lim D, Bang JK, Park JE, Shenoy SR, Liu F, Kang YH, Liao C, Soung NK, et al. Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1. Nat. Struct. Mol. Biol. 2009;16:876–882. doi: 10.1038/nsmb.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.