Abstract
Expression of the Cat-1 gene (cationic amino acid transporter-1) is induced in proliferating cells and in response to a variety of stress conditions. The expression of the gene is mediated via a TATA-less promoter. In the present study we show that an Sp1 (specificity protein 1)-binding site within a GC-rich region of the Cat-1 gene controls its basal expression and is important for induction of the gene during the UPR (unfolded protein response). We have shown previously that induction of Cat-1 gene expression during the UPR requires phosphorylation of the translation initiation factor eIF2α (eukaryotic initiation factor 2α) by PERK (protein-kinase-receptor-like endoplasmic reticulum kinase), one of the signalling pathways activated during the UPR. This leads to increased translation of the transcription factor ATF4 (activating transcription factor 4). We also show that a second signalling pathway is required for sustained transcriptional induction of the Cat-1 gene during the UPR, namely activation of IRE1 (inositol-requiring enzyme 1) leading to alternative splicing of the mRNA for the transcription factor XBP1 (X-box-binding protein 1). The resulting XBP1s (spliced XBP1) can bind to an ERSE (endoplasmic-reticulum-stress-response-element), ERSE-II-like, that was identified within the Cat-1 promoter. Surprisingly, eIF2α phosphorylation is required for accumulation of XBP1s. We propose that the signalling via phosphorylated eIF2α is required for maximum induction of Cat-1 transcription during the UPR by inducing the accumulation of both ATF4 and XBP1s.
Keywords: activating transcription factor 4 (ATF4), cationic amino acid transporter-1 (Cat-1), endoplasmic reticulum stress, specificity protein 1 (Sp1), unfolded protein response, X-box-binding protein 1 (XBP1)
INTRODUCTION
Cat-1 (cationic amino acid transporter-1) is a member of the CAT protein family, which mediates the Na+-independent transport of cationic amino acids. Cat-1 mediates bidirectional transport of arginine and lysine by facilitated diffusion [1]. It is expressed ubiquitously except in the adult liver. However, its expression varies in different tissues and cell types [2,3]. Transcription of the Cat-1 gene is modulated by ER (endoplasmic reticulum) stress, availability of nutrients, cell proliferation, growth factors and hormones [2,4]. Cat-1 supports vital metabolic functions, such as synthesis of proteins, polyamines and NO (reviewed in [1]).
During normal/unstressed conditions, low levels of Cat-1 are expressed by transcription of a TATA-less promoter within the 1.4 kb region upstream of the transcription start site [5]. Recently, Purα (purine-rich-binding protein A) was shown to bind to an INE (intronic enhancer element) within the first intron of the gene and to positively regulate promoter activity in the absence of stress [6]. However, the promoter sequence that drives transcription of this important gene remains unknown. The transcription start site of the Cat-1 gene has to be tightly controlled because it is important in generating the 5′-UTR (untranslated region) that regulates translation of the Cat-1 mRNA [7,8]. Therefore studies uncovering the mechanism of transcription start site selection in the TATA-less Cat-1 gene can be of great importance.
Many genes transcribed by Pol II (RNA polymerase II), especially housekeeping genes, lack a TATA box (reviewed in [9]). Some TATA-less promoters have GC-rich regions that initiate transcription at multiple sites. GC-rich elements serve as binding sites for members of the Sp (specificity protein) family, as well as Egr-1 (early growth response factor-1) [9,10]. Sp1 is a ubiquitously expressed protein belonging to the family of mammalian Sp/XKLF (X Krüppel-like factor) transcription factors characterized by their zinc-finger domains (reviewed in [11,12]). The consensus sequence for Sp1 binding is 5′-(G/T)GGGCGG(G/A)(G/A)(C/T)-3′ or 5′-(G/T)(G/A)-GGCG(G/T)(G/A)(G/A)(C/T)-3′. Sp1-binding sites are found in numerous genes including genes lacking TATA-box elements. Survivin, α1-soluble guanylate cyclase and neurogranin are examples of genes that have TATA-less promoters and require Sp1 for basal transcription [11,12]. Sp1 facilitates the binding of TFIID (transcription factor IID) to TATA-less promoters by interaction with TAFs (transcription-associated factors) or other transcription factors (reviewed in [9]).
In the present study, we identified Sp1 and XBP1s (spliced X-box-binding protein 1) as transcription factors that modulate expression of the Cat-1 gene. We characterized the minimal promoter sequence of the gene and the adaptive regulation of the promoter during ER stress by an ERSE-II (endoplasmic-reticulum-stress-element-II)-like element in the promoter. It is shown that a weak TATA-less promoter adapts to ER stress by recruiting ATF4 (activating transcription factor 4) early in the stress response (0–6 h) followed by recruitment of XBP1s for sustained induction during prolonged stress. All three factors, Sp1, ATF4 and XBP1s, are required for efficient transcription of the Cat-1 gene during ER stress. The Cat-1 promoter-regulatory unit described in the present paper is probably a key factor in the adaptation of Cat-1 gene expression to stress.
Synthesis, modification and folding of secretory or membrane-bound proteins occurs in the ER. The accumulation of unfolded or aggregated proteins in this organelle results in ER stress and activates the UPR (unfolded protein response) (reviewed in [13,14]). In mammalian cells, this response activates PERK [protein-kinase-receptor-like ER-localized eIF2α (eukaryotic initiation factor 2α) kinase], IRE1 (inositol-requiring enzyme 1) and ATF6. PERK activation phosphorylates the α subunit of eIF2 at Ser51, which causes a decrease in global mRNA translation initiation and an increase in translation of the ATF4 mRNA [15]. Upon activation, IRE1, a site-specific endoribonuclease in the ER membrane, cleaves the XBP1 mRNA leading to a spliced mRNA that encodes the potent bZIP (basic leucine zipper) transcription factor XBP1s [16,17]. ATF6 is released from the Golgi apparatus by specific proteases [18]. By binding to their respective target sequences, these transcription factors induce synthesis of ER resident proteins to assist in protein folding or ERAD (ER-associated protein degradation) to restore homoeostasis within the ER. However, under prolonged stress, cells may fail to recover and ultimately are directed towards the apoptotic pathway [19].
It has been demonstrated that Cat-1 mRNA increases during ER stress [5], but the mechanism of this induction is unknown. As cells within growing tumours experience ER stress, due to the hypoxic environment [20], and because actively proliferating tumour cells have elevated Cat-1 gene expression [21], we sought to identify the mechanism of Cat-1 gene transcriptional control during ER stress. We identified and characterized the Cat-1 minimal promoter and demonstrated the importance of Sp1 in Cat-1 gene transcription. We also identified a DNA element within the promoter that recruits XBP1s to sustain induction of Cat-1 mRNA levels during ER stress. Efficient induction mediated by XBP1s required eIF2α phosphorylation. We conclude that the signalling pathway of eIF2α phosphorylation and cis-DNA regulatory elements function in concert to ensure proper transcription of the Cat-1 gene during diverse stress conditions.
EXPERIMENTAL
Cell culture and DNA transfection
Cells were cultured in high-glucose DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine under a humidified atmosphere of 5 % CO2 at 37 °C. C6 rat glioma cells were cultured in medium supplemented with 5 % (v/v) heat-inactivated FBS (fetal bovine serum) and 5 % (v/v) calf serum. MEFs (mouse embryonic fibroblasts) with or without homozygous deletions of the ATF4 gene [22], XBP1 gene [23], WT (wild-type) eIF2α (S/S), and eIF2α-S51A (A/A) cells (gifts from R. Kaufman [24]) were grown in medium supplemented with 10 % (v/v) FBS. The S51A is a point mutation in the eIF2α protein at Ser51, which is a residue that is phosphorylated by stress-induced kinases. Embryonic stem cells with homozygous deletions of the Sp1 gene (a gift from J.M. Boss, Department of Microbiology and Immunology, Emory University School of Medicine, GA, U.S.A.) were grown in α-modified DMEM supplemented with 5 % (v/v) FBS, 1 mM L-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, 1 ng/ml basic fibroblast growth factor and 4 μg/ml insulin [25]. As a WT line was not available, MEFs from the same genetic background were used as a control and were cultured in the same conditions as the Sp1-null cells. Fugene 6 HD (Roche Applied Science) was used to transfect cultured cells according to the manufacturer’s instructions. Expression plasmids for β-gal (β-galactosidase) were co-transfected to monitor transfection efficiency [4]. LUC (firefly luciferase) and β-gal activities were measured as described previously [4]. Cells were starved of amino acids by incubating in KRB (Krebs–Ringer bicarbonate) solution (Sigma–Aldrich) supplemented with 10 %(v/v) dialysed FBS [4,26]. ER stress was induced in cells by incubating in the corresponding serum-containing medium in the presence of 400 nM thapsigargin as described previously [4,27].
Plasmid constructs
The CMVmin (cytomegalovirus minimal promoter) and PA1.4/Cat-1 5′-UTR LUC reporters (containing 1.4 kb of the Cat-1 promoter) were constructed as described previously [4,5]. The promoterless vector was generated by re-ligation of the vector remaining after digestion of CMVmin with XhoI and EcoRI. Promoterless/Cat-1 5′-UTR was generated by PCR-directed mutagenesis to remove the 1.4 kb sequence upstream of the Cat-1 exon1. The 5′-end truncation constructs were generated by PCR using PA1.4/Cat-1 5′-UTR as a template and were inserted between the XhoI and NcoI sites in the CMVmin plasmid. Mutations in these vectors were generated using PCR-directed mutagenesis. Regions of the Cat-1 promoter (from −63 to −1, relative to the transcription start site) without or with mutations in GC-rich regions were cloned in a vector lacking any promoter activity (pGL3-Basic; Promega), to generate constructs 13 and 13m respectively. Mutations of the ERSE-II-like element of construct 1 (from −25 to −14; 5′-ATTGGTGCCTGG-3′→5′-GCCAATAAATGG-3′) created construct 1m. The expression vector for Sp1 was from Dr Duna Massillon (Department of Nutrition, School of Medicine, Case Western Reserve University, Cleveland, OH, U.S.A.), that for ATF4 was from Dr David Ron (Kimmel Center for Biology and Medicine of the Skirball Institute, New York University School of Medicine, New York, U.S.A.), and that for XBP1u (unspliced XBP1), XBP1s, IRE1α and ATF6 were from Dr Randal Kaufman (Department of Biological Chemistry and Internal Medicine, University of Michigan Medical Center, Ann Arbor, MI, U.S.A.).
EMSA (electrophoretic mobility-shift assay)
Double-stranded DNA oligonucleotides containing the Cat-1 basal promoter sequence, Cat-1 (WT) and Cat-1 mutant (Sp1 MUT), 5′-GGTGTCCCCGCCCACAGGGGCGCGGCCGCG-3′ and 5′-GGTGTCCCCTTTAAATTTTTCGCGGCCGCG-3′ respectively (underlined sequences denote mutated nucleotides), were radiolabelled with [γ-32P]ATP using T4 polynucleotide kinase. For each binding reaction, 5 μg of C6 nuclear extract or 50 ng of recombinant Sp1 protein was incubated in 40 mM Tris/HCl, pH 7.5, containing 20 % (v/v) glycerol, 5 mM MgCl2, 100 mM NaCl, 0.01 % IGEPAL CA-630 (Sigma–Aldrich), 1 mM dithiothreitol and 20 mg/ml poly(dI-dC) · (dI-dC) for 1 h at 4 °C. Competition assays were performed using a 50-fold excess of unlabelled WT or Sp1 MUT oligonucleotides. Products were resolved on 4 % non-denaturing polyacrylamide gels, dried and analysed using the Storm Phosphorimager system (GE Healthcare).
ChIP (chromatin immunoprecipitation) analysis
ChIP analysis was performed on nuclear extracts as described in [26] using normal IgG or antibodies against RNA polymerase II (N-20), Sp1, ATF4 or XBP1 (Santa Cruz Biotechnology). Immunoprecipitated and purified DNA fragments were analysed by PCR. The primers used for were: Cat-1 promoter, 5′-TCGG-TTGGGGCTGCTGAGGACCAA-3′ (forward) and 5′-TTTCAT-CAGCCGCGCGCCGCCCT-3′ (reverse); Cat-1 exon 13, 5′-AG-CAAACCTGAGCAGTAAAGTGCT-3′ (forward) and 5′-CG-GACTTAATCTAATGTCATTGTA-3′ (reverse); and Cat-1 exon1 AARE (amino-acid-response element) and ERSE-II-like, 5′-TCGGTTGGGGCTGCTGAGGACCAA-3′ (forward) and 5′-TTTCATCAGCCGCGCGCCGCCCT-3′ (reverse).
RT (reverse transcription)–PCR and qRT-PCR (quantitative real-time PCR) analysis
cDNAs were synthesized from RNA samples using Superscript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen) as described previously [26,27]. Real-time PCR was performed using an iCycler (Bio-Rad) and SYBR GreenER qPCR SuperMix for the iCycler (Invitrogen) according to the manufacturer’s instructions. The primers used were: 18S, 5′-CAACAA-CTGGGCTAAGGGTCACTAC-3′ (forward) and 5′-CACCACA-TCCAAGACAGAGTCAACC-3′ (reverse); GAPDH (glyceralde-hyde-3-phosphate dehydrogenase), 5′-ACTTTGGCATCGTGG-AAGGG-3′ (forward) and 5′-TCATCATACTTGGCAGGTT-TCTCC-3′ (reverse); and Cat-1: 5′-CTTTGGATTCTCTGGTGT-CCTGTC-3′ (forward) and 5′-GTTCTTGACTTCTTCCCCT-GTGG-3′ (reverse).
Other methods
Total cell and nuclear extracts were prepared as described previously [7,27]. Proteins were detected via Western blotting with primary antibodies against ATF4 (cat. no. sc-200), XBP1 (cat. no. sc-7160) and CHOP [C/EBP (CCAAT/enhancer-binding protein)-homologous protein; (cat. no. sc-7351)] from Santa Cruz Biotechnology, tubulin (cat. no. T9026) from Sigma–Aldrich, eIF2α prepared by Quality Controlled Biochemicals and phospho-eIF2α (eIF2α-P; cat. no. 9721) from Cell Signaling.
RESULTS
Nucleotides from −63 to −25 of the Cat-1 gene promoter are essential for basal expression
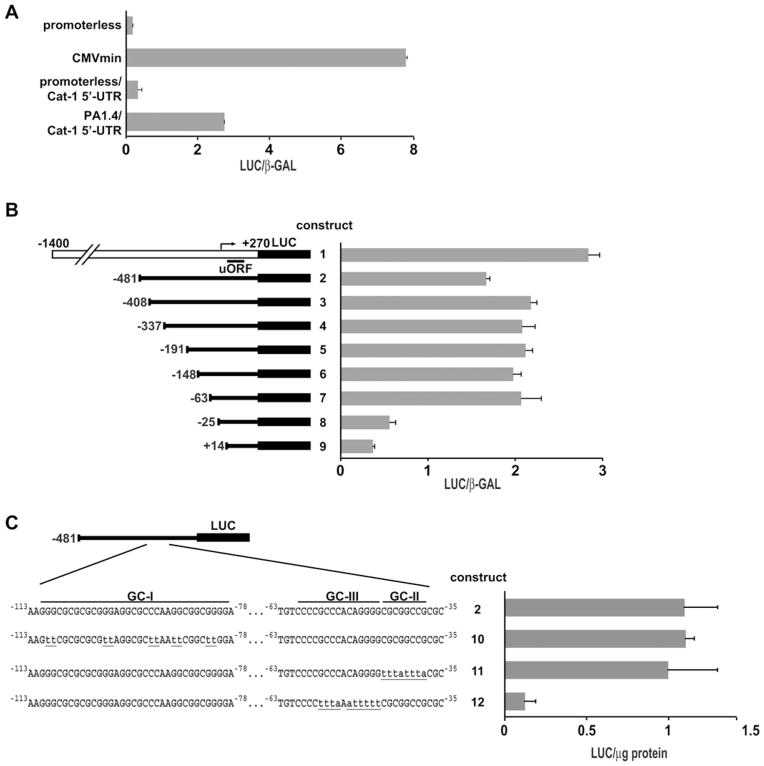
The presence of a dominant transcription start site for the rat Cat-1 gene and the absence of a TATA box [5] suggested that a cis-DNA element may mediate Cat-1 gene transcription initiation by binding a member of the Sp/XKLF family of transcription factors. To test this hypothesis, we first compared the strength of the Cat-1 gene promoter with the CMVmin-TATA box-containing promoter (Figure 1A) and utilized the chimaeric construct reported previously [26] that contains 1.4 kb of genomic DNA upstream of the Cat-1 transcription start site and 270 bp of the 5′-UTR containing the first three exons linked to a LUC reporter (Figures 1A and 1B, PA1.4/Cat-1 5′-UTR and construct 1). We showed previously that the first exon of the gene contains regulatory elements for transcriptional control during stress [26]. Promoter activities were determined by LUC assays in transiently transfected C6 rat glioma cells. LUC activity in cells transfected with the Cat-1 promoter construct was higher than with the promoterless or the promoterless/Cat-1 5′UTR-containing constructs (Figure 1A). However, LUC activity was 2.8-fold lower than with the CMVmin promoter harbouring a typical TATA box (Figure 1A). These results suggest that the 1.4 kb genomic region of the Cat-1 gene contains a weak TATA-less promoter, which is in agreement with the weak Cat-1 expression in fed cells [5].
Figure 1. A GC-rich motif within the Cat-1 gene promoter is required for transcription.
(A–C) The indicated vectors were transfected into C6 glioma cells along with an expression plasmid for β-gal (β-GAL) to normalize for transfection efficiency [except in (C) where protein content was used for normalization]. Enzymatic activities were measured in cell extracts 48 h post-transfection. Results are means ± S.E.M. for three independent experiments. uORF, upstream open reading frame.
To define the region of the Cat-1 promoter necessary for basal expression, we made a series of 5′-end truncation constructs and tested their activity in C6 cells. The promoter activity severely decreased in constructs that contained less than 63 nucleotides upstream of the transcription start site (Figure 1B, constructs 8 and 9). These results indicate that the GC-rich region between −63 and −25 is necessary for basal promoter activity. To further examine the relevance of this GC-rich region for promoter activity, we generated mutations in the three GC boxes within the (−481)/5′-UTR construct (Figure 1C, constructs 2, 10, 11 and 12). The promoter activity decreased in the construct with mutations in the sequence from −56 to −46 (GCCCACAGGGG; construct 12), but not with mutations in the other GC boxes. We conclude that the sequence between −63 and −35 constitutes the Cat-1 basal promoter and the GC-box at the 5′-end of this sequence is required to support basal transcription.
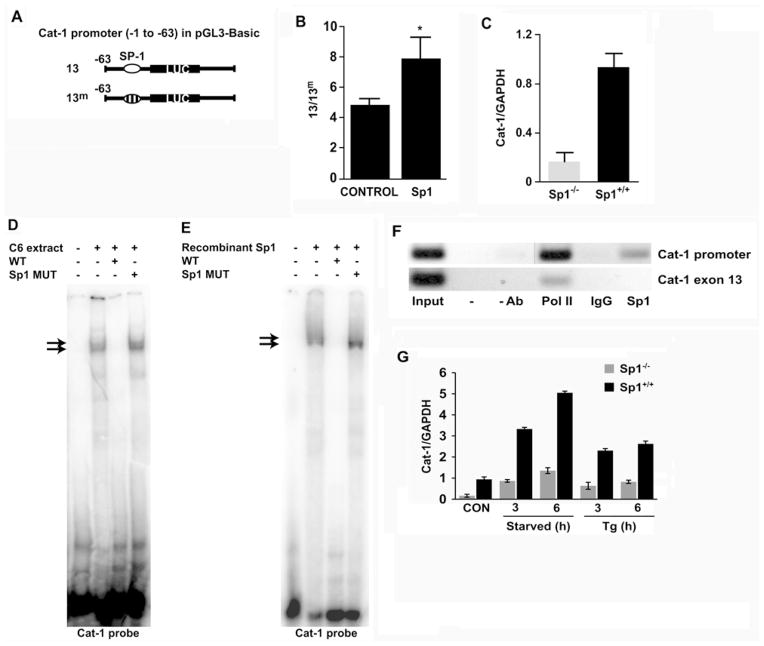
Sp1 binds the Cat-1 minimal promoter element both in vitro and in vivo
To determine the transcription factors that bind to the basal promoter, we scanned the −63 to −25 region using MatInspector software (Genomatix). A putative Sp1-binding site was identified in the promoter region corresponding to the GC-box that is required for promoter activity. To investigate the role of Sp1 in Cat-1 gene transcription driven by this sequence, we cloned the fragment (−63 to −1) into the promoterless pGL3-Basic vector (construct 13) and compared its activity with a construct with a mutation in the putative Sp1-binding site (construct 13m) (Figure 2A). In order to specifically examine the effect of the introduced sequences and not the sequences of the cloning vector, the results were expressed as the ratio of LUC activity from the cells transfected with construct 13 to 13m. C6 cells transfected with these constructs showed a ratio of approx. 5-fold difference between construct 13 and 13m (Figure 2B, Control). Furthermore, Sp1 overexpression in C6 cells caused an additional increase in the ratio of 13/13m (Figure 2B, Sp1). To further determine the significance of Sp1 in regulating Cat-1 gene transcription, we compared the levels of the Cat-1 mRNA in Sp1−/− and WT cells using qRT-PCR. The Cat-1 mRNA levels in the Sp1−/− cells were 17 %of that in the WT cells (Figure 2C); Western blotting for Cat-1 protein using cell extracts derived from these cell lines showed comparable results (results not shown). These results suggest that Sp1 is required for the basal transcription of the Cat-1 gene.
Figure 2. Efficient transcription in the Cat-1 gene promoter is mediated via a Sp1-binding site.
(A) Diagramatic representation of constructs 13 and 13m as used in the present study. (B) C6 cells were transfected with 100 ng of construct 13 or 13m without (CONTROL) or with 100 ng of Sp1 expression plasmids in cells plated in six-well plates. The total amount of transfected DNA was kept constant with the addition of non-specific DNA (pcDNA 3.1). LUC assays were performed 48 h post-transfection and values were normalized to protein content. Results are means ± S.E.M. expressed as the ratio of 13/13m. *P < 0.05 between Sp1 and CONTROL samples. (C) Cat-1 mRNA levels in Sp1−/− and Sp1+/+ cells. Results from qRT-PCR analysis of total RNA using gene-specific primers were normalized to the GAPDH mRNA signal. (D and E) EMSAs were performed by incubating a 32P-labelled double-stranded oligonucleotide containing the Sp1-binding site of the Cat-1 gene with nuclear extracts from (D) C6 cells or (E) cells expressing recombinant Sp1. Competition assays were performed with unlabelled Cat-1 (WT) or Cat-1 mutant (Sp1 MUT) oligonucleotides as indicated. (F) ChIP was performed using C6 cells with antibodies against Pol II and Sp1. Samples without antibody (-Ab) or with normal rabbit IgG were used as negative controls. PCR was performed with primer sets specific for the regions of interest (Cat-1 promoter or exon 13). (G) Cat-1 mRNA levels in Sp1−/− and Sp1+/+ either untreated amino-acid-fed (CON), amino-acid-starved (Starved) or thapsigargin (Tg)-treated for the indicated time. The CON condition involved serum-containing medium that was used to grow the Sp1 WT and Sp1−/− cells (see the Experimental section). Thapsigargin treatment was performed in the same medium, as described in the Experimental section. The amino-acid-starved condition was employed by incubating the cells in KRB supplemented with 10 % (v/v) dialyzed FBS. (B, C and G) Results are means ± S.E.M. for three independent experiments; (D–F) a representative gel from three independent experiments is shown.
We next determined the sequence-specific binding of Sp1 to a 32P-labelled double-stranded oligonucleotide containing the putative Sp1-binding site using EMSA (Figure 2D). Competing oligonucleotides were used to confirm the specificity of the complexes. The appearance of slowly migrating complexes was observed when nuclear extracts from C6 cells were incubated with an oligonucleotide containing the Cat-1 Sp1-binding site (Figure 2D). These complexes were effectively competed by an excess of unlabelled WT, but not mutant (Sp1 MUT), oligonucleotide (Figure 2D). In addition, similar complexes were observed when recombinant Sp1 protein was incubated with the WT oligonucleotide (Figure 2E, compare lane 2 with lane 2 of Figure 2D). Again, these complexes were effectively competed by an excess of unlabelled WT, but not Sp1 MUT oligonucleotide (Figure 2E).
To obtain direct proof of Sp1 binding to the Cat-1 promoter in vivo, we performed ChIP analysis. Chromatin from C6 glioma cells was immunoprecipitated with antibodies against Sp1, Pol II or IgG, and DNA fragments containing the Sp1-binding site within the promoter region or from exon 13 of the Cat-1 gene (used as control) were amplified by PCR (Figure 2F). In agreement with the EMSA results, Sp1 bound only the minimal promoter region and not DNA from exon 13 of the gene. Results from three independent ChIP experiments showed that DNA precipitated by Sp1 containing the minimal promoter region was 3.5 ± 0.1-fold that of the level of DNA precipitated by normal IgG (results not shown). Pol II was present within the Cat-1 exon 13 demonstrating that the absence of Sp1 within this region is not due to difficulty in PCR amplification. Furthermore, lack of PCR amplification in both regions from samples with no antibody or immunoprecipitated with normal IgGs confirmed the specificity of the immunoprecipitation. These results clearly demonstrate that the Cat-1 promoter contains a functional Sp1-binding site.
We also determined the importance of Sp1 in Cat-1 gene transcription during stress conditions that we have shown previously to induce expression of the Cat-1 gene [5]. Induction of Cat-1 mRNA during either thapsigargin-induced ER stress/UPR or amino acid deprivation were similar between WT and Sp1−/− cells when compared with the corresponding untreated control cells. However, the absolute amount of Cat-1 mRNA during stress conditions was at least 2-fold lower in cells lacking Sp1 compared with WT cells (Figure 2G). These results indicated that Sp1 is dispensable for Cat-1 mRNA induction during stress conditions, but is necessary for maximal induction of Cat-1 mRNA levels under these conditions.
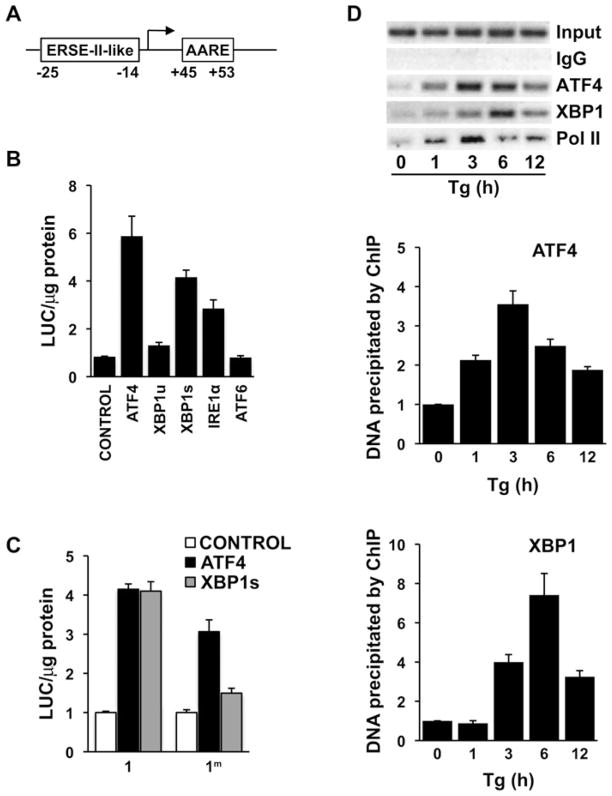
XBP1 confers regulation via a DNA element in the promoter
Our previous studies have shown that Cat-1 mRNA levels are induced during the UPR [5,6,27]. We sought to determine the transcription factors that control Cat-1 gene transcription during the UPR. As the Cat-1 gene contains a sequence that binds ATF4 (Figure 3A; AARE), we expected induction of the Cat-1 gene transcription during the UPR to involve ATF4. However, we also noticed the sequence from −25 to −14 (ATTGGTGCCTGG) within the minimal promoter that resembled the ERSE-II element (ATTGGNCCACG) of the human HERP (homocysteine-induced endoplasmic reticulum protein) gene, which is highly inducible by the UPR [28]. This DNA element mediates induction of HERP gene transcription by both ATF6 and XBP1 transcription factors during the UPR [29]. We therefore tested whether the Cat-1 gene promoter is regulated by ATF6 and XBP1 and if the putative ERSE-II-like element (ERSE-II-like) is involved in this regulation. Construct 1 (Figure 1B), containing the LUC reporter under the control of the Cat-1 gene promoter, was co-transfected with expression vectors for factors involved in the UPR: ATF4, XBP1 (both spliced and unspliced forms), active ATF6 and IRE1α, the protein that generates the spliced form of the XBP1 mRNA (XBP1s) from its unspliced counterpart (XBP1u). Their effects on reporter construct transcription were monitored by LUC activity (Figure 3B). LUC activities from construct 1 increased when cells were co-transfected with ATF4, XBP1s or IRE1α, but not with XBP1u or ATF6. These results suggest that the Cat-1 promoter contains response elements that bind ATF4 and XBP1s. We have shown previously that the AARE (Figure 3A) binds ATF4 and induces Cat-1 gene transcription during amino acid starvation [26]. We therefore determined whether XBP1s regulates the putative ERSE-II-like element. Construct 1 or a mutant lacking the ERSE-II-like sequence (construct 1m) was co-transfected with ATF4 or XBP1s expression vectors into C6 cells and LUC activities were measured (Figure 3C). As expected, both ATF4 and XBP1s increased LUC activity from the WT construct (Figure 3C, construct 1). However, mutations of the ERSE-II-like element abolished XBP1s-mediated induction and had only a minor effect on ATF4-mediated transcription. The sustained induction in the mutant construct by ATF4 could be explained by the presence of the AARE (Figure 3A). Therefore XBP1s and not ATF4 is responsible for the induction of Cat-1 gene transcription via the ERSE-II-like element.
Figure 3. XBP1s and ATF4 regulate Cat-1 gene transcription during ER stress.
(A) Representation of the ERSE-II-like and the AARE sequences within the Cat-1 gene. Numbers indicate the position of the element relative to the transcription start site. (B) C6 glioma cells were transiently transfected with 100 ng of construct 1 or (C) construct 1 or 1m and 100 ng of the indicated expression vectors in six-well plates. Control indicates the cloning vector which was used as a negative control. LUC assays were performed 48 h post-transfection as described in the Experimental section. Results were normalized to protein content and are means ± S.E.M. for three independent experiments. (D) ChIP was performed using C6 cells and the indicated antibodies. Thapsigargin (Tg)-treatment was performed in the growth medium for C6 cells as described in the Experimental section. Samples with normal rabbit IgG were used as negative controls. PCR was performed using the primer pair indicated in the Experimental section; a representative gel from three independent experiments is shown in the top panel. DNA isolated from ChIP analyses using anti-ATF4 (middle panel) and anti-XBP1 (bottom panel) antibodies from three independent experiments were also analysed by qRT-PCR. Values were obtained relative to input DNA and expressed as a fold change relative to untreated cells.
To demonstrate binding of ATF4 and XBP1s to the Cat-1 promoter in vivo during ER stress, a ChIP assay was performed using C6 cells treated with thapsigargin to induce the UPR (Figure 3D). Chromatin fragments immunoprecipitated with antibodies against ATF4, XBP1 and Pol II were amplified by PCR using primers flanking the region encompassing the AARE and the ERSE-II-like elements. ATF4 binding increased after 1 h of ER stress, showed maximum binding at 3 h and declined thereafter, while maintaining a level above unstressed conditions (Figure 3D, middle panel). Maximum binding of XBP1s was observed after 6 h of ER stress (Figure 3D, bottom panel). Pol II showed a binding pattern similar to ATF4. IgG was used as a negative control for the ChIP analysis and showed no detectable binding. Our results suggest that both transcription factors, ATF4 and XBP1s, contribute to Cat-1 gene transcriptional control during ER stress. Furthermore, the kinetics of recruitment of these transcription factors indicate that ATF4 bound first, followed by XBP1s.
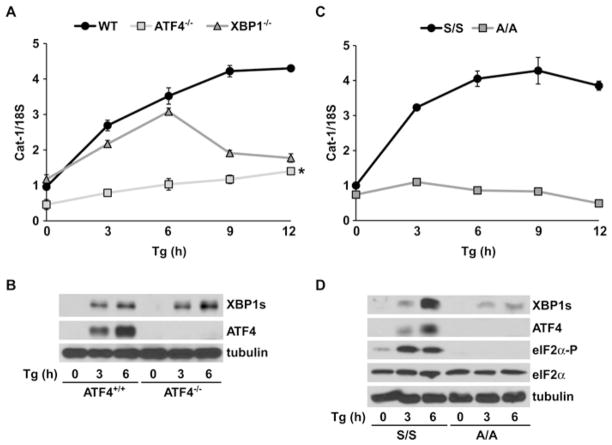
XBP1 is required for sustained induction of Cat-1 gene transcription during ER stress in a manner dependent on phosphorylation of eIF2α
To further support the role of ATF4- and XBP1s-mediated transcriptional induction of the Cat-1 gene during ER stress, we monitored the change in its mRNA level by qRT-PCR in MEFs lacking either ATF4 or XBP1 (Figure 4A). In WT MEFs, we observed the expected increase of 4.5-fold in Cat-1 mRNA levels after 6–12 h of ER stress [27]. In contrast, the levels of Cat-1 mRNA in ATF4−/− MEFs were induced less than 2-fold in the early hours of ER stress. This is consistent with the idea that binding of ATF4 to the AARE during ER stress is important for Cat-1 gene regulation (Figure 3C). Furthermore, the 3-fold induction of the Cat-1 mRNA levels after 6–12 h of stress in ATF4−/− MEFs was probably due to binding of XBP1 to the ERSE-II-like element of the Cat-1 gene promoter. In agreement with this idea, the levels of XBP1s protein in ATF4−/− MEFs were similar to WT MEFs (Figure 4B). In contrast, Cat-1 mRNA levels in XBP1−/− MEFs increased 3-fold by 6 h with a rapid decline thereafter, suggesting a role of XBP1 in sustaining transcription of the Cat-1 gene between 6 and 12 h of ER stress. As ATF4 levels are similar in WT and XBP1−/− MEFs (results not shown), we conclude that ATF4 stimulates transcription during early stress and XBP1 sustains transcription at later times.
Figure 4. Cat-1 mRNA accumulation during ER stress requires ATF4 and XBP1, the levels of which depend on eIF2α phosphorylation.
Quantification of Cat-1 mRNA levels in (A) Thapsigargin (Tg)-treated WT, ATF4−/− and XBP1−/− (C) or S/S (eIF2α-WT) and A/A (eIF2α-S51A) MEFs. Results are from qRT-PCR analysis of total RNA using gene-specific primers and were normalized to the 18S ribosomal RNA signal. *P < 0.05 between 0 and 12 h of Tg treatment for ATF4−/− MEFs compared with WT. Results are means ± S.E.M. for three independent experiments. (B and D) Western blot analysis of total cell extracts from the indicated Tg-treated MEFs using antibodies against the the indicated proteins. A -representative gel from three independent experiments is shown. All Tg-treatments were performed in DMEM supplemented with 10 % (v/v) FBS, as described in the Experimental section.
In order to further support the requirement of XBP1 for Cat-1 gene transcription during ER stress, we measured the Cat-1 mRNA levels in MEFs deficient in eIF2α phosphorylation. It is well known that during ER stress, phosphorylation of eIF2α at Ser51 by PERK induces ATF4 mRNA translation and accumulation of ATF4 protein. We therefore hypothesized that stress-induced Cat-1 transcription in A/A MEFs, which express an eIF2α that cannot be phosphorylated due to of a mutation in the phosphorylation site, should be similar to ATF4−/− MEFs. To test this hypothesis, we determined Cat-1 mRNA levels during ER stress in S/S (WT) and A/A MEFs (Figure 4C). As expected, Cat-1 mRNA levels were much lower in A/A than S/S MEFs during ER stress, in agreement with the absence of ATF4 induction in these cells (Figure 4D). Significantly, Cat-1 mRNA levels decreased in A/A cells at 6–12 h of stress, in contrast with the increase that was observed in ATF4−/− MEFs. This finding prompted us to examine the levels of XBP1s in A/A MEFs. To our surprise, ER stress induced XBP1s to lower levels in A/A than in S/S MEFs, suggesting regulation of the XBP1 gene expression during ER stress by the PERK/eIF2α pathway.
We have shown in the present study that eIF2α phosphorylation is required for induction of Cat-1 gene transcription during ER stress by mechanisms that involve increased levels of the transcription factors ATF4 and XBP1s. Although the mechanism of ATF4 induction by eIF2α phosphorylation has been described [15], the mechanism of regulation of XBP1 by the same pathway is novel and deserves further investigation.
DISCUSSION
In our previous studies we demonstrated that Cat-1 gene expression is under the control of a TATA-less promoter, consistent with its low expression in most tissues [5,26]. In the present study, we characterized the Cat-1 basal promoter and also identified an ERSE-II-like element near the basal promoter that functions during ER stress. These findings are supported by the following results: (i) the region from −63 to −25 upstream of the transcription start site contains a GC-rich motif that is essential for promoter activity; (ii) the transactivator Sp1 bound the GC-rich motif of the Cat-1 promoter both in vivo and in vitro, as demonstrated by ChIP and EMSA studies; (iii) induction of Cat-1 mRNA levels during ER stress required both the ERSE-II-like element that bound XBP1s and the exonic AARE that bound ATF4.
TATA-less genes (approx. 68 % of the protein-encoding human genes) have alternative mechanisms for recruitment of the polymerase and the subsequent transcription initiation [9] that involve GC- and CCAAT-boxes. Some GC boxes in TATA-less promoters have been shown to bind the transcriptional activator Sp1, which can interact with TFIID via direct binding of the TBP (TATA-box binding protein) or the TFIID subunits TAF4 and TAF7 [30]. Sp1 has also been implicated in chromatin remodelling through its interaction with HDAC1 (histone deacetylase 1), CBP/p300 [CREB (cAMP-response-element-binding protein)-binding protein] and the SWI/SNF complex, resulting in activation or inhibition of gene transcription. We have shown previously that the Cat-1 promoter is GC-rich and contains several putative binding sites for the Sp/XKLF transcription factor family [31]. Our finding that a single GC-box is the major determinant of Cat-1 basal promoter activity indicates the lack of synergism with other putative GC-boxes in the promoter. It also suggests that Sp3, which represses Sp1-mediated activation of promoters with more than two Sp1-binding sites [32], does not negatively regulate Cat-1. The single Sp1-binding site in the Cat-1 promoter is also consistent with our previous identification of a predominant transcription start site [5].
The importance of Sp1 in basal Cat-1 gene expression was demonstrated by the 6-fold decrease in Cat-1 mRNA levels in Sp1−/− cells (Figure 2C). Furthermore, although the extent of induction of Cat-1 mRNA levels by amino acid starvation and ER stress was similar in WT and Sp1−/− cells (Figure 2G), the absence of Sp1 resulted in lower Cat-1 mRNA levels under all conditions. This suggests that Sp1 is required for efficient Cat-1 transcription under basal and stress conditions, but is not involved in the induction by stress. The expression of Cat-1 in Sp1−/− cells may be due to binding of the functionally similar but weaker transactivator Sp3 to GC-boxes [11]. The binding of Sp proteins to GC-boxes in stress-induced gene expression has also been reported for the TATA-containing asparagine synthase gene [33].
The levels and activity of Sp proteins are regulated during stress [11]. For example, oxidative stress increases both the level and the DNA-binding activity of Sp1 and Sp3 in cortical neurons [34]. This induction may promote neuronal survival by activating Sp1-mediated transcription of anti-apoptotic genes [34]. Cat-1 is expressed in cortical neurons and arginine transport is an important defence mechanism against oxidative stress [35]. It is therefore possible that the Cat-1 gene is part of the Sp1-mediated survival response during oxidative stress.
The significance of Sp1 in Cat-1 gene expression is highlighted by the regulation of this gene during cell growth and physiological stress [1]. We have previously shown that the stress-induced transcription factor ATF4 enhances Cat-1 expression by binding to the AARE [26]. This regulatory mechanism may function during various physiological states and in development. Cat-1−/− mice develop severe anaemia and die within a few hours of birth; the major deficiency is impairment in erythrocyte maturation [36]. Interestingly, a similar phenotype was observed in Sp1+/−, Sp3+/− and ATF4−/− mice [22,37]. Because of these similarities, the Cat-1 gene may be a target of Sp1 and ATF4 in fetal liver that is important for haemopoiesis.
We have shown previously that transcription of the Cat-1 gene increases early in the ER-stress response and the LIP (liver-enriched inhibitory protein) isoform of C/EBPβ subsequently attenuated this induction [26,27]. The binding of heterodimers consisting of both C/EBPβ isoforms [LAP (liver-enriched activating protein) and LIP], or ATF4 and LIP, to the AARE played a role in this attenuation [26]. Furthermore, CHOP is also involved in this transcriptional attenuation via binding to the INE within the first intron of the gene [6]. In the present study, we expanded our understanding of the induction of Cat-1 gene transcription during the early phase of ER stress. We demonstrated that induction of Cat-1 gene transcription by ER stress is mediated by ATF4 via binding to the AARE in exon 1 and XBP1s via binding to the putative ERSE-II-like element near the Sp1-binding sequence. ERSE-II elements interact either with XBP1s or ATF6, either with or without the transcription factor NF-Y (nuclear factor Y) [29]. NF-Y has been shown to bind the ATTGG part of the sequence and both ATF6 and XBP1s bind the CCACG part. The binding of ATF6 requires NF-Y whereas XBP1 binding does not [38]. The rat Cat-1 ERSE-II-like element (−25 to −14; ATTGGTGCCTGG) contains a consensus NF-Y-binding site (5′-ATTGG-3′) and the sequence, 5′-CCTGG-3′, which in the reverse orientation is similar to the ATF6/XBP1s-binding site of the ERSE-II sequence. As the Cat-1 ERSE-II-like element is regulated by XBP1s and not ATF6, we also considered the presence of a UPRE (UPR element) within the putative ERE-II-like element. UPREs (consensus sequence TGACGTGG/A) have very low affinity for ATF6 and high affinity for XBP1s [29] and transcription driven by these elements requires XBP1s [39]. The CCTGG sequence in the Cat-1 ERSE-II-like element is similar to the core sequence of the UPRE [39]. To our knowledge, there is one report of a target gene (HRD1; 3- hydroxy-3-methylglutaryl-CoA reductase degradation 1) regulated by a UPRE that binds XBP1s and not ATF6 [40]. However, in contrast with the Cat-1 gene, which is regulated exclusively by XBP1s, the HRD1 gene is regulated by both ATF6 and XBP1s via the use of an additional ERSE within its promoter region [40,41]. Using genome-wide approaches, it was also shown that XBP1s-targeted genes function in many biological pathways [42]; the same authors, using computational analysis, identified six different sequence motifs that bound XBP1 [42]. However, none of these motifs was identical with the Cat-1 ERSE-II-like element. The ERSE-II-like element in the Cat-1 gene promoter may be the first natural XBP1s target sequence that is not a target of ATF6 during ER stress. The induction of Cat-1 transcription after 6–12 h of ER stress is also consistent with XBP1s being the regulating transcription factor; active ATF6 levels increase early in the stress response whereas XBP1s levels accumulate later.
An interesting finding of the present study was that the induction of Cat-1 transcription by ATF4 and XBP1s during ER stress required eIF2α phosphorylation. Our results suggest an unrecognized communication between the PERK/eIF2α pathway and the IRE1/XBP1 pathway during the UPR. In the absence of a phosphorylatable eIF2α, the production of the XBP1s protein is compromised. Although the mechanism of this regulation was not studied here, it may involve transcriptional control of the XBP1 gene by the ATF6 pathway. It has been reported that ATF6 induced XBP1 gene transcription [17]. Therefore the PERK/eIF2α signalling may involve activation of the ATF6 pathway and the subsequent induction of the IRE1/XBP1 pathway. Cells deficient in both XBP1 and ATF6 failed to induce several stress-response genes during the UPR, pointing to the importance of these transcription factors in the stress response [39]. However, there are no extensive studies on the regulation of the same genes in eIF2α phosphorylation-deficient cells. We are currently studying this mechanism.
We have shown in the present study that induction of Cat-1 gene transcription during ER stress involves ATF4 and XBP1s. As binding of ATF4 to the Cat-1 gene promoter precedes binding of XBP1s, we conclude that XBP1s mediates the sustained transcriptional activation of the gene during prolonged stress. Furthermore, modification of histones around the Cat-1 promoter by ATF4-associated factors may be required for efficient binding of XBP1s. In fact, such a mechanism was shown to enhance CHOP transcription during amino acid starvation via ATF4 association with the histone acetyltransferase PCAF (p300/CBP-associated factor) [43].
Among the three UPR-induced signalling pathways, the ATF6 pathway targets genes for the immediate response to stress whereas the IRE1/XBP1 pathway sustains this response [29]. The eIF2α/ATF4 pathway functions in both the early and late responses, due to ATF4’s ability to dimerize with different members of the bZIP family of transcription factors that are induced at various phases of the UPR [44]. This sophisticated and carefully timed transcription programme has both pro-survival and pro-apoptotic functions. The early pro-survival cellular response to stress is the inhibition of general protein synthesis, exit from the cell cycle and enhanced chaperone-mediated protein folding (reviewed in [14,20]). This is mediated by PERK phosphorylation of eIF2α and by proteins whose expression is stimulated by ATF6. Cells also increase expression of the ERAD protein degradation machinery in order to eliminate unfolded and misfolded proteins; XBP1s-target genes, such as EDEM (ER degradation enhancer) and HRD1 mediate this process [39,40]. During prolonged stress, cells will commit to the apoptotic pathway if restoration of homoeostasis or adaptation fails. Our results indicate that the transcription factors ATF4 and XBP1s are involved in transcriptional induction of the Cat-1 gene during early and late ER stress by binding the Cat-1 promoter region sequentially to induce and sustain transcription of the gene (Figure 5). Although the importance of Cat-1 during ER stress has not been studied, it is likely that Cat-1 functions to supply cationic amino acids in anticipation of relief from stress and re-entry into the cell cycle. These possible roles of Cat-1 during ER stress are currently under investigation. Understanding the interplay between Cat-1 and ER stress may be important when considering strategies to increase the survival of pancreatic β-cells under pathological conditions that involve ER stress-induced apoptosis [45].
Figure 5. Model of Cat-1 gene transcriptional control in ER stress by ATF4 and XBP1s downstream of eIF2α phosphorylation.
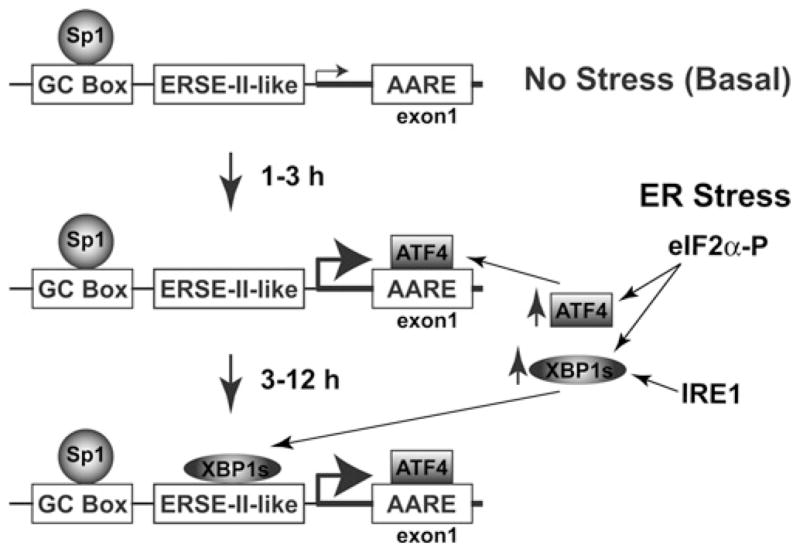
Top panel: when under no stress/basal conditions, Sp1 binds the GC box within the Cat-1 promoter to maintain a low level of transcription. Middle panel: early in the UPR, eIF2α phosphorylation causes increased ATF4 levels, which induces Cat-1 transcription by binding to the AARE in the first exon. Bottom panel: later in the UPR, a novel cross-talk between the PERK/eIF2α and the IRE1/XBP1 pathways of the UPR induces XBP1s expression. Cat-1 transcription is sustained by XBP1s binding to the ERSE-II-like element along with ATF4 bound to the AARE.
Acknowledgments
We thank Dr David Ron, Dr Duna Massillon, and Dr Jeremy M. Boss for providing expression plasmids and cell lines.
FUNDING
This work was supported by the National Institutes of Health [grant numbers R01DK060596 and R01DK053307 (to M.H.), R01CA103867 and R01CA124760 (to C.-M.C.), and DK042394, HL052173 and HL057346 (to R.J.K.)]. R.J.K. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used
- AARE
amino-acid-response element
- ATF
activating transcription factor
- β-gal
β-galactosidase
- bZIP
basic leucine zipper
- C/EBP
CCAAT/enhancer-binding protein
- Cat-1
cationic amino acid transporter-1
- CBP/p300
CREB (cAMP-response-element-binding protein)-binding protein
- ChIP
chromatin immunoprecipitation
- CHOP
C/EBP homologous protein
- CMVmin
cytomegalovirus minimal promoter
- DMEM
Dulbecco’s modified Eagle’s medium
- eIF2α
eukaryotic initiation factor 2α
- EMSA
electrophoretic mobility-shift assay
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- ERSE-II
ER-stress-element-II
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HERP
homocysteine-induced endoplasmic reticulum protein
- HRD1
3-hydroxy-3-methylglutaryl-CoA reductase degradation 1
- INE
intronic enhancer element
- IRE1
inositol-requiring enzyme 1
- KRB
Krebs–Ringer bicarbonate
- LAP
liver-enriched transcriptional activating protein
- LIP
liver-enriched transcriptional inhibitory protein
- LUC
firefly luciferase
- MEF
mouse embryonic fibroblast
- NF-Y
nuclear factor Y
- PCAF
p300/CBP-associated factor
- PERK
protein-kinase-receptor-like ER kinase
- Pol II
RNA polymerase II
- Purα
purine-rich-binding protein A
- qRT-PCR
quantitative real-time PCR
- Sp
specificity protein
- TAF
transcription-associated factor
- TFIID
transcription factor IID
- UPR
unfolded protein response
- UPRE
UPR element
- UTR
untranslated region
- WT
wild-type
- XBP1
X-box-binding protein 1
- XBP1s
spliced XBP1
- XBP1u
unspliced XBP1
- XKLF
X Krüppel-like factor
Footnotes
AUTHOR CONTRIBUTION
Charlie Huang, Yi Li and Alex Lopez performed the experiments and participated in their design. Cheng-Ming Chiang helped with experimental design and provided expertise of TATA-less promoters. Randal Kaufman provided essential reagents for the studies on the UPR. Martin Snider helped with data analysis and presentation. Maria Hatzoglou conceived and co-ordinated the project. Charlie Huang, Martin Snider and Maria Hatzoglou prepared the manuscript with input from all authors.
References
- 1.Hatzoglou M, Fernandez J, Yaman I, Closs E. Regulation of cationic amino acid transport: the story of the CAT-1 transporter. Annu Rev Nutr. 2004;24:377–399. doi: 10.1146/annurev.nutr.23.011702.073120. [DOI] [PubMed] [Google Scholar]
- 2.Aulak KS, Liu J, Wu J, Hyatt SL, Puppi M, Henning SJ, Hatzoglou M. Molecular sites of regulation of expression of the rat cationic amino acid transporter gene. J Biol Chem. 1996;271:29799–29806. doi: 10.1074/jbc.271.47.29799. [DOI] [PubMed] [Google Scholar]
- 3.Wu JY, Robinson D, Kung HJ, Hatzoglou M. Hormonal regulation of the gene for the type C ecotropic retrovirus receptor in rat liver cells. J Virol. 1994;68:1615–1623. doi: 10.1128/jvi.68.3.1615-1623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez J, Yaman I, Mishra R, Merrick WC, Snider MD, Lamers WH, Hatzoglou M. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J Biol Chem. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez J, Lopez AB, Wang C, Mishra R, Zhou L, Yaman I, Snider MD, Hatzoglou M, Hatzolgou M. Transcriptional control of the arginine/lysine transporter, cat-1, by physiological stress. J Biol Chem. 2003;278:50000–50009. doi: 10.1074/jbc.M305903200. [DOI] [PubMed] [Google Scholar]
- 6.Huang CC, Chiribau CB, Majumder M, Chiang CM, Wek RC, Kelm RJ, Jr, Khalili K, Snider MD, Hatzoglou M. A bifunctional intronic element regulates the expression of the arginine/lysine transporter Cat-1 via mechanisms involving the purine-rich element binding protein A (Purα) J Biol Chem. 2009;284:32312–32320. doi: 10.1074/jbc.M109.024471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez J, Bode B, Koromilas A, Diehl JA, Krukovets I, Snider MD, Hatzoglou M. Translation mediated by the internal ribosome entry site of the cat-1 mRNA is regulated by glucose availability in a PERK kinase-dependent manner. J Biol Chem. 2002;277:11780–11787. doi: 10.1074/jbc.M110778200. [DOI] [PubMed] [Google Scholar]
- 8.Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, Kaufman RJ, Hatzoglou M. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 9.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 10.Gidoni D, Dynan WS, Tjian R. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. Nature. 1984;312:409–413. doi: 10.1038/312409a0. [DOI] [PubMed] [Google Scholar]
- 11.Wierstra I. Sp1: emerging roles–beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 12.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 14.Mori K. Signaling pathways in the unfolded protein response: development from yeast to mammals. J Biochem (Tokyo) 2009;146:743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 15.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 18.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 19.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 20.Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6:55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimoto T, Yoshimoto E, Meruelo D. Enhanced gene expression of the murine ecotropic retroviral receptor and its human homolog in proliferating cells. J Virol. 1992;66:4377–4381. doi: 10.1128/jvi.66.7.4377-4381.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuoka HC, Townes TM. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood. 2002;99:736–745. doi: 10.1182/blood.v99.3.736. [DOI] [PubMed] [Google Scholar]
- 23.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 24.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 25.Guo Z, Boekhoudt GH, Boss JM. Role of the intronic enhancer in tumor necrosis factor-mediated induction of manganous superoxide dismutase. J Biol Chem. 2003;278:23570–23578. doi: 10.1074/jbc.M303431200. [DOI] [PubMed] [Google Scholar]
- 26.Lopez AB, Wang C, Huang CC, Yaman I, Li Y, Chakravarty K, Johnson PF, Chiang CM, Snider MD, Wek RC, Hatzoglou M. A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem J. 2007;402:163–173. doi: 10.1042/BJ20060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Bevilacqua E, Chiribau CB, Majumder M, Wang C, Croniger CM, Snider MD, Johnson PF, Hatzoglou M. Differential control of the CCAAT/enhancer-binding protein β (C/EBPβ) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J Biol Chem. 2008;283:22443–22456. doi: 10.1074/jbc.M801046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokame K, Kato H, Miyata T. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J Biol Chem. 2001;276:9199–9205. doi: 10.1074/jbc.M010486200. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem (Tokyo) 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 30.Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 31.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Yu B, Datta PK, Bagchi S. Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 2003;31:5368–5376. doi: 10.1093/nar/gkg706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung-Pineda V, Kilberg MS. Role of Sp1 and Sp3 in the nutrient-regulated expression of the human asparagine synthetase gene. J Biol Chem. 2002;277:16585–16591. doi: 10.1074/jbc.M110972200. [DOI] [PubMed] [Google Scholar]
- 34.Ryu H, Lee J, Zaman K, Kubilis J, Ferrante RJ, Ross BD, Neve R, Ratan RR. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J Neurosci. 2003;23:3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Kang BN, Tian J, Liu Y, Luo HR, Hester L, Snyder SH. The cationic amino acid transporters CAT1 and CAT3 mediate NMDA receptor activation-dependent changes in elaboration of neuronal processes via the mammalian target of rapamycin mTOR pathway. J Neurosci. 2007;27:449–458. doi: 10.1523/JNEUROSCI.4489-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perkins CP, Mar V, Shutter JR, del Castillo J, Danilenko DM, Medlock ES, Ponting IL, Graham M, Stark KL, Zuo Y, et al. Anemia and perinatal death result from loss of the murine ecotropic retrovirus receptor mCAT-1. Genes Dev. 1997;11:914–925. doi: 10.1101/gad.11.7.914. [DOI] [PubMed] [Google Scholar]
- 37.Kruger I, Vollmer M, Simmons DG, Elsasser HP, Philipsen S, Suske G. Sp1/Sp3 compound heterozygous mice are not viable: impaired erythropoiesis and severe placental defects. Dev Dyn. 2007;236:2235–2244. doi: 10.1002/dvdy.21222. [DOI] [PubMed] [Google Scholar]
- 38.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto K, Suzuki N, Wada T, Okada T, Yoshida H, Kaufman RJ, Mori K. Human HRD1 promoter carries a functional unfolded protein response element to which XBP1 but not ATF6 directly binds. J Biochem (Tokyo) 2008;144:477–486. doi: 10.1093/jb/mvn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneko M, Yasui S, Niinuma Y, Arai K, Omura T, Okuma Y, Nomura Y. A different pathway in the endoplasmic reticulum stress-induced expression of human HRD1 and SEL1 genes. FEBS Lett. 2007;581:5355–5360. doi: 10.1016/j.febslet.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 42.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Cherasse Y, Maurin AC, Chaveroux C, Jousse C, Carraro V, Parry L, Deval C, Chambon C, Fafournoux P, Bruhat A. The p300/CBP-associated factor (PCAF) is a cofactor of ATF4 for amino acid-regulated transcription of CHOP. Nucleic Acids Res. 2007;35:5954–5965. doi: 10.1093/nar/gkm642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su N, Kilberg MS. C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J Biol Chem. 2008;283:35106–35117. doi: 10.1074/jbc.M806874200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, Wada I, Akira S, Araki E, Mori M. Nitric oxide-induced apoptosis in pancreatic β cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci USA. 2001;98:10845–10850. doi: 10.1073/pnas.191207498. [DOI] [PMC free article] [PubMed] [Google Scholar]