Abstract
Transcription activator-like effectors (TALEs) are a class of naturally occurring DNA binding proteins found in the plant pathogen Xanthomonas sp. The DNA binding domain of each TALE consists of tandem 34-amino acid repeat modules that can be rearranged according to a simple cipher to target new DNA sequences. Customized TALEs can be used for a wide variety of genome engineering applications, including transcriptional modulation and genome editing. Here we describe a toolbox for rapid construction of custom TALE transcription factors (TALE-TFs) and nucleases (TALENs) using a hierarchical ligation procedure. This toolbox facilitates affordable and rapid construction of custom TALE-TFs and TALENs within one week and can be easily scaled up to construct TALEs for multiple targets in parallel. We also provide details for testing the activity in mammalian cells of custom TALE-TFs and TALENs using, respectively, qRT-PCR and Surveyor nuclease. The TALE toolbox described here will enable a broad range of biological applications.
Keywords: TALE, TAL effector, TALE-TF, TALE nuclease, TALEN, zinc finger, ZFN, transcription factor, gene activation, genome engineering, meganuclease, gene knockout, synthetic biology, homologous gene targeting
INTRODUCTION
Systematic reverse engineering of the functional architecture of the mammalian genome requires the ability to perform precise perturbations on gene sequences and transcription levels. Tools capable of facilitating targeted genome editing and transcription modulation are essential for elucidating the genetic and epigenetic basis of diverse biological functions and diseases. Recent discovery of the transcription activator-like effector (TALE) code1, 2 has enabled the generation of custom TALE DNA binding domains with programmable specificity3–12. When coupled to effector domains, customized TALEs provide a promising platform for achieving a wide variety of targeted genome manipulations3–5, 8, 11, 13, 14. Previously, we reported efficient construction of TALEs with customized DNA binding domains for activating endogenous genes in the mammalian genome3. Here we describe an improved protocol for rapid construction of customized TALEs and methods to apply these TALEs to achieve endogenous transcriptional activation3–5, 8 and site-specific genome editing4, 7, 9, 11–15. Investigators should be able to use this protocol to construct TALEs for targets of their choice in less than one week.
Transcription Activator-Like Effectors
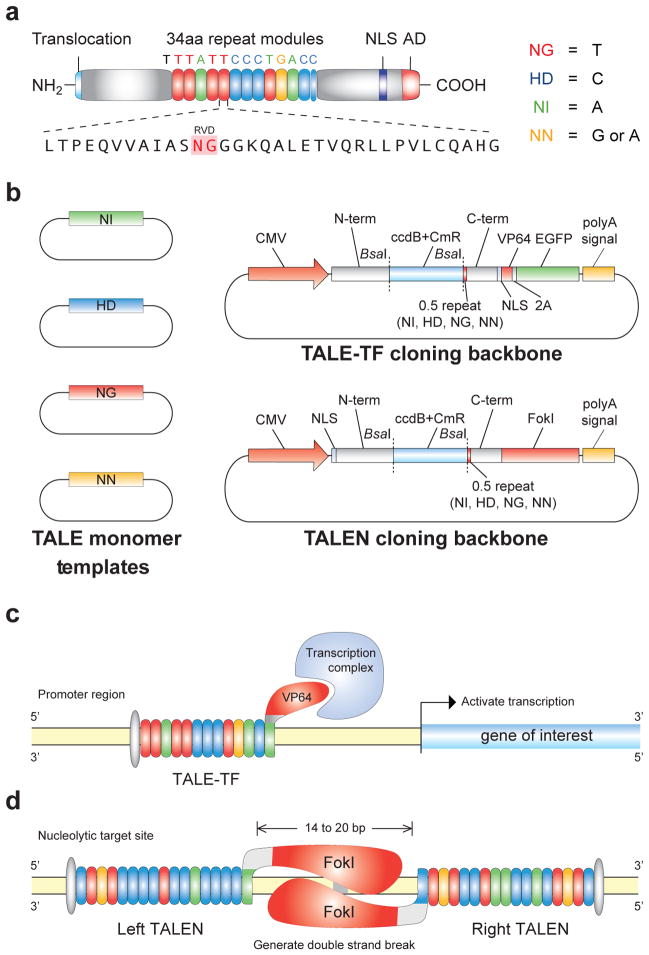
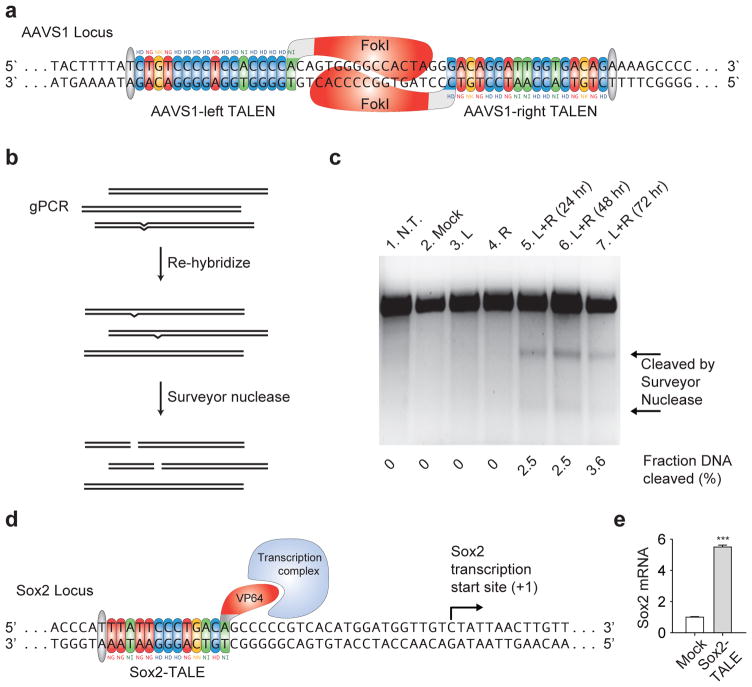
TALEs are natural bacterial effector proteins used by Xanthomonas sp. to modulate gene transcription in host plants to facilitate bacterial colonization16, 17. The central region of the protein contains tandem repeats of 34 amino acids sequences (termed monomers) that are required for DNA recognition and binding18–21 (Fig. 1a). Naturally occurring TALEs have been found to have a variable number of monomers, ranging from 1.5 to 33.5 (ref. 16). Although the sequence of each monomer is highly conserved, they differ primarily in two positions termed the repeat variable diresidues (RVDs, 12th and 13th positions). Recent reports have found that the identity of these two residues determines the nucleotide binding specificity of each TALE repeat and a simple cipher specifies the target base of each RVD (NI = A, HD = C, NG = T, NN = G or A)1, 2. Thus, each monomer targets one nucleotide and the linear sequence of monomers in a TALE specifies the target DNA sequence in the 5′ to 3′ orientation. The natural TALE binding sites within plant genomes always begin with a thymine1, 2, which is presumably specified by a cryptic signal within the non-repetitive N-terminus of TALEs. The tandem repeat DNA binding domain always ends with a half length repeat (0.5 repeat, Fig. 1a). Therefore, the length of DNA sequence being targeted is equal to the number of full repeat monomers plus two.
Figure 1. A TALE toolbox for genome engineering.
(a) Natural structure of TALEs derived from Xanthomonas sp. Each DNA binding module consists of 34 amino acids, where the repeat variable diresidues (RVDs) in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = T, HD = C, NI = A, and NN = G or A. The DNA binding modules are flanked by non-repetitive amino and carboxyl termini, which carry the translocation, nuclear localization (NLS), and transcription activation (AD) domains. A cryptic signal within the amino terminus specifies a thymine as the first base of the target site. (b) The TALE toolbox allows rapid and inexpensive construction of custom TALE-TFs and TALENs. The kit consists of 12 plasmids in total: 4 monomer plasmids to be used as templates for PCR amplification, 4 TALE-TF and 4 TALEN cloning backbones corresponding to 4 different bases targeted by the 0.5 repeat. CMV: cytomegalovirus promoter; N-term: non-repetitive amino terminus from the Hax3 TALE; C-term: non-repetitive carboxyl terminus from the Hax3 TALE; BsaI: type IIs restriction sites used for the insertion of custom TALE DNA binding domains; ccdB+CmR: negative selection cassette containing the ccdB negative selection gene and chloramphenicol resistance gene; NLS: nuclear localization signal; VP64: synthetic transcriptional activator derived from VP16 protein of herpes simplex virus; 2A: 2A self-cleavage linker; EGFP: enhanced green fluorescent protein; polyA signal: polyadenylation signal; FokI: catalytic domain from the FokI endonuclease. (c) TALEs can be used to generate custom transcription factors (TALE-TFs) and modulate the transcription of endogenous genes from the genome. This schematic shows a TALE-TF designed to target the SOX2 locus in the human genome. The SOX2 TALE-TF recognizes the sense strand of the SOX2 proximal promoter, and the recognition site begins with T. The TALE DNA-binding domain is fused to the synthetic VP64 transcriptional activator, which recruits RNA polymerase and other factors needed to initiate transcription. (d) TALE nucleases (TALENs) can be used to generate site-specific double strand breaks to facilitate genome editing through non-homologous repair or homology-directed repair. This schematic shows a pair of TALENs designed to target the AAVS1 locus in the human genome. Two TALENs target a pair of binding sites flanking a 16bp spacer. The left and right TALENs recognize the top and bottom strands of the target sites respectively. Each TALE DNA-binding domain is fused to the catalytic domain of FokI endonuclease; when FokI dimerizes, it cuts the DNA in the region between the left and right TALEN binding sites.
Comparison to other genome manipulation methods
For targeted gene insertion and knockout, there are several techniques that have been used widely in the past, such as homologous gene targeting22–24, transposases25, 26, site-specific recombinases27, meganucleases28, and integrating viral vectors29, 30. However most of these tools target a preferred DNA sequence and cannot be easily engineered to function at non-canonical DNA target sites. The most promising, programmable DNA-binding domain has been the artificial zinc finger (ZF) technology, which enables arrays of ZF modules to be assembled into a tandem array and target novel DNA binding sites in the genome. Each finger module in a ZF array targets three DNA bases31, 32. In comparison, TALE DNA binding monomers target single nucleotides and are much more modular than ZF modules. For instance, when two independent ZF modules are assembled into a new array, the resulting target site cannot be easily predicted based on the known binding sites for the individual finger modules. Perhaps the biggest caveat of ZFs is that most of the intellectual property surrounding the ZF technology platform is proprietary and expensive (>$10k per target site). A public effort for ZF technology development also exists through the Zinc Finger Consortium but the publicly available ZF modules can only target a subset of the 64 possible trinucleotide combinations33–35. TALEs theoretically can target any sequence and have already been deployed in many organisms with impressive success (see Table 1). Although TALEs seem superior in many ways, zinc fingers have a much longer track record in DNA-targeting applications32, including their use in human clinical trials36. Despite their relatively recent development, early results with TALEs have been promising and it seems that they can be applied in the same way as zinc fingers for many DNA-targeting applications (e.g. transcriptional modulator3–5, 8, nuclease4, 7, 9, 11–15, recombinase37–39, transposase40, 41).
Table 1.
Applications of custom TALEs on endogenous genome targets
Constructing Customized TALE-TFs and TALENs
Due to the repetitive nature of TALEs, construction of the DNA-binding monomers can be difficult. Previously, we and other groups have used a hierarchical ligation strategy to overcome the difficulty of assembling the monomers into ordered multimer arrays, taking advantage of degeneracy in codons surrounding the monomer junction and Type IIs restriction enzymes3, 6–10. In this protocol, we employ the same basic strategy that we previously used3 to construct TALE-TFs to modulate transcription of endogenous human genes. We have further improved the TALE assembly system with a few optimizations, including maximizing the dissimilarity of ligation adaptors to minimize misligations and combining separate digest and ligation steps into single Golden Gate42–44 reactions. Briefly, we first amplify each nucleotide-specific monomer sequence with ligation adaptors that uniquely specify the monomer position within the TALE tandem repeats. Once this monomer library is produced, it can conveniently be re-used for the assembly of many TALEs. For each TALE desired, the appropriate monomers are first ligated into hexamers, which are then amplified via PCR. Then, a second Golden Gate digestion-ligation with the appropriate TALE cloning backbone (Fig. 1b) yields a fully-assembled, sequence-specific TALE. The backbone contains a ccdB negative selection cassette flanked by the TALE N- and C-termini, which is replaced by the tandem repeat DNA-binding domain when the TALE has been successfully constructed. ccdB selects against cells transformed with an empty backbone, therefore yielding clones with tandem repeats inserted7.
Assemblies of monomeric DNA binding domains can be inserted into the appropriate TALE transcription factor (TALE-TF) or TALE nuclease (TALEN) cloning backbones to construct customized TALE-TFs and TALENs. TALE-TFs are constructed by replacing the natural activation domain within the TALE C-term with the synthetic transcription activation domain VP64 (ref. 3) (Fig. 1c). By targeting a binding site upstream of the transcription start site, TALE-TFs recruit the transcription complex in a site-specific manner and initiate gene transcription. TALENs are constructed by fusing a C-term truncation (+63aa) of the TALE DNA binding domain4 with the non-specific FokI endonuclease catalytic domain (Fig. 1d). The +63aa C-term truncation has also been shown to function as the minimal C-term sufficient for transcriptional modulation3. TALENs form dimers through binding to two target sequences separated by ~17 bases. Between the pair of binding sites, the FokI catalytic domains dimerize and function as molecular scissors by introducing double-strand breaks (DSBs) (Fig 1d). Normally, DSBs are repaired by the non-homologous end-joining45 (NHEJ) pathway, resulting in small deletions and functional gene knock-out. Alternatively, TALEN-mediated DSBs can stimulate homologous recombination, enabling site-specific insertion of an exogenous donor DNA template4, 13.
We also present a short procedure for verifying correct TALE assembly: using colony PCR to verify the correct insert length followed by DNA sequencing. With our cloning procedure, we routinely achieve high efficiency (correct length) and high accuracy (correct sequence). The cloning procedure is modular in several ways: We can construct TALEs to target DNA sequences of different lengths and the protocol is the same for producing either TALE-TFs or TALENs. The backbone vectors can be modified with different promoters to achieve cell-type specific expression.
Our protocol includes functional assays for evaluating TALE-TF and TALEN activity in human cells. This step is important because we have observed some variability in TALE activity on the endogenous genome, possibly due to epigenetic repression and/or inaccessible chromatin at certain loci. For TALE-TFs, we perform quantitative reverse-transcription polymerase chain reaction (qRT-PCR) to quantify changes in gene expression. For TALENs, we use the Surveyor mutation detection assay (i.e. the base-mismatch cleaving endonuclease Cel2) to quantify NHEJ. Although these assays are standard and have already been described elsewhere46, 47, we feel that the functional characterization is integral to TALE production and therefore have presented it here with the assembly procedure. Other functional assays such as plasmid-based reporter constructs3, 7, restriction sites destroyed by NHEJ48, or other enzymes that detect DNA mismatch49 may also be used to validate TALE activity.
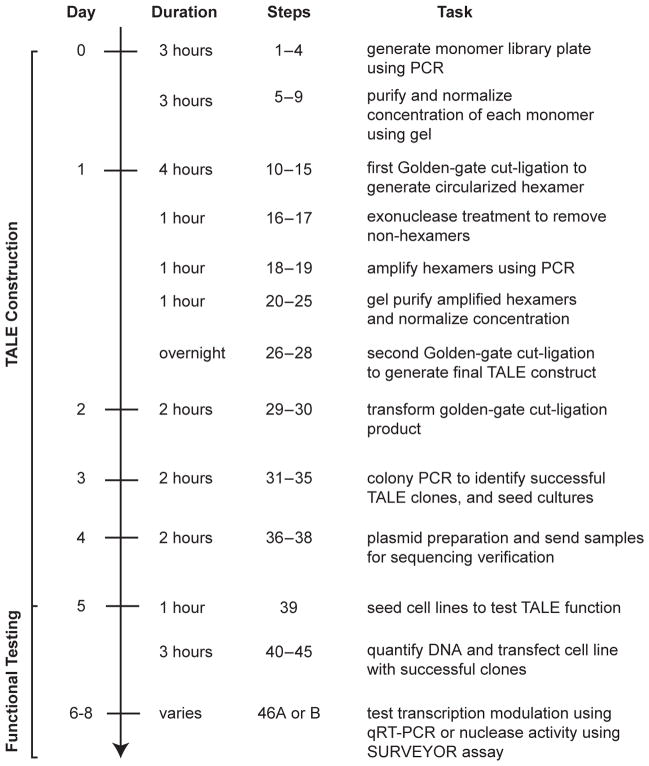
Our protocol (Fig 2) begins with the generation of a monomer library, which takes one day and can be re-used for building many TALEs. Using the monomer library, several TALEs can be constructed in a single day with an additional two days for transformation and sequence verification. To assess TALE function on the endogenous genome, we take ~3 days to go from mammalian cell transfection to qRT-PCR or Surveyor results.
Figure 2. Timeline for the construction of TALE-TFs and TALENs.
Steps for the construction and functional testing of TALE-TFs and TALENs are outlined. TALEs can be constructed and sequence verified in 5 days following a series of ligation and amplification steps. During the construction phase, samples can be stored at −20°C at the end of each step and continued at a later date. After TALE construction, functional validation via qRT-PCR (for TALE-TFs) and Surveyor nuclease assay (for TALENs) can be completed in 2–3 days.
Comparison with other TALE assembly procedures
A number of TALE assembly procedures have described the use of Golden-Gate cloning to construct customized TALE DNA binding domains3, 6–10. These methods rely on the use of a large collection of plasmids (typically over 50 plasmids) encoding repeat monomers and intermediate cloning vectors. Our PCR-based approach requires significantly less initial plasmid preparation, as our monomer library can be amplified on one 96-well PCR plate, and facilitates more rapid construction of custom TALEs. Plasmid-based amplification has a much lower mutation/error rate but, in our experience, the combination of a high-fidelity polymerase and the short length of the monomer template (~100 nt) results in accurate assembly. For building similar length TALEs to those presented in this protocol, the plasmid-based approaches also require an additional transformation and colony selection that extends the time needed to build TALEs. Thus, these alternative assembly protocols require a greater time investment both upfront (for monomer library preparation) and on a recurring basis (for each new TALE). For laboratories seeking to produce TALEs quickly, our protocol requires only a few hours to prepare a complete monomer library and less than a day to proceed from monomers to the final transformation into bacteria.
Targeting limitations
There are a few important limitations with the TALE technology. Although the RVD cipher is known, it is still not well understood why different TALEs designed according to the same cipher act on their target sites in the native genome with different levels of activity. It is possible that there are yet unknown sequence dependencies for efficient binding or site-specific constraints (e.g. chromatin state) that are responsible for differences in functional activity. Therefore we suggest constructing at least 2 or 3 TALE-TFs or TALEN pairs for each target locus. Also, it is possible that engineered TALEs can have off-target effects – binding unintended genomic loci – which can be difficult to detect without additional functional assays at these loci. Given the relatively early state of TALE technology development, these issues remain to be addressed in a conclusive manner.
Experimental design
TALE-TF target site selection
The programmable nature of TALEs allows for a virtually arbitrary selection of target DNA binding sites. As previously reported, the N-terminus of the TALE requires that the target site begin with a thymine nucleotide. For TALE-TFs, we have been successful targeting 14 to 20 bp sequences within 200 bp of the transcription start site (Fig. 1c). It can be advantageous to select a longer sequence to reduce off-target activation, as it is known from reporter activation assays that TALEs interact less efficiently with targets contain more than one mismatching base. In our assembly protocol, we describe ligation of 18 monomers into a backbone containing a nucleotide-specific final 0.5 monomer; combined with the initial thymine requirement, this yields a total sequence specificity of 20 nucleotides. Specifically, the TALE-TF binding site takes the form 5′-TN19-3′. When selecting TALE-TF targeting sites for modulating endogenous gene transcription, we recommend selecting multiple target sites within the proximal promoter region (can target either the sense or antisense strand), as epigenetic and local chromatin dynamics might impede TALE binding. Larger TALEs might be beneficial for TALE-TFs targeting genes with less unique regions upstream of their transcription start site.
TALEN target site selection
Since TALENs function as dimers, a pair of TALENs, referred to as the left and right TALENs, need to be designed to target a given site in the genome. The left and right TALENs target sequences on opposite strands of DNA (Fig. 1d). As with TALE-TF, we design each TALEN to target a 20 base pair sequence. TALENs are engineered as a fusion of the TALE DNA-binding domain and a monomeric FokI catalytic domain. To facilitate FokI dimerization, the left and right TALEN target sites are chosen with a spacing of ~14–20 bases. Therefore, for a pair of TALENs, each targeting 20 base pair sequences, the complete target site should have the form 5′-TN19N14–20N19A-3′, where the left TALEN targets 5′-TN19-3′ and the right TALEN targets the antisense strand of 5′-N19A-3′ (N = A, G, T, or C). TALENs should have fewer off-target effects due to the dimerization requirement for the FokI nuclease, although no significant off-target effects have been observed in limited sequencing verifications13. Because DSB formation only occurs if the spacer between the left and right TALEN binding sites (Fig. 1d) is ~14–20 bases, nuclease activity is restricted to genomic sites with both the specific sequences of the left TALEN and the right TALEN with this small range of spacing distances between those sites. These constraints should greatly reduce potential off-target effects.
TALE monomer design
To ensure that all synthesized TALEs are transcribed at a similar level, all of the monomers have been optimized to share identical DNA sequences except in the variable di-residues – and are codon-optimized for expression in human cells (see Supplementary Data 1). This should minimize any difference in translation due to codon availability.
Construction strategy
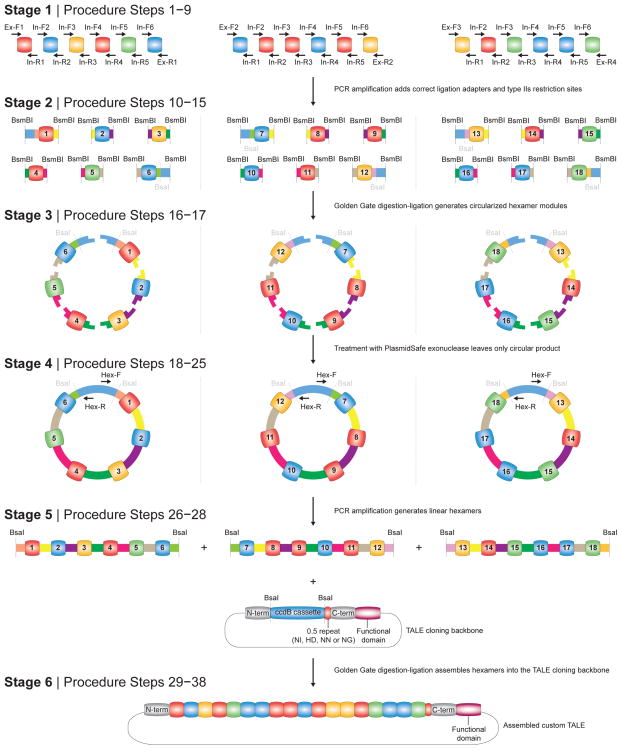
Synthesis of monomeric TALE DNA binding domains in a precise order is challenging due to their highly repetitive nature. Previously3, we took advantage of codon redundancy at the junctions between neighboring monomers and devised a hierarchical ligation strategy to construct ordered assemblies of multiple monomers. In this protocol, we describe a similar strategy but with several important improvements that make the procedure easier, more flexible, and more reliable (Fig. 3).
Figure 3. Construction of TALE DNA binding domains using hierarchical ligation assembly.
Schematic of the construction process for a custom TALE containing a 18-mer tandem repeat DNA binding domain. Stage 1: specific primers are used to amplify each monomer and add the appropriate ligation adapters (Procedure Steps 1–9). Stage 2: hexameric tandem repeats (1—6, 7—12, and 13—18) are assembled first using Golden Gate digestion-ligation. The 5′ ends of monomers 1, 7, and 13 and the 3′ ends of monomers 6, 12, and 18 are designed so that each tandem hexamer assembles into an intact circle (Procedure Steps 10–15). Stage 3: the Golden Gate reaction is treated with an exonuclease to remove all linear DNA, leaving only the properly assembled tandem hexamer (Procedure Steps 16–17). Stage 4: each tandem hexamer is amplified individually using PCR and purified (Procedure Steps 18–25). Stage 5: tandem hexamers corresponding to 1—6, 7—12, and 13—18 are ligated into the appropriate TALE-TF or TALEN cloning backbone using Golden Gate cut-ligation (Procedure Steps 26–28). Stage 6: The assembled TALE-TF or TALEN is transformed into competent cells and successful clones are isolated and sequence verified (Procedure Steps 29–38).
In our initial protocol3, the digestion and ligation steps were carried out separately with an intervening DNA purification step. This improved protocol adopts the powerful Golden Gate cloning technique42–44, requiring less hands-on time and resulting in a more efficient reaction. The Golden Gate procedure involves combining the restriction enzyme and ligase together in a single reaction with a mutually compatible buffer. The reaction is cycled between optimal temperatures for digestion and ligation. Golden Gate digestion-ligation capitalizes on Type IIs restriction enzymes, for which the recognition sequence is spatially separated from where the cut is made. During a Golden Gate reaction, the correctly ligated products no longer contain restriction enzyme recognition sites and cannot be further digested. In this manner, Golden Gate drives the reaction toward the correct ligation product as the number of cycles of digestion and ligation increases.
For the hierarchical ligation steps, we have optimized our previous cloning strategy for faster TALE production. The improved design takes advantage of a circularization step that allows only properly assembled hexameric intermediates to be preserved (Fig. 3). Correctly ligated hexamers consist of six monomers ligated together in a closed circle, and incomplete ligation products are left as linear DNA. After this ligation step, an exonuclease degrades all non-circular DNA, leaving intact only the complete circular hexamers. Without circularization and exonuclease treatment, the correct ligation product would need to be gel purified before proceeding. The combination of Golden Gate digestion-ligation and circularization reduces the overall hands-on time required for TALE assembly.
Primer design for monomer library preparation
Each monomer in the tandem repeat must have its position uniquely specified. The monomer primers are designed to add ligation adaptors that enforce this positioning. Our protocol uses a hierarchical ligation strategy: For the 18mer tandem repeat, we first ligate monomers into hexamers. Then, we ligate three hexamers together to form the 18mer. By breaking down the assembly into two steps, we do not need unique ligation junctions for each monomer in the 18mer. Instead the same set of ligation junctions internal to each hexamer are re-used in all three hexamers (first ligation step), whereas unique (external) ligation junctions are used to flank each hexamer (second ligation step). As shown in Figure 4, the internal primers used to amplify the monomers within each hexamer are the same, but the external primers differ between the hexamers. By re-using the same internal primers between different hexamers, our protocol minimizes the number of primers necessary for monomer amplification.
Figure 4. PCR plate setup used to generate a plate of monomers for constructing custom 18-mer TALE DNA binding domains.
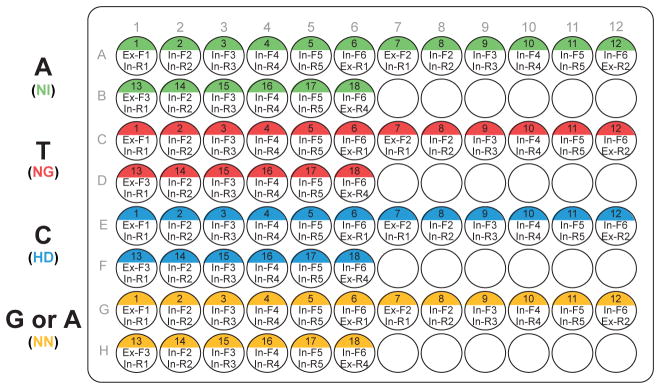
One 96-well plate can be used to carry out 72 reactions (18 for each monomer template). The position of each monomer and the primers used for the position is indicated in the well. Color coding in the well indicates the monomer used as the PCR template. Typically, 2–4 plates of 100 ul PCR reactions are pooled together and purified to generate a monomer library of sufficient quantity for production of many TALEs. During TALE construction, the corresponding monomer for each DNA base in the 18 bp target sequence can be easily picked from the plate.
Controls
As a negative control for Golden Gate assembly, we recommend performing a separate reaction with only the TALE-TF or TALEN backbone. Transformation of this negative control should result in few or no colonies due to the omission of the tandem repeats and resulting re-ligation of the toxic ccdB insert. After completing the TALE cloning, we use colony PCR or restriction digests to screen for correct length clones. For the final verification of proper assembly, we sequence the entire length of the tandem repeats. Due to limits in Sanger sequencing read length, other TALE assembly protocols have difficulty sequencing the entire tandem repeat region7, 9, 10. The similarity of the monomers within the region makes primer annealing to specific monomers impossible. We have overcome this problem by slightly modifying the codon usage at the 5′ end of monomer 7 to create a unique annealing site so that a TALE with a 18mer DNA binding array can be verified through a combination of three staggered sequencing reads. Specifically, during the monomer amplification, the codons for the first 5 amino acids in monomer 7 are mutated via PCR to use different but synonymous codons, creating a unique priming site without changing the encoded TALE protein. This modification allows each hexamer in the 18mer to be sequenced with a separate sequencing read and requires only a standard read length of ~700 bp for complete sequence verification. For TALEs containing more than 18 full monomer repeats, we introduced a third unique priming site for sequencing at the 3′ end of the 18th monomer using a similar approach. For construction of TALEs containing up to 24 full monomers with the entire tandem repeat region easily sequenced, see Box 1.
Box 1. Building TALEs that target DNA sequences of different lengths.
In the main protocol, we present a hierarchical ligation strategy for the construction of TALEs that contain 18 full monomer repeats; however the general approach can be easily adapted to construct TALEs of any length. These TALEs containing 18 full repeat monomers bind to 20 bp DNA sequences, where the first and last bases are specified by the N-terminus and the 0.5 repeat, respectively (Fig 1a). We chose this length because, empirically, we have observed that 20 bp sequences tend to be unique within the human genome. Nevertheless, for different species (eg. with larger or more repetitive genomes) or for repetitive regions within the human genome, it can be advantageous to construct longer or shorter TALEs. For certain genomic loci, it might also be difficult to identify TALEN target sites that satisfy the spacing constraints when the binding sites for both left and right TALENs are restricted to 20 bp sequences.
Our protocol is easily modified for the construction of TALEs containing up to 24 full monomer repeats by changing the order in which particular primers are used during the preparation of the monomer library plate (as described in Procedure Steps 1 – 9). All other steps remain essentially the same. A plate of monomer amplification primers (similar to Figure 4) can be prepared for building TALEs with 24 full monomer repeats, which bind to 26 bp DNA sequences, as illustrated below. In this case, a fourth circular hexamer, corresponding to monomers 19 through 24, is also built and treated identically as the other three circular hexamers (1 – 6, 7 – 12, and 13 – 18).
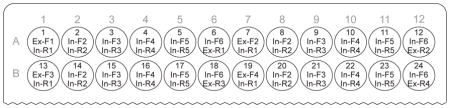
For building shorter TALEs, only a single change to monomer amplification is needed: The final monomer should be amplified with the Ex-R4 reverse primer. For example, to build TALEs with 17 monomers instead of 18, the monomers templates (NI, NG, NN, HD) should be amplified with the forward/reverse primer combination In-F5/Ex-R4. Note that during gel purification (Procedure Step 20) the desired PCR amplicon is a pentamer containing monomers 13 – 17 and will run faster than the hexamers (1 – 6, 7 – 12). After purification, ensure that the pentameric and hexameric intermediates are used at an equimolar ratio in the final Golden Gate digestion-ligation.
Design of functional validation assays
For TALE-TFs, qRT-PCR quantitatively measures the increase in transcription driven by the TALE-TF. For TALENs, the Surveyor assay provides a functional validation of TALEN cutting and quantifies the cutting efficiency of a particular pair of TALENs. These assays should be performed in the same cell type as intended for the TALE application, as TALE efficacy can vary between cell types, presumably due to differences in chromatin state or epigenetic modifications.
For qRT-PCR, we use commercially-available probes to measure increased transcription of the TALE-TF-targeted gene. For most genes in the human or mouse genomes, specific probes can be purchased (e.g. TaqMan Gene Expression Probes from Applied Biosystems). There are a wide variety of qRT-PCR protocols and, although we describe one of them here, others can be substituted. For example, a more economical option is to design custom, transcript-specific primers (e.g. with NCBI Primer-BLAST) and use a standard fluorescent dye to detect amplified dsDNA (e.g. SYBR Green).
For Surveyor, we follow the recommendations given by the assay manufacturer when designing specific primers for genomic PCR. We typically design primers that are ~30 nucleotides long and with melting temperatures of ~65 °C. The primers should flank the TALEN target site and generate an amplicon of ~300–800 bp with the TALEN target site near the middle. During the design, we also check to make sure the primers are specific over the intended genome using NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Before using the primers for Surveyor, the primers and specific PCR cycling parameters should be tested to ensure that amplification results in a single clean band. In difficult cases where a single band product cannot be achieved, it is acceptable to gel extract the correct length band before proceeding with heteroduplex re-annealing and Surveyor nuclease digest.
MATERIALS
REAGENTS
TALE construction
-
TALE monomer template plasmids:
pNI_v2
pNG_v2
pNN_v2
pHD_v2
-
TALE transcriptional activator (TALE-TF) plasmids:
pTALE-TF_v2 (NI)
pTALE-TF_v2 (NG)
pTALE-TF_v2 (NN)
pTALE-TF_v2 (HD)
-
TALE nuclease (TALEN) backbone plasmids:
pTALEN_v2 (NI)
pTALEN_v2 (NG)
pTALEN_v2 (NN)
pTALEN_v2 (HD)
These plasmids can be obtained individually or bundled together as a single kit from the Zhang Lab plasmid collection at Addgene (http://www.addgene.org/TALE_Toolbox). See Supplementary Data 1 for plasmid sequences.
PCR primers for TALE construction (Table 2, Integrated DNA Technologies, custom DNA oligonucleotides)
Herculase II Fusion polymerase (Agilent Technologies, cat. no. 600679)
Table 2.
Primer sequences for TALE construction
| Name | Sequence | Purpose |
|---|---|---|
| Ex-F1 | TGCGTCcgtctcCGAACCTTAAACCGGCCAACATACCggtctcCTGACCCCAGAGCAGGTCGTG | monomer amplification |
| Ex-F2 | TGCGTCcgtctcCGAACCTTAAACCGGCCAACATACCggtctcGACTTACACCCGAACAAGTCGTGGCAATTGCGAGC | |
| Ex-F3 | TGCGTCcgtctcCGAACCTTAAACCGGCCAACATACCggtctcGCGGCCTCACCCCAGAGCAGGTCG | |
| Ex-F4 | TGCGTCcgtctcCGAACCTTAAACCGGCCAACATACCggtctcGTGGGCTCACCCCAGAGCAGGTCG | |
| Ex-R1 | GCTGACcgtctcCGTTCAGTCTGTCTTTCCCCTTTCCggtctcTAAGTCCGTGCGCTTGGCAC | |
| Ex-R2 | GCTGACcgtctcCGTTCAGTCTGTCTTTCCCCTTTCCggtctcAGCCGTGCGCTTGGCACAG | |
| Ex-R3 | GCTGACcgtctcCGTTCAGTCTGTCTTTCCCCTTTCCggtctcTCCCATGGGCCTGACATAACACAGGCAGCAACCTCTG | |
| Ex-R4 | GCTGACcgtctcCGTTCAGTCTGTCTTTCCCCTTTCCggtctcTGAGTCCGTGCGCTTGGCAC | |
| In-F2 | CTTGTTATGGACGAGTTGCCcgtctcGTACGCCAGAGCAGGTCGTGGC | |
| In-F3 | CCAAAGATTCAACCGTCCTGcgtctcGAACCCCAGAGCAGGTCGTG | |
| In-F4 | TATTCATGCTTGGACGGACTcgtctcGGTTGACCCCAGAGCAGGTCGTG | |
| In-F5 | GTCCTAGTGAGGAATACCGGcgtctcGCCTGACCCCAGAGCAGGTCGTG | |
| In-F6 | TTCCTTGATACCGTAGCTCGcgtctcGGACACCAGAGCAGGTCGTGGC | |
| In-R1 | TCTTATCGGTGCTTCGTTCTcgtctcCCGTAAGTCCGTGCGCTTGGCAC | |
| In-R2 | CGTTTCTTTCCGGTCGTTAGcgtctcTGGTTAGTCCGTGCGCTTGGCAC | |
| In-R3 | TGAGCCTTATGATTTCCCGTcgtctcTCAACCCGTGCGCTTGGCACAG | |
| In-R4 | AGTCTGTCTTTCCCCTTTCCcgtctcTCAGGCCGTGCGCTTGGCACAG | |
| In-R5 | CCGAAGAATCGCAGATCCTAcgtctcTTGTCAGTCCGTGCGCTTGGCAC | |
| Hex-F | CTTAAACCGGCCAACATACC | hexamer amplification |
| Hex-R | AGTCTGTCTTTCCCCTTTCC | |
| TALE-Seq-F1 (aka colony PCR forward) | CCAGTTGCTGAAGATCGCGAAGC | sequencing forward primer used to check monomers 1–6; also used as colony PCR forward primer |
| TALE-Seq-F2 | ACTTACACCCGAACAAGTCG | sequencing forward primer used to check monomers 7–12 |
| TALE-Seq-R1 (aka colony PCR reverse) | TGCCACTCGATGTGATGTCCTC | sequencing primer used to check monomers 13–18 for TALEs with less than 18 full monomer repeats, and used to check monomers 19–24 for TALEs with more than 18 monomers (use TALE-Seq-R2 to check monomers 13–18 in this case); also used as colony PCR reverse primer |
| TALE-Seq-R2 | CCCATGGGCCTGACATAA | sequencing reverse primer used to check monomers 13–18 in TALEs with more than 18 full monomer repeats |
CRITICAL Standard Taq polymerase, which lacks 3′-5′ exonuclease proofreading activity, has lower fidelity and can lead to errors in the final assembled TALE. Herculase II is a high-fidelity polymerase (equivalent fidelity to Pfu) that produces high yields of PCR product with minimal optimization. Other high-fidelity polymerases may be substituted.
5x Herculase II reaction buffer (Agilent Technologies, included with polymerase)
Taq-B polymerase (Enzymatics, cat. no. P725L)
10x Taq-B buffer (Enzymatics, included with polymerase)
25 mM (each) dNTP solution mix (Enzymatics, cat. no. N205L)
MinElute Gel Extraction Kit (Qiagen, cat. no. 28606)
CRITICAL MinElute columns should be stored at 4°C until use.
QIAprep Spin Miniprep Kit (Qiagen, cat. no. 27106)
QIAquick 96 PCR Purification (Qiagen, cat. no. 28181)
UltraPure DNase/RNase-Free Distilled Water (Invitrogen, cat. no. 10977-023)
UltraPure 10X TBE Buffer (Invitrogen, cat. no. 15581-028)
SeaKem LE agarose (Lonza, cat. no. 50004)
10,000x SYBR Safe DNA stain (Invitrogen, cat. no. S33102)
Low DNA Mass Ladder (Invitrogen, cat. no. 10068-013)
1 kb Plus DNA Ladder (Invitrogen, cat. no. 10787-018)
TrackIt™ Cyan/Orange Loading Buffer (Invitrogen, cat. no. 10482-028)
-
Restriction enzymes:
BsmBI (Esp3I) (Fermentas/ThermoScientific cat. no. ER0451)
BsaI-HF (New England Biolabs, cat. no. R3535L)
AfeI (New England Biolabs, cat. no. R0652S)
Fermentas Tango Buffer and 10x NEBuffer 4 (included with enzymes)
100x Bovine Serum Albumin (New England Biolabs, included with BsaI-HF)
DL-Dithiothreitol (DTT) (Fermentas/ThermoScientific cat. no. R0862)
T7 DNA ligase, 3,000 U/ul (Enzymatics, cat. no. L602L)
CRITICAL Do not substitute the more commonly-used T4 ligase. T7 ligase has 1000-fold higher activity on sticky ends than blunt ends and higher overall activity than commercially available concentrated T4 ligases.
10 mM Adenosine 5′-Triphosphate (New England Biolabs, cat. no. P0756S)
Plasmid-Safe™ ATP-Dependent DNase (Epicentre, cat. no. E3101K)
One Shot® Stbl3™ Chemically Competent E. coli (Invitrogen, cat. no. C7373-03)
SOC medium (New England Biolabs, cat. no. B9020S)
LB medium (Sigma, cat. no. L3022)
LB agar medium (Sigma, cat. no. L2897)
100 mg/ml ampicillin, sterile-filtered (Sigma, cat. no. A5354)
TALEN and TALE-TF functional validation in mammalian cells
HEK293FT cells (Invitrogen, cat. no. R700-07)
Dulbecco’s Minimum Eagle Medium (DMEM) (1X), high glucose (Invitrogen, cat. no. 10313-039)
Dulbecco’s Phosphate Buffered Saline (DPBS) (1X) (Invitrogen, cat. no. 14190-250)
Fetal bovine serum, qualified and heat inactivated (Invitrogen, cat. no. 10438-034)
Opti-MEM® I reduced-serum medium (Invitrogen, cat. no. 11058-021)
GlutaMAX™-I (100X) (Invitrogen, cat. no. 35050079)
Penicillin-streptomycin (100X) (Invitrogen, cat. no. 15140-163)
Trypsin, 0.05% (1X) with EDTA•4Na (Invitrogen, cat. no. 25300-062)
Lipofectamine 2000 ™ transfection reagent (Invitrogen, cat. no. 11668027)
QuickExtract ™ DNA extraction solution (Epicentre, cat. no. QE09050)
Herculase II Fusion polymerase (Agilent Technologies, cat. no. 600679)
CRITICAL Since Surveyor assay is sensitive to single-base mismatches, it is important to use only a high-fidelity polymerase. Other high-fidelity polymerases can be substituted; refer to the Surveyor manual for PCR buffer compatibility details.
5x Herculase II reaction buffer (Agilent Technologies, included with polymerase)
Surveyor Mutation Detection Kit for Standard Gel Electrophoresis (Transgenomic, cat. no. 706025)
CRITICAL The Surveyor assay includes the Cel2 base-mismatch nuclease. Alternatives include the Cel1, T7, mung bean, and S1 nucleases50, 51. Of these, Cel1 has been applied extensively for mutation detection52–54 and established protocols are available for its purification52, 54.
Primers for Surveyor assay of TALEN cutting efficiency (see Experimental design for further information on Primer design,, Integrated DNA Technologies, custom DNA oligonucleotides)
RNeasy Mini Kit (Qiagen, cat. no. 74104)
QIAshredder (Qiagen, cat. no. 79654)
-
2-mercaptoethanol (Sigma, cat. no. 63689)
! CAUTION Wear appropriate personal protective equipment and work in a fume hood when handling 2-mercaptoethanol, which is acutely toxic and corrosive.
RNAseZAP (Applied Biosystems, cat. no. AM9780)
iScript cDNA synthesis kit (BioRad, cat. no. 170-8890)
TaqMan® Universal Master Mix (Applied Biosystems, cat. no. 4364341)
TaqMan® Gene Expression Assay Probes for the TALE-TF-targeted gene (Applied Biosystems, http://bioinfo.appliedbiosystems.com/genomic-products/gene-expression.html)
EQUIPMENT
96-well thermocycler with programmable temperature stepping functionality (Applied Biosystems Veriti, cat no. 4375786)
CRITICAL Programmable temperature stepping is needed for the TALEN (Surveyor) functional assay. Other steps only require a PCR-capable thermocycler.
96-well qPCR system (Applied Biosystems StepOnePlus™ Real-Time PCR System, Cat. No. 4376600)
96-well optical plates (Applied Biosystems MicroAmp, cat. no. N801-0560)
96-well PCR plates (Axygen, cat. no. PCR-96-FS-C)
8-well strip PCR tubes (Applied Biosystems, cat. no. N801-0580)
QIAvac 96 vacuum manifold (Qiagen, cat. no. 19504)
Gel electrophoresis system (BioRad PowerPac Basic Power Supply, cat no. 164-5050, and BioRad Sub-Cell GT System gel tray, cat. no. 170-4401).
Digital gel imaging system (BioRad GelDoc EZ, cat. no. 170-8270, and BioRad Blue Sample Tray, cat. no. 170-8273)
Blue light transilluminator and orange filter goggles (Invitrogen SafeImager 2.0, cat. no. G6600)
Sterile 20 ul pipette tips for colony picking
Gel quantification software (BioRad ImageLab, included with GelDoc EZ, or open-source NIH ImageJ, available at http://rsbweb.nih.gov/ij/)
TALE online sequence verification software (Zhang Lab: http://taleffectors.com/tools/)
60 mm × 15 mm petri dishes (BD Biosciences, cat. no. 351007)
Incubator for bacteria plates (Quincy Lab Inc, cat. no. 12-140E)
Shaking incubator for bacteria suspension culture (Infors HT Ecotron)
6-well, cell culture-treated polystyrene plates (Corning, cat. no. 3506)
UV spectrophotometer (ThermoScientific, cat. no. NanoDrop 2000c)
REAGENT SETUP
Tris-borate EDTA (TBE) electrophoresis solution
Dilute in distilled water to 1x working solution for casting agarose gels and as a buffer for gel electrophoresis. Buffer can be stored at room temperature indefinitely.
10X BSA
Dilute 100x bovine serum albumin (BSA, supplied with BsaI-HF) to 10x concentration and store at −20 °C for at least 1 year in 20 ul aliquots.
10 mM ATP
Divide 10 mM ATP into 50 ul aliquots and store at −20 °C for up to 1 year; avoid repeated freeze-thaw cycles.
10 mM DTT
Prepare 10 mM DTT solution in distilled water and store 20 ul aliquots at −70 °C for up to 2 years; for each reaction, use a new aliquot since DTT is easily oxidized.
D10 culture medium
For culture of HEK293FT cells, prepare D10 culture medium by supplementing DMEM with 1X GlutaMAX and 10% fetal bovine serum. As indicated in the protocol, this medium can also be supplemented with 1X penicillin-streptomycin. D10 medium can be made in advance and stored at 4 °C for up to 1 month.
PROCEDURE
Amplification and normalization of monomer library with ligation adaptors for 18mer TALE DNA binding domain construction
TIMING 6 hr
-
1|
Prepare diluted forward and reverse monomer primer mixes. In a 96-well PCR plate, prepare primer mixes for amplifying a TALE monomer library (Figure 3, stage 1). Mix forward and reverse primers for each of the 18 positions according to the first two rows (A and B) of Figure 4 and achieve a final concentration of 10 uM for each primer. If using multi-channel pipettes, arrange the oligonucleotide primers in the order indicated in Figure 4 to allow for easy pipetting. Typically, prepare 50 ul mixes for each primer pair (40 ul ddH2O, 5 ul 100 uM forward primer, 5 ul 100 uM reverse primer).
-
2|
Set up two 96-well monomer library plates following the organization shown in Figure 4; each plate will contain a total of 72 PCR reactions (18 positions for each monomer × 4 types of monomers). Although it is acceptable to have smaller volume PCR reactions, we typically make the monomer set in larger quantities since one monomer library plate can be used repeatedly for the construction of many TALEs. Each PCR reaction should be made up as follows to a total volume of 200 ul, and then split between the two 96-well plates so that each well contains a 100 ul PCR reaction:
Component Amount Final concentration Monomer template plasmid (5 ng/ul) 2 ul 50 pg/ul 100 mM dNTP (25 mM each) 2 ul 1 mM 5X Herculase II PCR buffer 40 ul 1x 20 uM primer mix (10 uM forward primer and 10 uM reverse primers from Step 1) 4 ul 200 nM Herculase II Fusion polymerase 2 ul Distilled water 150 ul Total 200 ul (for 2 reactions) -
3|
Perform PCR on the reactions from Step 2 using the following cycling conditions:
Cycle number Denature Anneal Extend 1 95°C, 2 min 2–31 95°C, 20 s 60°C, 20 s 72°C, 10 s 32 72°C, 3 min -
4|
After the reaction has completed, use gel electrophoresis to verify that monomer amplification was successful. Cast a 2% agarose gel in 1x TBE electrophoresis buffer with 1x SYBR Safe dye. The gel should have enough lanes to run out 2 ul of each PCR product from Step 3. Run the gel at 15 V/cm for 20 minutes. It is not necessary to check all 72 reactions at this step; it is sufficient to check all 18 reactions for one type of monomer template. Successful amplification should show a ~100 bp product. Monomers positioned at the ends of each hexamer (monomers 1, 6, 7, 12, 13 and 18) should be slightly longer than the other monomers due to the length difference of the longer external primers.
? TROUBLESHOOTING
-
5|
Pool both of the 100 ul PCR plates into a single deep-well plate. Purify the combined reactions using the QIAquick 96 PCR Purification kit following the manufacturer’s directions. Elute the DNA from each well using 100 ul of Buffer EB (included with kit) pre-warmed to 55°C. Alternatively, PCR products can also be purified using individual columns found in standard PCR cleanup kits.
◆ CRITICAL STEP Before eluting the DNA, let the 96-well column plate air dry, preferably at 37°C, for 30 minutes on a clean Kimwipe so that all residual ethanol has enough time to evaporate.
-
6|
Normalization of monomer concentration. Cast a 2% agarose gel. The gel should have enough lanes to run out 2 ul of each purified PCR product from Step 5. Include in one lane 10 ul of the quantitative DNA ladder. Run the gel at 20 V/cm for 20 minutes.
-
7|
Image the gel using a quantitative gel imaging system. Monomers 1, 6, 7, 12, 13, and 18 are ~170 bp in size, whereas the other monomers are ~150 bp size (Fig. 5a, lanes 1–6). Make sure the exposure is short enough so that none of the bands are saturated.
-
8|
Quantify the integrated intensity of each PCR product band using ImageJ or other gel quantification software. Use the quantitative ladder with known concentrations (5, 10, 20, 40, 100 ng) to generate a linear fit and quantify the concentration of each purified PCR product.
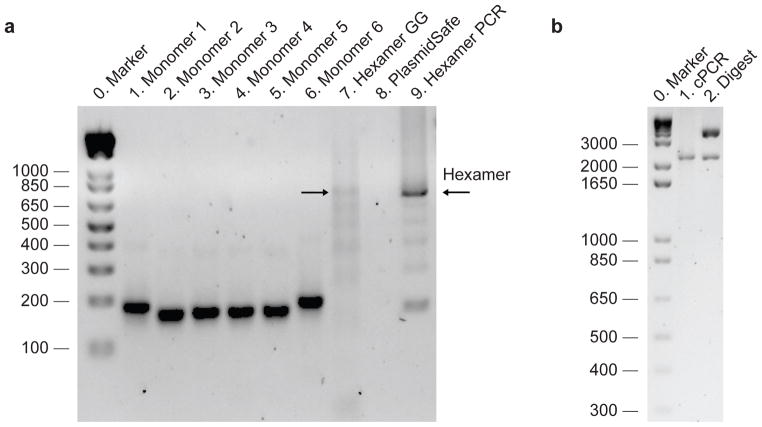
Figure 5. Example gel results from the TALE construction procedure.
(a) Lanes 1—6: products from the monomer PCR reaction (Stage 1 in Figure 3) after purification and gel normalization (Procedure Steps 8–9). The molar concentrations of samples shown on this gel have been normalized so that equal moles of monomers are mixed for downstream steps. Monomers 1 and 6 are slightly longer than monomers 2, 3, 4, and 5 due to the addition of sequences used for circularization. Lane 7: result of the hexamer Golden Gate cut-ligation (Procedure Step 15). A series of bands with size ~700 bp and lower can be seen. Successful hexamer Golden Gate assembly should show a band ~700 bp (as indicated by arrow). Lane 8: hexamer assembly after PlasmidSafe exonuclease treatment (Procedure Step 17). Typically the amount of circular DNA remaining is difficult to visualize by gel. Lane 9: result of hexamer amplification (Procedure Step 20). A ~700 bp band should be clearly visible. The hexamer gel band should be gel-purified to remove shorter DNA fragments. (b) Properly assembled TALE-TFs and TALENs can be verified using bacterial colony PCR (2175 bp band, lane 1) (Procedure Step 35) and restriction digest with AfeI (2118 bp band for correctly assembled 18-mer in either backbone; other bands for TALE-TF are 165 bp, 3435 bp, 3544 bp; other bands for TALEN are 165 bp, 2803 bp, 3236 bp; digest shown is for TALE-TF backbone vector, lane 2) (Procedure Step 35).
? TROUBLESHOOTING
-
9|
Adjust the plate of purified PCR products by adding Buffer EB so that each monomer has the same molar concentration. Since monomers 1, 6, 7, 12, 13, and 18 are longer than the other monomers, it is necessary to adjust them to a slightly higher concentration. For example, we adjust monomers 1, 6, 7, 12, 13, and 18 to 18 ng/ul and the other monomers to 15 ng/ul.
◆ CRITICAL STEP For subsequent digestion and ligation reactions, it is important that all monomers are at equimolar concentrations.
PAUSE POINT Amplified monomers can be stored at −20 °C for several months and can be reused for assembling additional TALEs.
Construction of custom 20bp-targeting TALEs
TIMING 1.5 days (5 hr hands-on time)
-
10|
Select target sequence(s). Typical TALE recognition sequences are identified in the 5′ to 3′ direction and begin with a 5′ thymine. The procedure below describes the construction of TALEs that bind a 20 bp target sequence (5′-T0N1N2N3N4N5N6N7N8N9N10N11N12N13N14N15N16N17N18N19-3′, where N = A, G, T, or C), where the first base (typically a thymine) and the last base are specified by sequences within the TALE backbone vector. The middle 18 bp are specified by the RVDs within the middle tandem repeat of 18 monomers according to the cipher NI = A, HD = C, NG = T, and NN = G or A. For targeting shorter or longer sequences, see Box 1.
-
11|
Divide target sequences into hexamers. Divide N1-N18 into sub-sequences of length 6 (N1N2N3N4N5N6,N7N8N9N10N11N12, and N13N14N15N16N17N18). For example, a TALE targeting 5′-TGAAGCACTTACTTTAGAAA-3′ can be divided into hexamers as (T) GAAGCA CTTACT TTAGAA (A), where the initial thymine and final adenine (in parenthesis) are encoded by the appropriate backbone. In this example, the three hexamers will be: hexamer 1 = NN-NI-NI-NN-HD-NI, hexamer 2 = HD-NG-NG-NI-HD-NG, hexamer 3= NG-NG-NI-NN-NI-NI. Due to the adenine in the final position, we will use one of the NI backbones: pTALE-TF_v2(NI) or pTALEN_v2(NI).
-
12|
Assembling hexamers using Golden Gate digestion-ligation (Fig. 3, stage 2). Prepare one reaction tube for each hexamer. Using the monomer plate schematic (Fig. 4), pipette 1 ul of each normalized monomer into the corresponding hexamer reaction tube. Repeat this for all hexamers. For example, for the target from Step 10, set up tube 1 (1 ul from each of G1, A2, A3, G4, E5, and A6), tube 2 (1 ul from each of E7, C8, C9, A10, E11, C12), and tube 3 (1 ul from each of D1, D2, B3, H4, B5, B6). To construct a TALE with 18 full repeats, 3 separate hexamer tubes are used.
◆ CRITICAL STEP Pay close attention when pipetting the monomers; it is very easy to accidentally pipette from the wrong well during this step.
-
13|
To perform a simultaneous digestion-ligation (Golden Gate) reaction to assemble each hexamer (Fig. 3, stage 2) add the following reagents to each hexamer tube:
Component Amount Final concentration Esp3I (BsmBI) 5 U/ul 0.75 ul 0.375 U/ul Tango Buffer 10X 1 ul 1x Dithiothreitol (DTT) 10 mM 1 ul 1 mM T7 Ligase 3000 U/ul 0.25 ul 75 U/ul ATP 10 mM 1 ul
1 mM 4 ul 6 monomers 6 × 1 ul Total 10 ul
◆ CRITICAL STEP Dithiothreitol (DTT) is easily oxidized in air. It should be freshly made or thawed from aliquots stored at −70 °C and used immediately.
-
14|
Place each hexamer tube in a thermocycler to carry out the Golden Gate reactions using the following cycling conditions for ~ 3 hours:
Cycle number Digest Ligate 1–15 37°C, 5 20°C, 5 Hold at 4°C. min min
PAUSE POINT This reaction can be left to run overnight.
-
15|
Run out the ligation product on a gel to check for ~700 bp bands corresponding to the hexamer products (Fig. 5a, lane 7). Cast a 2% agarose gel in 1x TBE electrophoresis buffer with 2x SYBR Safe dye. The additional dye helps to visualize faint bands. The gel should have enough lanes to run out each Golden Gate reaction from Step 14; load 3 ul of each ligation product in separate lanes. Include in one lane 1 ug of the 1 kb Plus DNA ladder. Run the gel at 15 V/cm until there is separation of the 650 bp ladder band from neighboring bands.
? TROUBLESHOOTING
-
16|
Exonuclease treatment to degrade non-circular ligation products (Fig. 3, stage 3). During the Golden Gate reaction, only fully-ligated hexamers should be able to circularize. PlasmidSafe exonuclease selectively degrades non-circular (incomplete) ligation products. Add the following reagents to each hexamer reaction tube:
Component Amount Final concentration PlasmidSafe DNAse 10U/ul 1 ul 0.66 U/ul Plasmid-Safe 10X Reaction Buffer 1 ul 1x ATP 10 mM 1 ul
1 mM 3 ul Golden gate reaction from Step 14 7 ul Total 10 ul -
17|
Incubate each hexamer reaction tube with PlasmidSafe at 37°C for 30 minutes followed by inactivation at 70°C for 30 minutes.
PAUSE POINT After completion, the reaction can be frozen and continued later. The circular DNA should be stable for at least a week.
-
18|
Hexamer PCR (Fig. 3, stage 4). Amplify each PlasmidSafe-treated hexamer in a 50 ul PCR reaction using high-fidelity Herculase II polymerase and the hexamer forward and reverse primers (Hex-F and Hex-R; Table 2). Add the following reagents to each PCR reaction:
Component Amount Final concentration 100 mM dNTP (25 mM each) 0.5 ul 1 mM 5X Herculase II reaction buffer 10 ul 1x 10 uM each Hex-F and Hex-R primers 1 ul 200 nM Herculase II Fusion DNA polymerase 0.5 ul 1x Distilled water 37 ul
49 ul PlasmidSafe-treated hexamer from Step 17 1 ul Total 50 ul -
19|
Perform PCR on the reactions in Step 18 using the following cycling conditions:
Cycle number Denature Anneal Extend 1 95°C, 2 min 2–36 95°C, 20 s 60°C, 20 s 72°C, 30 s 37 72°C, 3 min -
20|
Gel purification of amplified hexamers. Due to the highly repetitive template, it is necessary to purify the amplified hexamer product from the other amplicons. Cast a 2% agarose gel in 1x TBE electrophoresis buffer with 1x SYBR Safe dye. The gel should have enough lanes to run out each PCR product from Step 19 and the comb size should be big enough to load 40–50 ul of PCR product. Include in one lane 1 ug of the 1 kb Plus DNA ladder. Run the gel at 15 V/cm until there is separation of the 650 bp ladder band from neighboring bands. Using a clean razor blade, excise each hexamer band, which should be nearly aligned with the 650 bp band from the ladder (Fig. 5, lane 9).
◆ CRITICAL STEP Avoid any cross-contamination by ethanol sterilization of work surfaces, razor blades, etc. during the gel extraction and between each individual band excision.
! CAUTION Wear appropriate personal protective equipment, including a facemask, when performing gel stabs to minimize risks associated with prolonged light or mutagenic DNA dye exposure.
? TROUBLESHOOTING
-
21|
Purify the hexamer gel bands from Step 20 using the MinElute Gel Extraction kit following the manufacturer’s directions. Elute the DNA from each reaction using 20 ul of Buffer EB prewarmed to 55°C.
-
22|
Gel normalization of purified hexamer concentrations. Cast a 2% agarose gel in 1x TBE electrophoresis buffer with 1x SYBR Safe dye. The gel should have enough lanes to run out 2 ul of each purified hexamer from Step 21. Include in one lane 10 ul of the quantitative DNA ladder. Run the gel at 15V/cm until all lanes of the quantitative ladder are clearly separated. Each hexamer lane should contain only a single (purified) band.
-
23|
Image the gel using a quantitative gel imaging system. Each lane should have only the ~700 bp hexamer product. Make sure the exposure is short enough so that none of the bands are saturated.
-
24|
Quantify the integrated intensity of each hexamer band using ImageJ or other gel quantification software. Use the quantitative ladder with known concentrations (5, 10, 20, 40, 100 ng) to generate a linear fit and quantify the concentration of each purified hexamer.
? TROUBLESHOOTING
-
25|
Adjust the concentration of each hexamer to 20 ng/ul by adding Buffer EB.
-
26|
Golden Gate assembly of hexamers into TALE backbone (Fig. 3, stage 5). Combine the hexamers and the appropriate TALE backbone vector (transcription factor or nuclease) in a Golden Gate digestion-ligation. For example, we will use a TALE backbone with NI as the 0.5 repeat for the target sequence in Step 10 since N19=A. For this ligation, a 1:1 molar ratio of insert:vector works well. Set up one reaction tube for each TALE. Also, prepare a negative control ligation by including the TALE backbone vector without any hexamers.
Component TALE Negative control Final concentration TALE backbone vector (100 ng/ul) 1 ul 1 ul 10 ng/ul BsaI-HF (20 U/ul) 0.75 ul 0.75 ul 1.5 U/ul 10x NEBuffer 4 1 ul 1 ul 1x 10x Bovine serum albumin 1 ul 1 ul 1x ATP 10 mM 1 ul 1 ul 1 mM T7 Ligase (3000 U/ul) 0.25 ul 0.25 ul 75 U/ul
5 ul 5 ul 3 purified hexamers (20 ng/ul) 3 ul (1 ul each) 2 ng/ul each Distilled water 2 ul 5 ul Total 10 ul 10 ul
◆ CRITICAL STEP As a negative control, set up a separate reaction omitting the purified hexamers (i.e. including only the TALEN or TALE-TF backbone).
-
27|
Place the tubes from Step 26 in a thermocycler to carry out the Golden Gate reactions using the following cycling conditions for ~4 hours:
Cycle number Digest Ligate Inactivate 1–20 37°C, 5 min 20°C, 5 min 21 80°C, 20 min
PAUSE POINT Ligation products can be frozen at −20 °C and stored at least one month for transformation into bacteria at a later time.
-
28|
Although it is not necessary, it is possible to run out the ligation product on a gel to check for ~1.8 kbp band corresponding to the properly assembled 18mer tandem repeat. To check the ligation product, cast a 2% agarose gel in 1x TBE electrophoresis buffer with 2x SYBR Safe dye. The additional dye helps to visualize faint bands. Load 5 ul of the ligation product from Step 27. Include in one lane 1 ug of the 1 kb Plus DNA ladder. Run the gel at 15 V/cm until there is clear separation of the 1650 bp and 2000 bp ladder bands. Alternatively, proceed directly to transformation (Step 29) without running a gel; transformation is very sensitive and, even when a clear band cannot be visualized on the gel, there is often enough plasmid for transformation of high competency cells.
? TROUBLESHOOTING
Verifying correct TALE repeat assembly
TIMING 3 days (4 hr hands-on time)
-
29|
Transformation. Transform the ligation products from Step 27 into a competent E. coli; in our lab, we use Stbl3 for routine transformation. Transformation can be done following the protocol supplied with the cells. Briefly, add 5 ul of the ligation product to 50 ul of ice-cold chemically competent Stbl3 cells, incubate on ice for 5 min, incubate at 42°C for 45 sec, return immediately to ice for 5 min, add 250 ul of SOC medium, incubate at 37°C for 1 hr on a shaking incubator (250 rpm), plate 100 ul of transformation on a LB plate containing 100 ug ml−1 ampicillin and incubate overnight at 37°C.
-
30|
Inspect all plates from Step 29 for bacterial colony growth. Typically, we see few colonies on the negative control plates (only backbone in the Golden Gate digestion-ligation) and tens to hundreds of colonies on the complete TALE ligation plates.
? TROUBLESHOOTING
-
31|
For each TALE plate, pick 8 colonies to check the assembly fidelity. Using a sterile 20 ul pipette tip, touch the tip to a single colony, streak onto a single square on a pre-warmed, new gridded LB-ampicillin plate to save the colony, and then swirl the tip in 100 ul of distilled water to dissolve the colony for colony PCR. Repeat this procedure for all colonies to be checked, streaking each new colony into a separate square on the gridded LB-ampicillin plate. After finishing, incubate the gridded plate at 37°C for at least 4 hours to grow up the colony streaks.
-
32|
Colony PCR. Using the colonies selected in Step 31 as templates, set up colony PCR to verify that the correctly assembled tandem 18mer repeat has been ligated into the TALE backbone. We have found that the colony PCR reaction is sensitive to excessive template concentration, therefore we typically use 1 ul of the 100 ul colony suspension from Step 31. For colony PCR, use primers TALE-Seq-F1 and TALE-Seq-R1 for amplification (Table 2). Set up the following colony PCR reaction:
Component Amount Final concentration Colony suspension from Step 31 1 ul 100 mM dNTP (25 mM each) 0.25 ul 1 mM 10x Taq-B polymerase buffer 2.5 ul 1x 10 uM each TALE-Seq-F1 and TALE-Seq-R1 primers 0.25 ul 100 nM Taq-B polymerase (5 U/ul) 0.1 ul 0.02 U/ul Distilled water 20.9 ul
Total 25 ul -
33|
Perform colony PCR on the reactions in Step 32 using the following cycling conditions:
Cycle number Denature Anneal Extend 1 94°C, 3 min 2–31 94°C, 30 s 60°C, 30 s 68°C, 2 min 32 68°C, 5 min -
34|
To check the colony PCR result, cast a 1% agarose gel in 1x TBE electrophoresis buffer with 1x SYBR Safe dye. The gel should have enough lanes to run out 10 ul of each PCR product from Step 33. Include in one lane 1 ug of the 1 kb Plus DNA Ladder. Run the gel at 15 V/cm until there is clear separation of the 1650 bp and 2000 bp ladder bands.
-
35|
Image the gel and identify which colonies have the correct insert size. For an insert of 18 monomers (3 hexamers ligated into the TALE backbone vector), the product should be a single band of size 2175 bp (Fig. 5b, lane 1). Incorrect ligation products will show bands of different sizes. In place of colony PCR, plasmid DNA from prepared clones can be digested with AfeI. In both backbones (TALE-TF and TALEN), AfeI cuts 4 times. For both backbones, one fragment contains the entire tandem repeat region and should be of size 2118 bp for a correctly assembled 18mer. For the TALE-TF backbone, the correct clone will produce 4 bands with sizes: 165bp, 2118bp, 3435bp and 3544bp (Fig. 5b, lane 2). The 3435bp and 3544bp bands are difficult to separate on a 1% agarose gel and therefore a correct clone will show three bands with the middle 2118bp band indicating an intact tandem 18mer repeat (Fig. 5b, lane 2). For the TALEN backbone, the correct clone will produce 4 bands with sizes: 165 bp, 2118 bp, 2803 bp, 3236 bp.
? TROUBLESHOOTING
-
36|
Miniprep and sequencing. For each clone with the correct band size, inoculate a colony from the gridded plate into 3 ml of LB media with 100 ug ml−1 ampicillin and incubate at 37°C in a shaking incubator overnight.
-
37|
Isolate plasmid DNA from overnight cultures using a QIAprep Spin Miniprep Kit following the manufacturer’s instructions.
-
38|
Verify the sequence of each clone by sequencing the tandem repeat region using sequencing primers (see Table 2) TALE-Seq-F1 (forward primer annealing just before the first monomer), TALE-Seq-F2 (forward primer annealing at the beginning of the seventh monomer) and TALE-Seq-R1 (reverse primer annealing after the final 0.5 monomer). For most TALEs, reads from all 3 primers are necessary to unambiguously verify the entire sequence. Verify the sequencing result using our online, freely-available TALE software (http://taleffectors.com/tools/) or using standard sequence alignment methods (e.g. ClustalW). After entering the target site sequence, our software generates a TALE-TF or TALEN reference sequence in either FASTA format or as an annotated GenBank vector map (*.gb file) that can be viewed using standard plasmid editor software (e.g. everyVECTOR, VectorNTI, or LaserGene SeqBuilder). The software also aligns sequencing reads (entered in FASTA format) to the generated reference sequence to allow for easy clone verification. Detailed instructions can be found on our website.
? TROUBLESHOOTING
Transfection of TALE-TF and TALEN into HEK293FT cells
TIMING 2 days (1 hour hands-on time)
-
39|
Plate HEK293FT cells onto 6-well plates in D10 culture medium without antibiotics approximately 24h prior to transfection at a seeding density of around 1×106 cells per well and a seeding volume of 2 mL. Scale up and down the culture according to the manufacturer’s manual provided with the 293FT cells if needed.
-
40|
Prepare DNA for transfection. Quantify the DNA concentration of the TALE plasmids used for transfection using reliable methods (such as UV spectrophotometry or gel quantification).
◆ CRITICAL STEP: The DNA concentration of the TALE plasmids should be quantified to guarantee that an accurate amount of TALE DNA will be used during the transfection.
-
41|
Prepare the DNA-Opti-MEM mix as follows using option A if testing transcriptional modulation, or option B if testing nuclease activity:
A. DNA-Opti-MEM mix for testing transcriptional modulation
i) Mix 4 μg of TALE-TF plasmid DNA with 250 μl of Opti-MEM medium. Include controls (e.g. RFP plasmid or mock transfection) to monitor transfection efficiency and cell health respectively.
B. DNA-Opti-MEM mix for testing nuclease activity
i) Mix 2 μg of the Left and 2 μg of the Right TALEN (Figure 1d) plasmid DNA with 250 μl of Opti-MEM medium. Control transfections should be done by omitting one or both of the TALENs. Also include controls (e.g. an RFP plasmid or mock transfection) to monitor transfection efficiency and cell health respectively. For all transfections, make sure the total amount of DNA transfected is the same across conditions – when omitting one or both TALENs, supplement with empty vector DNA to maintain the same total DNA amount.
-
42|
Prepare the Lipofectamine-Opti-MEM solution by diluting 10 μl of Lipofectamine 2000 with 250 μl of Opti-MEM. Mix the solution thoroughly by tapping the tube and incubating for 5 minutes at room temperature.
-
43|
Add the Lipofectamine-Opti-MEM solution to the DNA-Opti-MEM solution to form the DNA-Lipofectamine complex. Mix well by gently pipetting up and down. Incubate for 20 minutes at room temperature.
◆ CRITICAL STEP Make sure the complex is thoroughly mixed. Insufficient mixing results in lower transfection efficiency.
◆ PAUSE POINT The transfection complex will remain stable for 6 hours at room temperature.
-
44|
Add 500 μl of the DNA-Lipofectamine complex to each well of the 6-well plate from Step 39 directly. Mix gently by rocking the plates back and forth.
-
45|
Incubate cells at 37°C with 5% CO2 for 24 hours. At this point, determine the transfection efficiency by estimating the fraction of fluorescent cells in the positive control transfection (e.g. RFP plasmid) using a fluorescence microscope.
CRITICAL STEP If incubation beyond 48 hours is needed, change the culture medium with fresh D10 supplemented with antibiotics on a daily basis. This will not affect the transfection efficiency.
? TROUBLESHOOTING
TALE functional characterization
-
46|
To measure TALEN cutting efficiency using Surveyor nuclease follow option A, or to measure TALE-TF transcriptional activation using qRT-PCR follow option B:
A. Measuring TALEN cutting efficiency using Surveyor nuclease
TIMING 6 hr (3 hr hands-on time)
-
i)
Remove culture medium from each well from Step 45 and add 100 μl of QuickExtract DNA Extraction Solution to each well and pipette thoroughly to lyse cells. Transfer the lysate to a PCR tube.
-
ii)
Extract DNA from the lysate from Step 46Ai using the following cycling conditions:
Cycle number Condition 1 68°C, 15 min 2 95°C, 8 min -
iii)
PCR amplification of region surrounding TALEN target site. Prepare the following PCR reaction using the genomic DNA from Step 46Aii:
Component Amount Final concentration gDNA from Step 46Aii 0.5 ul 100 mM dNTP (25 mM each) 0.5 ul 1 mM 5X Herculase II reaction buffer 10 ul 1x 10 uM each of target-specific Surveyor forward and reverse primers (see Experimental Design) 1 ul 200 nM Herculase II Fusion DNA polymerase 0.5 ul 1x Distilled water 37.5 ul
Total 50 ul
◆ CRITICAL STEP Surveyor procedure (Steps 46Aiii–xv) is carried out according to the manufacturer’s protocol and is described in greater detail in the Surveyor manual. We provide brief details here since mutation detection by mismatch endonuclease is not a common procedure for most laboratories.
◆ CRITICAL STEP When performing the Surveyor assay for the first time, we suggest carrying out the positive control reaction included with the Surveyor nuclease kit.
-
iv)
Perform PCR using the following cycling conditions:
Cycle number Denature Anneal Extend 1 95°C, 3 min 2–36 95°C, 30 s 55°C, 15 s 72°C, 30s 37 72°C, 5 min -
v)
Check the PCR result by running 5 ul of PCR product on a 2% agarose gel in 1x TBE electrophoresis buffer with 1x SYBR Safe dye. Include in one lane 10 ul of the quantitative DNA ladder. Run the gel at 15 V/cm until all bands are clearly separated. For all templates, it is important to make sure that there is only a single band corresponding to the intended product for the primer pair. The size of this band should be the same as calculated from the distance between the two primer annealing sites in the genome.
◆ CRITICAL STEP If multiple amplicons are generated from the PCR reaction, re-design primers and re-optimize the PCR conditions to avoid off-target amplification.
? TROUBLESHOOTING
-
vi)
Image the gel using a quantitative gel imaging system. Make sure the exposure is short enough so that none of the bands are saturated. Quantify the integrated intensity of each PCR product using ImageJ or other gel quantification software. Use the quantitative ladder with known concentrations (5, 10, 20, 40, 100 ng) to generate a linear fit. Adjust the DNA concentration of the PCR product by diluting with 1x Herculase II reaction buffer so that it is in the range of 25 – 80 ng/μl.
-
vii)
DNA heteroduplex formation. At this point, the amplified PCR product includes a mixture of both modified and unmodified genomic DNA (TALEN-modified DNA will have a few bases of sequence deletion near the TALEN cut site due to exonuclease activity during NHEJ). For Surveyor mismatch detection, this mixture of products must first be melted and re-annealed such that heteroduplexes are formed. DNA heteroduplexes contain strands of DNA that are slightly different but annealed (imperfectly) together. Given the presence of both unmodified and modified DNA in a sample, a heteroduplex may include one strand of unmodified DNA and one strand of TALEN-modified DNA. Heteroduplexes can also be formed from re-annealing of two different TALEN-modified products as NHEJ exonuclease activity can produce different mutations. To cross-hybridize wild type and TALEN-modified PCR products into hetero- and homoduplexes, all strands are melted and then slowly re-annealed (Figure 6a). Place 300 ng of the PCR product from Step 46Avi in a thermocycler tube and bring to a total volume of 20 μl with 1x Herculase II reaction buffer.
-
viii)
Perform cross-hybridization on the diluted PCR amplicon from Step 46Avii using the following cycling conditions:
Cycle number Condition 1 95°C, 10 min 2 95°C to 85°C, −2°C/s 3 85°C, 1 min 4 85°C to 75°C, −0.3°C/s 5 75°C, 1 min 6 75°C to 65°C, −0.3°C/s 7 65°C, 1 min 8 65°C to 55°C, −0.3°C/s 9 55°C, 1 min 10 55°C to 45°C, −0.3°C/s 11 45°C, 1 min 12 45°C to 35°C, −0.3°C/s 13 35°C, 1 min 14 35°C to 25°C, −0.3°C/s 15 25°C, 1 min -
ix)
Surveyor Nuclease S digestion. To treat the cross-hybridized homo- and hetero-duplexes using Surveyor Nuclease S to determine TALEN cleavage efficiency (Figure 6a), add the following components together on ice and mix by pipetting gently:
Component Amount Final concentration 0.15 M MgCl2 solution 2 ul 15 mM Surveyor Nuclease S 1 ul 1x Surveyor Enhancer S 1 ul
1x 4 ul Re-annealed duplexes from Step 46Aviii 16 ul
Total 20 ul -
x)
Incubate the reaction from Step 46Aix at 42°C for 1 hour.
-
xi)
Add 2 ul of the Stop Solution from the Surveyor kit.
Figure 6. Examples of TALE-TF and TALEN activity in 293FT cells.
(a) Schematic of the Surveyor nuclease assay used to determine TALEN cleavage efficiency. First, genomic PCR is used to amplify the TALEN target region from a heterogeneous population of TALEN-modified and unmodified cells, and the gPCR products are re-annealed slowly to generate heteroduplexes. The re-annealed heteroduplexes are cleaved by Surveyor nuclease while homoduplexes are left intact. TALEN cleavage efficiency is calculated based on the fraction of cleaved DNA. (b) Gel showing the Surveyor nuclease result from the AAVS1 TALEN pair (from Fig. 1d). Lanes 1—4: controls from un-transfected (N.T.) cells and cells transfected with a plasmid carrying GFP (Mock), AAVS1 left TALEN only (L), and AAVS1 right TALEN only (R). Lanes 5—7: cells transfected with AAVS1 Left and Right TALENs (L+R) for 24, 48, and 72 hours. The two lower bands indicated by the arrows are Surveyor-cleaved DNA products. (c) 293FT cells transfected with the SOX2 TALE-TF (from Fig. 1c) exhibited a 5 fold increase in the amount of SOX2 mRNA compared with mock transfected cells. Error bars indicate s.e.m.; n = 3. *** indicates P < 0.005. Panel c was modified with permission from Nature Biotechnology3 (Nature Biotechnology (c) 2011, Macmillian Publishers Ltd.).
◆ PAUSE POINT: The digestion product can be stored at −20°C for analysis at a later time.
-
xii)
Cast a 2% agarose gel in 1x TBE electrophoresis buffer with 1x SYBR Safe dye. When casting the gel, it is preferable to use a thin comb size (<1 mm) for the sharpest possible bands. The gel should have enough lanes to run out 20 ul of each digestion product band from Step 46Axi. Include in one lane 1 ug of the 1 kb Plus DNA ladder. Run the gel at 5 V/cm until the Orange G loading dye has migrated 2/3rds of the way down the gel.
-
xiii)
Image the gel using a quantitative gel imaging system. Make sure the exposure is short enough so that none of the bands are saturated. Each lane from samples transfected with both left and right TALENs should have a larger band corresponding to the uncut genomic amplicon (the same size as in Step46Av) and smaller bands corresponding to the DNA fragments resulting from the cleavage of the genomic amplicon by Surveyor nuclease. Controls (no transfection, control plasmid transfection, or transfection omitting one of the TALENs) should only have the larger band corresponding to the uncut genomic amplicon.
? TROUBLESHOOTING
-
xiv)
Quantify the integrated intensity of each band using ImageJ or other gel quantification software. For each lane, calculate the fraction of the PCR product cleaved (fcut) using the following formula: fcut = a/(a+b), where a = the integrated intensity of both of the cleavage product bands, and b = the integrated intensity of uncleaved PCR product band. A sample Surveyor gel for TALENs targeting human AAVS1 is shown in Figure 6b.
-
xv)
Estimate the percentage of TALEN-mediated gene modification using the following formula47:
This calculation can be derived from the binomial probability distribution given a few conditions: that strand reassortment during the duplex formation is random, that there is a negligible probability of the identical mutations reannealing during duplex formation, and that the Surveyor nuclease digestion is complete.
B. Measuring TALE-TF transcriptional activation using qRT-PCR
TIMING 5 hr (3 hr hands-on time)
-
i)
RNA extraction. Aspirate the medium in each well of the 6-well plates from Step 45 at 72 hours after transfection.
◆ CRITICAL STEP Use proper RNA handling techniques to prevent RNA degradation, including cleaning bench surfaces and pipettes with RNAseZAP. Use RNAse-free consumables and reagents.
-
ii)
Wash the cells in each well twice with 1 ml of DPBS.
-
iii)
Harvest ~1×106 cells for subsequent total RNA extraction by trypsinizing the cells with 500 μl trypsin with EDTA. Incubate for 1–2 minutes to let the cells detach from the bottom of the wells.
◆ CRITICAL STEP Do not leave the cells in trypsin for longer than a few minutes.
-
iv)
Neutralize the trypsin by adding 2 ml of D10 medium.
-
v)
In a 15 ml centrifuge tube, centrifuge the cell suspension at 300×g for 5 min. Carefully aspirate all of the supernatant.
◆ CRITICAL STEP Incomplete removal of the supernatant can result in inhibition of cell lysis. PAUSE POINT: Cells can be frozen at −80°C for 24 hours.
-
vi)
Extract and purify RNA from the cells in Step 46Bv using the RNeasy Mini Kit and QIAshredder following the manufacturer’s directions. Elute the RNA from each column using 30 ul of nuclease-free water.
-
vii)
Measure the RNA concentration using a UV spectrophotometer.
-
viii)
cDNA reverse-transcription. Generate cDNA using the iScript cDNA Synthesis Kit following the manufacturer’s directions. For matched negative controls, perform the reverse transcription without the reverse-transcriptase enzyme.
-
ix)
Quantitative PCR. Thaw on ice the appropriate TaqMan probe for the target gene and for an endogenous control gene.
◆ CRITICAL STEP Protect the probes from light and do not allow the thawed probes to stay on ice for an extended time.
-
x)
Following the TaqMan Universal PCR Master Mix manufacturer’s directions, prepare 4 technical replicate qPCR reactions for each sample in optical thermocycler strip tubes or 96-well plates. Set up negative controls for non-specific amplification as indicated in the directions: namely, RNA template processed without reverse transcriptase (“no RT”) and a no-template control.
-
xi)
Briefly centrifuge the samples to remove any bubbles and amplify them in a TaqMan-compatible qRT-PCR machine with the following cycling parameters.
Cycle number Denature Anneal and Extend 1 95°C, 20 s 2–41 95°C, 1 s 60°C, 20 s -
xii)
Analyze data and calculate the level of gene activation using the ΔΔCT method46, 55. TALE-TF results from qRT-PCR assay of SOX2 activation in HEK293 cells are shown in Figure 6c.
◆ CRITICAL STEP The ΔΔCT method assumes that amplification efficiency is 100% (ie. number of amplicons doubles after each cycle). For new probes (such as custom TaqMan probes), amplification from a template dilution series (spanning at least 5 orders of magnitude) should be performed to characterize amplification efficiency. For standard TaqMan Gene Expression Assay probes, this is not necessary as they are designed to have 100±10% amplification efficiency.
? TROUBLESHOOTING
● TIMING
Steps 1–9, Monomer library amplification and normalization: 6 hr
Steps 10–28, TALE hierarchical ligation assembly: 1.5 days (5 hr hands-on time)
Steps 29–38, TALE transformation and sequence verification: 3 days (4 hr hands-on time)
Steps 39–45, Transfection of TALE-TF and TALEN into HEK293FT cells: 2 days (1 hr hands-on time)
Steps 46A and B, TALE functional characterization with RT-qPCR or Surveyor: 6 hr (3 hr hands-on time)
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
Table 3.
Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 4 | Uneven amplification across monomers | Not using Herculase 2 Fusion polymerase | Optimize annealing temperature and Mg2+ and DMSO concentrations |
| 8 | Low DNA concentration after elution | Residual ethanol on purification column | Air dry columns before elution at 37°C for a longer period of time |
| Incorrect vacuum pressure during DNA binding | Adjust vacuum pressure according to the manufacturer’s suggestions | ||
| 15 | No visible hexamer band (~700 bp) | Not adding equimolar amounts of monomers | Gel normalize monomer concentration |
| Degraded DTT or ATP | Use fresh stocks of DTT and ATP, which degrade easily | ||
| No visible hexamer band (~700 bp) but smaller bands present | Wrong monomer(s) added during pipetting | Re-select monomers | |
| Monomer concentration is too low | Increase the number of Golden Gate digestion-ligation cycles and/or increase the concentration of monomers to >20 ng/ul; there is no detrimental effect to using more monomers in an equimolar ratio | ||
| 20 | No visible hexamer band (~700 bp) | Unsuccessful Golden Gate digestion-ligation | Verify on a gel that the Golden Gate digestion-ligation product from Step 15 is visible; increase monomer concentration |
| 24 | Low concentration for purified hexamers | Unsuccessful gel extraction | Ensure that there is no residual ethanol during elution or increase PCR reaction volume |
| 28 | No visible 18mer band (~1.8 kbp) | Unsuccessful Golden Gate digestion-ligation | Increase hexamer concentration in Golden Gate digestion-ligation in Step 26 or proceed directly to transformation in Step 29 |
| 30 | More than a few colonies on negative control plate | Compromised TALE backbone | Perform a restriction digest of the backbone to verify integrity |
| 35 | Colony PCR bands are smeared | Too much template | Dilute colony suspension 10x–100x |
| 38 | Monomers assembled in incorrect order | Misligation | Misligation occurs at a very low frequency; analyze two additional clones |
| 45 | Low transfection efficiency | Low DNA quality | Prepare DNA using high quality plasmid preparation |
| Suboptimal DNA to Lipofectamine2000 ratio | Titrate DNA to Lipofectamine2000 ratio to determine optimal transfection condition | ||
| 46Av | Multiple amplicons | Nonspecific primers | Design new primers and verify specificity using PrimerBLAST; use touchdown PCR |
| No amplification | Suboptimal PCR condition | Optimize annealing temperature and Mg2+ and DMSO concentrations | |
| 46Axiii | No cleavage bands visible | TALEN unable to cleave target site | Design new TALEN pairs targeting nearby sequences |
| 46Bxii | No increase in transcription in target mRNA | TALE-TF unable to access target site | Design new TALE-TFs targeting nearby sequences |
ANTICIPATED RESULTS
TALE-TFs and TALENs can facilitate site-specific transcriptional modulation3–5, 8 and genome editing4, 7, 9, 11–15 (Table 1). TALENs can be readily designed to introduce double-stranded breaks at specific genomic loci with high efficiency. In our experience, a pair of TALENs designed to target the human AAVS1 locus is able to achieve up to 3.6% cutting efficiency in 293FT cells as determined by Surveyor nuclease assay (Fig. 6a–b). TALE-TFs can also robustly increase the mRNA levels of endogenous genes. For example, a TALE-TF designed to target the proximal promoter region of SOX2 in human cells is able to elevate the level of endogenous SOX2 gene expression by up to 5 fold3 (Fig. 6c). The ability for TALE-TFs and TALENs to act at endogenous genomic loci is dependent on the chromatin state as well as yet-to-be-determined mechanisms regulating TALE DNA binding56, 57. For these reasons we typically build several TALE-TFs or TALEN pairs for each genomic locus we aim to target. These TALE-TFs and TALENs are designed to bind neighboring regions around a specific target site since some binding sites might be more accessible than others. The reason why some TALEs exhibit significantly lower levels of activity remains unknown, though it is likely due to position- or cell state-specific epigenetic modifications preventing access to the binding site. Due to differences in epigenetic states between different cells, it is possible that TALEs that fail to work in a particular cell type might work in a different cell type.
Supplementary Data 1. Nucleotide sequences of the 4 monomer plasmids, 4 TALE-TF cloning backbone plasmids, and 4 TALEN cloning backbone plasmids.
Acknowledgments
We thank the entire Zhang laboratory for their support. L.C. is supported by a HHMI International Student Research Fellowship. Y.Z. is supported by a Simons Foundation Fellowship. M.M.C. is supported by a MIT Undergraduate Research Opportunities scholarship. F.Z. is supported by a NIH Transformative R01, the McKnight and Simons Foundations, Robert Metcalfe, and Michael Boylan.
Footnotes
Author contributions
N.E.S., L.C., Y.Z., and F.Z. wrote the manuscript. M.M.C. designed the online TALE sequence verification software. F.Z. and G.F. supervised the research.
Competing financial interest
The authors declare that they have no competing financial interests.
References
- 1.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 2.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 5.Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of Designer TAL Effectors by Golden Gate Cloning. PLoS One. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiβler R, et al. Transcriptional Activators of Human Genes with Programmable DNA-Specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011 doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahfouz MM, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci U S A. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 17.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Romer P, et al. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- 19.Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- 20.Kay S, Hahn S, Marois E, Wieduwild R, Bonas U. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 2009;59:859–871. doi: 10.1111/j.1365-313X.2009.03922.x. [DOI] [PubMed] [Google Scholar]
- 21.Romer P, et al. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 2009;150:1697–1712. doi: 10.1104/pp.109.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinnen A, Hicks JB, Fink GR. Transformation of yeast. Proc Natl Acad Sci U S A. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 24.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 25.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akagi K, et al. Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res. 1997;25:1766–1773. doi: 10.1093/nar/25.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epinat JC, et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 30.Khan IF, Hirata RK, Russell DW. AAV-mediated gene targeting methods for human cells. Nat Protoc. 2011;6:482–501. doi: 10.1038/nprot.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 32.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 33.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JS, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods. 2010;7:91. doi: 10.1038/nmeth0210-91b. author reply 91–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keenholtz RA, Rowland SJ, Boocock MR, Stark WM, Rice PA. Structural basis for catalytic activation of a serine recombinase. Structure. 2011;19:799–809. doi: 10.1016/j.str.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF., 3rd Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF., 3rd Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc Natl Acad Sci U S A. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 41.Wilson MH, Kaminski JM, George AL., Jr Functional zinc finger/sleeping beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 47.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 48.Zhang F, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci U S A. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buzdin AA. In: Nucleic Acids Hybridization. Buzdin A, Lukyanov S, editors. Springer; 2007. pp. 211–239. [Google Scholar]
- 50.Till BJ, Burtner C, Comai L, Henikoff S. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 2004;32:2632–2641. doi: 10.1093/nar/gkh599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babon JJ, McKenzie M, Cotton RG. The use of resolvases T4 endonuclease VII and T7 endonuclease I in mutation detection. Mol Biotechnol. 2003;23:73–81. doi: 10.1385/MB:23:1:73. [DOI] [PubMed] [Google Scholar]
- 52.Yang B, et al. Purification, cloning, and characterization of the CEL I nuclease. Biochemistry. 2000;39:3533–3541. doi: 10.1021/bi992376z. [DOI] [PubMed] [Google Scholar]
- 53.Kulinski J, Besack D, Oleykowski CA, Godwin AK, Yeung AT. CEL I enzymatic mutation detection assay. Biotechniques. 2000;29:44–46. 48. doi: 10.2144/00291bm07. [DOI] [PubMed] [Google Scholar]
- 54.Oleykowski CA, Bronson Mullins CR, Godwin AK, Yeung AT. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998;26:4597–4602. doi: 10.1093/nar/26.20.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murakami MT, et al. The repeat domain of the type III effector protein PthA shows a TPR-like structure and undergoes conformational changes upon DNA interaction. Proteins. 2010;78:3386–3395. doi: 10.1002/prot.22846. [DOI] [PubMed] [Google Scholar]
- 57.Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Huang P, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 59.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tesson L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]