Abstract
The expression, localization and activity of the serum- and glucocorticoid- induced protein kinase, Sgk-1, are regulated by multiple hormonal and environmental cues including cellular stress. Biochemical fractionation and indirect immunofluorescence demonstrated that sorbitol induced hyperosmotic stress stimulated expression and triggered the localization of endogenous Sgk-1 into the mitochondria of NMuMG mammary epithelial cells. The immunofluorescence pattern of endogenous Sgk-1 was similar to that of a green fluorescent linked fusion protein linked to the N-terminal Sgk-1 fragment that encodes the mitochondrial targeting signal. In the presence or absence of cellular stress, exogenously expressed wild type Sgk-1 efficiently compartmentalized into the mitochondria demonstrating the mitochondrial import machinery per se is not stressed regulated. Co-immunoprecipitation and GST-pull down assays identified the IF-1 mitochondrial matrix inhibitor of the F1F0-ATPase as a new Sgk-1 binding partner, which represents the first observed mitochondrial target of Sgk-1. The Sgk-1/IF-1 interaction requires the 122-176 amino acid region within the catalytic domain of Sgk-1 and is pH dependent, occurring at neutral pH but not at slightly acidic pH, which suggests that this interaction is dependent on mitochondrial integrity. An in vitro protein kinase assay showed that the F1F0-ATPase can be directly phosphorylated by Sgk-1. Taken together, our results suggest that stress-induced Sgk-1 localizes to the mitochondria, which permits access to physiologically appropriate mitochondrial interacting proteins and substrates, such as IF-1 and the F1F0-ATPase, as part of the cellular stressed induced program.
Keywords: Serum and glucocorticoid induced protein kinase, Cellular stress response, Mitochondria localization, Protein-protein interactions, IF-1
1. Introduction
We originally identified the Serum- and glucocorticoid-induced protein kinase, Sgk (denoted as Sgk-1) as a novel serine/threonine protein kinase under transcriptional control by serum and glucocorticoids [1, 2]. The SGK-1 gene encodes a 50 kDa protein that is a member of the “AGC” family of serine/threonine protein kinases that includes Akt/PKB and protein kinase C (PKC) [1, 3, 4]. Sgk-1 has two other protein homologues (Sgk-2 and Sgk-3), and there are four Sgk-1 isoforms that are products of alternate translation initiation and can localize to a variety of cellular compartments [5]. The catalytic domains of the Sgk homologues are most closely related to the catalytic domain of the Akt/PKB protein kinase, which are directly phosphorylated and activated by phosphoinositide-dependent kinase 1 (PDK1) [6-8] and mTOR complex 2 [9] after activation through the phosphatidylinositol 3-kinase (PI3-kinase) signaling pathway. The expression, enzymatic activity, protein stability and subcellular localization of Sgk-1 can be regulated in a stimulus-dependent manner in many normal and transformed cell types [1, 8, 10], allowing the selective accessibility of this protein kinase to its substrates and interacting proteins in order to mediate its physiologically appropriate cellular response to different environmental cues.
The transcriptional regulation of Sgk-1 is a particularly distinguishing feature of this protein kinase compared to other closely related AGC kinases [1, 4, 8], which are constitutively expressed [4]. Sgk-1 is a key component of the cellular stress response that promotes survival of cells exposed to several types of extracellular stress [3, 8, 11, 12]. Depending on the cell and tissue type, Sgk -1 expression is stimulated by changes in osmolarity and cell volume [10, 13, 14], as well as by cellular stress such as hyperosmotic stress, oxidative stress, heat shock, and UV irradiation [11, 12]. The stimulus control of Sgk-1 promoter activity through specific response elements in the Sgk-1 promoter to glucocorticoids [1, 15], p53 [16] and hyperosmotic stress [12] accounts for the increased transcript expression in several cell systems. The kinetics, magnitude and duration of the induced Sgk-1 expression vary with each stimulus [1, 8, 11, 12], which implicates a diverse role for this protein kinase in the maintenance of physiological homeostasis.
The subcellular compartmentalization and selective access to compartment-specific substrates of Sgk-1 is also regulated in a stimulus-dependent manner in a variety of cell types [8, 10, 17, 18]. However, the precise localization and the protein targets of Sgk-1 after exposure to cellular stress stimuli are not well understood. We now report that endogenous, hyperosmotic stress-induced Sgk-1 localizes to the mitochondria, and that Sgk-1 interacts with the IF-1 stress-regulated inhibitor of mitochondrial F1F0-ATPase through a specific region within the catalytic domain of Sgk-1. We also show that Sgk-1 can phosphorylated the F1F0-ATPase in vitro, which together with the IF-1 binding results implicate a role for Sgk-1 and its interaction with IF-1 and phosphorylation of the F1F0-ATPase in the overall mitochondrial cell stress response.
2. Materials and Methods
2.1 Cell Culture
Cell culture reagents such as DMEM, fetal bovine serum, calcium- and magnesium-free phosphate-buffered saline (PBS), and trypsin/EDTA were supplied by BioWhittaker, Inc. (Walkersville, MD). Insulin and D-sorbitol were purchased from Sigma (St. Louis, MO). NMuMG nontumorigenic mouse mammary epithelial cells (ATCC# CRL-1636) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 50 units/ml penicillin (BioWhittaker, Inc), as well as 50 μg/ml streptomycin and 10 μg/ml insulin, which were both purchased from Sigma (St. Louis, MO). Human embryonic kidney (HEK) 293T cells (ATCC# CRL-11268) were cultured this same medium without the addition of insulin. All cells were propagated at 37 °C in humidified air containing 5% CO2. To induce hyperosmotic stress, cells were treated with 0.3 M sorbitol in DMEM for 24 hours. Vehicle control cells were incubated with an equal volume of DMEM.
2.2 Generation of Sgk-1 Constructs
Wild type rat Sgk-1 was cloned into pFLAG-CMV5a (Sigma, St. Louis, MO) in order to express Sgk-1 with a FLAG epitope at the carboxyl terminus in mammalian cells. Sgk-1 was amplified by PCR from CMV-HA-Sgk [6] with the following oligonucleotide primers: forward primer: 5′-CCGGAATTCCGGACCA TGACCGTCAAAACCGAG-3′, reverse primer: 5′-CGCGGATCCGAGGAAGGAGTCCATAGGAG-3′. The forward primer contains a restriction site for EcoRI (underlined sequence) and a Kozak sequence (bold), and the reverse primer contains a restriction site for BamHI (underlined). The amplified PCR product of Sgk was cloned between the EcoRI and BamHI restriction sites of pFLAG-CMV5a.
The production of constructs encoding full length wild-type Sgk-1 (WT Sgk), N- and C-terminal deleted Sgk-1 (ΔN Sgk and ΔC Sgk, respectively), and the catalytic domain of Sgk-1 (Cat 60-355 Sgk) in pcDNA3 or pCITE-4a was described previously [18, 19]. Different deletions of Sgk-1 were cloned between the EcoRI and XhoI restriction sites of pCITE-4a as described above, using the following oligonucleotide primers: forward primer targeting Sgk-1 from amino acid 63 (F Sgk 63): 5′-GCAAAGAATTCACCATGCCTCAGGAGCCCGAACTTATG-3′, reverse primer targeting Sgk up to amino acid 122 (R Sgk 122): 5′-GCAAACTCGAGTCATGCT TCTTCTGCCTTGTGCCTTGC-3′, (R Sgk 156): 5′-GCAAACTCGAGTCAAGGGTGC TTCACATTCTTCAACAG-3′, (R Sgk 176): 5′-GCAAACTCGAGTCATAGGACGAAG TAGAGTTTGTCAGC-3′, (R Sgk 289): 5′-GCAAACTCGAGTCACATCTCATACAAGACAGCCCCG-3′
2.3 Transfections
NMuMG cells were transfected with Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions as we previously described [11]. Transfected cells were kept in the tissue culture incubator for 5 hours before the transfection medium was replaced with the culture medium for NMuMG cells. HEK 293T cells were transfected with FuGene 6 reagent (Roche, Madison WI) according to manufacturer’s instructions. Approximately 3 × 105 cells were plated into 35 mm tissue culture dishes in DMEM supplemented only with 10% FBS. After 24 hours, a transfection mixture was added directly to the cells in a drop wise fashion. The transfection mixture was prepared by mixing 4 μL FuGene with 2 μg DNA in DMEM and then incubated at room temperature for 30 minutes. No change of medium was required. All transfected cells were kept in the tissue culture incubator for a minimum of 24 hours before being treated or harvested.
2.4 Biochemical fractionation of stress-treated and untreated cells
Cells washed twice with cold phosphate buffered saline (PBS) were scraped into PBS and collected by centrifugation at 1,000 rpm for 3 minutes in a tabletop Beckman GPR centrifuge. The pelleted cells were gently resuspended in two volumes of hypotonic lysis buffer (5 mM Tris, pH 7.4, 5 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, pH 8.0), and swelled on ice for 30 minutes. The cells were then lysed by hand with 50 strokes in a Potter-Elvehjem Homogenizer. Both before and after lysis, cells were examined by trypan blue (Sigma, St. Louis MO) exclusion to verify initial integrity of the swollen cells and completion of homogenization. Lysates were centrifuged for 10 minutes at 600 x g in an SE-12 Sorvoll rotor. The pellet represented crude nuclei and other cellular debris (Nuc). The 600 x g supernatant was then centrifuged for 10 minutes at 10,000 x g, yielding the heavy membrane fraction (HMP) in the pellet and the 10,000 x g supernatant (Sup).
2.5 Western Blot Analysis
Protein samples were boiled in SDS-sample buffer, resolved on 7.8% SDS polyacrylamide gels in SDS-PAGE, and transferred to nitrocellulose membranes according to standard protocols. Where applicable, membranes were stained with Ponceau-S dye (Sigma, St. Louis MO) to verify equal loading of total protein levels. Membranes were then blocked in 10% non-fat dried milk (Apex Bioresearch Products, San Diego CA) in 0.5 M NaCl, 10 mM Tris, pH 7.4 (ST) for 30 minutes at room temperature. Primary antibodies were diluted in 3% milk in ST, and they included the following: affinity purified rabbit polyclonal Sgk-1 antibody (1:2500) [1], 2.5 μg/mL cytochrome oxidase subunit IV (COX IV) antibody (Molecular Probes, Eugene OR), Sp1 (PEP2) antibody (1:2000) (Santa Cruz Biotechnology, Santa Cruz CA), 12CA5 HA antibody (1:1000) (Roche, Madison WI), and FLAG antibody (1:2000) (Sigma, St. Louis MO).
Membranes were incubated in primary antibody on a room temperature rocker for 2 hours or on a rocker at 4°C overnight. Membranes were then thoroughly washed and then incubated in goat anti-rabbit or goat anti-mouse secondary antibodies (BioRad, Hercules CA), diluted 1:5,000 in 3% milk in ST, for 1 hour on a room temperature rocker. Membranes were again washed and then exposed to a 1:1 mixture of reagents in the Renaissance developing kit (PerkinElmer Life Sciences, Waltham MA) for 1 minute before being exposed to x-ray film.
2.6 Indirect Immunofluorescence of endogenous Sgk-1
NMuMG cells were plated onto 8-well LabTek chamber slides (Nalgene, Rochester NY). After 24 hours, the cells were treated with 0.3 M sorbitol for 24 hours to induce Sgk-1 expression. Prior to fixing cells, 250 nM of MitoTracker Orange CMTMRos (Molecular Probes, Eugene OR), diluted in plain DMEM, was added directly to the medium of cells and incubated in the dark (foil wrap) in the tissue culture incubator for 30 minutes. All following steps were performed under restricted light conditions due to the photosensitivity of the MitoTracker dye. Cells gently washed three times with cold PBS were fixed with freshly prepared 3.7% formaldehyde, 0.1% glutaraldehyde in PBS on ice for 15 minutes, then washed three times with cold PBS. Cells were permeabilized with ice cold 50% acetone, 50% methanol for 1 minute, and then washed three times with cold PBS. The cells were then blocked in 3% non-fat dried milk in PBS for 30 minutes at room temperature on a slow rocker, then 30 μL of Sgk-1 antibody (1:150) or PBS alone was added to cells, which were then incubated on a rocker at room temperature for 2 hours. Cells were washed three times with 3% non-fat dried milk in PBS, then 30 μL of Alexa 488 secondary antibodies (1:300) (Molecular Probes, Eugene OR) was added to all cells which were then incubated on a rocker at room temperature for 1 hour. Cells were washed four times with PBS, the gaskets were removed from the slides, and coverslips were mounted onto cells in Vectashield (Vector Laboratories, Burlingame CA) with 1 μM DAPI. Cells were visualized on a Zeiss Laser-scanning confocal microscope. Red signals represented staining of the mitochondria by MitoTracker dye, green signals represented indirect immunostaining of Sgk-1 with Alexa 488, and blue signals represented staining of cell nuclei by the DAPI in the mounting solution.
2.7 Generation and Expression of Recombinant GST and GST-IF-1 Fusion Proteins
IF-1 isolated from the yeast two-hybrid screen [19], encoding the full-length protein of 107 amino acids, was subcloned between EcoRI and XhoI sites within the glutathione-S-transferase (GST) vector pGEX-4T1 (Amersham Pharmacia Biotech, Uppsala Sweden) to yield GST-IF-1. The GST-IF-1 fusion proteins and GST alone were isolated from the XL1-Blue competent bacterial cells (Stratagene, La Jolla CA) transformed with the GST-IF-1 construct and the pGEX-4T1 vector alone. Bacteria were grown from 1:10 diluted inoculations at 37°C for 2 hours (O.D. = 0.5-0.7) and subsequently induced with 0.1 mM IPTG (isopropyl-1-thio-beta-D-galactpyranoside) for 6 h at 37°C. Cells were lysed using the French Press (three times) in lysis buffer (PBS containing 0.05% Tween 20, 2 mM EDTA, 1 mM DTT, and 0.1% beta-mercapto-ethanol). The GST and GST-IF-1 fusion proteins were purified on glutathione sepharose beads (Pharmacia, Uppsala Sweden) according to manufacturer’s instructions.
2.8 In vitro-translation and IF-1 GST Pull Down Assays
In vitro transcription and translation of Sgk-1, Jun NH2-terminal kinase (JNK) and PDK-1 in pcDNA3 or pCITE-4a expression vectors was accomplished using the TNT coupled rabbit reticulocyte kit (Promega, Madison WI) according to manufacturer’s instructions. Expression plasmids encoding JNK and PDK1 were provided by Dr. J. S. Gutkind (National Institute of Dental Research, Bethesda, MD) and Dr. B. A. Hemmings (Friedrich Miescher-Institut, Basel, Switzerland).
To test the specific binding of the various forms of in vitro translated Sgk-1 to IF-1, GST alone or GST-IF-1 were immobilized on glutathione sepharose beads (Amersham Pharmacia Biotech, Uppsala Sweden) and incubated with 2-5 μl of [35S]Sgk-1 in vitro translation products in 180 μl of binding buffer (20 mM HEPES-KOH, pH 7.9, 50 mM KCl, 2.5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.2% NP-40, 1.5 mM PMSF, and 3 μl of normal goat serum/180 μl binding buffer). The slurry was incubated on a nutator at 4°C overnight, then the beads were washed five times in wash buffer (200 mM NaCl, 0.2%Tween 20, 10 mM Tris, pH 7.5, and 0.5% nonfat dry milk). After removing the supernatant fraction in the final wash, samples were resuspended in 25 μl of 2× SDS sample buffer, boiled, and resolved on 7.8% SDS-PAGE. Binding was compared with 10% of the in vitro-translated products added to the binding reactions. Gels were dried at 60°C and autoradiography carried out at 70°C.
2.9 Generation of HA-IF-1 construct and Co-immunoprecipitation of IF-1 with Sgk-1
Co-immunoprecipitations of HA-IF-1 and Sgk-1-FLAG were performed in HEK 293T cells. Using standard PCR cloning protocols, the full-length IF-1 coding sequence was subcloned between EcoRI and XhoI sites within the pCMV4 vector, modified to include a sequence encoding a HA-epitope tag upstream of the multiple cloning site [19]. HEK 293T cells were transfected with Sgk-1-FLAG plus HA-IF-1 or CMV-FLAG vector plus HA-IF-1. After 36 hours, cells were washed with and scraped into cold PBS and centrifuged at 1000 rpm for 3 minutes in a tabletop Beckman GPR centrifuge. Cell pellets were then divided and lysed in either pH 6.7 or pH 7.4 lysis buffers (50 mM Tris, pH 6.7 or pH 7.4, 150 mM NaCl, 1 mM EDTA, 1.0% Triton X-100, 10 μg/mL Sigma Protease Inhibitor Cocktail) for 15 minutes on ice. Lysates were sheared through a 26-gauge needle five times and then clarified of insoluble membranes by centrifugation at 5,000 rpm at 4°C for 10 minutes. Approximately 1-5 mg of total protein from the different lysates was used for the co-immunoprecipitation assays. Lysates were precleared with plain Sepharose beads (Amersham Pharmacia Biotech, Uppsala Sweden) on a rocker at 4°C for 1 hour. Beads were separated from supernatants by centrifugation at 5,000 rpm at 4°C for 2 minutes. Lysate supernatants from the preclear step were transferred to new tubes containing 20 μL of M2 anti-FLAG affinity bead slurry (Sigma, St. Louis MO), prewashed with corresponding lysis buffers, and incubated on a nutator at 4°C for 2 hours. The beads were then washed four times with 0.5 mL of corresponding lysis buffers and aspirated with a 30-gauge needle. FLAG peptide (30 μL of 200 μg/mL in TBE) was added to the beads and incubated at room temperature with frequent tapping for 30 minutes. The peptide elutions were collected by inserting holes in the tops and bottoms of the tubes of beads, inserting these tubes into new tubes, and centrifuging the stacked tubes at 5,000 rpm for 2 minutes at room temperature. Elutions were then boiled in SDS sample buffer, resolved on 7.8% SDS-PAGE, and examined by Western blot analysis for the presence of both Sgk-FLAG and HA-IF-1 using Sgk-specific and the HA epitope-specific antibodies as described in Section 2.5.
2.10 Construction of Sgk-GFP and analysis of subcellular localization by fluorescence microscopy
Amino terminal sequences of Sgk-1 (aa 1-36 or aa 1-60) were cloned using standard procedures into pEGFP-N3 (Clontech. Mountain View CA) in order to express a fusion protein of N-terminal Sgk-1 sequences linked to an enhanced green fluorescent protein (EGFP). The N-terminal sequences of Sgk-1 were amplified by PCR from CMV-HA-Sgk [17] with the following oligonucleotide primers: common forward primer for Sgk-1: 5′-GAA TTC GCC ACC ATG ACC GTC AAA ACC GAG-3′ (EcoRI restriction site underlined; followed by Kozak sequence in bold); reverse primer for 1-36 peptide: 5′-GGA TCC GTT CAG GCC CAT CCT TCT CTG-3′ (BamHI restriction site underlined); reverse primer for 1-60 peptide: 5′-GGA TCC GAT TTT CAA ATA GGA TTG AAC-3′ (BamHI restriction site underlined).
NMuMG cells were transfected on glass coverslips in 6-well plates using lipofectamine transfection reagent (Invitrogen, Carlsbad CA) according to package instructions, with 0.4 μg of DNA and 1.0 μL of lipofectamine added to each well. The DNA/lipofectamine mixture was allowed to incubate at room temperature for 30 minutes prior to adding the mixture to the cells in serum-free medium. The transfection reaction was stopped after 6-7 hours of incubation at 37°C by changing the cell medium. After approximately 24 hours, cells were incubated with 250 nM MitoTracker red dye (Molecular Probes, Eugene OR) for 30 minutes, then rinsed and fixed onto the coverslips with cold 4% paraformaldehyde in PBS. After rinsing, cells were permeabilized with ice cold acetone/methanol (50/50%) at -20°C for 1-5 minutes. Cells were washed and then mounted onto slides (without polish) into 20 μL aliquots of a mixture of Antifade and DAPI. A confocal microscope was used to view and capture images of the EGFP (488 nm/507 nm) and MitoTracker (579 nm/599 nm).
3. Results
3.1 Sgk-1 interacts specifically with the IF-1 inhibitor of mitochondrial F1F0-ATPase
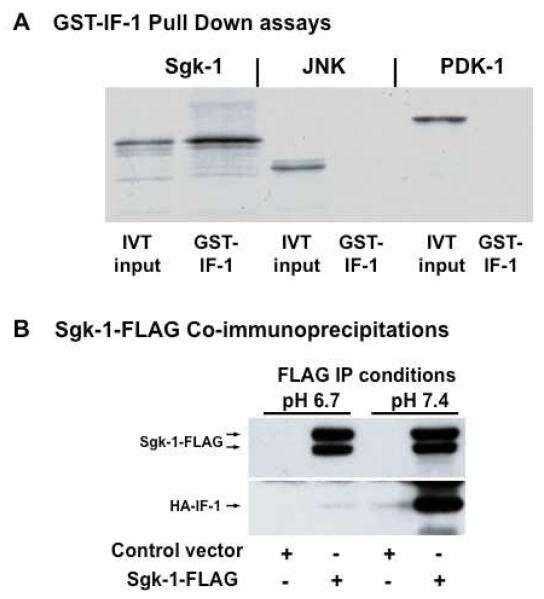
IF-1 is a mitochondria-associated low molecular weight (9.5 kDa) protein inhibitor of mitochondrial F1F0-ATPase, or ATP synthase [20], although little is known about IF-1 physiological interactions outside its inhibitory activity on the ATP synthase. Using a yeast-two-hybrid screen for Sgk-interacting proteins [19], we identified IF-1 from the protein expression library as a potential Sgk-1 interacting partner. In vitro GST-pull down assays were performed to examine the specificity of the interaction between Sgk-1 and IF-1. GST-IF-1 was immobilized on glutathione agarose beads and incubated with in vitro translated [35S]-labeled Sgk-1, or two other kinases JNK and PDK-1. Electrophoretic analysis of specifically bound proteins demonstrated that [35S]-labeled Sgk-1 can be recovered from the fractions bound to GST-IF-1 (Figure 1A) but not from GST control beads (data not shown), thus confirming the interaction between Sgk and IF-1 in vitro. Neither [35S]-labeled JNK nor [35S]-labeled PDK-1 bound to GST-IF-1, indicating the specificity of the interaction observed between Sgk-1 and GST-IF-1 (Fig 1A).
Fig 1.
Specificity of Sgk-1 interaction with IF-1 and the pH-dependent co-immunoprecipitation of Sgk-1 IF-1. Panel A: Sgk-1, PDK-1 and JNK kinases were expressed as [35S]labeled in vitro translated proteins in reticulocyte lysates and examined for binding to GST-IF-1 immobilized on glutathione beads. Electrophoretic fractionation of the glutathione eluted protein as well as ten percent of input protein used for the full glutathione elutions are shown in the panel. Panel B: NMuMG cells were co-transfected with HA-IF-1 and CMV-FLAG (control vector) or with HA-IF-1 and Sgk-1-FLAG. Sgk-1-FLAG was immunoprecipitated from lysates at either pH 7.4 or pH 6.7 using anti-FLAG affinity sepharose beads, eluted from the beads with FLAG peptide, electrophoretically fractionated and examined by Western blots analyzed for Sgk-1 using Sgk-1 specific antibodies or for HA-IF-1 using HA-specific antibodies.
3.2 Sgk-1 interacts with HA-IF-1 in mammalian cell lysates in a pH-dependent manner
A co-precipitation analysis was used to verify and characterize the Sgk-1 interactions with IF-1 in a cellular context. HA-epitope-tagged IF-1 (HA-IF-1) was co-expressed in 293T human embryonic kidney cells transfected with either Sgk-FLAG or with the CMV-FLAG empty vector. The 293T cells were used for this analysis because of their high efficiency of co-transfection and that exogenously expressed Sgk-1-FLAG localizes to the mitochondria (see later sections). Because the oligomeric and functional states of IF-1 are dependent on pH [21-23], transfected cells were lysed in either pH 7.4 or pH 6.7 buffers to test the potential significance of pH on the interaction between Sgk and IF-1. Sgk-FLAG was immunoprecipitated from the cell lysates using an anti-FLAG M2 agarose affinity gel, and the recovered Sgk-FLAG protein complexes were eluted from the beads with the FLAG peptide. Western blot analysis of the eluted protein complexes using antibodies specific for the HA epitope (to detected HA-IF-1) or for Sgk-1 showed that similar levels of Sgk-1-FLAG can be immunoprecipitated from both the pH 6.7 and the pH 7.4 lysates (Fig 1B). Showing the fidelity of the anti-FLAG immunoprecipitations, no Sgk-1-FLAG was detected in lysates of cells transfected with the CMV-FLAG empty control expression vector. HA-IF-1 specifically co-immunoprecipitated with Sgk-FLAG at pH 7.4, but failed to co-immunoprecipitate with Sgk-1 at pH 6.7 (Figure 1B). These data demonstrate that Sgk-FLAG binds HA-IF-1 in a pH-dependent manner, and because IF-1 exists in cells as homotetramers at pH 7.4, Sgk-1 most likely interacts with IF-1 when IF-1 is in a tetrameric configuration.
3.3 Mutagenic identification of a discrete IF-1 binding region within Sgk-1
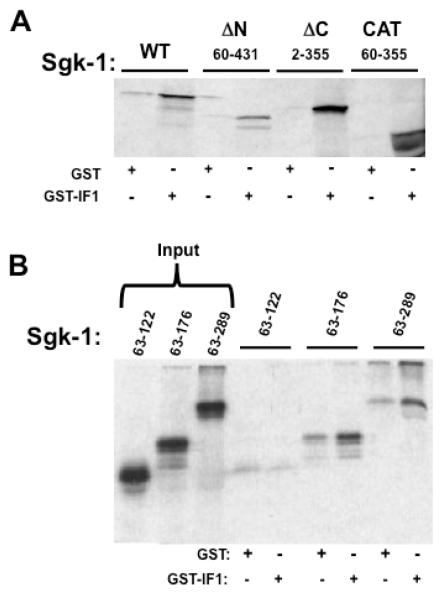
To identify the putative region within the Sgk-1 protein that binds IF-1, various in vitro translated [35S]-labeled deletions of Sgk-1 were tested for binding to IF-1 using GST-IF-1 pull down assays. Initial IF-1 binding studies were carried out with the wild type Sgk-1 (WT), the amino terminal deleted ΔN-Sgk-1 (60-431 aa), the carboxyterminal ΔC-Sgk-1 (2-355 aa) and the catalytic domain Sgk-1 (60-355 aa). The relative electrophoretic migration pattern of the ΔN-Sgk-1 (60-431 aa) and the ΔC-Sgk-1 (2-355 aa) fragments do not reflect their predicted size differences, which is likely due to secondary structures that attenuate SDS binding and therefore alter the optimal charge:mass ratio. As shown in Fig 2A, all three of the deletion constructs specifically bound to GST-IF-1, indicating that the IF-1 binding domain is located within the catalytic domain of Sgk-1. To further narrow down the boundaries of the IF-1 binding domain, several additional truncated versions of the Sgk-1 catalytic domain were assayed in GST-IF-1 pull down assays. As shown in Fig 2B, the Sgk-1 63-289 aa and 63-176 aa fragments bind specifically to GST-IF-1, whereas the 63-122 aa fragment does not bind to GST-IF-1. Another Sgk-1 region between 63-156 aa binds non-specifically to GST alone (data not shown). Taken together, these results suggest the presence of a discrete IF-1 binding domain in Sgk-1 between 122-176 aa, although this region has not been directly tested for specific binding. This newly defined functional region within Sgk-1 (see Fig 4 diagram) is highly charged (10 basic and 3 acidic residues), and it is predicted to reside on a solvent-accessible surface, which is consistent with this region being involved in protein-protein interactions.
Fig 2.
In vitro analysis of Sgk-1 interaction with GST-IF-1. Panel A: Full length Sgk-1 (WT), Sgk-1 aa 60-431 (ΔN), Sgk-1 aa 2-355 (ΔC) and Sgk-1 aa 60-355 (Cat) were expressed as [35S]labeled proteins by in vitro translation in reticulocyte lysates, and examined for binding to GST-IF-1 or GST alone immobilized on glutathione beads. Glutathione eluted proteins were electrophoretically fractioned. Panel B: Sgk-1 domains of amino acids 63-122, 63-176 and 63-289, along with full length Sgk (WT) were expressed as [35S]labeled proteins by in vitro translation in reticulocyte lysates, and examined for to GST-IF-1 or GST alone immobilized on glutathione beads. Protein extracts from glutathione elutions were electrophoretically fractionated and detected by autoradiography.
Fig 4.
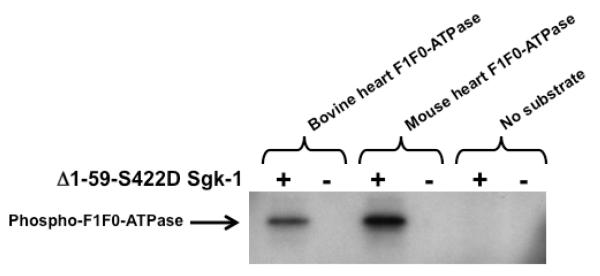
In vitro phosphorylation of the F1F0-ATPase by Sgk-1. In vitro kinase reactions contained [γ-32P]ATP as well as either purified bovine heart F1F0-ATPase, mouse heart F1F0-ATPase or no added substrate. The reactions were initiated in the presence or absence of added constitutively active Δ1-59-S422D Sgk-1 and the incubations continued for 1 hour. The in vitro reactions were terminated by heating each mixture in SDS-PAGE sample buffer and then electrophoretically fractionated in polyacrylamide gels. The presence of phosphorylated F1F0-ATPase was determined by autoradiographic analysis of the electrophoretic gels.
3.4 Stress induced endogenous Sgk-1 localizes to the mitochondria in NMuMG mammary epithelial cells
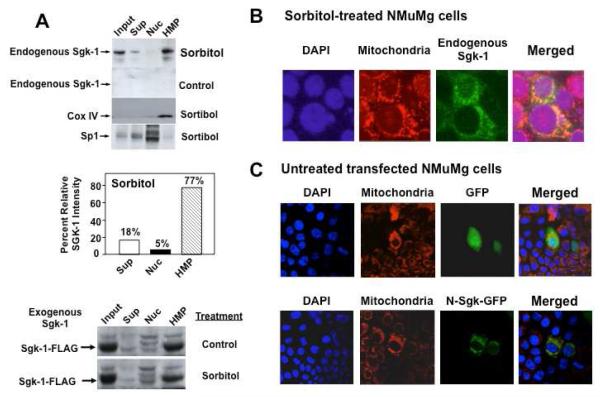
The specific interactions of Sgk-1 with IF-1 predict that stress induced endogenous Sgk-1 should localize to the mitochondria. To test this possibility, NMuMG mouse mammary epithelial cells were treated with or without 0.3 mM sorbitol to stimulate Sgk-1 expression by hyperosmotic stress [11, 12]. Cell extracts were differentially fractionation into nuclear (600 x g, pellet), heavy membrane (10,000 x g, pellet: HMP) and cytoplasmic fractions (10,000 x g, supernate), and the enrichment of Sgk-1 protein in each fraction was determined by western blot analysis. As shown in Fig 3A, after exposure to 0.3 mM sorbitol, Sgk-1 was highly enriched in the heavy membrane pellet. Densitometry of the western blots revealed that a relatively large percentage (~80%) of Sgk-1 protein induced by hyperosmotic shock localized to the heavy membrane fraction and much less to the cytoplasmic fractions (Fig 3A, bar graphs). Under these hyperosmotic stress conditions, little or no Sgk-1 was detected in the nuclear fraction. The mitochondrial marker cytochrome oxidase IV (COX IV) was detected only in the heavy membrane fractions, demonstrating an enrichment of mitochondria in this biochemical fraction (Fig. 3A). As previously observed for NMuMG cells [38, 41], in the absence of hyperosmotic stress (control conditions) Sgk-1 cannot be detected in the total cell extracts or in any of the subcellular fractions (Fig 3A).
Fig 3.
Mitochondria localization of stress-induced endogenous Sgk-1 and of exogenously expressed Sgk-1 or an exogenous N-terminal Sgk-1 linked GFP fusion protein. Panel A: NMuMG cells were treated with or without sorbitol (hyperosmotic shock), and isolated cell lysates were fractionated by differential centrifugation into subcellular fractions as described in the methods section. Equal amount of protein from input lysates (Input), 10,000 x g supernatants (Sup), nuclear extracts (Nuc) and heavy membrane fractions (HMP) were electrophoretically fractionated and examined by Western blot analysis for production of endogenous Sgk-1, for the cytochrome oxidase IV (COX IV) mitochondria marker protein, and for the Sp-1 transcription factor nuclear marker protein. The relative level of Sgk-1 localized in the supernatant, nuclear and heavy membrane fractions after sorbitol treatment was quantified by densitometry of the western blots and the values shown as the percentage of Sgk-1 localization for each of stress conditions. Cells transfected with Sgk-1-FLAG were treated with or without sorbitol and fractionated into 10,000 x g supernatants (Sup), nuclear extracts (Nuc) and heavy membrane pellets (HMP) and analyzed by western blots probed for the FLAG epitope. Panel B: The localization of endogenous Sgk-1 was analyzed in sorbitol-treated NMuMG cells by confocal immunofluorescence microscopy. Sgk-1 was visualized as a green signal by Alexa 488 secondary antibody against polyclonal Sgk-1 antibody, mitochondria were stained red with the vital dye, MitoTracker Orange CMTMRos, and nuclei were stained blue by DAPI. Panel C: NMuMG cells were transiently transfected with expression vectors for either the Green Fluorescence Protein (GFP) or a fusion protein comprised of the N-terminal 60 amino acids of Sgk-1 linked to the GFP forming N-Sgk-GFP. The nuclei were stained by DAPI, the mitochondria were stained red with the vital dye MitoTracker Orange CMTMRos, and the subcellular localization of GFP or the N-Sgk-GFP fusion protein was determined by confocal microscopy. All results in this figure are representative of three independent experiments.
Control untreated and sorbitol-treated NMuMG cells expressing exogenous Sgk-1-FLAG were biochemically fractionated into supernatant (Sup), nuclear (Nuc) and heavy membrane pellets (HMP) as described earlier. Western blots were probed for exogenous Sgk-1-FLAG using FLAG epitope specific antibodies. As shown in Fig 3A (lower panels), similar levels of exogenous Sgk-1-FLAG were highly enriched in the heavy membrane fractions containing mitochondria in the presence or absence of stress conditions. Therefore, the cellular localization of Sgk-1 is not stress induced, but rather, hyperosmotic stress induces the level of endogenous Sgkp-1, which is then compartmentalized into heavy membrane fraction.
Indirect immunofluorescence of Sgk-1 was employed to determine a more precise subcellular localization of the stress-induced Sgk-1. Staining of the mitochondria with a vital dye marker MitoTracker Orange CMTMRos in sorbitol-treated NMuMG cells was used to determine whether stress-induced Sgk-1 resides in this organelle. Co-staining of fixed, permeabilized cells revealed that the sorbitol-induced Sgk-1 co-localizes with the mitochondria dye marker (Fig 3B, Merged) in addition to being partially dispersed in the cytoplasm (Fig 3B, Endogenous Sgk-1). In untreated cells, endogenous Sgk-1 protein could not be detected by an immunofluorescence signal (data not shown), which is consistent with the lack of expressed endogenous Sgk-1 in untreated NMuMG cells. This result with the endogenous rat Sgk-1 is consistent with previous studies showing that the mitochondrial localization of exogenously expressed human Sgk-1 is driven by an intrinsic mitochondria localization domain [24, 25].
Based on amino acid sequences, publically available prediction software programs, MitoProt II 1.0a4 [26] and PSORT II [27, 28] assigned probabilities of rat Sgk (NCBI # Q06226), human Sgk (NCBI# NP_005618), and mouse Sgk (NCBI#NP_035491) being imported into the mitochondria as 97.1%, 94.7%, and 97.7%, respectively. Furthermore, sequence analysis using MitoProt II revealed a mitochondrial targeting sequence with a net positive charge within the first 33 amino terminal amino acids (MTVKTEAAKGTLTYSRMRGMVAILIAFMKQRRM), which includes a potential cleavage site at residue 33 between methionine and glycine in the sequence: RRM/GL. COILS prediction software [29] predicts a helical structure that spans amino acids 14-33, which ends at the predicted cleavage site. Therefore, to confirm the mitochondrial localization of stress induced endogenous Sgk-1, the NMuMG cells were transfected either with the enhanced Green Fluorescence Protein (GFP) or with a Sgk-GFP fusion protein with the Sgk-1 1-60 amino terminal peptide linked as an amino terminal extension to GFP (N-Sgk-1-GFP aa 1-60), which includes the rat Sgk-1 mitochondrial targeting signal as well as a proline-rich region. Fluorescence microscopy revealed that most of the expressed N-Sgk-GFP co-localized with the mitochondria vital dye marker MitoTracker Orange CMTMRos, whereas, the control GFP protein remained excluded from the mitochondria (Fig. 3C). The subcellular location pattern of endogenous Sgk-1 was essentially identical to that of the N-terminal-Sgk-1-GFP fusion protein (Fig 3B vs Fig 3C), which demonstrates that the stress induced endogenous Sgk-1 is localized to the mitochondria and likely accessible to IF-1 and other potential mitochondrial-associated targets.
3.5 In vitro Sgk-1 phosphorylation of the F1F0-ATPase/ATP synthase
The stress induced localization of Sgk-1 to the mitochondria and its interaction with IF-1 suggests that Sgk-1 may recognize specific mitochondrial components as substrates. Sequence analysis revealed that the beta subunit of the bovine, mouse, and human F1F0-ATPases contains a conserved RARVALT amino acid sequence at 279-285 (human sequence) that represents a putative consensus sequence for Sgk-1. Therefore, an in vitro protein kinase assay was used to determine if the F1F0-ATPase could be directly phosphorylated by Sgk-1. The reaction mixtures were formed with or without the Δ1-59-S422D Sgk-1, which is constitutively active, and contained [γ32P]ATP and either purified bovine heart F1F0-ATPase, purified mouse heart F1F0-ATPase, or no added substrate. The reaction mixtures were electrophoretically fractioned and analyzed for phosphorylated substrates by autoradiography of the electrophoretic gels. As shown in Fig 4, both the bovine and mouse heart F1F0-ATPases were phosphorylated in vitro by Sgk-1, the 50 kDa size of the phosphorylated protein band is consistent it being the beta-subunit of the F1F0-ATPase [30]. Phosphorylation of the purified F1F0-ATPases required the presence Sgk-1 in that no phosphorylated protein bands were detected in the absence of substrate.
4. Discussion and conclusion
We have identified the mitochondria matrix protein IF-1 as a new Sgk-1 binding partner, which we propose plays an important role in the cellular response to hyperosmotic stress and other stress stimuli (such as heat and oxidative stress; data not shown) that promote Sgk-1 localization into the mitochondria. IF-1 is the first documented Sgk-1 binding partner that resides primarily as a mitochondrial component. We propose that the IF-1 interaction with Sgk-1 plays a role in sequestering this protein kinase within the mitochondria, likely in the matrix or inner membrane where IF-1 functions. In this regard, IF1 was reported to be tethered to the inner mitochondrial membrane by a membrane receptor distinct from the F1F0-ATPase beta subunit [31]. The cellular components that facilitate Sgk-1 localization to the mitochondria are not known. Conceivably, mitochondrial import proteins could potentially direct Sgk-1 localization to distinct compartments within mitochondria, such as the matrix and the outer membrane, to interact with different substrate and non-substrate protein targets such as IF-1. The mitochondrial localization of exogenously expressed Sgk-1 in the absence of cell stress demonstrated that the ability of the mitochondrial transport machinery to recognize and import Sgk-1 is not stress regulated per se.
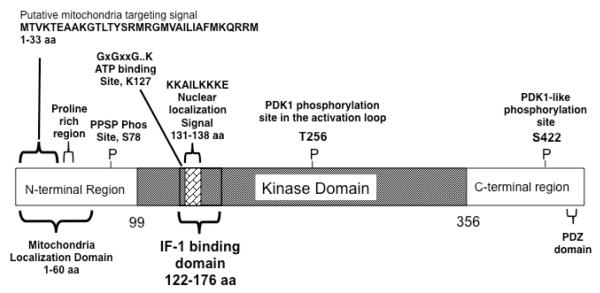
As summarized in the Fig 5 diagram, Sgk-1 has multiple regulatory domains within its structure that account for its activation, compartmentalization and many cellular functions in a variety of different cell types. Our studies have uncovered a new functional domain between 122-176 amino acids in the central catalytic domain that mediate its protein-protein interaction with IF-1. Consistent with previously reported studies [24, 25], the aminoterminal 60 amino acids of Sgk-1 is sufficient to drive the mitochondrial localization of the green fluorescence protein under non-stress conditions. Furthermore, another report showed that deletion of the aminoterminal 60 amino acids of human Sgk-1 results in a homogenously dispersed localization pattern throughout the cell [32]. Sequence analysis predicts a mitochondria targeting signal within the first 33 aminoterminal amino acids of Sgk-1, and one study has shown that amino acids 17-32 function as an anchor for the outer mitochondrial membrane [25]. Interestingly, Sgk-1 contains a proline rich region near the aminoterminal 36 amino acids (see Fig 5) that conceivably plays a role in mediating the stable accumulation of Sgk-1 in the mitochondria or cellular other compartments. We propose that the aminoterminal 60 amino acids of Sgk-1 functions as a Mitochondrial Localization Domain that includes a mitochondrial targeting signal as well as a proline-rich region.
Fig 5.
Locations of functional domains, interaction sites and phosphorylation sites within the Sgk-1 protein. Sgk-1 is a multidomain protein that contains protein interaction motifs (proline rich region and the PDZ domain), phosphorylation sites in the amino terminal and carboxy terminal domains as well as in the central kinase domain, an ATP binding site, and nuclear and mitochondrial localization signals. We propose that the IF-1 binding domain is comprises amino acids 122-176, which is located within the central kinase domain, and that amino terminal sequences 1-60 present a mitochondrial localization domain that contains a mitochondrial targeting signal and a proline rich region.
Mitochondrial IF-1 can be stress regulated and blocks the hydrolytic activities of F1F0-ATPase by binding to the beta subunits of the F1 moiety of the enzyme complex [33]. IF-1, which exists as an inactive homotetramer or homo-oligomer at neutral pH, is activated by the acidification of the mitochondrial matrix that occurs during de-energizing cellular conditions [34]. The resulting decreased pH activates IF-1 by the protonation of histidine residues that causes an activating conformational change in IF-1. This conformational change leads to the formation of IF-1 homodimers that are capable of binding F1 and inactivating the F1F0-ATPase [21, 35]. Co-immunoprecipitation assays in cellular lysates showed that Sgk-1 strongly binds IF-1 at pH 7.4, but does not bind IF-1 at pH 6.7. Our observation in combination with the location of the IF-1 interacting domain within the catalytic domain of Sgk-1 suggests a pH-dependent switch mechanism that may regulate Sgk-1 function within the mitochondria. IF-1 is most likely a nonsubstrate target of Sgk-1 because IF-1 lacks a consensus Sgk-1 phosphorylation sequence, and IF-1 failed to be phosphorylated by Sgk-1 in in vitro kinase assays (data not shown).
Consistent with the notion that the IF-1 interaction with Sgk-1 may serve to tether Sgk-1 to a functionally relevant position in the mitochondria, we observed, for the first time, that the F1F0-ATPase is a substrate of Sgk-1. The approximately 50 kD molecular weight of the phosphorylated protein band that is phosphorylated in vitro by Sgk-1 is consistent in size with the beta subunit of the F1F0-ATPase [30]. Biochemical and structural studies show that IF-1 inhibits the F1F0-ATPase (or ATP synthase) by binding to the beta-subunit of the enzyme [36, 37], which suggests that in mitochondria IF-1 could potentially tether Sgk-1 close enough for recognition of the F1F0-ATPase as a substrate. In addition, sequence analysis reveals that the beta subunit of the bovine, mouse, and human F1F0-ATPases contains a conserved RARVALT amino acid sequence at 279-285 (human sequence) that represents a putative consensus sequence for Sgk-1/AKT. Akt/PKB, which is homologous to Sgk-1 in the catalytic domain and has similar substrate specificities and cell survival functions [1, 3, 4], has been shown to localize to the mitochondria in its active state in human neuroblastoma cells under conditions in which the beta subunit of the F1F0-ATPase is phosphorylated, although a direct role of Akt/PKB in the phosphorylation of F1F0-ATPase was not established [38]. Furthermore, Akt activation has been shown to promote mitochondrial respiration and ATP production in toxicant-injured renal proximal tubular cells [39]. Because Sgk-1 interacts with the inactive, tetrameric IF-1 and is released at acidic pH, a tethering of Sgk-1 by IF-1 may potentially prime the kinase for action to phosphorylate F1F0-ATPase under low energy mitochondrial conditions that also activate the activity of IF-1. Future studies will focus on whether the stimulus induced Sgk-1, through its specific interactions with IF-1, regulates F1F0-ATPase activity and energy metabolism in the context of the cellular stress response.
Highlights.
Cell stress response triggers expression of a mitochondria localized protein kinase
IF-1 is the first established mitochondria target for Sgk-1
Sgk-1 interaction with IF-1 is a new potential cell survival response to stress
Sgk-1 can directly phosphorylate F1F0-ATPase in an in vitro assay
Acknowledgements
This work is dedication to the memory of Dr. Anita C. Maiyar, friend and colleague, who originally discovered that IF-1 and Sgk-1 are binding partners. The research described in this paper was supported by National Institute of Health Grant DK-42799 (GLF). SC was a postdoctoral trainee supported by a National Research Service Grant (CA-09041) awarded by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures The authors declare that there are no financial and personal relationships with other people or organizations that could inappropriately influence or bias their work.
References
- [1].Webster MK, Goya L, Ge Y, Maiyar AC, Firestone GL. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol. Cell Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Webster MK, Goya L, Firestone GL. Immediate-early transcriptional regulation and rapid mRNA turnover of a putative serine/threonine protein kinase. J. Biol. Chem. 1993;268:11482–11485. [PubMed] [Google Scholar]
- [3].Bruhn MA, Pearson RB, Hannan RD, Sheppard KE. Second AKT: the rise of SGK in cancer signaling. Growth Factors. 2010;28:394–408. doi: 10.3109/08977194.2010.518616. [DOI] [PubMed] [Google Scholar]
- [4].Parker PJ, Parkinson SJ. AGC protein kinase phosphorylation and protein kinase C. Biochem. Soc. Trans. 2001;29:860–863. doi: 10.1042/0300-5127:0290860. [DOI] [PubMed] [Google Scholar]
- [5].Arteaga MF, Alvarez de la Rosa D, Alvarez JA, Canessa CM. Multiple translational isoforms give functional specificity to serum- and glucocorticoid-induced kinase 1. Mol. Biol. Cell. 2007;18:2072–2080. doi: 10.1091/mbc.E06-10-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Park J, Leong ML, Buse P, Maiyar AC, Firestone GL, Hemmings BA. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. Embo. J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- [8].Firestone GL, Giampaolo JR, O’Keeffe BA. Stimulus-dependent regulation of serum and glucocorticoid inducible protein kinase (SGK) transcription, subcellular localization and enzymatic activity. Cell Physiol. Biochem. 2003;13:1–12. doi: 10.1159/000070244. [DOI] [PubMed] [Google Scholar]
- [9].García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- [10].Lang F, Bohmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V. (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol. Rev. 2006;86:1151–1178. doi: 10.1152/physrev.00050.2005. [DOI] [PubMed] [Google Scholar]
- [11].Leong ML, Maiyar AC, Kim B, O’Keeffe BA, Firestone GL. Expression of the serum- and glucocorticoid-inducible protein kinase, Sgk, is a cell survival response to multiple types of environmental stress stimuli in mammary epithelial cells. J. Biol. Chem. 2003;278:5871–5882. doi: 10.1074/jbc.M211649200. [DOI] [PubMed] [Google Scholar]
- [12].Bell LM, Leong ML, Kim B, Wang E, Park J, Hemmings BA, Firestone GL. Hyperosmotic stress stimulates promoter activity and regulates cellular utilization of the serum- and glucocorticoid-inducible protein kinase (Sgk) by a p38 MAPK-dependent pathway. J. Biol. Chem. 2000;275:25262–25272. doi: 10.1074/jbc.M002076200. [DOI] [PubMed] [Google Scholar]
- [13].Fillon S, Warntges S, Matskevitch J, Moschen I, Setiawan I, Gamper N, Feng YX, Stegen C, Friedrich B, Waldegger S, Broer S, Wagner CA, Huber SM, Klingel K, Vereninov A, Lang F. Serum- and glucocorticoid-dependent kinase, cell volume, and the regulation of epithelial transport. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2001;130:367–376. doi: 10.1016/s1095-6433(01)00422-6. [DOI] [PubMed] [Google Scholar]
- [14].Waldegger S, Barth P, Raber G, Lang F. Cloning and characterization of a putative human serine/threonine protein kinase transcriptionally modified during anisotonic and isotonic alterations of cell volume. Proc. Natl. Acad. Sci. U S A. 1997;94:4440–4445. doi: 10.1073/pnas.94.9.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Maiyar AC, Phu PT, Huang AJ, Firestone GL. Repression of glucocorticoid receptor transactivation and DNA binding of a glucocorticoid response element within the serum/glucocorticoid-inducible protein kinase (sgk) gene promoter by the p53 tumor suppressor protein. Mol. Endocrinol. 1997;11:312–329. doi: 10.1210/mend.11.3.9893. [DOI] [PubMed] [Google Scholar]
- [16].Maiyar AC, Huang AJ, Phu PT, Cha HH, Firestone GL. p53 stimulates promoter activity of the sgk. serum/glucocorticoid-inducible serine/threonine protein kinase gene in rodent mammary epithelial cells. J. Biol. Chem. 1996;271:12414–12422. doi: 10.1074/jbc.271.21.12414. [DOI] [PubMed] [Google Scholar]
- [17].Buse P, Tran SH, Luther E, Phu PT, Aponte GW, Firestone GL. Cell cycle and hormonal control of nuclear-cytoplasmic localization of the serum- and glucocorticoid-inducible protein kinase, Sgk, in mammary tumor cells. A novel convergence point of anti-proliferative and proliferative cell signaling pathways. J. Biol. Chem. 1999;274:7253–7263. doi: 10.1074/jbc.274.11.7253. [DOI] [PubMed] [Google Scholar]
- [18].Failor KL, Desyatnikov Y, Finger LA, Firestone GL. Glucocorticoid-induced degradation of GSK3 protein is triggered by Sgk and Akt signaling and controls beta-catenin dynamics and tight junction formation in mammary epithelial tumor cells. Molecular Endocrinology. 2007;21:2403–2415. doi: 10.1210/me.2007-0143. [DOI] [PubMed] [Google Scholar]
- [19].Maiyar AC, Leong ML, Firestone GL. Importin-alpha mediates the regulated nuclear targeting of serum- and glucocorticoid-inducible protein kinase (Sgk) by recognition of a nuclear localization signal in the kinase central domain. Mol. Biol. Cell. 2003;14:1221–1239. doi: 10.1091/mbc.E02-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Walker JE. The regulation of catalysis in ATP synthase. Curr. Opin. Struct. Biol. 1994;4:912–918. doi: 10.1016/0959-440x(94)90274-7. [DOI] [PubMed] [Google Scholar]
- [21].Lebowitz MS, Pedersen PL. Protein inhibitor of mitochondrial ATP synthase: relationship of inhibitor structure to pH-dependent regulation. Arch. Biochem. Biophys. 1996;330:342–354. doi: 10.1006/abbi.1996.0261. [DOI] [PubMed] [Google Scholar]
- [22].Cabezon E, Butler PJ, Runswick MJ, Walker JE. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J Biol Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]
- [23].Sah JF, Kumar C, Mohanty P. pH dependent conformational changes modulate functional activity of the mitochondrial ATPase inhibitor protein. Biochem. Biophys. Res. Commun. 1993;194:1521–1528. doi: 10.1006/bbrc.1993.1997. [DOI] [PubMed] [Google Scholar]
- [24].Cordas E, Naray-Fejes-Toth A, Fejes-Toth G. Subcellular localization of serum- and glucocorticoid-induced kinase-1 in renal and mammary epithelial cells. Am J. Physiol Cell Physiol. 2007;292:C1971–C1981. doi: 10.1152/ajpcell.00399.2006. [DOI] [PubMed] [Google Scholar]
- [25].Engelsberg A, Kobelt F, Kuhl D. The N-terminus of the serum- and glucocorticoid-inducible kinase Sgk1 specifies mitochondrial localization and rapid turnover. Biochem. J. 2006;399:69–76. doi: 10.1042/BJ20060386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- [27].Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- [28].Horton P, Nakai K. A probabilistic classification system for predicting the cellular localization sites of proteins. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1996;4:109–115. [PubMed] [Google Scholar]
- [29].Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- [30].Gaballo A, Zanotti F, Papa S. Structures and interactions of proteins involved in the coupling function of the protonmotive F(o)F(1)-ATP Synthase. Curr. Protein Pept. Sci. 2002;3:451–460. doi: 10.2174/1389203023380558. [DOI] [PubMed] [Google Scholar]
- [31].Lopez-Mediavilla C, Vigny H, Godinot C. Docking the mitochondrial inhibitor IF-1 to a membrane receptor different from the F1-ATPase beta subunit. Eur J. Biochem. 1993;215:487–496. doi: 10.1111/j.1432-1033.1993.tb18058.x. [DOI] [PubMed] [Google Scholar]
- [32].Brickley DR, Mikosz CA, Hagan CR, Conzen SD. Ubiquitin modification of serum and glucocorticoid-induced protein kinase-1 (SGK-1) J. Biol. Chem. 2002;277:43064–43070. doi: 10.1074/jbc.M207604200. [DOI] [PubMed] [Google Scholar]
- [33].Campanella M, Parker N, Tan CH, Hall AM, Duchen MR. IF(1): setting the pace of the F(1)F(o)-ATP synthase. Trends Biochem. Sci. 2009;34:343–350. doi: 10.1016/j.tibs.2009.03.006. [DOI] [PubMed] [Google Scholar]
- [34].Cabezon E, Runswick MJ, Leslie AG, Walker JE. The structure of bovine IF(1), the regulatory subunit of mitochondrial F-ATPase. Embo J. 2001;20:6990–6996. doi: 10.1093/emboj/20.24.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gledhill JR, Montgomery MG, Leslie AG, Walker JE. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. U S A. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gledhill JR, Montgomery MG, Leslie AG, Walker JE. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. U S A. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Green DW, Grover GJ. The IF(1) inhibitor protein of the mitochondrial F(1)F(0)-ATPase. Biochim. Biophys. Acta. 2000;1458:343–355. doi: 10.1016/s0005-2728(00)00085-2. [DOI] [PubMed] [Google Scholar]
- [38].Bijur GN, Jope RS. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J. Neurochem. 2003;87:1427–1435. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shaik ZP, Fifer EK, Nowak G. Akt activation improves oxidative phosphorylation in renal proximal tubular cells following nephrotoxicant injury. Am J Physiol Renal Physiol. 2008;294:F423–F432. doi: 10.1152/ajprenal.00463.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]