Abstract
Recently in Cell, Jia et al. (2012) reported novel Utf1-controlled mechanisms of maintaining pluripotency and self-renewal in embryonic stem cells (ESCs). Utf1 buffers bivalent gene expression by competitive binding with polycomb repressive complex 2 and initiation of mRNA degradation.
In 1998, Okuda et al. discovered a protein expressed specifically in two pluripotent cell lines: mouse embryonic carcinoma cells (mECs) and mouse embryonic stem cells (mESCs). Because of its restricted expression pattern, the protein was named Undifferentiated Embryonic Cell Transcription Factor 1 (Utf1) (Okuda et al., 1998). Utf1 is quickly downregulated in mESCs during differentiation and the protein is not expressed in somatic cells. Van den Boom et al. (2007) later identified Utf1 as a chromatin-associated factor that is required for proper differentiation of both mECs and mESCs. Now Jia et al. (2012) have made a significant advance in our understanding of not only Utf1 itself, but also the regulatory machinery controlling ESC pluripotency and self-renewal, cellular processes in which Utf1 plays integral roles.
Surprisingly, despite its name invoking a surmised role as a transcription factor, the activities of Utf1 in ESCs identified to date do not appear to involve action as a classical transcription factor. Jia et al. (2012) found instead that Utf1 contributes to regulation of pluripotency by acting as an epigenetic and translational factor. In the former role, Utf1 fine-tunes the effects of ESC-specific bivalent domains on gene expression. Bivalent domains are epigenetically regulated chromatin regions that allow key developmental genes to be poised in preparation for differentiation, while concurrently maintaining low levels of actual expression in the undifferentiated state (Bernstein et al., 2006). Bivalent domains are characterized by a combination of large regions of repressive H3 lysine 27 trimethylation (H3K27me3) epigenetic marks, and smaller regions of active H3 lysine 4 trimethylation (H3K4me3) epigenetic marks. Jia et al. (2012) found that Utf1 contributes to regulation of pluripotency by fine-tuning the effects of these ESC-specific bivalent domains, in particular via H3K27me3 and also by posttranscriptional mRNA pruning.
Using ChIP-Seq to map global genomic DNA binding sites of Utf1 in mESCs, Jia et al. (2012) reported approximately 75,000 chromatin regions bound by Utf1 including a large cohort of genes related to organ/system development and cell differentiation. RNA-Seq of Utf1 null versus control mESCs identified almost 800 genes influenced by Utf1 levels by at least 1.5-fold. Integrating the genomic binding and RNA-Seq data sets revealed that Utf1 directly bound most genes that exhibited altered expression in ESCs with Utf1 loss of function. Intriguingly, the majority of these Utf1 target genes were both bivalent and targets of polycomb repressive complex 2 (PRC2) including its Suz12 subunit. They further identified the mechanism of Utf1-mediated gene upregulation, which involves competing with PRC2 to bind an AG-rich motif within CpG islands. It was already known that PRC2 is required for establishing silenced and poised states of bivalent genes in ESCs (reviewed in Zhou et al., 2011). Jia et al. (2012) propose a model in which Utf1 prevents excessive inhibition of bivalent genes by blocking PRC2 binding and subsequent silencing via H3K27me3.
Utf1 also operates on a second, unexpected level to buffer bivalency. Jia et al. (2012) found that Utf1 fine-tunes bivalent gene expression by tagging newly transcribed mRNAs in the nucleus for cytoplasmic degradation. Biotin-Utf1 pull-down of mESC nuclear extracts identified interactions between Utf1 and three members of the mRNA decapping complex: Dcp1a, Ddx6, and Edc3. Sequential ChIP-qPCR with Utf1 and Dcp1a antibodies of randomly selected bivalent genes demonstrated that Utf1 is necessary to recruit Dcp1a to bivalent gene promoter regions. These findings establish an exciting, new area of research for mRNA pruning in ESCs because mRNA pruning has only been studied previously in fungi and animal somatic cells (reviewed in Ling et al., 2011). Through its opposing functions of inhibiting PRC2 binding and initiating mRNA degradation via recruitment and interaction with the mRNA decapping complex, Utf1 maintains a precise level of bivalent gene expression that is “just right” for maintaining pluripotency of ESC chromatin. In this sense, Utf1 is the youngest bear from the Goldilocks tale, ensuring that the conditions are precisely right for maintaining pluripotency.
Clues as to how Utf1 is involved in the differentiation process are also suggested by Utf1 loss of function and changes in bivalent gene expression levels during ectoderm, mesoderm, and trophectoderm differentiation. Loss of Utf1 expression in ectoderm and mesoendoderm by shRNA results in an increase in PRC2 binding of key developmental genes Olig2, Nestin, and T as well as an increase in H3K27me3 at these promoters. On the other hand, the expression level of Hoxa1 significantly increases after loss of Utf1 due to a decrease in mRNA pruning activity; Dcp1a knockdown recapitulates this phenotype. When Utf1 null mESCs are differentiated by embryoid-body formation Olig2, Nestin, T, Hoxa1, and other important mesoderm development genes, Gata6 and Bmp4 are misregulated. From extensive in vivo assays of trophectoderm differentiation and Cdx2 expression, Jia et al. (2012) found that Utf1 deletion does not block trophectoderm differentiation, but rather, disrupts the connection between the cells’ differentiation and proliferation cycles.
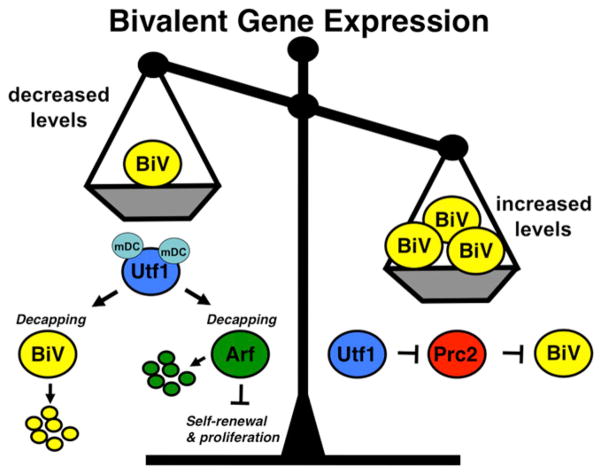
Jia et al. (2012) report that Utf1, in addition to fine-tuning bivalency to preserve pluripotency, also maintains self-renewal in mESCs. Utf1 acts directly to downregulate the levels of Arf, a known inhibitor of ESC proliferation, by initiating Arf mRNA degradation. Arf is also known to be directly upregulated by Myc in ESCs. Oct3/4 and Sox2 directly upregulate Utf1 transcription (Nishimoto et al., 1999) and Utf1 in turn translationally inhibits the Myc-Arf feedback loop within mESCs. Thus, Utf1 links together key pathways in ESCs in a novel way. We highlight the seemingly opposing mechanisms of Utf1 function in mESCs as observed by Jia et al. (2012) that together balance bivalent gene expression (Figure 1).
Figure 1. A Model Showing Opposing Functions of Utf1 on Bivalent Gene Expression in Pluripotent Stem Cells.
Yellow circles labeled “BiV” represent bivalent genes. Other factors are presented as indicated with Utf1 in blue, Arf in green, and PRC2 in red. mDc is the mRNA decapping complex. Smaller circles represent mRNAs being degraded. In the balance, decreased BiV levels are reflected by the scale pan going up and being less weighty, while the reverse is the case on the right with increasing BiV levels.
The indirect connection between Myc and Utf1 raised by Jia et al. (2012) suggests possible mechanisms underlying the observation made in 2008 that replacing Myc with Utf1 increases reprogramming efficiency (relative to Oct4, Sox2, and Klf4 alone) (Zhao et al., 2008). One interesting notion is that in the absence of exogenous Myc, which suppresses differentiation-associated gene expression (Varlakhanova et al., 2010), Utf1 switches to suppressing differentiation-associated gene expression. However, the mechanisms by which Utf1 stimulates reprogramming in the absence of exogenous Myc remain unknown. It is also notable that Utf1 is an early stage indicator of successful reprogramming (Buganim et al., 2012).
A number of important open questions remain about Utf1. Does Utf1 have an effect on global chromatin structure as suggested by its 75,000 binding sites on ESC chromatin? Is there a more direct relationship between Utf1 and Myc? What specific roles does Utf1 have during ESC differentiation? How does Utf1 function to enhance cellular reprogramming, and do any of the mechanisms by which Utf1 functions in ESCs manifest in iPSCs as well? Does Utf1 function in some contexts as a transcription factor as its name implies? Do the functions outlined for Utf1 in mESCs also manifest in human ESCs? The Jia et al. (2012) paper also raises exciting broader questions about the potential role of mRNA decapping in cellular reprogramming. Future studies tackling these questions, in part inspired by these recent studies of Utf1, will greatly advance our understanding of self-renewal and pluripotency.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zheng X, Hu G, Cui K, Zhang J, Zhang A, Jiang H, Lu B, Yates J, 3rd, Liu C, et al. Cell. 2012;151:576–589. doi: 10.1016/j.cell.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SH, Qamra R, Song H. Wiley Interdiscip Rev RNA. 2011;2:193–208. doi: 10.1002/wrna.44. [DOI] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, Muramatsu M. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, Kuro-o M, Nabeshima Y, Boon K, Keaveney M, et al. EMBO J. 1998;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom V, Kooistra SM, Boesjes M, Geverts B, Houtsmuller AB, Monzen K, Komuro I, Essers J, Drenth-Diephuis LJ, Eggen BJ. J Cell Biol. 2007;178:913–924. doi: 10.1083/jcb.200702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, et al. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]