Abstract
Peptide nucleic acids have emerged over the past two decades as a promising class of nucleic acid mimics because of their strong binding affinity and sequence selectivity toward DNA and RNA, and resistance to enzymatic degradation by proteases and nucleases. While they have been shown to be effective in regulation of gene expression in vitro, and to a small extent in vivo, their full potential for molecular therapy has not yet been fully realized due to poor cellular uptake. Herein, we report the development of cell-permeable, guanidine-based peptide nucleic acids targeting the epidermal growth factor receptor (EGFR) in preclinical models as therapeutic modality for head and neck squamous cell carcinoma (HNSCC) and nonsmall cell lung cancer (NSCLC). A GPNA oligomer, 16 nucleotides in length, designed to bind to EGFR gene transcript elicited potent antisense effects in HNSCC and NSCLC cells in preclinical models. When administered intraperitoneally in mice, EGFRAS-GPNA was taken-up by several tissues including the xenograft tumor. Systemic administration of EGFRAS-GPNA induced antitumor effects in HNSCC xenografts, with similar efficacies as the FDA-approved EGFR inhibitors: cetuximab and erlotinib. In addition to targeting wild-type EGFR, EGFRAS-GPNA is effective against the constitutively active EGFR vIII mutant implicated in cetuximab resistance. Our data reveals that GPNA is just as effective as a molecular platform for treating cetuximab resistant cells, demonstrating its utility in the treatment of cancer.
Efforts including the Human Genome and the Cancer Genome Projects have led to the identification of numerous molecular targets critical for cancer growth and progression.1 Molecular targeted therapies against oncogenic proteins may be more effective and induce less side-effects than conventional chemo- and radiotherapies. Typically, this is accomplished through the use of small-molecules or antibody antagonists, which, upon binding, inactivate the physiological functions of the protein (or receptor) targets. Many anticancer drugs have been developed on the basis of this mode of action. Although effective in treating cancer in preclinical models, these agents demonstrate low clinical response rates in patients with solid tumors due to primary or acquired drug resistance.2 An antisense approach, involving the use of oligonucleotide molecules to target the gene transcripts and thereby blocking protein production, may be more effective because of the specificity of recognition and the flexibility in sequence design. Since drug resistance, in many instances, occurs as the result of genetic mutations of the targeted genes, the ability to modify the oligonucleotide’s sequence to match that of the emerging clones is essential to countering drug resistance. Despite their promising outlook, oligonucleotide agents are rarely employed in molecular therapy because of their susceptibility to enzymatic degradation, nonspecific binding,3 and poor cellular uptake.4
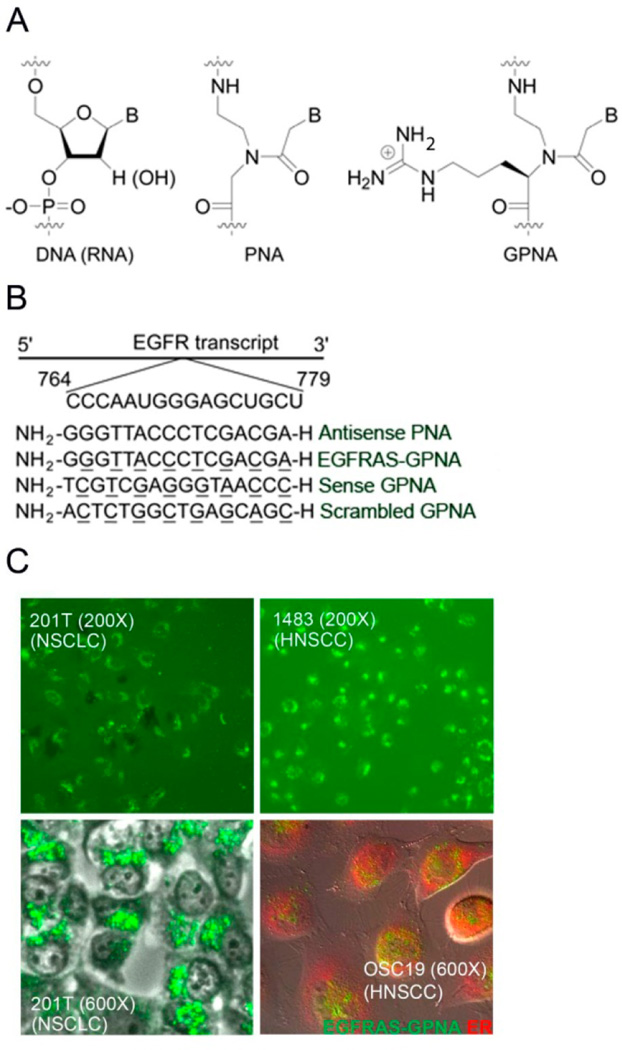
Attempts to improve enzymatic stability have led to the development of several classes of synthetic oligonucleotides;5 one such promising class is peptide nucleic acids (PNAs).6 PNAs are DNA and RNA analogues in which the sugar phosphodiester backbone is replaced by N-(2-aminoethyl) glycine units to which the nucleobases are attached through a flexible carboxymethylene linker.7 The charge-neutral backbone enables PNAs to form highly stable duplexes with complementary DNA and RNA strands,8 while the unnatural polyamide linkage renders them impervious to recognition and degradation by proteases and nucleases.9 Together, these properties make PNAs attractive as antisense reagents for molecular therapy. Though their ability to regulate gene expression has been demonstrated in cell culture,10−15 and to a small extent in vivo,16−19 their potential application as therapeutics have not yet been fully realized due to poor cellular uptake.20 To overcome this limitation, we have prepared a chiral class of PNAs called guanidine-based peptide nucleic acids (GPNAs).21 GPNA contains an arginine side-chain with an R-configuration at the α-backbone (Figure 1A). The choice of the guanidinium group was based on the work of Wender22 and Dowdy,23 which showed that this chemical functionality is essential for the uptake of many cell-penetrating peptides and proteins. The R-configuration was chosen because prior studies revealed that this particular stereochemistry has minimal effect on the hybridization properties of PNAs.24 The ability of GPNAs to traverse the cell membrane and selectively inhibit gene expression has been demonstrated in cell culture;25 however, whether they could be used as molecular therapeutics to regulate gene expression in vivo or treat cancer in animal models has not yet been explored. As a proof-of-concept, we selected the epidermal growth factor receptor (EGFR) as a target for our study because of its involvement in growth and progression of several types of cancer.26
Figure 1.
(A) Chemical structure of DNA, PNA, and GPNA unit. (B) Target site within the EGFR gene and sequence of PNA and GPNA oligomers employed in this study; underlined are GPNA units. (C) Fluorescent images of live NSCLC and HNSCC cells following incubation with 1 μM FITC-EGFRAS-GPNA (green) in a complete medium for 24 h. Bottom right panel: an image of live cells costained with an ER dye (red), demonstrating colocalization (yellow) in the perinuclear regions. Three independent experiments were carried out showing similar results.
High levels of EGFR in epithelial tumors have been associated with advanced stage, large tumor size, invasion, decreased survival, and poor prognosis.27,28 EGFR inhibition can be achieved by either blocking EGFR autophosphorylation or downmodulating total EGFR levels within the cell. Several inhibitors that block EGFR activation and downstream signaling have been developed and shown to be effective in suppressing cancer growth; however, the therapeutic effects are often short-lived due to the emergence of resistant clones.29,30 Likewise, antisense EGFR therapy has also been shown to be effective;31 but because of their enzymatic lability and poor cellular uptake, these agents require intratumoral delivery. Such a delivery route is suitable for treating localized tumors but is not practical for curtailing metastatic tumors at hard-to-reach sites. In an attempt to overcome the cellular delivery issue, we examined the cellular uptake of GPNAs in cell culture and in vivo and assessed their antitumor effects in a xenograft model. The results presented here have important implications for in vivo gene regulation and for the future treatment of head and neck cancer, as well as a number of other cancer types including lung and stomach, associated with overexpression of EGFR.
RESULTS
Cellular Uptake and Localization of EGFRAS-GPNA
Despite the charge-neutral backbone, PNA oligomers are not readily taken-up by cells.20 We have shown that PNA oligomers, 10 to 20 nucleotides in length, containing GPNA units at every other position are readily taken-up by mammalian cells.24,32 Further, we showed that internal placement of the guanidinium groups is less toxic to cells than the conventional head-to-head or head-to-tail conjugation,25 presumably due to the reduction in the amphipathic character of the former. We previously demonstrated that phosphorothioate-modified oligonucleotides specific to the EGFR mRNA sequence (NCBI accession NM_005228.3) from site 829 to 844, effectively reduced EGFR levels and had antitumor efficacy in vivo.31 The target sequence lies in the extracellular domain of EGFR, which is responsible for ligand binding. On the basis of this design concept, a 16-nucleotide, anti-EGFR GPNA oligomer (EGF-RAS-GPNA), along with the sequence-scrambled control, was generated (Figures 1A,B and S1−S3, Supporting Information). To assess cellular uptake in live cells, an N-terminal FITC-tagged EGFRAS-GPNA oligomer was incubated with NSCLC and HNSCC cells in 10% serum-containing medium and imaged with confocal fluorescent microscopy without fixing (Figures 1C and S4, Supporting Information). No autofluorescence was observed in untransfected cells in the FITC channel (data not shown). Transfection efficiencies of greater than 99% were achieved with a dosage as low as 1 μM (Figure 1C, top and bottom left panels). The fluorescent signals were observed primarily in the peri-nuclear region. Endoplasmic reticulum (ER) is located in the perinuclear regions, the sites at which mRNA molecules are translated into protein. To determine if EGFRAS-GPNA localizes in the ER, we stained live cells with an ER specific dye (red) and assessed the localization of FITC-tagged EGFRAS-GPNA (green). Our data demonstrate that EGFRAS-GPNA colocalizes with the ER (Figure 1C, bottom right panel). EGFRAS-GPNA localization was also examined in HNSCC cell lines UMSCC-22B and PCI-15B (Supplementary Figure S5A,B). Further, we tested the uptake and localization of FITC-tagged EGFRAS-GPNA in normal oral epithelial tissue (Het 1A) cells and found the uptake efficiency to be similar to that seen in the HNSCC cells (Supplementary Figure S5C). We carried out a line-scan analysis of Het 1A cells stained with the ER marker and transfected with FITC-tagged EGFRAS-GPNA and found that the two fluorescent signals localized to the same regions within the cells (Supplementary Figure S6). Together, the data suggest that GPNA was taken-up by HNSCC, NSCLC, and normal epithelial cells, and localized in the ER where mRNA molecules are most abundant.
EGFRAS-GPNA Suppressed EGFR Expression and Reduced Cell Growth
We and others have previously shown that EGFR antisense oligonucleotides were effective in downmodulating EGFR protein levels.33 However, transfecting agents were used to facilitate transport of the oligonucleotides into cells. While this is a viable strategy for in vitro cell culture work, it is not practical for in vivo applications or for the treatment of genetic diseases. GPNAs offer a solution to this problem because they are cell permeable and therefore could be delivered into live cells and intact organisms without the aid of transfecting reagents or other mechanical or electrical transduction means. We selected EGFR as a model gene target because of the critical role it plays in promoting tumor growth. Cetuximab is the only FDA-approved EGFR targeted agent for clinical use in HNSCC, but the clinical response rates have been low, in the range of 10−13%.34 In addition to cetuximab, a small molecule tyrosine kinase inhibitor erlotinib extensively tested in HNSCC and NSCLC patients also demonstrated a low clinical response rate of 4%.35
To determine whether EGFRAS-GPNA could be used to inhibit the expression of EGFR without the assistance of the transfecting reagents, we incubated HNSCC and NSCLC cells with 3 μM of GPNA in a complete DMEM medium. After 72 h of incubation, cells were harvested and lysed as described previously.36 We chose a dose at which there would be minimal cytotoxic effects of EGFRAS GPNA. Examination of cell lysates by RT-PCR and immunoblotting revealed that treatment with EGFRAS-GPNA resulted in the reduction of EGFR mRNA and protein levels compared to the negative controls (Figure 2A,B, respectively). Since the expression of EGFR has been shown to correlate with cell growth, we further tested the ability of EGFRAS-GPNA to inhibit the proliferation of HNSCC and NSCLC cells in culture. Cells were treated with GPNA for 72 h in a complete medium, and the viability was assessed using CellTiter-Glo luminescent assay. Our results demonstrate that cells treated with 10 μM EGFRAS-GPNA had greater than 50% growth inhibition compared to the scrambled sequence, for both HNSCC 1483 and NSCLC 201T lines (Figures 2C,D). This indicates that squamous cell carcinomas are sensitive to EGFRAS-GPNA treatment. Lesser growth inhibition was observed at a lower concentration (5 μM).
Figure 2.
Effects of EGFRAS-GPNA on (A,B) gene expression and (C,D) cancer cell growth. (A) RT-PCR and (B) immunoblotting analyses of HNSCC and NSCLC cells treated with 3 μM oligomers for 72 h in complete medium. (C) HNSCC cell line 1483 and (D) NSCLC cell line 201T treated with EGFRAS-GPNA or scrambled control GPNA at 5 and 10 μM concentrations for 72 h. The y-axis depicts the relative light units (RLU) determined by a luminescence-based assay that estimates the ATP levels in metabolically active cells. The experiment was repeated three times with similar results.
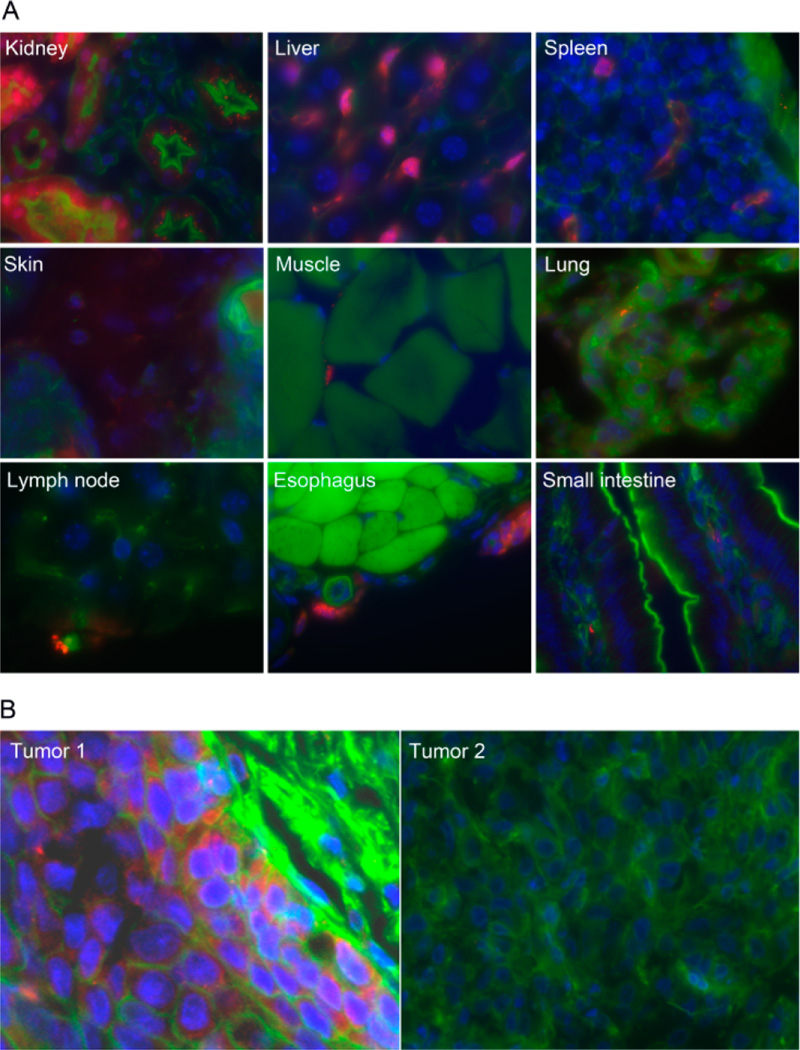
Systemic delivery of EGFRAS-GPNA in a xenograft tumor model. Next, we assessed the uptake of EGFRAS-GPNA in a xenograft model to determine whether GPNA could be delivered into intact organisms. Athymic nude mice were treated with saline alone, as a negative control, 5 mg/kg body weight of red fluorescent-TAMRA-tagged EGFRAS-GPNA, or the corresponding TAMRA-tagged standard PNA by intraperitoneal injection (IP). Animals were sacrificed at 4 and 24 h postinjection. Fluorescence confocal imaging of tissue from various organs and the HNSCC xenograft revealed uptake of EGFRAS-GPNA by the liver and kidney (Figure 3A) as well as by the tumor (Figure 3B) 4 h postinjection, as evidenced from the red fluorescent signals. No fluorescent signals, however, were detected from samples treated with unmodified PNA except for the liver and kidney, which showed signals slightly above the background (data not shown), indicating that uptake was strongly enhanced for GPNA. Further, the primary routes of elimination of EGFRAS-GPNA are likely hepatic and renal.
Figure 3.
(A) Tissue distribution of EGFRAS-GPNA in intact mice following IP injection. UMSCC-22B xenograft-bearing nude mice (on the back) were injected IP with 5 mg/kg of EGFRAS-GPNA-TAMRA or PNA-TAMRA. After 4 h, tissues were excised, fixed, sectioned, stained, and imaged at 200×. The nuclei were stained blue with DAPI, actin stained green with phalloidin, and GPNA stained red with TAMRA. (B) Image of xenograft tumors treated with EGFRAS-GPNA-TAMRA (tumor 1) and PNA-TAMRA (tumor 2). Note that only tumor 1 showed red-stains indicating systemic uptake of EGFRAS-GPNA-TAMRA but not PNA-TAMRA.
EGFRAS-GPNA Demonstrates Antitumor Effects in Vivo
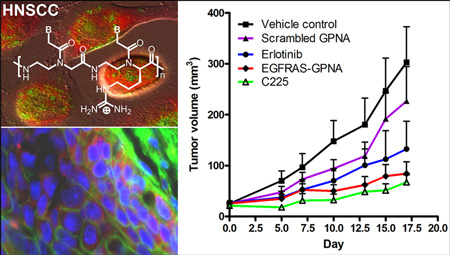
Because of the short half-life in plasma, antisense gene therapy for HNSCC tumors has previously been administered intratumorally.37 However, intralesional administration of antitumor agents precludes treatment of metastatic tumors at distal and difficult to access sites. To determine the optimal method for administration of GPNA, we treated tumor bearing mice with intratumoral or intraperitoneal injections of EGFRAS-GPNA. As a control, an EGFR sense GPNA was administered intratumorally. HNSCC xenograft tumors were treated daily with GPNAs. The tumors were measured twice a week. As expected from the result of downmodulation of EGFR, the tumors administered with EGFRAS-GPNA grew at a much slower rate compared to those administered with the controls, and more importantly, there was no difference in the volumes of the tumors treated with intraperitoneal or intratumoral injections (Figure 4A). This showed that GPNA can be systemically delivered into live organisms. The antitumor effect is sequence-specific; it was only observed with EGFRAS-GPNA, whereas the scrambled control GPNA was indistinguishable from the vehicle control at day 10 (p = 0.005) (Figure 4B). We attribute the improvement in antitumor efficacy to enhancements in cellular uptake and enzymatic stability of GPNAs.
Figure 4.
Antitumor effects of EGFRAS-GPNA in a xenograft mouse model (A) delivered by IT vs IP and (B) for the perfectly matched vs scrambled sequence. (C) Comparative antitumor effect of EGFRAS-GPNA, scrambled GPNA, FDA-approved cetuximab (C225, monoclonal antibody), and erlotinib (small-molecule kinase inhibitor). See the Materials and Methods section for detailed experimental procedures.
Antitumor Efficacy of EGFR-GPNA Is Comparable with EGFR Inhibitors
Several EGFR-specific tyrosine kinase inhibitors are currently in clinical trials.29 Cetuximab (C225) is an FDA-approved EGFR-specific antibody that binds the extracellular ligand-binding domain of EGFR, which prevents receptor activation. It has been reported that treatment with cetuximab in combination with radiation increased the survival rate in HNSCC patients.38 Results from a phase II trial with a small molecule EGFR inhibitor erlotinib demonstrated a 4% response rate but 38% disease stabilization for 16 weeks.39 Here, we compared the antitumor effects of cetuximab and erlotinib to that of the EGFRAS-GPNA in vivo. There was a statistically significant difference in tumor volumes across the erlotinib, cetuximab, and EGFRAS-GPNA groups when compared to the vehicle control group over time (p = 0.02) (Figure 4C). However, there was no significant difference in the response among erlotinib, cetuximab, and EGFRAS-GPNA groups indicating that the antitumor effects were comparable. There is a significant difference in tumor volumes of EGFRAS-GPNA and scrambled GPNA treated mice indicating a sequence specific antitumor effect of EGFRAS-GPNA (p = 0.05). The reduction in tumor volume is correlated with reduced EGFR mRNA and protein levels (Supplementary Figure S7). Although the outcomes of the treatment are similar, GPNA-based therapy has the potential to overcome drug-resistance due to mutation in the targeting gene by modifying the nucleobase sequence to match that of the mutant, which is not feasible with small-molecule and antibody inhibitors such as erlotinib and cetuximab.
EGFRAS-GPNA Inhibits Expression of Mutant EGFR vIII
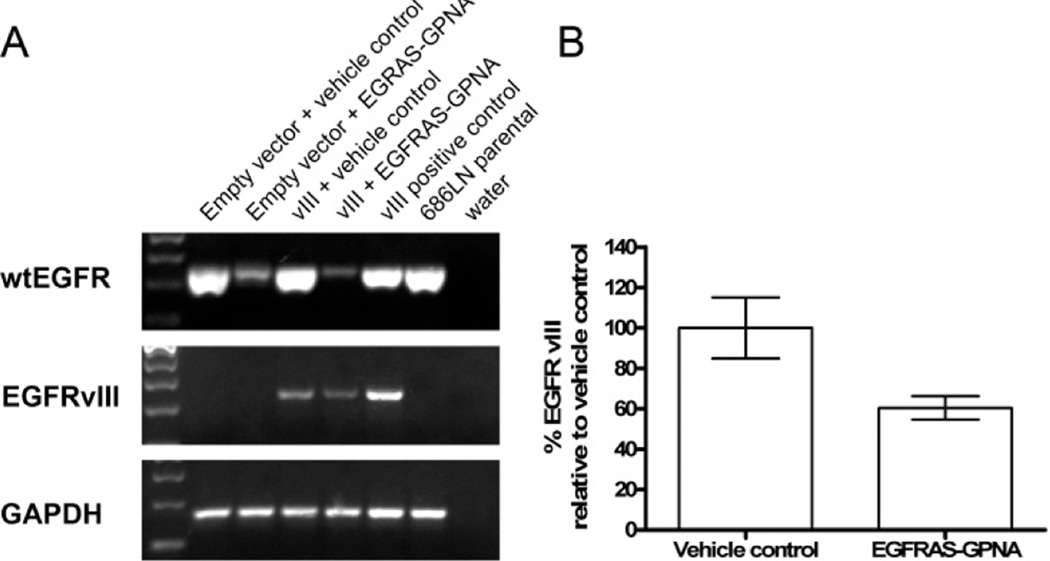
Mechanisms of resistance to EGFR tyrosine kinase inhibitors are currently being investigated in order to develop strategies to improve the sensitivity of tumors to these agents.40 We have previously reported that HNSCC cells expressing the constitutively active EGFR vIII mutant are resistant to the effects of EGFR-specific antibody cetuximab.41 To determine whether EGFRAS-GPNA is effective against cancer cells expressing a mutated form of EGFR, we treated HNSCC cell line 686LN engineered to transiently express mutant EGFR vIII. RT-PCR densitometric determinations showed a 40% reduction in the levels of EGFR vIII mRNA upon 3 μM EGFRAS-GPNA treatment (Figure 5A,B). This result demonstrates that EGFRAS-GPNA can be used to target both EGFR and EGFRvIII expressing lines, further demonstrating the flexibility and versatility of GPNA therapeutics.
Figure 5.
Effects of EGFRAS-GPNA on the expression of EGFR vIII. HNSCC cells expressing the vIII receptor were treated with 3 μM EGFRAS-GPNA for 72 h. (A) Cell lysates were subjected to RT-PCR to determine the levels of EGFR and EGFR vIII, and (B) EGFR vIII levels from cells treated with vehicle control or EGFRAS-GPNA from 2 independent experiments were plotted and demonstrate a 40% reduction in EGFR vIII levels on EGFRAS-GPNA treatment.
DISCUSSION
A major challenge in the field of cancer therapy is the identification and selective inhibition of oncogenic proteins (or signal ligands) that are differentially expressed in cancer cells compared to normal cells. Several studies have demonstrated that more than 90% of patients with head and neck squamous cell carcinoma (HNSCC) and more than 50% of patients with nonsmall cell lung carcinoma (NSCLC) overexpress the epidermal growth factor receptor (EGFR). Further, preclinical studies have demonstrated that inhibition of EGFR reduces tumor growth. EGFR inhibition can be achieved at the protein level, where agents abrogate EGFR activation, or at the mRNA level, where EGFR protein synthesis is inhibited. Several clinical trials with small molecules such as erlotinib or EGFR-specific antibodies such as cetuximab that block the receptor activity have demonstrated low-level antitumor effects in HNSCC patients. Antisense oligonucleotides designed to bind to the EGFR gene transcript, thereby blocking the production of EGFR protein, may be more effective in suppressing tumor growth than agents that inhibit receptor activation.42 A 29% (5/17 patients) response rate has been achieved with intratumoral administration of EGFR antisense gene therapy.37 This success rate is higher than the EGFR inhibitors clinically tested. Thus, it may be more efficacious to downmodulate EGFR levels. However, systemic delivery of DNA-based agents remains a challenge because of the poor cellular uptake and susceptibility to degradation by nucleases in the serum and in the cellular milieu. Administration of these agents intratumorally is technically challenging, especially for tumors that are difficult to access or for metastatic lesions. To circumvent these challenges, we have developed GPNAs, which are resistant to enzymatic degradation by proteases and nucleases and are taken-up by both somatic and embryonic stem cells.24 Our present data reveals that EGFRAS-GPNA is taken-up by HNSCC and NSCLC cells. Further, EGFRAS-GPNA effectively downregulated EGFR mRNA and protein levels and reduced tumor cell growth.
HNSCC xenograft-bearing nude mice treated with either systemically delivered EGFR-GPNA, via either intraperitoneal or intratumoral, showed a similar response. We have previously demonstrated that a phosphorothioate modification of EGFRAS oligonucleotides also has antitumor effects, but they need to be delivered intratumorally, and the effect is less dramatic and occurred with a long delay-period as compared to GPNA.43 EGFRAS-GPNA elicits antitumor effects in less than one week post-treatment as compared to three weeks for the phosphorothioate oligonucleotides, indicating that it is more potent.43 The enhancement in antitumor efficacy is likely due to improvements in enzymatic stability and cellular uptake. Our studies show that GPNA can be detected in HNSCC tumors within 4 h postadministration. GPNA uptake was also seen in the liver and kidney at 4 h indicating that these may be the main routes of elimination. A limitation of this study is the paucity of an efficient method to quantify GPNA oligonucleotides in serum or tumor tissue. Further studies are underway to develop such techniques.
To assess the relative efficacy of EGFRAS-GPNA, we compared the antitumor effect of EGFRAS-GPNA to that of a small molecule EGFR inhibitor erlotinib and an EGFR-specific antibody cetuximab. Our finding indicates that, in animal models of HNSCC, EGFRAS-GPNA has comparable antitumor effects as the other EGFR-targeting agents. It is worth noting that EGFR inhibitors that are FDA-approved have been very effective in preclinical models but have poor responses as single agents with the exception of NSCLC patients who harbor EGFR activating mutations. However, xenograft tumors expressing the EGFR vIII mutation, which is a constitutively active receptor lacking the extracellular ligand binding domain of EGFR and resistant to EGFR-specific antibody cetuximab were also inhibited with concomitant regulation of the levels of EGFR vIII mRNA.41 Because of the flexibility in the sequence design, GPNAs could potentially be used to target other genes that are important for tumor growth and progression, and for combating drug resistance due to mutations in the original targets. Improvements in binding affinity and sequence specificity, along with reduction in the cost of the monomer production, could further be made by installing the guanidinium group at the γ-backbone.44 Other classes of oligonucleotides that have been developed with a similar design concept include deoxyribonucleic guanidine45 and positively charged phosphorodiamidate morpholino (PMOplus).46,47 The work reported herein provides compelling evidence for EGFRAS-GPNA as a therapeutic agent for the treatment of HNSCC and possibly NSCLC. Cumulative data demonstrate that GPNA oligomers targeting unique sequences can be used to effectively reduce the levels of proteins that are implicated in cancer progression and survival.
MATERIALS AND METHODS
Synthesis of GPNA Monomers and Oligomers
GPNA monomers were prepared according to published procedures.21 EGFRAS-specific GPNA oligomers, with and without the fluorescein (FITC) probe, along with the scrambled control were synthesized on solid-support using standard Boc-chemistry.48 Upon cleavage from the resin by treating with m-cresol/thioanisole/TFSMA/TFA (1:1:2:6) cocktail, under which condition the protecting groups were also removed, the oligomers were precipitated with ethyl ether, purified by reverse-phase HPLC, and characterized by MALDI-TOF mass spectrometry. EGFRAS-GPNA oligomers were designed to bind to the EGFR mRNA transcript spanning nucleotides 829−844 (NCBI accession NM_005228.3). Antisense PNA, scrambled GPNA, and sense GPNA sequences were also prepared and used as a controls. All sequences are listed in Figure 1B.
Cell Lines and Reagents
Well-characterized HNSCC cell lines 1483, UMSCC-22B, PCI-15B, and OSC19 were used in this study.49 HNSCC cell line 686LN was a kind gift from Dr. Zhou (Georgia) Chen, Emory University. Cells were transiently transfected with a vector control or EGFR vIII expressing plasmid DNA as previously reported.50 All HNSCC cells except OSC19 were maintained in DMEM with 10% fetal bovine serum (FBS). OSC19 cells were maintained in MEM with 1% nonessential amino acids and 10% FBS. Het 1A is a transformed esophageal epithelial line obtained from American Type Culture Collection (ATCC) and was maintained in airway epithelial cell culture medium with supplements and 2% FBS. Nonsmall cell lung carcinoma (NSCLC) line 201T was a gift from Dr. Jill Siegfried, University of Pittsburgh Cancer Institute. NSCLC cells were maintained in minimal Eagle’s medium (MEM) with 10% FBS. For immunoblotting, Anti-EGFR antibody was obtained from Millipore and anti-β-tubulin antibody was purchased from Abcam. For immunohistochemistry, anti-EGFR antibody was obtained from Sigma Aldrich. Cetuximab was obtained from Imclone Systems and erlotinib from OSI Pharmaceuticals.
In Vitro Uptake Studies
Cells (2 × 103 cells/well) were plated on the coverslip in the slide chamber overnight. The next day, cells were treated with FITC-labeled EGFRAS-GPNA for 24 h. Live cells were co-stained with the endoplasmic reticulum-specific marker (ER tracker, Molecular Probes) and imaged on a Nikon Eclipse TE200-E inverted microscope at 600× magnification. Co-localization was analyzed using MetaMorph software (Molecular Devices).
In vitro cytotoxicity assay
HNSCC and NSCLC cells were plated at a density of 2 × 104 cells per well in a 24-well plate. The next day, cells were treated with either EGFRAS or scrambled control GPNA. Cells were treated daily for 3 days followed by either cell counting or CellTiter-Glo luminescent cell viability assay (Promega) to measure the amounts of ATP in the viable cells according to the manufacturer’s instructions.
RT-PCR and Immunoblotting Analysis of EGFR Levels
HNSCC and NSCLC cells were plated at a density of 4 × 104 cells per well in a 6-well plate for RNA extraction or at 5 × 105 cells per 100 mm dish for protein extraction and treated with 3 μM GPNA for 72 h. Total mRNA was isolated using Qiagen RNeasy Mini kit and reverse transcribed using the Superscript First-Strand synthesis kit with an input of 2.5 μg of RNA. All PCR reactions were run using 2× PCR Mastermix from Promega with 0.7 μM forward and reverse primers and 120 ng of cDNA. EGFRvIII specific primers, Forward [5′-ATGCGACCCTCCGGGACG-3′] and Reverse [5′-ATTCCGTTACACACTTTGCGGC-3′], and GAPDH specific primers, Forward [5′-TGGAATTTGCCATGGGTG- 3 ′] and Reverse [5′-GTGAAGGTCGGAGTCAAC-3′], were used.41 Thermocycling conditions were as follows: 95 °C for 5 min, 35 cycles each at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min, followed by reaction termination at 72 °C for 10 min. Wild-type EGFR (wtEGFR) primers were designed against exons 2−7: Forward primer [5′-CCTGCCCTGTGCAACGTGGA-3′] and Reverse primer [5′-CACTGGGGGACTTGCCACGG-3′]. The PCR reaction for wtEGFR was run as follows: 94 °C for 5 min, 35 cycles each at 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s, followed 7 min at 72 °C. PCR products were separated on a 1.5% agarose gel, stained with GelRed from Biotium, and imaged on a Kodak Image Station 4000MM. Cell lysates were subjected to protein extraction and immunoblotting as previously described.36
In Vivo Studies
All in vivo studies were carried out in compliance with the University of Pittsburgh Institutional Animal Use and Care Committee (IACUC). Athymic nude mice were inoculated with head and neck squamous cell carcinoma (HNSCC) cell line 1483 (106 cells per site). Tumors were allowed to establish for 10 days. The tumor volumes were measured in two dimensions using a vernier caliper. Mice were randomized into groups such that the average tumor volume across the groups was the same. To determine whether systemic delivery of EGFRAS-GPNA was a viable strategy for treating HNSCC, we administered EGFRAS-GPNA to 2 mice with intraperitoneal (IP) injections. Three mice were treated with intratumoral (IT) injections of EGFRAS-GPNA and 2 mice were treated with EGFR sense oligonucleotides as a control. The mice were treated once daily with 2.5 mg/kg of EGFRAS-GPNA or EGFR sense oligonucleotides IT or 5 mg/kg of EGFRAS-GPNA IP. Treatment was carried out for 16 days. To determine the specificity of the EGFRAS-GPNA, HNSCC tumor-bearing mice were randomized into 3 groups of 10 mice each. Mice were treated with EGFRAS-GPNA (5 mg/kg once daily), scrambled control GPNA (5 mg/kg once daily), or saline. Treatments were administered intraperitoneally for 10 days. To compare the antitumor efficacy of EGFRAS-GPNA to EGFR targeting agents that are currently approved by the FDA or under clinical investigation for HNSCC, tumor-bearing mice were randomized into 5 groups including vehicle control, EGFRAS-GPNA, scrambled control GPNA, erlotinib, and cetuximab. The EGFRAS and scrambled GPNA had 10 mice per group. Other groups had 8 to 9 mice per group. Treatment was initiated on day 10 post-tumor inoculation. Vehicle control mice were administered saline intraperitoneally and also the 20% trappsol in saline (vehicle for erlotinib) via oral gavage daily. EGFRAS-GPNA or scrambled control GPNA was administered by IP at 5 mg/kg daily. Erlotinib was administered as a suspension via oral gavage once daily at 90 mg/kg body weight. Cetuximab was administered by IP at 0.8 mg/mouse twice a week. The treatment was carried out for 2 weeks. For all in vivo experiments, tumor volumes were measured thrice a week in 2 dimensions with a vernier caliper, and volumes were determined in mm3 using the formula 0.52lb2 (where l is the larger diameter and b is the smaller diameter of the tumor). At the end of the study, animals were euthanized, and the tumors were harvested. A part of the tumors were snap frozen on dry ice and the other part fixed in 10% buffered formalin for paraffin embedding. Frozen tumors were processed for protein and total RNA extraction. Sections of paraffin embedded tumors were subjected to immunohistochemical staining for EGFR and counter stained with hematoxylin and eosin. Percent positive EGFR staining and intensity of the stain were assessed by a board certified pathologist. Intensity of stain was graded on a scale of 0 to 3 with 0 being no stain, 1 being weak, 2 being moderate, and 3 being strong EGFR stain. The product of the percent positive cells and intensity was used to calculate the composite score.
Statistical Analyses
In vivo antitumor efficacy at each time point was assessed using the Kruskal−Wallis nonparametric test. Repeated measure ANOVA was used to compare the antitumor effect over time across different treatment groups.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by P50 CA097190 and a Pilot project in Lung cancer 5P50 CA 090440-07 (to S.M.T.), EY08098, R01 GM076251 (to D.H.L.), and DSF Charitable Foundation (to D.H.L. and B.A.A.), and American Cancer Society Clinical Research Professorship (to J.R.G.). The authors are grateful to L. G. M. Trevison for technical assistance with immunofluorescence imaging.
ABBREVIATIONS USED
- EGFR
epidermal growth factor receptor
- HNSCC
head and neck squamous cell carcinoma
- NSCLC
nonsmall cell lung cancer
- PNA
peptide nucleic acid
- GPNA
guanidine-based peptide nucleic acid
- FITC
fluorescein isothiocyanate
- TAMRA
tetramethylrhodamine
- ER
endoplasmic reticulum
- TFSMA
trifluoromethanesulfonic acid
- TFA
trifluoroacetic acid
Footnotes
ASSOCIATED CONTENT
Supporting Information
Molecular structures, method for monomer and oligomer synthesis, purification and analyses are included. Uptake of EGFRAS-GPNA by HNSCC cell lines (UMSCC-22B and PC-15B) and normal oral epithelial tissue (Het 1A) cells are presented. EGFR levels in animal tumors post-treatment are depicted. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Taramelli R, Acquati F. The human genome project and the discovery of genetic determinants of cancer susceptibility. Eur. J. Cancer. 2004;40:2537–2543. doi: 10.1016/j.ejca.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 2.Tan DS, Gerlinger M, Teh BT, Swanton C. Anti-cancer drug resistance: understanding the mechanisms through the use of integrative genomics and functional RNA interference. Eur. J. Cancer. 2010;46:2166–2177. doi: 10.1016/j.ejca.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Stein CA. Keeping the biotechnology of antisense in context. Nat. Biotechnol. 1999;17:209. doi: 10.1038/6909. [DOI] [PubMed] [Google Scholar]
- 4.Juliano RL, Yoon H. Aspect of the transport and delivery of antisense oligonucleotides. Curr. Opin. Mol. Ther. 2000;2:297–303. [PubMed] [Google Scholar]
- 5.De Mesmaeker A, Haner R, Martin P, Moser HE. Antisense oligonucleotide. Acc. Chem. Res. 1995;28:366–374. [Google Scholar]
- 6.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen PE. Peptide nucleic acid. A molecule with two identities. Acc. Chem. Res. 1999;32:624–630. [Google Scholar]
- 8.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 9.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sonnichsen SH, Nielsen PE. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 10.Ray A, Norden B. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J. 2000;14:1041–1060. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- 11.Chin JY, Kuan JY, Lonkar PS, Krause DS, Seidman MM, Peterson KR, Nielsen PE, Kole R, Glazer PM. Correction of a splice-site mutation in the beta-globin gene stimulated by triplex-forming peptide nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13514–13519. doi: 10.1073/pnas.0711793105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Satsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allelespecific silencing of mutant huntington and ataxia-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominski Z, Kole R. Restoration of correct splicing in thalassemic premessenger RNA by antisense oligonucleotides. Proc. Nat. Acad. Sci. U.S.A. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutrona G, Carpaneto EM, Ulivi M, Roncella S, Landt O, Ferrarini M, Boffa LC. Effects in live cells of a c-myc anti-gene PNA linked to a nuclear localization signal. Nat. Biotechnol. 2000;18:300–303. doi: 10.1038/73745. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen PE. PNA technology. Mol. Biotechnol. 2004;26:233–248. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 16.Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 2001;19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- 17.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 18.Amirkhanov NV, Zhang K, Aruva MR, Thakur ML, Wickstrom E. Imaging human pancreatic cancer xenograft by targeting mutant KRAS2 mRNA with [111In]DOTAn-poly-(diamidopropanoyl)m-KRAS2 PNA-D(Cys-Ser-Lys-Cys) nanoparticles. Bioconjugate Chem. 2010;21:731–740. doi: 10.1021/bc900523c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sazani P, Gemignani F, Kang S-H, Maier MA, Manoharan M, Persmark M, Bortner D, Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 20.Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA) Adv. Drug Delivery Rev. 2003;55:267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhou P, Wang M, Du L, Fisher GW, Waggoner A, Ly DH. Novel binding and efficient cellular uptake of guanidine-based peptide nucleic acids (GPNA) J. Am. Chem. Soc. 2003;125:6878–6879. doi: 10.1021/ja029665m. [DOI] [PubMed] [Google Scholar]
- 22.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 24.Dragulescu-Andrasi A, Zhou P, He G, Ly DH. Cell-permeable GPNA with appropriate backbone stereochemistry and spacing binds sequence-specifically to RNA. Chem. Commun. 2005:244–246. doi: 10.1039/b412522c. [DOI] [PubMed] [Google Scholar]
- 25.Dragulescu-Andrasi A, Rapireddy S, He G, Bhattacharya B, Hyldig-Nielsen JJ, Zon G, Ly DH. Cell-permeable peptide nucleic acid designed to bind to the 5′-untranslated region of e-cadherin transcript induces potent and sequence-specific antisense effects. J. Am. Chem. Soc. 2006;128:16104–16112. doi: 10.1021/ja063383v. [DOI] [PubMed] [Google Scholar]
- 26.Thomas S, Grandis J. Importance of epidermal growth factor receptor in head and neck cancer. Head Neck Cancer Updates. 2001;1:1–11. [Google Scholar]
- 27.Grandis JR, Tweardy DJ. TGF-alpha and EGFR in head and neck cancer. J. Cell. Biochem. 1993;(Suppl. 17F):188–191. doi: 10.1002/jcb.240531027. [DOI] [PubMed] [Google Scholar]
- 28.Kruser TJ, Wheeler DL. Mechanisms of resistance to HER family targeting antibodies. Exp. Cell Res. 2010;316:1083–1100. doi: 10.1016/j.yexcr.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Thomas SM, Grandis JR. Pharmacokinetic and pharmacodynamic properties of EGFR inhibitors under clinical investigation. Cancer Treat. Rev. 2004;30:255–268. doi: 10.1016/j.ctrv.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Chai RL, Grandis JR. Advances in molecular diagnostics and therapeutics in head and neck cancer. Curr. Treat. Options Oncol. 2006;7:3–11. doi: 10.1007/s11864-006-0027-4. [DOI] [PubMed] [Google Scholar]
- 31.Niwa H, Wentzel AL, Li M, Gooding WE, Lui VW, Grandis JR. Antitumor effects of epidermal growth factor receptor antisense oligonucleotides in combination with docetaxel in squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2003;9:5028–5035. [PubMed] [Google Scholar]
- 32.Zhou P, Dragulescu-Andrasi A, Bhattacharya B, O’Keefe H, Vatta P, Hyldig-Nielsen JJ, Ly DH. Synthesis of cell-permeable peptide nucleic acids and characterization of their hybridization and uptake properties. Bioorg. Med. Chem. Lett. 2006;16:4931–4935. doi: 10.1016/j.bmcl.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 33.Cassell A, Grandis JR. Investigational EGFR-targeted therapy in head and neck squamous cell carcinoma. Expert Opin. Invest. Drugs. 2010;19:709–722. doi: 10.1517/13543781003769844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vermorken JB, Herbst RS, Leon X, Amellal N, Baselga J. Overview of the efficacy of cetuximab in recurrent and/or metastatic squamous cell carcinoma of the head and neck in patients who previously failed platinum-based therapies. Cancer. 2008;112:2710–2719. doi: 10.1002/cncr.23442. [DOI] [PubMed] [Google Scholar]
- 35.Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu LL. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J. Clin. Oncol. 2004;22:77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- 36.Lui VW, Thomas SM, Zhang Q, Wentzel AL, Siegfried JM, Li JY, Grandis JR. Mitogenic effects of gastrinreleasing peptide in head and neck squamous cancer cells are mediated by activation of the epidermal growth factor receptor. Oncogene. 2003;22:6183–6193. doi: 10.1038/sj.onc.1206720. [DOI] [PubMed] [Google Scholar]
- 37.Lai SY, Koppikar P, Thomas SM, Childs EE, Egloff AM, Seethala RR, Branstetter BF, Gooding WE, Muthukrishnan A, Mountz JM, Lui VW, Shin DM, Agarwala SS, Johnson R, Couture LA, Myers EN, Johnson JT, Mills G, Argiris A, Grandis JR. Intratumoral epidermal growth factor receptor antisense DNA therapy in head and neck cancer: first human application and potential antitumor mechanisms. J. Clin. Oncol. 2009;27:1235–1242. doi: 10.1200/JCO.2008.17.8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, Ove R, Kies MS, Baselga J, Youssoufian H, Amellal N, Rowinsky EK, Ang KK. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 39.Loeffler-Ragg J, Schwentner I, Sprinzl GM, Zwierzina H. EGFR inhibition as a therapy for head and neck squamous cell carcinoma. Expert Opin. Invest. Drugs. 2008;17:1517–1531. doi: 10.1517/13543784.17.10.1517. [DOI] [PubMed] [Google Scholar]
- 40.Bianco R, Troiani T, Tortora G, Ciardiello F. Intrinsic and acquired resistance to EGFR inhibitors in human cancer therapy. Endocr.-Relat. Cancer. 2005;12(Suppl 1):S159–S171. doi: 10.1677/erc.1.00999. [DOI] [PubMed] [Google Scholar]
- 41.Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, Freilino ML, Graner MW, Wikstrand CJ, Bigner DD, Gooding WE, Furnari FB, Grandis JR. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin. Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 42.Rubin Grandis J, Chakraborty A, Melhem MF, Zeng Q, Tweardy DJ. Inhibition of epidermal growth factor receptor gene expression and function decreases proliferation of head and neck squamous carcinoma but not normal mucosal epithelial cells. Oncogene. 1997;15:409–416. doi: 10.1038/sj.onc.1201188. [DOI] [PubMed] [Google Scholar]
- 43.Thomas SM, Ogagan MJ, Freilino ML, Strychor S, Walsh DR, Gooding WE, Grandis JR, Zamboni WC. Antitumor mechanisms of systemically administered epidermal growth factor receptor antisense oligonucleotides in combination with docetaxel in squamous cell carcinoma of the head and neck. Mol. Pharmacol. 2008;73:627–638. doi: 10.1124/mol.107.041160. [DOI] [PubMed] [Google Scholar]
- 44.Sahu B, Chenna V, Lathrop KL, Thomas SM, Zon G, Livak KJ, Ly DH. Synthesis of conformationally preorganized and cell-permeable guanidine-based gamma-peptide nucleic acids (gammaGPNAs) J. Org. Chem. 2009;74:1509–1516. doi: 10.1021/jo802211n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browne KA, Dempcy RO, Bruice TC. Binding studies of cationic thymidyl deoxyribonucleic guanidine to RNA homopolynucleotides. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7051–7055. doi: 10.1073/pnas.92.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swenson DL, Warfield KL, Warren TK, Lovejoy C, Hassinger JN, Ruthel G, Blouch RE, Moulton HM, Weller DD, Iversen PL, Bavari S. Chemical modifications of antisense morpholino oligomers enhance their efficacy against Ebola virus infection. Antimicrob. Agents Chemother. 2009;53:2089–2099. doi: 10.1128/AAC.00936-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warren TK, Warfield KL, Wells J, Swenson DL, Donner KS, Van Tongeren SA, Garza NL, Dong L, Mourich DV, Crumley S, Nichols DK, Iversen PL, Bavari S. Advanced antisense therapies for postexposure protection against lethal filovirus infections. Nat. Med. 2010;16:991–994. doi: 10.1038/nm.2202. [DOI] [PubMed] [Google Scholar]
- 48.Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, Egholm M, Buchardt O, Nielsen PE, Coull J, Berg RH. Solid-phase synthesis of peptide nucleic acids. J. Pept. Sci. 1995;1:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 49.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, Ferris RL, Lai SY. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 50.Wheeler SE, Suzuki S, Thomas SM, Sen M, Leeman-Neill RJ, Chiosea SI, Kuan CT, Bigner DD, Gooding WE, Lai SY, Grandis JR. Epidermal growth factor receptor variant III mediates head and neck cancer cell invasion via STAT3 activation. Oncogene. 2010;29:5135–5145. doi: 10.1038/onc.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.