Abstract
While breast cancer mortality rate has seen a steady decline in the last few decades, advances in better treatment and diagnostic tools remain important as we come into the age of personalized therapy. In this report, we describe our studies of SGK3’s role in breast cancer. SGK3 (also known as CISK) is a member of the AGC family of kinases. Our previous work indicates that SGK3 functions downstream of the PI 3-kinase cascade and shares molecular and biochemical similarities with Akt. Here we show that SGK3 expression is linked to estrogen receptor (ER) both in breast caner cell lines and in primary tumor samples. Our analysis also indicated a positive correlation between SGK3 expression and tumor prognosis. Importantly, our immunochemistry analysis of human tumor samples established a clinical link between SGK3 expression and ER+ tumors. These findings implicate SGK3 as an additional component to a complex and heterogeneous disease, and point to the potential benefits of incorporating SGK3 into the process of breast cancer diagnosis and treatment.
Keywords: SGK3, CISK, estrogen receptor, breast cancer, immunohistochemistry, PI 3-kinase
Introduction
Improved screening and treatment has significantly reduced the mortality rates of breast cancer, the most commonly occurring cancer among women [1, 2]. Hereditary breast cancers can be attributed mostly to high-penetrance breast cancer susceptibility genes, for example, tumor suppressor genes BRCA1, BRCA2, p53, ATM, BARD1, and PTEN [3–12]. Sporadic breast cancer cases, which make up the majority of all breast cancers, are likely the results of low-penetrance risk factors working together with endogenous (e.g., hormones) and exogenous risk factors (e.g., pollution and diet). The inherent heterogeneity and complexity of breast cancer will continue to challenge researchers and clinicians in diagnosing and treating the disease
Among the many genetic factors associated with cancer, the growth factor receptor (GFR)/PI 3-kinase cascade has been one of the most widely studied. Mutations of various components of the PI 3-kinase signaling pathway have been found in many types of cancers including breast cancer. These include the oncogenic amplification and over-activation of human EGFR family (HER) of receptor tyrosine kinases such as EGFR [13, 14] and HER2 [15, 16], activating mutations and amplification of the gene that encodes the catalytic subunit of PI 3-kinase (p110) [17–19], overexpression of PDK1 [20, 21], activation of Akt genes [22, 23], and germ-line and somatic mutations of the tumor suppressor PTEN [24, 25]. These observations indicate that players of the PI 3-kinase pathway are part of the cancer molecular signature and represent prime targets for potential anti-cancer drugs and therapies.
Clinically, ER is one of the most important markers in breast cancer diagnosis and treatment. Nearly 70% of all breast cancers express ER where higher ER expression is often associated with better outcome for endocrine therapy. More recently, breast cancer research has benefited from laboratory technological advances that enable more nuanced classification of breast tumors both clinically and molecularly. For example, mature technologies such as microarray and more recently deep-sequencing have enabled breast tumors to be divided based on an ever-expanding list of potential diagnostic and prognostic markers [29–34]. Some of the most commonly seen tumors fall into the luminal A or B categories. Luminal A tumors are characterized by high ER/PR levels along with low expression of proliferative markers such as Ki-67, whereas luminal B tumors are often low for ER/PR but higher for proliferative markers [35]. Molecularly, crosstalks between ER and PI 3-kinase/Akt pathways have also been shown to play a role in the development of drug resistance [36–39].
During our previous studies, we carried out an Enhanced Retroviral Mutagen (ERM)-mediated genetic screen and identified the ser/thr kinase SGK3 (also known as CISK) as a survival kinase that functions downstream of the PI 3-kinase cascade [26, 27]. It is a member of the serum-glucocorticoid-regulated (SGK) family protein kinases, as well as the AGC superfamily. We showed that SGK3 could protect cells from apoptosis induced by factor withdrawal [28]. Importantly, activated SGK3 can promote estrogen/estrogen receptor (ER) dependent transcription and cell survival [28]. Recent studies using the MCF-7 breast cancer cell line support the model that SGK3 and ER form a feedback loop [46]. In this report, we provide further evidence for the feedback regulation between SGK3 and ER. Furthermore, this model was corroborated by our analysis of human breast tumor samples. Our data establish a clinical link between SGK3 and ER, and underline the importance of incorporating SGK3 as a new component in the assessment of breast cancer.
Materials and Methods
Cell lines, constructs, and antibodies
MCF-7L [28, 40, 41] cells were used for qRT-PCR, western blotting, and microarray analysis. Cells were cultured in DMEM supplemented with 10% FBS. Alternatively, MCF-7 cells were maintained in phenol red-free DMEM media containing 10% charcoal-stripped (CS) serum for 48 hours, before the addition of 10nM 17β-estradiol (E2) (Sigma-Aldrich). Cells were then collected at the indicated time points after addition of E2 for mRNA or protein expression analysis. Parental HEK293T cells and those transiently expressing activated SGK3 [26, 27] or SGK3 shRNA sequences [28] were used for antibody testing for immunohistochemistry analysis.
Real-time qRT-PCR and western blotting
Real-time quantitative PCR was performed as described previously [42]. Total RNA was isolated with the RNeasy Mini Kit (Qiagen, Valencia, CA) for reverse transcription using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA). Real-time qPCR amplification reactions were performed using the SYBR green master mix and an ABI StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA).
Cells were freshly lysed in lysis buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.15 unit/ml aprotinin, 20 µM leupeptin, 1 mM sodium vanadate, 1 µM β-glycerol phosphate, and 1 µM dithiothreitol). Cell lysates were resolved by 8% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad). Rabbit polyclonal antibodies for SGK3 (against the PX domain) were generated by Bethyl Laboratories (Montgomery, TX). Anti-Rabbit-HRP was obtained from Bio-Rad.
Establishment of endocrine resistance breast cancer cell models
Two endocrine resistance cell models of T47D and BT483 were established in this study. Briefly, the ER+ T47D and BT483 cells growing in full medium (RPMI-1640, 10% FBS) were adapted in estrogen-deprived (ED) medium (phenol red-free RPMI-1640, 10% CS-FBS), with or without the addition of 10−7 M of 4-OH-tamoxifen (Tam) (Sigma-Aldrich) for >6 months. In contrast to parental cell lines that are sensitive to endocrine treatment (ED or Tam), the derivatives developed resistance phenotypes to ED (EDR) or Tam (TamR) and exhibited regular growth rate (passaged twice/week with a ratio of 1:3). The endocrine resistant MCF-7 xenograft models have been previously published [40, 41].
Patient tumor samples
A total of 133 patients, who underwent surgical excision between 2006 and 2011 at MDACC and had available formalin-fixed, paraffin-embedded tissue blocks of primary invasive breast carcinoma, were identified from the surgical pathology files in the Department of Pathology and included in this study. Patient age, tumor histologic type, pathologic tumor size, tumor histologic grade, prognostic/predictive marker status, lymph node status, and the history of neoadjuvant chemotherapy were retrospectively recorded from the medical records. The American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guideline recommendations were followed for scoring estrogen receptor (ER), progesterone receptor (PR), and HER2 [43, 44], and performed as part of the routine pathology evaluation. Briefly, ER and PR status were determined by immunohistochemical (IHC) staining with monoclonal antibodies against ER (6F11, Novacastra laboratories Ltd, Burlingame, CA) and PR (1A6, Novacastra Laboratories Ltd). Positive staining was defined as nuclear staining in at least 1% of invasive cancer cells. HER2 status was tested by fluorescence in situ hybridization (FISH) with the PathVision kit (Vysis, Downers Grove, IL), or by IHC staining with a monoclonal antibody (AB8, Neomarkers, Fremont, CA). Triple-negative (ER negative, PR negative, and HER2 negative) was defined accordingly. In addition, Ki67 proliferation index performed as part of routine pathology evaluation by IHC (MIB-1, Dako, Carpenteria, CA) was recorded from the medical records whenever available. This study was approved by the institutional review board of MDACC.
SGK3 immunohistochemistry staining and evaluation
In each case, one whole slide of unstained tissue section (4-μm thick) that had been prepared from a representative paraffin block of a surgically excised primary invasive breast carcinoma was deparaffinized in xylene and rehydrated to 50% ethanol. Antigen retrieval was achieved by pressure cooking the slide in 10 mM citric buffer (pH 6.0) for 5 minutes. Endogenous peroxidase activity was inactivated by treating the slide with 3% H2O2 for 5 minutes at room temperature. The slide was then washed and blocked for 1 hour in 1xPBS containing 3% normal non-immune goat serum to reduce nonspecific staining. This was followed by incubating the slide with rabbit polyclonal anti-CISK PX antibody (5µg/ml) overnight at 4°C, washing 3 times with 1xTBST, and incubation with EnVision+ System-HRP labeled anti-Rabbit polymer (DAKO) for 30 minutes at room temperature. Visualization was achieved by incubating slides with Liquid DAB+ Substrate Chromogen System (DAKO) for 10 minutes at room temperature.
SGK3 expression was reviewed and recorded as the proportion of cells stained and staining intensity in the invasive carcinoma in each case. The results were evaluated using the Q score method [45]. The proportion of stained cells was scored as follows: < 1%, score 0; 1%–25%, score 1; 26%–50%, score 2; 51%–75%, score 3; >75%, score 4. The staining intensity was scored as follows: no staining, score 0; weak average staining, score 1; moderate average staining, score 2; strong average staining, score 3. The Q score was generated from adding the proportion score and the intensity score, which led to a range of 0 to 7. A Q score of >2 was considered positive for SGK3.
Statistical analysis
All statistical tests were performed using R version 2.12.0 (http:www/r-project.org). For analyzing the correlation between two categorical variables, Fisher’s exact test was used. Logistic regression was used to analyze the association between SGK3 positivity as defined above and factors of interest. A P value of <0.05 indicated statistical significance. Cox proportional hazards regression analysis was used to assess the association between SGK3 expression and patient survival.
Results
SGK3 functions as a classic ER-dependent gene
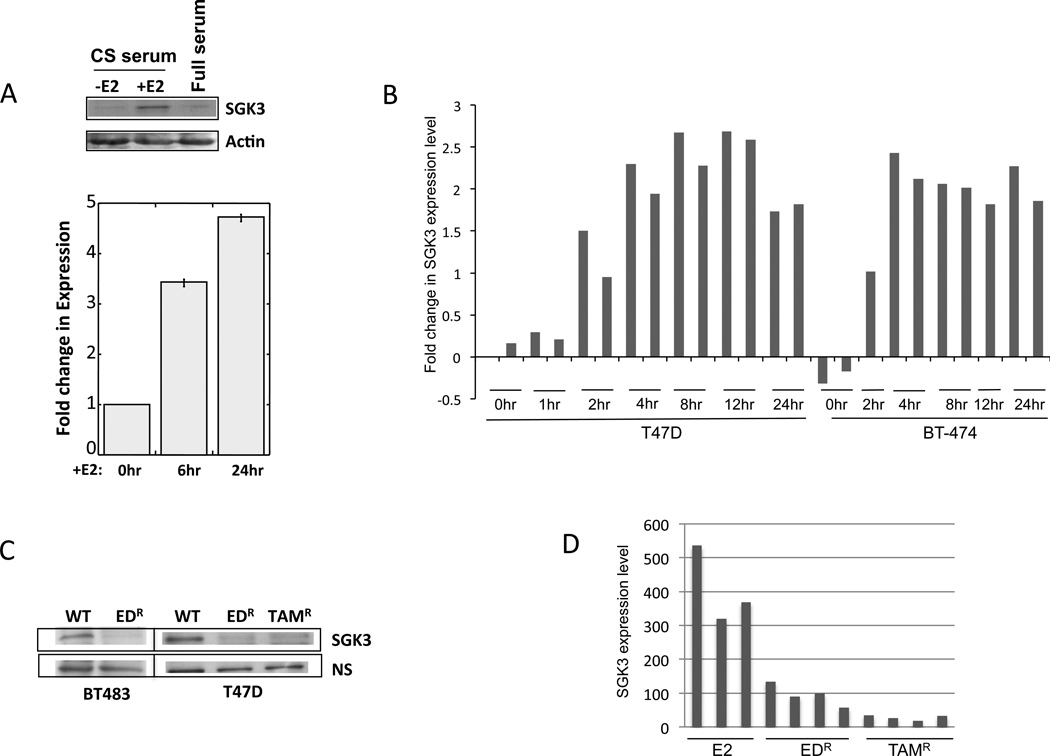
SGK3 appears to be widely expressed in a variety of tissues both in mice [27] and in human (suppl Fig.S1). Previously we have shown that SGK3 is crucial to estrogen-dependent growth of the ER+ breast cancer cell line MCF-7 [28], suggesting that SGK3 potentiates the transcriptional activation function and proliferative effect of estrogen/ER signaling. Using the MCF-7 cell line model, Wang et al provided evidence that SGK3 is an E2 dependent gene [46]. To further explore the regulation of SGK3 by estrogen/ER, we examined the mRNA as well as protein expression of SGK3 in our MCF-7 cells [28]. Consistent with published data, when these cells were stimulated with the addition of estrogen, we observed increased expression of SGK3 proteins as well as mRNAs (Figure 1a). This estrogen/ER-dependent transcriptional activation could also be observed in additional ER+ breast cancer cell lines (BT474 and T47D), when we analyzed gene expression data from our previously-published studies (Figure 1b) [47]. These findings support the model that SGK3 forms a feedback regulatory loop with ER.
Figure 1.
SGK3 expression is controlled by estrogen/ER signaling. A) Parental MCF-7 cells pretreated with charcoal-stripped (CS) serum for 2 days were stimulated with 10nM β-estradiol (+E2). Cells grown in full serum were also included. Whole cell lysates were collected 24 hours after E2 addition for western blot analysis. An anti-actin antibody was used as loading control. mRNA was collected at the indicated time points for qPCR. B) Up-regulated SGK3 mRNA expression in response to estrogen treatment is a general feature of ER+ breast cancer cell lines. SGK3 mRNA expression following estrogen treatment in T47D and BT-474 cells from a previously published study [47] were plotted. C) SGK3 protein expression was suppressed when estrogen receptor was inhibited in breast cancer cells. Anti-SGK3 antibodies were used to western blot whole cell lysate from wildtype (WT) T47D and BT483 cells and their derivatives. NS, non-specific band was used as loading control. EDR, resistant clones from estrogen deprivation treatment. TamR, resistant clones of tamoxifen-treated cells. D) SGK3 mRNA expression was suppressed when estrogen receptor was inhibited in MCF-7 endocrine resistance cells. E2, estrogen treatment.
Further corroboration came from our analysis of SGK3 expression under conditions that inhibited ER signaling. Here, we established two endocrine resistance cell models (T47D and BT483) (see Materials and Methods). As shown in Figure 1c, protein expression of SGK3 was down-regulated in these derivative cells (TamR and EDR) compared to their parental counterparts. Furthermore, in our previously reported MCF-7 xenograft mouse models of estrogen deprivation or tamoxifen treatment [40, 41], the mRNA level of SGK3 was also significantly decreased (Figure 1d). When endocrine-resistant tumors eventually developed in these xenografted mice, SGK3 expression remained blocked (data not shown). Collectively, these data indicate that SGK3 bears the hallmarks of a classic ER-dependent gene that functions downstream of estrogen/ER.
SGK3 expression in breast cancer patient samples
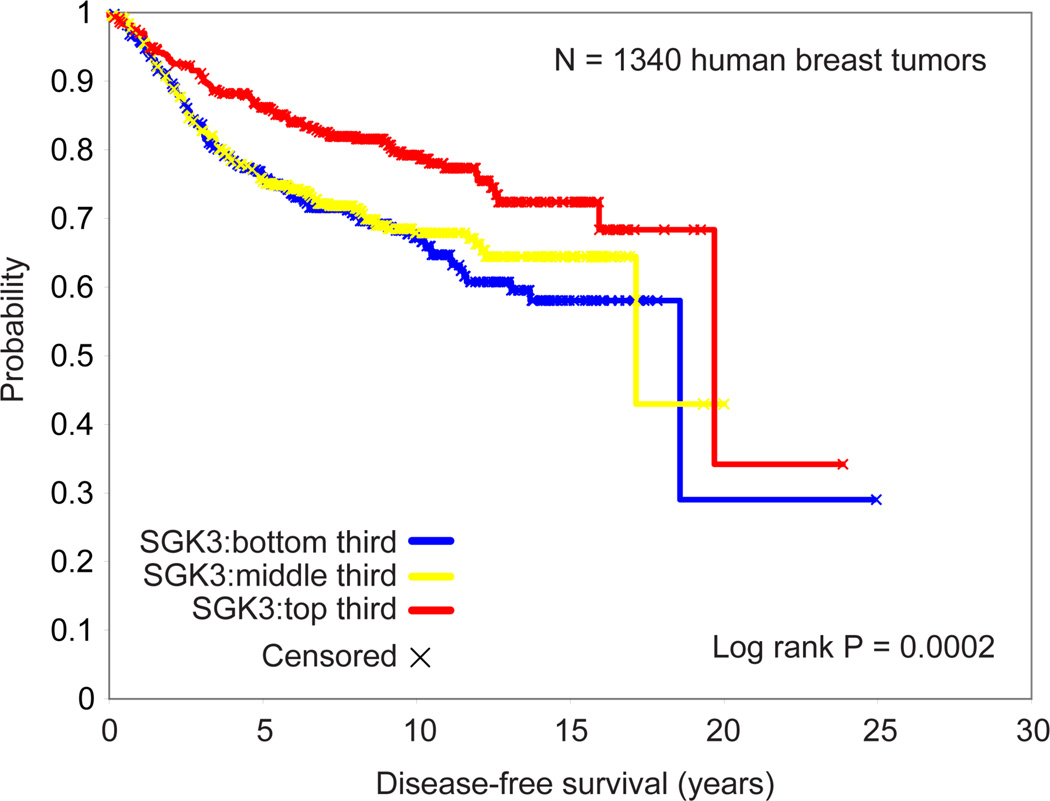
The analysis thus far suggests a strong correlation of SGK3 and ER expression. We then analyzed SGK3 expression in patients using the Oncomine Research database (http://www.oncomine.org) for gene expression studies involving ≥200 patients with known molecular subtypes based on PAM50 classification. Patients from the Hatzis dataset were found to have significantly higher SGK3 expression in the luminal A and B subtypes when compared to those in HER2, basal, or normal subtypes (suppl. Fig.S2)[48]. When we examined SGK3 levels in a compendium of expression profiles (Affymetrix U133A) on 1340 human breast tumors (which were compiled previously from published studies [49]) we found lower SGK3 expression to associate with poorer prognosis (Figure 2) (P<0.0001, univariate Cox). When we only examined ER+ tumors (N=686), an association between higher SGK3 level and better prognosis could still be observed, though the trend here was not as strong (P=0.04, univariate Cox).
Figure 2.
SGK3 mRNA expression is associated with better outcome in breast cancer, based on data from a compendium of gene expression profiles from published studies (N=1340).
Validation of anti-SGK3 antibody for immunohistochemistry (IHC) analysis
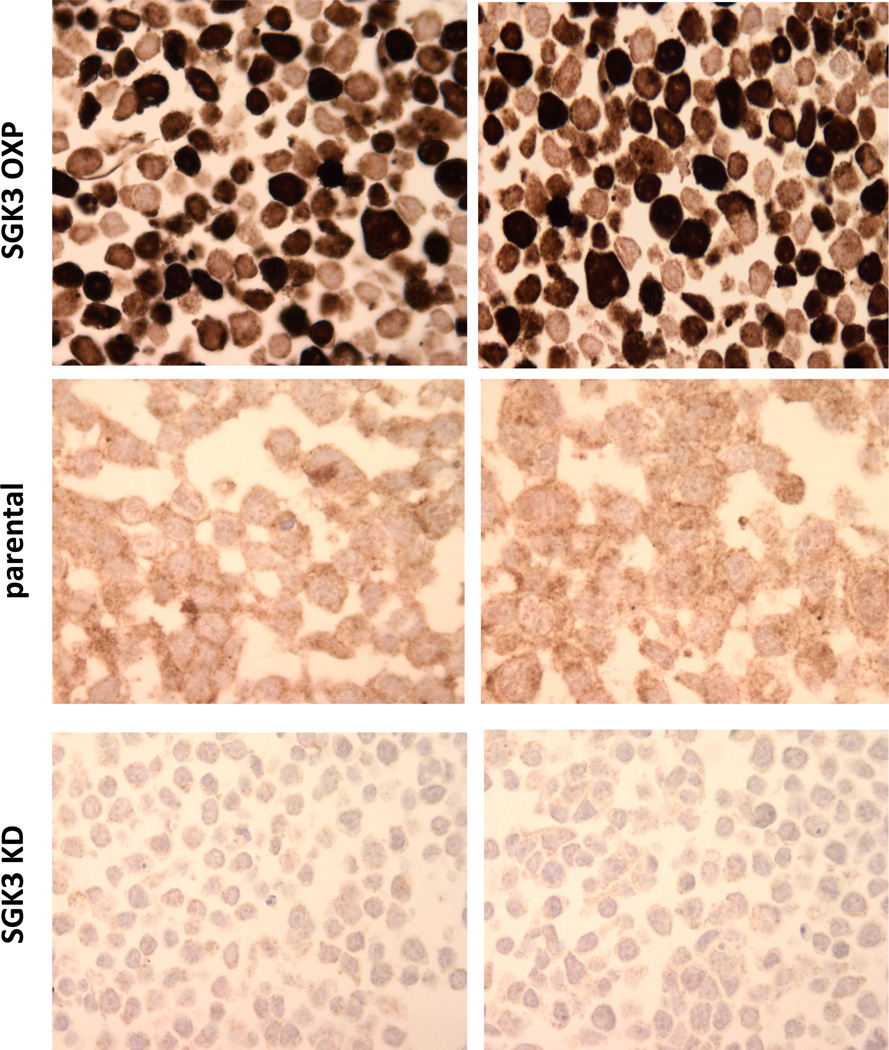
To probe the link between SGK3 and ER expression in breast tumors, we next established a SGK3 IHC assay for clinical samples. We have previously generated an antibody against the unique N-terminal PX domain of SGK3, and successfully used it to western blot and immunoprecipitate SGK3 using cultured cells (Figure 1a) [28]. To characterize the antibody for IHC, we generated 293T cells transiently expressing full-length activated SGK3 (SGK3 OXP) [26, 27], or shRNA sequences against SGK3 (SGK3 KD) [28], and made formalin-fixed, paraffin embedded pellets of these cells. As shown in Figure 3, our antibody could recognize both endogenous and exogenous SGK3 for IHC. More importantly, SGK3 antibody staining was significantly reduced in cells expressing SGK3 shRNA sequences. These results indicate that the antibody is suitable for IHC analysis of clinical breast cancer samples.
Figure 3.
Analysis of anti-SGK3 antibody for IHC. Parental 293T cells and those transiently expressing activated SGK3 (SGK3 OXP, top) or an shRNA sequence against SGK3 (SGK3 KD, bottom) were harvested and used for IHC analysis.
Distinct SGK3 expression patterns in breast tumor samples
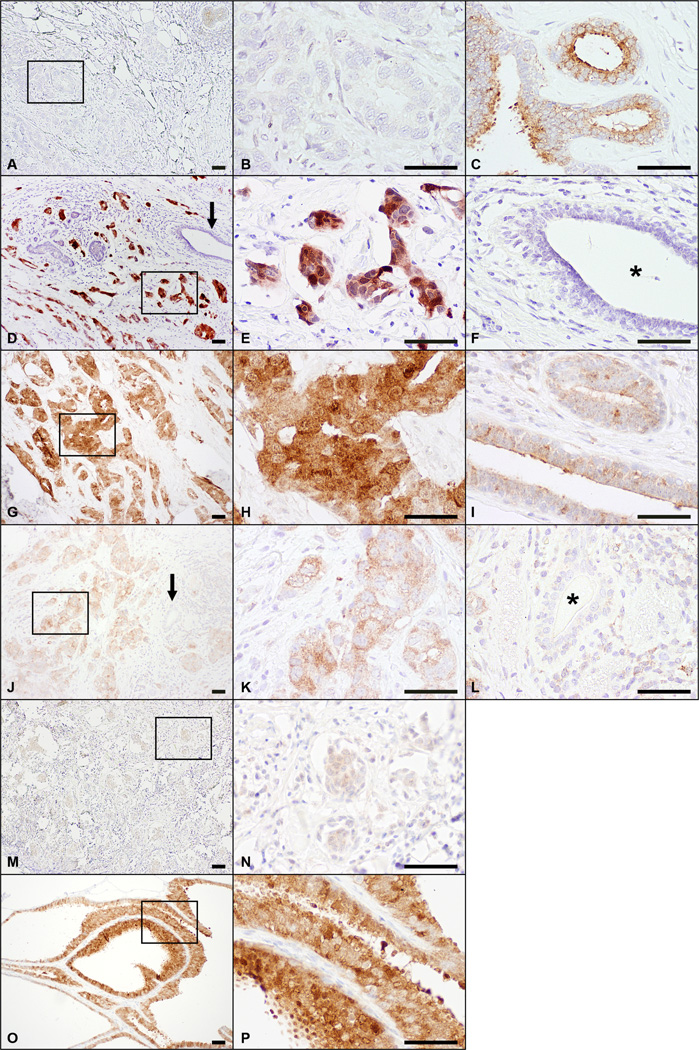
To further investigate the role of SGK3 in breast cancer, we examined the expression of SGK3 in 133 clinical primary invasive breast carcinomas from 133 patients. All patients were women between the ages of 27 and 88 years (median, 55 years). The clinicopathologic features of these patients are summarized in Table 1. The Q score distribution for SGK3 staining is summarized in Table 2. In tumors with strong staining, simultaneous cytoplasmic and nuclear staining was noted (Figure 4). In tumors with weak or moderate staining, SGK3 expression appeared to be predominantly in the cytoplasm, where simultaneous staining of the nucleus was only seen in rare cases. Scattered non-neoplastic breast epithelium was present at the periphery of the invasive carcinoma in 105 tumor samples that stained for SGK3. SGK3 expression was predominantly cytoplasmic in the surrounding non-neoplastic epithelium with rare cases showing both cytoplasmic and nuclear staining. Using a Q score of >2 as cutoff, positive staining for SGK3 was observed in 71 tumors (53%) and in the surrounding non-neoplastic epithelium in 64 samples (61%). As summarized in Table 1, in univariate analyses, there was substantial evidence of an association between SGK3 staining and ER status, PR status, and triple-negative status of the tumor (p<0.0001). Patients with ER-positive, PR-positive, or non-triple-negative tumors were more likely to have positive SGK3 staining. In addition, there was a statistically significant association between SGK3 staining and tumor grade (p=0.008) where patients with lower grade tumors were more likely to have positive SGK3 staining. An association between SGK3 staining and axillary lymph node status (p=0.02) was also observed. Other clinicopathologic features were not significantly associated with SGK3 staining.
Table 1.
SGK3 IHC staining in association with clinicopathologic features. %* indicates the percentage of patients out of each corresponding subgroup from the preceding column.
| Clinicopathologic features | Number of patients (%) (Total=133) |
Number of positive SGK3 staining (%*) |
P-value (Fisher’s exact test) |
|---|---|---|---|
| Patient age | 1.00 | ||
| ≤ 55 yrs | 68 (51%) | 36 (53%) | |
| > 55 yrs | 65 (49%) | 35 (54%) | |
| Tumor type | 0.83 | ||
| IDC/IMC | 118 (89%) | 62 (53%) | |
| ILC | 11 (8%) | 7 (63%) | |
| Metaplastic carcinoma | 4 (3%) | 2 (50%) | |
| Tumor size | 0.31 | ||
| ≤2 cm | 75 (56%) | 37 (49%) | |
| >2 cm and ≤ 5 cm | 41 (31%) | 26 (63%) | |
| >5 cm | 17 (13%) | 8 (47%) | |
| Tumor Grade | 0.008 | ||
| 1 | 16 (12%) | 12 (75%) | |
| 2 | 58 (44%) | 36 (62%) | |
| 3 | 59 (44%) | 23 (39%) | |
| Axillary lymph node status | 0.02 | ||
| Negative | 70 (53%) | 36 (51%) | |
| Micrometastasis | 2 (2%) | 1 (50%) | |
| 1–3 positive | 36 (27%) | 14 (39%) | |
| 4–9 positive | 16 (12%) | 13 (81%) | |
| >10 positive | 9 (7%) | 7 (78%) | |
| Neoadjuvant chemotherapy | 0.16 | ||
| Yes | 34 (26%) | 22 (65%) | |
| No | 99 (74%) | 49 (50%) | |
| ER status | <0.0001 | ||
| Positive (≥1%) | 80 (60%) | 56 (70%) | |
| Negative | 52 (39%) | 14 (27%) | |
| Unknown | 1 (1%) | 1 (100%) | |
| PR status | <0.0001 | ||
| Positive (≥1%) | 74 (56%) | 51 (69%) | |
| Negative | 58 (44%) | 19 (33%) | |
| Unknown | 1 (1%) | 1 (100%) | |
| HER2 status | 0.68 | ||
| Positive | 6 (5%) | 4 (67%) | |
| Negative | 126 (95%) | 66 (52%) | |
| Unknown | 1 (1%) | 1 (100%) | |
| Ki67 status | 0.09 | ||
| Low (≤17%) | 21 (16%) | 15 (71%) | |
| Intermediate (>17% and ≤ 35%) | 12 (9%) | 7 (58%) | |
| High (>35%) | 19 (14%) | 7 (37%) | |
| Unknown | 81 (61%) | 42 (52%) | |
| Triple-negative status | <0.0001 | ||
| Yes | 47 (36%) | 12 (26%) | |
| No | 85 (64%) | 58 (68%) | |
| Unknown | 1 (1%) | 1 (100%) | |
Table 2.
Q-score distribution in patient samples from Table 1
| Q Score | 0 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
|
Number of samples |
31 | 31 | 17 | 17 | 15 | 15 | 7 |
Figure 4.
SGK3 expression in human breast tumor samples. A-C, A sample with negative SGK3 staining in invasive carcinoma (A, B) and moderate staining in the surrounding non-neoplastic breast epithelium (C). D-F, A sample with strong SGK3 staining in the invasive carcinoma (D, E) and negative staining in the surrounding non-neoplastic breast epithelium (arrow in D, and F). Both cytoplasmic and nuclear staining was seen in the tumor cells. G-I, A sample with strong SGK3 staining in the invasive carcinoma (G, H) with predominantly cytoplasmic staining. The neighboring nonneoplastic breast epithelium had weak to moderate staining (I). J-K, A sample with moderate SGK3 staining in the invasive carcinoma (J, K) and negative staining in the surrounding breast epithelium (arrow in J, and L). The staining in the tumor cells was cytoplasmic. M-N, A sample with weak SGK3 staining in the invasive carcinoma. O-P, Examples of strong SGK3 staining in non-neoplastic breast epithelium. The middle panels represent boxed higher magnification images of the boxed area in the left panel. Non-neoplastic breast epithelium in D and J as indicated by arrows is shown in F and L respectively. * indicates the lumen of the normal gland. Scale bars, 50 μm. Left panel, ×100. Middle and right panels, ×400.
Factors that were significantly associated with SGK3 expression were fit in a multivariate model. Because ER and PR are highly correlated, and ER, PR, and HER2 are highly correlated with triple negativity, two separate multivariate models were considered: one with ER status and another with triple-negative status. In these models, ER status and triple negativity remained statistically highly significant factors associated with SGK3 expression (Table 3 and Table 4). ER positive patients were more likely to have positive SGK3 expression (P=0.0002, OR=6.18) and triple-negative patients were more likely to have negative SGK expression (P=0.0007, OR=0.19). Lymph node status was of marginal significance in both models (P=0.05 and P=0.08, respectively). Taken together, the findings indicate that SGK3 expression is significantly associated with ER status in human breast tumor samples.
Table 3.
Multivariate Logistic Regression Model #1 (including tumor grade, ER status, and lymph node status).
| Parameter | Category | SGK 3 negative |
SGK3 positive |
OR | 95% CI (OR) |
P-value |
|---|---|---|---|---|---|---|
| Grade | 1 | 4 | 11 | ----- | ----- | 0.91 |
| 2 | 22 | 36 | 0.81 | 0.21, 3.09 | ||
| 3 | 36 | 23 | 0.71 | 0.16, 3.24 | ||
| ER | Negative | 38 | 14 | ----- | ----- | 0.0002 |
| Positive | 24 | 56 | 6.18 | 2.26, 16.9 | ||
| Lymph node | 0 | 34 | 36 | ----- | ----- | 0.05 |
| Micro | 1 | 1 | 0.42 | 0.02, 7.38 | ||
| 1–3 | 22 | 14 | 0.65 | 0.26, 1.61 | ||
| 4–9 | 3 | 12 | 3.76 | 0.86, 16.4 | ||
| 10+ | 2 | 7 | 5.05 | 0.82, 31.0 |
Table 4.
Multivariate Logistic Regression Model #2 (including tumor grade, triple-negative status, and lymph node status).
| Parameter | Category | SGK3 negative |
SGK3 positive |
OR | 95% CI (OR) |
P-value |
|---|---|---|---|---|---|---|
| Grade | 1 | 4 | 11 | ----- | ----- | 0.77 |
| 2 | 22 | 36 | 0.75 | 0.20, 2.81 | ||
| 3 | 36 | 23 | 0.59 | 0.13, 2.58 | ||
| Triple-Negative | No | 27 | 58 | ----- | ----- | 0.0007 |
| Yes | 35 | 12 | 0.19 | 0.07, 0.52 | ||
| Lymph node | 0 | 34 | 36 | ----- | ----- | 0.08 |
| Micro | 1 | 1 | 0.43 | 0.02, 7.66 | ||
| 1–3 | 22 | 14 | 0.65 | 0.27, 1.60 | ||
| 4–9 | 3 | 12 | 3.38 | 0.79, 14.5 | ||
| 10+ | 2 | 7 | 4.30 | 0.71, 26.1 |
Discussion
In this study, we explored estrogen/ER-dependent regulation of SGK3 in multiple ER+ cell line models, and in derivative cells that were resistant to tamoxifen or estrogen-deprivation treatment. Furthermore, we established for the first time the link between SGK3 upregulation and ER status in clinical breast cancer samples.
We have shown previously that SGK3 can positively regulate ER transcriptional activation in vitro [28]. This function depends on SGK3 kinase activity, which can be assessed by its phosphorylation status. Due to the lack of an anti-phosphoSGK3 antibody suitable for IHC analysis, we were unable to assess the activation status of SGK3 in patient tumor samples. However, accumulating data support the model of a positive feedback loop between ER transcriptional activity and SGK3 expression and possibly enzymatic activation. Crosstalks between GFR/PI 3-kinase signaling and ER pathway have been well documented [36–39]. It has been shown that PI 3-kinase pathway activation is inversely associated with ER expression and patient outcome [50–59]. As a result, combination therapies that also target the GFR pathways may prove effective in combating endocrine therapy resistance. The expression profile of SGK3, however, appears to positively correlate with ER expression and better prognosis. Such observations underline the uniqueness of individual components of the PI 3-kinase cascade.
We initially identified SGK3 in a genome-wide screen for cell survival genes [27]. As a survival kinase downstream of the PI 3-kinase pathway, SGK3 shares both sequence and functional similarity with Akt, another AGC family member. However, structurally and molecularly, there is clear non-redundancy between SGK3 and Akt. In place of the PH domain, SGK3 contains a PX domain that binds phospholipids with specificities distinct from the PH domain [27, 60]. One of the substrates shared between Akt and SGK3 is the pro-apoptotic protein BAD [61, 62]. BAD contains multiple potential phosphorylation sites for ser/thr kinases, and their phosphorylation is important for Bad function [63]. Even though the kinase domains for SGK3 and Akt exhibit extensive homology, the SGK3 phosphorylation sites on BAD differ from those for Akt [27]. Furthermore, proteomic analysis revealed that the SGK3 protein complex in cells differed from the Akt protein complex [64], suggesting unique interaction, activity, and signaling to each of the two kinases. These observations underline the unique properties for SGK3 and Akt, both biochemically and at the cellular level.
While the role of Akt in breast cancer has been extensively studied, the function of SGK3 in breast cancer remains unclear. Our analysis of >1,000 tumor data revealed a positive correlation between SGK3 level and patient survival and prognosis. In comparison, no correlation between Akt mRNA expression and tumor prognosis was found in a previous analysis of breast tumors [50]. Such differences cannot be easily explained by our current knowledge of SGK3 and Akt, and are supported by findings from Vasudevan et al who demonstrated that SGK3 represents a non-redundant alternative pathway that is downstream of PI 3-kinase signaling and distinct from Akt in breast cancer cells [65]. More in-depth studies that dissect the intrinsic signaling pathways mediated by these kinases may shed more light on this subject, and offer better insight into the complex molecular properties of different types of breast cancer.
Supplementary Material
Acknowledgement
We would like to thank Drs. Yi Li and Zhou Songyang for technical help and Kim-Anh Vu for her assistance with the figures. We would also like to acknowledge the support of SU2C Breast Cancer programs, the Breast Cancer Research Foundation, P50 CA58183 (Breast Cancer SPORE), the Dan L. Duncan Cancer Center (P30CA125123), and the Administrative and Genome-wide RNAi Screens Cores (IDDRC P30HD024064). This research is supported in part by MD Anderson institutional Start-Up Funds (to L.H.) and the National Institutes of Health through MD Anderson's Cancer Center Support Grant CA016672.
Footnotes
Conflict of interest statement
The authors declare no conflicts of interest.
References
- 1.Dumitrescu RG, Cotarla I. Understanding breast cancer risk -- where do we stand in 2005? J Cell Mol Med. 2005;9(1):208–221. doi: 10.1111/j.1582-4934.2005.tb00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin DM. International variation. Oncogene. 2004;23(38):6329–6340. doi: 10.1038/sj.onc.1207726. [DOI] [PubMed] [Google Scholar]
- 3.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 4.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266(5182):120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 5.Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12(3):333–337. doi: 10.1038/ng0396-333. [see comments] [DOI] [PubMed] [Google Scholar]
- 6.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789–792. doi: 10.1038/378789a0. [see comments] [published erratum appears in Nature 1996 Feb 22;379, (6567):749] [DOI] [PubMed] [Google Scholar]
- 7.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268(5218):1749–1753. doi: 10.1126/science.7792600. [see comments] [DOI] [PubMed] [Google Scholar]
- 8.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer [see comments] Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 9.Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 10.Ellisen LW, Haber DA. Hereditary breast cancer. Annu Rev Med. 1998;49:425–436. doi: 10.1146/annurev.med.49.1.425. [DOI] [PubMed] [Google Scholar]
- 11.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108(2):171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 12.Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 13.Sauter G, Maeda T, Waldman FM, Davis RL, Feuerstein BG. Patterns of epidermal growth factor receptor amplification in malignant gliomas. Am J Pathol. 1996;148(4):1047–1053. [PMC free article] [PubMed] [Google Scholar]
- 14.Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res. 2002;62(22):6764–6769. [PubMed] [Google Scholar]
- 15.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26(45):6469–6487. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O'Meara S, Smith R, Parker A, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431(7008):525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 17.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7(6):561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 19.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peifer C, Alessi DR. New anti-cancer role for PDK1 inhibitors: preventing resistance to tamoxifen. Biochem J. 2009;417(1):e5–e7. doi: 10.1042/BJ20082243. [DOI] [PubMed] [Google Scholar]
- 21.Kim SH, Miller FR, Tait L, Zheng J, Novak RF. Proteomic and phosphoproteomic alterations in benign, premalignant and tumor human breast epithelial cells and xenograft lesions: biomarkers of progression. Int J Cancer. 2009;124(12):2813–2828. doi: 10.1002/ijc.24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 23.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448(7152):439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 24.Pendaries C, Tronchere H, Plantavid M, Payrastre B. Phosphoinositide signaling disorders in human diseases. FEBS Lett. 2003;546(1):25–31. doi: 10.1016/s0014-5793(03)00437-x. [DOI] [PubMed] [Google Scholar]
- 25.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27(41):5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 26.Liu D, Songyang Z. ERM-mediated genetic screens in mammalian cells. Methods Enzymol. 2008;446:409–419. doi: 10.1016/S0076-6879(08)01624-8. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, Yang X, Songyang Z. Identification of CISK, a new member of the SGK kinase family that promotes IL-3-dependent survival. Curr Biol. 2000;10(19):1233–1236. doi: 10.1016/s0960-9822(00)00733-8. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Liao L, Qin J, Xu J, Liu D, Songyang Z. Identification of Flightless-I as a Substrate of the Cytokine-independent Survival Kinase CISK. J Biol Chem. 2009;284(21):14377–14385. doi: 10.1074/jbc.M807770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220(2):263–280. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 30.Reis-Filho JS, Weigelt B, Fumagalli D, Sotiriou C. Molecular profiling: moving away from tumor philately. Sci Transl Med. 2010;2(47):47ps43. doi: 10.1126/scitranslmed.3001329. [DOI] [PubMed] [Google Scholar]
- 31.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 32.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61(16):5979–5984. [PubMed] [Google Scholar]
- 34.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 35.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YL, Law PY, Loh HH. Inhibition of PI3K/Akt signaling: an emerging paradigm for targeted cancer therapy. Curr Med Chem Anticancer Agents. 2005;5(6):575–589. doi: 10.2174/156801105774574649. [DOI] [PubMed] [Google Scholar]
- 37.Miller TW, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3- kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13(6):224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26(9):1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- 39.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 40.Creighton CJ, Massarweh S, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Shou J, Malorni L, Schiff R. Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts. Cancer Res. 2008;68(18):7493–7501. doi: 10.1158/0008-5472.CAN-08-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, Weiss H, Rimawi M, Schiff R. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68(3):826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 42.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 43.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131(1):18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 45.Barnes DM, Harris WH, Smith P, Millis RR, Rubens RD. Immunohistochemical determination of oestrogen receptor: comparison of different methods of assessment of staining and correlation with clinical outcome of breast cancer patients. Br J Cancer. 1996;74(9):1445–1451. doi: 10.1038/bjc.1996.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Zhou D, Phung S, Masri S, Smith D, Chen S. SGK3 is an estrogen-inducible kinase promoting estrogen-mediated survival of breast cancer cells. Mol Endocrinol. 2011;25(1):72–82. doi: 10.1210/me.2010-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Creighton CJ, Cordero KE, Larios JM, Miller RS, Johnson MD, Chinnaiyan AM, Lippman ME, Rae JM. Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors. Genome Biol. 2006;7(4):R28. doi: 10.1186/gb-2006-7-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, et al. A genomic predictor of response and survival following taxaneanthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335(6066):348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Creighton CJ. A gene transcription signature of the Akt/mTOR pathway in clinical breast tumors. Oncogene. 2007;26(32):4648–4655. doi: 10.1038/sj.onc.1210245. [DOI] [PubMed] [Google Scholar]
- 51.Tokunaga E, Kimura Y, Mashino K, Oki E, Kataoka A, Ohno S, Morita M, Kakeji Y, Baba H, Maehara Y. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13(2):137–144. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- 52.Tokunaga E, Kataoka A, Kimura Y, Oki E, Mashino K, Nishida K, Koga T, Morita M, Kakeji Y, Baba H, et al. The association between Akt activation and resistance to hormone therapy in metastatic breast cancer. Eur J Cancer. 2006;42(5):629–635. doi: 10.1016/j.ejca.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 53.Shin I, Arteaga CL. Expression of active Akt protects against tamoxifen-induced apoptosis in MCF-7 Cells. IUBMB Life. 2006;58(11):664–669. doi: 10.1080/15216540601001681. [DOI] [PubMed] [Google Scholar]
- 54.Vestey SB, Sen C, Calder CJ, Perks CM, Pignatelli M, Winters ZE. Activated Akt expression in breast cancer: correlation with p53, Hdm2 and patient outcome. Eur J Cancer. 2005;41(7):1017–1025. doi: 10.1016/j.ejca.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 55.Kirkegaard T, Witton CJ, McGlynn LM, Tovey SM, Dunne B, Lyon A, Bartlett JM. AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J Pathol. 2005;207(2):139–146. doi: 10.1002/path.1829. [DOI] [PubMed] [Google Scholar]
- 56.Stal O, Perez-Tenorio G, Akerberg L, Olsson B, Nordenskjold B, Skoog L, Rutqvist LE. Akt kinases in breast cancer and the results of adjuvant therapy. Breast Cancer Res. 2003;5(2):R37–R44. doi: 10.1186/bcr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi W, Zhang X, Pintilie M, Ma N, Miller N, Banerjee D, Tsao MS, Mak T, Fyles A, Liu FF. Dysregulated PTEN-PKB and negative receptor status in human breast cancer. Int J Cancer. 2003;104(2):195–203. doi: 10.1002/ijc.10909. [DOI] [PubMed] [Google Scholar]
- 58.Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL, et al. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol. 1999;155(4):1253–1260. doi: 10.1016/S0002-9440(10)65227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia JM, Silva JM, Dominguez G, Gonzalez R, Navarro A, Carretero L, Provencio M, Espana P, Bonilla F. Allelic loss of the PTEN region (10q23) in breast carcinomas of poor pathophenotype. Breast Cancer Res Treat. 1999;57(3):237–243. doi: 10.1023/a:1006273516976. [DOI] [PubMed] [Google Scholar]
- 60.Xu J, Liu D, Gill G, Songyang Z. Regulation of cytokine-independent survival kinase (CISK) by the Phox homology domain and phosphoinositides. J Cell Biol. 2001;154(4):699–705. doi: 10.1083/jcb.200105089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278(5338):687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 62.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell- intrinsic death machinery. Cell. 1997;91(2):231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 63.Gajewski TF, Thompson CB. Apoptosis meets signal transduction: elimination of a BAD influence. Cell. 1996;87:589–592. doi: 10.1016/s0092-8674(00)81377-x. [DOI] [PubMed] [Google Scholar]
- 64.Ding Z, Liang J, Lu Y, Yu Q, Songyang Z, Lin SY, Mills GB. A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc Natl Acad Sci U S A. 2006;103(41):15014–15019. doi: 10.1073/pnas.0606917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16(1):21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.