Abstract
Background
RNA-dependent RNA polymerases (RDRs) function in anti-viral silencing in Arabidopsis thaliana and other plants. Salicylic acid (SA), an important defensive signal, increases RDR1 gene expression, suggesting that RDR1 contributes to SA-induced virus resistance. In Nicotiana attenuata RDR1 also regulates plant-insect interactions and is induced by another important signal, jasmonic acid (JA). Despite its importance in defense RDR1 regulation has not been investigated in detail.
Methodology/Principal Findings
In Arabidopsis, SA-induced RDR1 expression was dependent on ‘NON-EXPRESSER OF PATHOGENESIS-RELATED GENES 1’, indicating regulation involves the same mechanism controlling many other SA- defense-related genes, including pathogenesis-related 1 (PR1). Isochorismate synthase 1 (ICS1) is required for SA biosynthesis. In defensive signal transduction RDR1 lies downstream of ICS1. However, supplying exogenous SA to ics1-mutant plants did not induce RDR1 or PR1 expression to the same extent as seen in wild type plants. Analysing ICS1 gene expression using transgenic plants expressing ICS1 promoter:reporter gene (β-glucuronidase) constructs and by measuring steady-state ICS1 transcript levels showed that SA positively regulates ICS1. In contrast, ICS2, which is expressed at lower levels than ICS1, is unaffected by SA. The wound-response hormone JA affects expression of Arabidopsis RDR1 but jasmonate-induced expression is independent of CORONATINE-INSENSITIVE 1, which conditions expression of many other JA-responsive genes. Transiently increased RDR1 expression following tobacco mosaic virus inoculation was due to wounding and was not a direct effect of infection. RDR1 gene expression was induced by ethylene and by abscisic acid (an important regulator of drought resistance). However, rdr1-mutant plants showed normal responses to drought.
Conclusions/Significance
RDR1 is regulated by a much broader range of phytohormones than previously thought, indicating that it plays roles beyond those already suggested in virus resistance and plant-insect interactions. SA positively regulates ICS1.
Introduction
RNA silencing refers to a set of gene regulation mechanisms occurring in most eukaryotes, whereby transcript stability or translatability is suppressed in a sequence-specific manner, guided by small 19–24 nt RNA molecules [1], [2]. RNA silencing is an important component of anti-viral defense in plants [3], [4]. Double-stranded structures within viral RNA can be cleaved by dicer-like (DCL) nucleases to generate double-stranded small interfering (si)RNAs. In Arabidopsis thaliana, there are four DCL enzymes, of which DCL4 and DCL2 are the most important in the generation of virus-derived siRNAs [5], [6], [7], [8]. After further processing, single-stranded forms of virus-derived siRNA molecules associate with Argonaute (AGO) nucleases and direct AGO-catalysed slicing of complementary viral RNA molecules [9]. Of the ten AGOs encoded by the Arabidopsis genome, AGO1 is the primary ‘antiviral’ AGO, with secondary roles for AGO2, and in certain instances for AGO7 [10], [11], [12].
Another important feature of the anti-viral RNA silencing pathway in plants is referred to as amplification, whereby more virus-specific dsRNA substrates for DCLs are generated de novo by cellular RNA-dependent RNA polymerases (RDRs) [13]. The Arabidopsis thaliana genome encodes six RDRs, characterized by the DFDGD catalytic domain, of which RDRs 1, 2 and 6 are known to be involved in biogenesis of siRNAs [14]. In Arabidopsis and other plants, RDRs 1 and 6 contribute to antiviral RNA silencing, whilst RDR2 is involved in establishment of transcriptional gene silencing [8], [13], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]. RDRs also contribute to silencing mediated turnover of transcripts encoded by endogenous plant genes and transgenes [1], [25].
Xie and colleagues [15] reported that in tobacco (Nicotiana tabacum) RDR1 gene expression is induced by the defensive phytohormone salicylic acid (SA). This was a notable finding because it provided for the first time a possible connection between RNA silencing and two well-studied resistance phenomena that are dependent upon SA-mediated signal transduction: (i) the hypersensitive response, a genetically defined and highly pathogen-specific defense; and (ii) systemic acquired resistance (SAR) a broad-spectrum resistance to pathogens that is often triggered by a hypersensitive response [26].
However, it was also reported by Xie et al. [15] that although knockdown of NtRDR1 expression in transgenic tobacco enhanced the susceptibility of these plants to infection by tobacco mosaic virus (TMV) and potato virus X, resistance to these viruses could still be induced by treatment of the plants with exogenous SA. Subsequently, it was shown that Arabidopsis mutants compromised in AtRDR1 expression showed normal responses to bacterial infection and normal SA-induced expression of pathogenesis-related protein 1 (PR1: a marker for SA-induced resistance to bacteria, oomycetes and fungi), while no effect on SA-induced virus resistance in these Atrdr1 mutants was reported [16]. Constitutive expression of the Medicago truncatula RDR1 in N. benthamiana (a natural rdr1 mutant: [18]) did not enhance SA-induced resistance or rescue chemically-induced resistance in plants compromised in induced resistance by expression of a mutant form of alternative oxidase [27]. Curiously, expression of NtRDR1 in transgenic N. benthamiana plants enhanced, rather than ameliorated, infection by plum pox virus [28]. Thus, although it is possible that RDR1 may contribute to SA-induced resistance to certain viruses, it is not an indispensible component of anti-viral resistance, and in some cases its expression may enhance susceptibility.
The regulation of RDR1 gene expression is not well understood. It was reported that TMV infection triggered increased NtRDR1 transcript accumulation in the tobacco cultivar Xanthi (nn genotype) [15] but this could not be due to increased levels of SA, since infection of this cultivar with TMV does not induce SA accumulation [29]. Interestingly, in N. attenuata, RDR1 was induced by jasmonic acid (JA) [30], a phytohormone that is often assumed to be antagonistic to SA-mediated defensive signaling [31]. JA regulates induced resistance to herbivorous insects, and experiments with transgenic N. attenuata plants deficient in RDR1 expression showed that NaRDR1 regulates inducible genes conferring resistance to insect herbivory [32]. Diminishing NtRDR1 expression in transgenic tobacco decreased the expression of several endogenous transcripts related to virus resistance including Alternative Oxidase 1a and NtRDR6 [33]. Thus, in addition to its hypothesized role in enhancing antiviral RNA silencing [15], [16], RDR1 may play indirect roles in plant defense via silencing-mediated regulation of cellular mRNAs encoding resistance factors. In this study we have investigated in more detail the regulation of AtRDR1 gene expression by SA and JA, and found that other phytohormones, including abscisic acid (ABA) and ethylene, trigger its expression.
Results
SA-induced AtRDR1 Expression is NPR1-dependent
Wild-type Arabidopsis plants (ecotype Col-0) were treated with 1 mM SA, then samples were taken over a time course spanning 72 hours (h) and were analysed by reverse transcription coupled with quantitative PCR (RTqPCR) for RDR1 and PR1 expression (Figure 1). In agreement with previous studies using northern blotting [16], AtRDR1 transcript accumulation increased following SA treatment. In this study SA treatment peaked (at approximately four-fold basal level) between 2 and 6 h post-treatment before decreasing to approximately two-fold at 24 h post-treatment and returning to near starting levels by 72 h post-treatment (Figure 1A). The increase in accumulation of AtPR1 transcripts confirmed that the treatment with SA had been effective (Figure 1). This is in contrast to the work of Yu and colleagues, who reported that AtRDR1 induction took longer to become detectable (4 to 8 h) with no diminution of AtRDR1 expression apparent at 24 h post-treatment, the point at which the analysis was terminated [16]. The current work shows that, in contrast to SA-induced AtPR1 gene expression, the effect of SA on AtRDR1 gene expression is transient (Figure 1).
Figure 1. SA treatment causes transient induction of AtRDR1 expression.
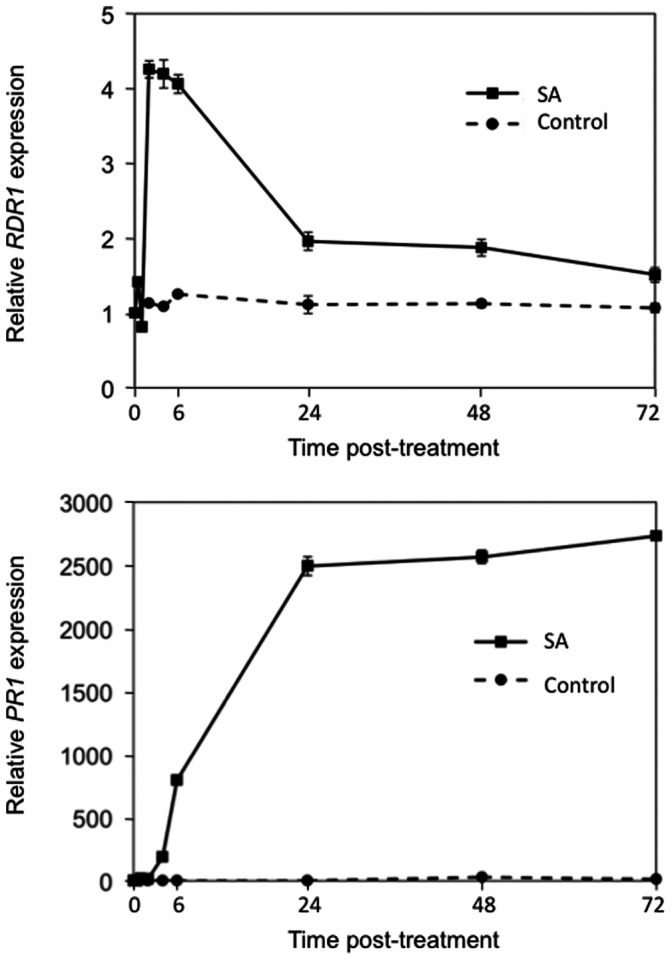
AtRDR1 expression and AtPR1 expression in control and SA-treated Arabidopsis (Col-0) plants sampled at immediately before treatment (‘0’ time) and over a time course of 72 h. Error bars represent standard error of the mean.
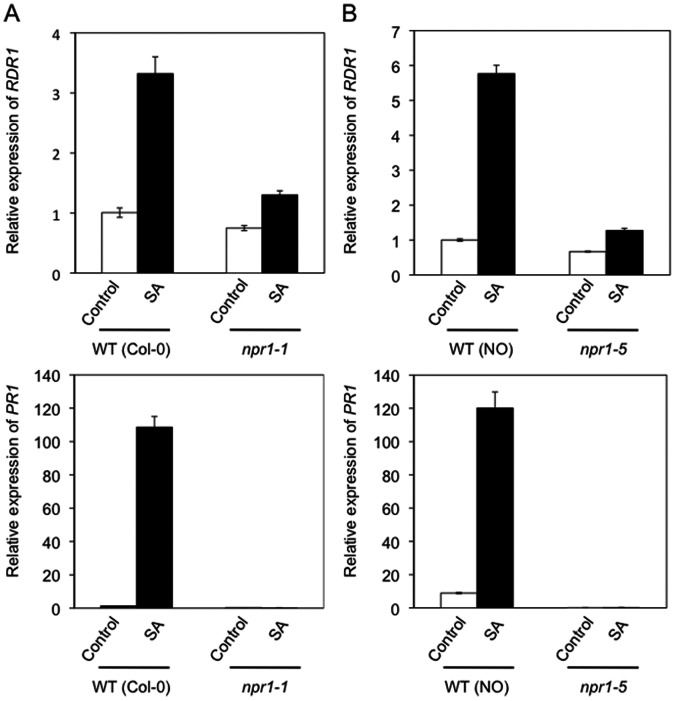
The transcriptional activator ‘Non-Expressor of PR proteins 1’ (NPR1) is required for PR gene induction and SAR against a wide range of microbial pathogens [34], although it is not required for SA-induced resistance to viruses [35], [36]. To determine if RDR1 gene expression is NPR1-dependent, we used two independent mutant lines: npr1-1 (Col-0 background: [37]) and npr1-5, which was originally named salicylic acid insensitive 1 (sai1) (Nössen background: [38]), and examined AtRDR1 transcript accumulation in plants at 6 h post-treatment with SA. As a control, the induction of AtPR1 by SA, which is dependent upon NPR1, was also examined in wild-type and npr1-mutant plants. SA-induced AtRDR1 expression in both npr1 mutant lines was markedly lower than that in wild-type plants (Figure 2), indicating that it is NPR1-dependent. It was noted that in all experiments AtRDR1 expression was consistently higher at 6 h post-treatment in plants of the Nössen ecotype than in plants of the Col-0 ecotype (Figure 2).
Figure 2. SA induces AtRDR1 expression in an NPR1-dependent manner.
(A) AtRDR1 and AtPR1 expression in ecotype Col-0 wild type and npr1-1 control and SA treated plants 6 h after treatment. (B) AtRDR1 and AtPR1 expression in Nössen (NO) wild-type and npr1-5 control and SA treated plants 6 h after treatment. Error bars represent standard error of the mean.
ICS1 Expression is Auto-regulated by SA and is Required for Maximal SA-Induced Expression of RDR1
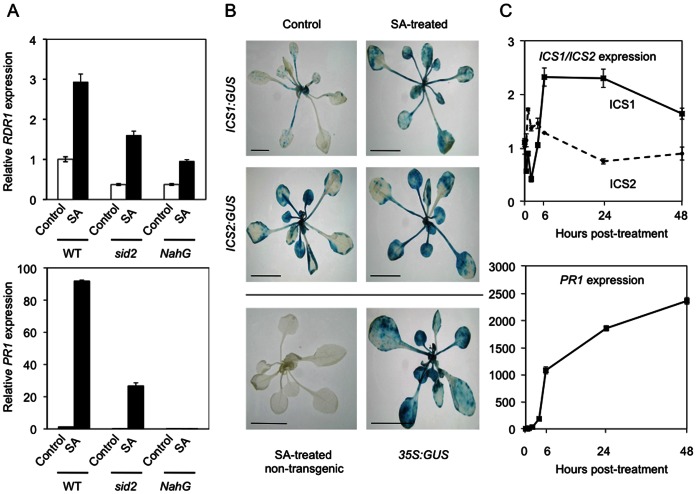
Plants of the sid2 line are impaired in their ability to synthesize SA due to a lesion in the gene encoding the isochorismate synthase isozyme, ICS1, upon which Arabidopsis is dependent for the bulk of its stress-induced SA biosynthesis, and which is a key factor in the induction of SAR in this species [39], [40], [41]. It was observed during initial experiments on the role of NPR1 in AtRDR1 induction that SA treatment of sid2 mutant plants caused induction of less AtRDR1 and PR1 expression than was seen in wild-type plants (data not shown and Figure 3A). This result was unexpected since although these plants are compromised in their ability to produce SA, it was anticipated that addition of exogenous SA would rescue expression of the two SA-inducible transcripts. As expected, in plants of the transgenic NahG line, which expresses a bacterial salicylate hydroxylase [42], SA-induced accumulation of both transcripts was greatly diminished (Figure 3A).
Figure 3. AtICS1 expression is positively regulated by SA and is required for optimal expression of AtRDR1 and PR1 in response to exogenous SA treatment.
(A) RTqPCR analysis of transcript accumulation for AtRDR1 and AtPR1 expression in Arabidopsis ecotype Col-0 wild type (WT), sid2-mutant (compromised in expression of ICS1) and NahG-transgenic plants 6 h post SA treatment. (B) Transgenic Arabidopsis plants harboring the promoter: reporter constructs ICS1:GUS and ICS2:GUS stained for GUS activity 24 h after control (water) or SA treatment. SA treated Col-0 WT and 35S:GUS plants, 24 h after treatment are included as controls. (C) RTqPCR analysis of AtICS1, AtICS2, and AtPR1 transcript accumulation in wild-type plants treated with SA, over a 48 h time course. Error bars represent standard error of the mean.
We investigated the effect of exogenous SA application on expression of the AtICS1 gene, as well as the other Arabidopsis ICS ortholog, ICS2. Transgenic plants harboring ICS1:β-glucuronidase (GUS) and ICS2:GUS promoter:reporter gene fusion constructs were treated with SA. ICS1:GUS-transgenic plants consistently exhibited increased GUS activity 24 and 48 h after SA treatment (histochemical analysis of 24 h samples are shown in Figure 3B). In contrast, GUS activity was already detectable in untreated ICS2:GUS–transgenic plants, and showed no induction at either time-point.
The responsiveness of ICS gene expression was investigated further by examining transcript accumulation for ICS1 and ICS2 using RTqPCR (Figure 3C). ICS2 transcript accumulation increased transiently after SA treatment but decreased again by 6 h post-treatment. ICS1 transcript accumulation increased by 2–2.5 fold within 6 h of SA treatment but this elevated level was sustained over 24 hours, followed by a gradual decline (Figure 3C). These results were consistent over three biological replicates. Therefore, ICS1 gene expression appears to be positively auto-regulated by SA. ICS1 is the isozyme responsible for the bulk of SA biosynthesis [40], [43], [44] and unlike the gene for ICS2, the ICS1 gene is stimulated in a sustained fashion by SA (Figure 3C). This positive auto-regulation of ICS1 expression by SA appears to explain why in sid2 mutant plants the increase in PR1 and RDR1 gene expression triggered by exogenous SA was weaker than in wild-type plants (Figure 3A).
Mock Inoculation Induced RDR1 Expression
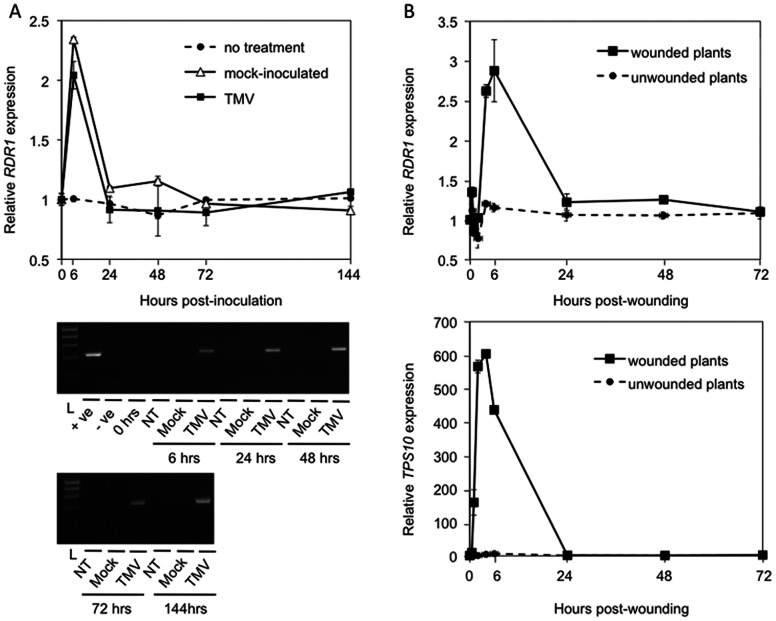
TMV infection was reported to increase accumulation of RDR1 transcripts in the inoculated leaves of Arabidopsis [16]. However, we found that the kinetics of RDR1 transcript accumulation were similar in mock-inoculated and TMV-inoculated leaves (Figure 4A). In both cases, increased RDR1 expression was transient, peaking and declining during the first 24 h following treatments and in the TMV-inoculated leaves there was no obvious relationship between RDR1 expression and the kinetics of viral RNA accumulation (Figure 4A). This suggests that the process of inoculation, involving abrasion of the adaxial surfaces of the leaves with Carborundum, rather than virus infection per se, was responsible for increased AtRDR1 expression.
Figure 4. Wounding and mock-inoculation induces AtRDR1 expression.
(A) AtRDR1 expression in TMV-infected, mock-inoculated (Mock) and untreated control (no treatment) Arabidopsis (Col-0) plants over the course of 144 h monitored by RTqPCR (Upper panel). Lower panel shows confirmation by RT-PCR of infection in TMV-inoculated compared with mock-inoculated and untreated (NT, no treatment). (B) RTqPCR of expression of AtRDR1 and the wounding- and JA-responsive gene AtTPS10 over 72 h following wounding. Error bars represent standard error of the mean.
To explore the possibility that RDR1 expression was triggered by abrasion, further experiments were carried out to follow the expression of both RDR1 and a well-characterized wound-induced gene, terpene synthase 10 (TPS10), over a period of 72 h following mock inoculation (Figure 4B). Although the response of RDR1 to mock-inoculation in terms of fold-increase in expression was at least two orders of magnitude less than the response of TPS10, the timing of expression following mock-inoculation was similar for both transcripts, supporting the idea that wounding had triggered increased RDR1 gene expression in TMV-inoculated plants (Figure 4A,B).
JA Induces RDR1 Expression in a COI1-independent Manner
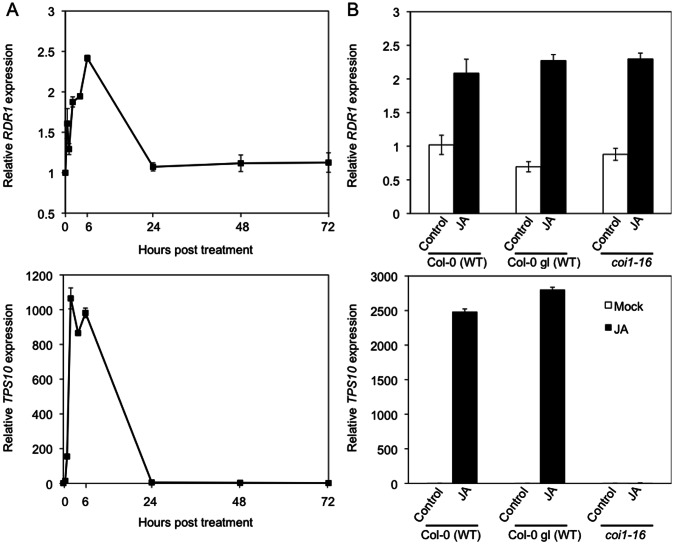
JA-mediated signaling co-ordinates a large proportion of wound-induced gene expression [45]. Indeed, expression of the wound-inducible TPS10 transcript is stimulated by treatment with methyl-JA [46]. In N. attenuata, NaRDR1 expression was shown to be induced by JA [30], suggesting that the abrasion-induced expression of AtRDR1 is regulated by JA-dependent signaling. To examine this further, wild type Col-0 plants were treated with 250 µM methyl-JA and RNAs were extracted at various times over a 72 h time-course for analysis of gene expression by Q-RT-PCT (Figure 5A). Methyl-JA treatment induced transient increases in expression of both AtRDR1 and TPS10, used here as a positive control for JA-induced gene expression (Figure 5A). AtRDR1 expression peaked at 6 h post-treatment, and by 24 h had decreased to pre-treatment levels (Figure 5A). The results show that regulation of RDR1 gene expression by JA is conserved between N. attenuata and Arabidopsis.
Figure 5. JA-induced AtRDR1 expression is COI1-independent.
(A) RTqPCR analysis of transcript accumulation for AtRDR1 and the JA-responsive, COI1-dependent gene AtTPS10 over 72 h following treatment of Arabidopsis (ecotype Col-0) plants with methyl-JA. (B) AtRDR1 and AtTPS10 transcript accumulation in methyl-JA (JA) or control-treated coi1-16 mutant plants and wild-type Col-0 or Col-0 gl (the coi1-16 background) at 6 h post-treatment. Error bars represent standard error of the mean.
To investigate further the relationship of JA-mediated signal transduction to AtRDR1 expression, plants of the mutant line coronatine insensitive 1–16 (coi1-16), which is compromised in perception of the active form of JA, JA-Ile, were treated with methyl-JA and samples were harvested at 6 h post-treatment for RNA extraction and analysis of expression of AtRDR1 and TPS10 by RTqPCR (Figure 5B).
It has been shown previously that the induction by methyl-JA of increased TPS10 expression is COI1-dependent [45] and our data were consistent with this. Thus, we found that induction of TPS10 transcript accumulation by methyl-JA was inhibited in coi1-16 mutant plants (Figure 5B). In contrast, AtRDR1 expression following methyl-JA treatment was similar in wild-type and coi1-16 mutant plants (Figure 5B), demonstrating that JA-induced AtRDR1 expression is COI1-independent.
Ethylene and ABA Induce RDR1 Expression
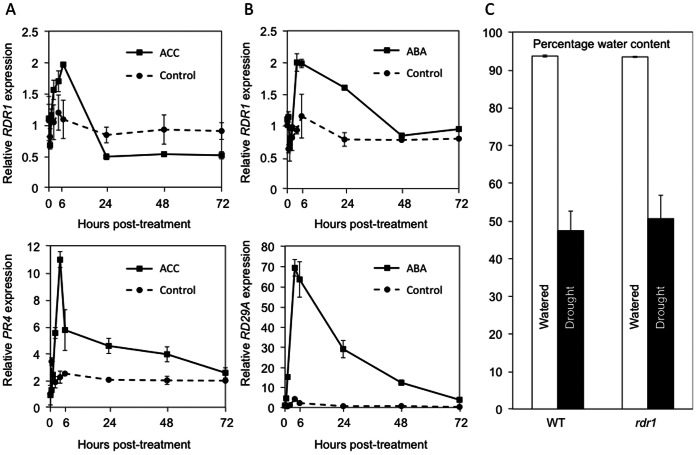
There is significant cross-talk between JA- and SA-regulated signaling and signaling mediated by ethylene and ABA [47]. Therefore, we investigated if these other stress-related phytohormones affected RDR1 expression. Arabidopsis plants were treated with solutions of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) or ABA. ACC treatment induced the expression of AtRDR1 in parallel with induction of the ethylene-responsive AtPR4 gene peaking at 6 h post treatment (Figure 6A). ABA treatment induced AtRDR1 expression and expression of the ABA-responsive gene RD29A, also with a peak in expression at 6 h post-treatment (Figure 6B).
Figure 6. Ethylene and ABA induce AtRDR1 expression but RDR1 is not required for drought resistance.
RTqPCR analysis of expression of AtRDR1 and the ethylene-regulated gene AtPR4 expression in Arabidopsis thaliana Col-0 plants sprayed with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) at 1 mM or water (Control) (A), or of AtRDR1 and the ABA-inducible gene RD29A after treatment with ABA (B) over time courses of 72 h. (C) Analysis of water content in wild-type (WT) or rdr1-mutant Arabidopsis plants watered normally (Watered) or deprived of water for 9 days (Drought). In the experiment shown, 40 well-watered plants were divided into two groups of 20, one group watered, the other subjected to drought. There was no significant difference (t-test: p = 0.693) in water content of water-deprived WT and rdr1-transgenic plants. Error bars represent standard error of the mean.
ABA is an important regulator of drought responses [48]. As AtRDR1 is induced by ABA treatment, this suggested the possibility that RDR1 might play a role in resistance to drought stress. However, when plants were subjected to drought by 9 days of water deprivation, there was no significant difference between the percentage water content of wild-type or rdr1 mutant plants (Figure 6C). The same result was seen in three independent experiments.
Discussion
Although often viewed as being SA-inducible, RDR1 expression displays some intriguing differences, as well as similarities too, to the behaviour of other SA-regulated plant genes. As is the case with the well-studied PR genes, SA-induced AtRDR1 expression was shown to be NPR1-dependent, which was confirmed using two independent npr1 mutant lines from different Arabidopsis ecotypes. However, unlike the transcript for the PR1 protein, the increase in AtRDR1 transcript accumulation was transient and its peak expression was markedly lower than that for AtPR1. This suggests that AtRDR1 transcript accumulation is under tighter transcriptional and post-transcriptional control than AtPR1. Interestingly, SA-induced accumulation of both AtPR1 and AtRDR1 transcripts was diminished in plants of the SA biosynthetic mutant line sid2 (which is compromised in ICS1). This led to our finding that the ICS1 gene but not the ICS2 gene is under a form of positive feedback from SA, the ultimate end product of ICS1 activity. The results also confirmed the primacy of ICS1 over ICS2 in facilitating SA biosynthesis in Arabidopsis [44].
The induction by SA of resistance to viruses is not dependent upon NPR1 [35], [36]. The dependence on NPR1 of SA-induced AtRDR1 expression provides additional evidence, along with previous studies with transgenic plants [15], [27], that the major contribution of RDR1 to virus resistance lies in its role in basal defense, and that it is not essential for SA-induced resistance to viruses. The role of RDR1 in stress tolerance and defense via the silencing of endogenous genes is something that has been suggested previously by Pandey and Baldwin (2007) as an explanation for the susceptibility observed in rdr1 mutant N. attenuata lines to herbivory [30]. Furthermore, tobacco lines deficient in RDR1 have been shown to have altered expression of other defense related genes, suggested by the authors that RDR1 plays a role in regulating other endogenous defense-related genes by suppressing the expression of regulatory molecules [33].
The JA-mediated and SA-mediated defensive signaling pathways are to a great extent antagonistic, and few transcripts are positively regulated by both [49]. Thus, the responsiveness of RDR1 to both of these phytohormones seen in N. attenuata [30] and in Arabidopsis (this study) sets this ‘SA-responsive’ gene apart from typical SA-responsive genes like PR1. RDR1 is also not a typical JA-responsive gene, since its induction by methyl-JA was not dependent upon COI1, a F-box protein responsible for degradation of JASMONATE ZIM-domain proteins that negatively regulate most JA-responsive genes [50], [51], [52]. Although most JA-responsive genes are dependent upon COI1 for induction, several, including genes involved in plant defense, have been discovered to be COI1-independent [53]. Perhaps the independence from COI1-mediated jasmonate perception allows RDR1 regulation to be outside the typical SA-JA antagonism, and may allow the gene to be similarly responsive to such a wide range of distinct stress signals as JA, SA, ethylene and ABA.
In N. attenuata, simulating herbivory by wounding leaves and applying oral secretions from leaf-chewing larvae caused NaRDR1 expression to increase, due to the JA-responsiveness of the gene [30]. We found that gentle wounding, specifically the abrasion used during mechanical inoculation with virus, is sufficient to induce AtRDR1 transcript accumulation and it was wounding, rather than an effect of the virus, that caused AtRDR1 induction in directly-inoculated leaves. The finding is reminiscent of findings of induction by abrasion of host RDR enzyme activity in plant tissues in early studies of plant viral RNA synthesis [54]. In previous work it was suggested that induction of RDR1 expression in inoculated and systemically infected tissues of virus-infected Arabidopsis was due to effects of the virus [15], [16]. However, our results indicate that these findings should be re-assessed and that RDR1 induction in inoculated tissue was most likely due to wounding, while induction in systemically infected leaves is probably attributable to localised induction of RNA silencing, such as that which occurs during ‘green-island’ formation [55], rather than as a direct effect of the virus or its gene products.
The responsiveness of AtRDR1 expression to a wide range of stress related hormones might imply a role in co-ordination of resistance to both biotic and abiotic insult. The responsiveness to ABA suggested that one of the stresses that RDR1 may help protect against is drought. However, rdr1-mutant plants were neither more nor less resistant to water loss than wild-type plants. In one way this was a surprising result because in a number of studies it has been shown that small RNA pathways affect drought responses. For example, where RNA silencing pathways have been compromised through mutation of AGO1 or DCLs 1–4, the mutant plants showed increased resistance to water loss [56], [57], [58]. Hence, although RDR1 is a component of the silencing pathway it does not appear to play a critical role in drought resistance, unlike the DCLs and AGO1. Thus, the biological implications of the ABA-responsiveness of the RDR1 gene remain to be discovered.
Materials and Methods
Arabidopsis Mutants and Growth Conditions
Arabidopsis thaliana wild type Col-0, Col-0gl, and Nössen (NO) ecotypes were used in this study, either alone or alongside mutant lines with the corresponding ecotype. Arabidopsis mutants used included the previously characterised NahG, sid2, npr1-1, npr1-5, coi1-16 and rdr1, ICS1:GUS, ICS2:GUS, and 35S:GUS lines. All plants were grown in short-day condition growth chambers (Conviron Ltd., Winnipeg, Manitoba, Canada): 8 h of light at 200 micromol (photons).m−2.s−1 and 22°C, with 60% humidity.
Hormone Treatments
Hormone treatments were conducted on plants four weeks of age. The plant hormone treatments were SA (1 mM), methyl-JA (250 µM), ABA (50 µM) and ACC (the precursor of ethylene; 1 mM) dissolved in water. Hormone concentrations were selected on the basis of previous optimization for SA and methyl-JA [46], ACC [59], and ABA [58], [60]. Control treatment used water only. Plants were sprayed until surface run-off and samples (aerial tissues of six plants) were taken and frozen in liquid nitrogen at time points of 0.5, 1, 2, 4, 6, 24, 48 and, in some experiments, 72 h post treatment. All experiments were carried out at least three times.
Wounding Treatment
Four-week-old, wild-type Col-0 plants were wounded by squeezing leaves twice with a pair of tweezers. Two leaves per plant were wounded. Aerial plant tissue (from six plants per time point) was harvested and immediately frozen in liquid nitrogen at subsequent time points of 0.5, 1, 2, 4, 6, 24, 48 and 72 h.
TMV Inoculation
TMV strain U1 (20 µg.ml−1 purified virions in sterile water) was mechanically inoculated onto Carborundum-dusted leaves. Mock-inoculation used sterile water only. Arabidopsis plants were inoculated at the 4- to 6-true-leaf stage. Successful inoculation was confirmed by RT-PCR on extracted plant RNA, using primers for the TMV coat protein gene (forward primer 5′-TTCTTGTCATCAGCGTGGGCCG-3′; reverse primer 5′-GCAGGACCAGAGGTCCAGACCAA-3′.
Reverse Transcription-coupled Quantitative Polymerase Chain Reaction
Total RNA for RTqPCR analysis was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Total RNA was then further purified by a phenol-chloroform extraction and subsequently treated with TURBO-DNase (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. First strand cDNA synthesis was carried out on 0.5 µg total RNA using GoScript (Promega, Madison, WI, USA) with random hexamer primers according to the manufacturer’s instructions. The cDNA produced was diluted 1 to 5 and RTqPCR performed using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich, St Louis, MO, USA) in 15 µl reactions according to the manufacturer’s instructions. Reactions were conducted in triplicate. Primers sequences are given in Table 1. The gene glyceraldehyde-3-phosphate dehydrogenase was used as the reference gene as its expression was identified as being stable under the experimental conditions. The instrument used was a BioRad C1000 thermal cycler connected to a CFX96 Real-Time PCR Detection System and a PC running on CFX manager software (BioRad). The data was analysed using LinRegPCR [61] to give Ct and amplification efficiency values. Relative gene expression was calculated using efficiency adjusted ΔΔCt methodology, incorporating the reference transcript to control for variation in loading. Gene expression was expressed relative to mock-treated wild-type plants.
Table 1. Primers used for quantitative PCR.
| Transcript | Locus | Forward primer 5′-3′ | Reverse primer 5′-3′ |
| AtGAPDH | AT1G13440 | AGGCTGGGATTGCATTGAGCGA | ACACACAAACTCTCGCCGGTGT |
| AtRDR1 | AT1G14790 | AAGAGCGGTTCGGGCGTTGA | AGCCGAAGCCTTTGCTGACTCA |
| AtPR1 | AT2G14610 | CGAAAGCTCAAGATAGCCCA | AAGGCCCACCAGAGTGTATG |
| AtTPS10 | AT2G24210 | CTGGTGGATGGAGACAGGTT | TGAGGCTCTTGGATTTGTCC |
| AtRD29A | AT5G52310 | ACCGATTCATCATCCTCTGTCCGAA | ACGTTATCGGGGTCTCGACGTT |
| AtPR4 | AT3G04720 | TGTTCTCCGACCAACAACTG | TGGAGCAATAAGCACTCACG |
| AtICS1 | AT1G74710 | CTTTTCAGTCCCTCAGGTTG | AGTTCATCATCCCAAGCAAT |
| AtICS2 | AT1G18870 | TGCAGTGTGAAGGACAAGAC | GAAGAGTCTCTCAGGCGTGT |
AtGAPDH was used as the ‘housekeeping’ reference transcript. All primers (used at a final concentration of 10 pmol.µl−1) were designed to have an annealing temperature of 57°C.
Generation of ICS:GUS Transgenic Lines
Generation of ICS1:GUS-transgenic lines was previously described by Lewsey et al. [46]. PCR amplification of the ICS2 promoter region for cloning was performed using oligonucleotides designed to incorporate 5′ XbaI (forward primer, target sequence underlined) and 3′ XmaI (reverse primer, target sequence underlined) restriction sites (forward primer 5′-ATATCTAGATTAATTGTTACGAGACG-3′ and reverse primer 5′-TATCCCGGGTAGAGAGACTACGAAG-3′). The 1.5-kb product was cloned into pGEM-T Easy (Promega), sequenced to check for mutations and sub-cloned into pGreen-GUS [62] using XbaI and XmaI. The resulting plasmid was introduced into Agrobacterium tumefaciens GV3101::pMP90::pSOUP by electroporation and used to transform A. thaliana Col-0 by floral dipping [63].
Detection of GUS Activity
Plants of four-week-old ICS1:GUS and ICS2:GUS lines were sprayed with 1 mM SA until surface run-off. As a control plants of the same age were treated with water. Aerial plant tissue was harvested at time points of 24 and 48 h post treatment for both control and SA-treated plants. Immediately after harvesting, plant tissue was stained with 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-Gluc) by submerging the rosettes in 5 ml of X-Gluc solution and infiltrating them under a vacuum for 15 min. Samples were incubated at 37°C overnight. The indigo stain develops and indicates regions where the GUS reporter gene has been expressed. Samples were then soaked in 70% ethanol to remove chlorophyll.
Drought Experiments
Water content analysis was performed according to the methods of Xu et al. [64]. At least 20 four-week-old plants were drenched in water for 30 min to achieve 100% soil saturation. The plants were divided into equal numbers, half receiving watering as a control, whilst the other half did not receive any more water. The position of individual plants was randomized and plants were observed daily. After 9 days without water, the aerial tissues of well-watered and drought-stressed plants were harvested and fresh weights recorded. Samples were then dried over a period of 5 days at 50°C. Dry weight was recorded and the weight loss for each plant, which is equal to water weight, was calculated. Percentage water content of each rosette was calculated by dividing the fresh weight by the water weight for each sample. This experiment was done three times.
Acknowledgments
The authors are grateful to Alex Murphy, Kristina Gruden and Simon Groen for useful discussions and to Adrienne Pate for excellent technical support.
Funding Statement
First author (LJRH) supported by a studentship co-funded by the James Hutton (formerly Scottish Crop Research) Institute and the UK Biotechnological and Biological Sciences Research Council (BBSRC). Work in the JPC lab is funded by The Leverhulme Trust (RPG-2012-667), BBSRC (BB/D014376/1, BB/J011762/1) and the Cambridge University Newton Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- 2. Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- 3. Csorba T, Pantaleo V, Burgyán J (2009) RNA silencing: an antiviral mechanism. Advances in Virus Research 75: 35–71. [DOI] [PubMed] [Google Scholar]
- 4. Palukaitis P (2011) The road to RNA silencing is paved with plant-virus interactions. Plant Pathology Journal 27: 197–206. [Google Scholar]
- 5. Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, et al. (2005) Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. The Journal of Virology 79: 7812–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, et al. (2006) Four plant dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Research 34: 6233–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, et al. (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71. [DOI] [PubMed] [Google Scholar]
- 8. Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, et al. (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 22: 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of the National Academy of Sciences of the United States of America 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qu F, Ye X, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proceedings of the National Academy of Sciences of the United States of America 105: 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, et al. (2010) Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen encoded GW repeat protein. Genes and Development 24: 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harvey JJW, Lewsey MG, Patel K, Westwood J, Heimstädt S, et al. (2011) An antiviral defense role of AGO2 in plants. Plos ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542. [DOI] [PubMed] [Google Scholar]
- 14. Wassenegger M, Krczal G (2006) Nomenclature and functions of RNA-directed RNA polymerases. Trends in Plant Science 11: 142–151. [DOI] [PubMed] [Google Scholar]
- 15. Xie Z, Fan B, Chen C, Chen Z (2001) An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proceedings of the National Academy of Sciences of the United States of America 98: 6516–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu D, Fan B, MacFarlane SA, Chen Z (2003) Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Molecular Plant-Microbe Interactions 16: 206–216. [DOI] [PubMed] [Google Scholar]
- 17. Muangsan N, Beclin C, Vaucheret H, Robertson D (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. The Plant Journal 38: 1004–1014. [DOI] [PubMed] [Google Scholar]
- 18. Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS (2004) A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana . Proceedings of the National Academy of Sciences of the United States of America 101: 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwach F, Vaistij FE, Jones L, Baulcombe DC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by Potato Virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiology 138: 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diaz-Pendon J, Li F, Li W-X, Ding S-W (2007) Suppression of Antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. The Plant Cell 19: 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Donaire L, Barajas D, Martinez-Garcia B, Martinez-Priego L, Pagan I, et al. (2008) Structural and genetic requirements for the biogenesis of Tobacco Rattle Virus-derived small interfering RNAs. Journal of Virology 82: 5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi X, Bao FS, Xie Z (2009) Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS.One 4: 3, e4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He J, Dong Z, Jia Z, Wang J, Wang G (2010) Isolation, expression and functional analysis of a putative RNA-dependent RNA polymerase gene from maize (Zea mays L.). Molecular Biology Reports 37: 865–874. [DOI] [PubMed] [Google Scholar]
- 24. Yang H, Wang M, Gao Z, Zhu C, Guo X (2011) Isolation of a novel RNA-dependent RNA polymerase 6 from Nicotiana glutinosa, NgRDR6, and analysis of its response to biotic and abiotic stresses. Molecular Biology Reports 38: 929–937. [DOI] [PubMed] [Google Scholar]
- 25. Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO Journal 22: 4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carr JP, Lewsey MG, Palukaitis P (2010) Signalling in induced resistance. Advances in Virus Research 76: 57–121. [DOI] [PubMed] [Google Scholar]
- 27. Lee WS, Fu SF, Verchot-Lubicz J, Carr JP (2011) Genetic modification of alternative respiration in Nicotiana benthamiana affects basal and salicylic acid-induced resistance to potato virus X. BMC Plant Biology. 11: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ying X-B, Dong L, Zhu H, Duan C-G, Du Q-S, et al. (2010) RNA-Dependent RNA Polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana . The Plant Cell 22: 1358–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic Acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004. [DOI] [PubMed] [Google Scholar]
- 30. Pandey SP, Baldwin IT (2007) RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature. Plant Journal 50: 40–53. [DOI] [PubMed] [Google Scholar]
- 31. Thaler JS, Owen B, Higgins VJ (2004) The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiology 135: 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandey SP, Shahi P, Gase K, Baldwin IT (2008) Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata . Proceedings of the National Academy of Sciences of the United States of America 105: 4559–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rakhshandehroo F, Takeshita M, Squires J, Palukaitis P (2009) The influence of RNA-dependent RNA polymerase 1 on Potato virus Y infection and on other antiviral response genes. Molecular Plant-Microbe Interactions 22: 1312–1318. [DOI] [PubMed] [Google Scholar]
- 34. Durrant WE, Dong X (2004) Systemic acquired resistance. Annual Review of Phytopathology 42: 185–209. [DOI] [PubMed] [Google Scholar]
- 35. Kachroo P, Yoshioka K, Shah J, Dooner HK, Klessig DF (2000) Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell 12: 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wong CE, Carson RA, Carr JP (2002) Chemically induced virus resistance in Arabidopsis thaliana is independent of pathogenesis-related protein expression and the NPR1 gene. Molecular Plant Microbe Interactions 15: 75–81. [DOI] [PubMed] [Google Scholar]
- 37. Cao H, Scott A, Bowling A, Gordon S, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shah J, Tsui F, Klessig DF (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Molecular Plant-Microbe Interactions 10: 69–78. [DOI] [PubMed] [Google Scholar]
- 39. Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y, Xu S, Ding P, Wang D, Cheng YT, et al. (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proceedings of the National Academy of Sciences of the United States of America 107: 18220–18225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250. [DOI] [PubMed] [Google Scholar]
- 43. Strawn MA, Marr SK, Inoue K, Inada K, Zubieta C, et al. (2007) Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. Journal of Biological Chemistry 282: 5919–5933. [DOI] [PubMed] [Google Scholar]
- 44. Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, et al. (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiology. 147: 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Devoto A, Ellis C, Magusin A, Chang H-S, Chilcott C, et al. (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Molecular Biology 58: 497–513. [DOI] [PubMed] [Google Scholar]
- 46. Lewsey MG, Murphy AM, MacLean D, Dalchau N, Westwood JH, et al. (2010) Disruption of two defensive signaling pathways by a viral RNA silencing suppressor. Molecular Plant-Microbe Interactions 23: 835–845. [DOI] [PubMed] [Google Scholar]
- 47. Koornneef A, Pieterse CMJ (2008) Cross talk in defense signaling. Plant Physiology 146: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robertson FC, Skeffington AW, Gardner MJ, Webb AAR (2009) Interactions between circadian and hormonal signalling in plants. Plant Molecular Biology 69: 419–427. [DOI] [PubMed] [Google Scholar]
- 49. Pieterse CMJ, Leon-Reyes A, van der Ent S, van Wees SCM (2009) Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5: 308–316. [DOI] [PubMed] [Google Scholar]
- 50. Chini A, Fonseca S, Fernández G, Adie B, Chico JM, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448: 666–673. [DOI] [PubMed] [Google Scholar]
- 51. Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, et al. (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiology 146: 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signaling. Nature 448: 661–666. [DOI] [PubMed] [Google Scholar]
- 53. Stotz HU, Jikumaru Y, Shimada Y, Sasaki E, Nadja Stingl N (2011) Jasmonate-dependent and COI1-independent defense responses against Sclerotinia sclerotiorum in Arabidopsis thaliana: Auxin is part of COI1-independent defense signaling. Plant Cell Physiology 52: 1941–1956. [DOI] [PubMed] [Google Scholar]
- 54. Zaitlin M, Hull R (1987) Plant Virus-Host Interactions. Annual Review of Plant Physiology and Plant Molecular Biology 38: 291–315. [Google Scholar]
- 55. MacDiarmid R (2005) RNA silencing in productive virus infections. Annual Review of Phytopathology 43: 523–544. [DOI] [PubMed] [Google Scholar]
- 56. Earley K, Smith M, Weber R, Gregory B, Poethig R (2010) An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana . Silence 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li W, Cui X, Meng Z, Huang X, Xie Q (2012) Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiology 158: 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Westwood JH, McCann L, Naish M, Dixon H, Murphy AM, et al. (2013) A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana . Molecular Plant Pathology 14: 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hase S, Van Pelt JA, van Loon LC, Pieterse CMJ (2003) Colonization of Arabidopsis roots by Pseudomonas fluorescens primes the plant to produce higher levels of ethylene upon pathogen infection. Physiological and Molecular Plant Pathology 62: 219–226. [Google Scholar]
- 60. Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, et al. (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences of the United States of America 103: 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339: 62–6. [DOI] [PubMed] [Google Scholar]
- 62. Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology 42: 819–832. [DOI] [PubMed] [Google Scholar]
- 63. Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 64. Xu P, Chen F, Mannas JP, Feldman T, Sumner LW (2008) Virus infection improves drought tolerance. New Phytologist 180: 911–921. [DOI] [PubMed] [Google Scholar]