Abstract
The biosynthetic gene cluster for the pyralomicin antibiotics has been cloned and sequenced from Nonomuraea spiralis IMC A-0156. The 41-kb gene cluster contains 27 ORFs predicted to encode all of the functions for pyralomicin biosynthesis. This includes non-ribosomal peptide synthetases (NRPS) and polyketide synthases (PKS) required for the formation of the benzopyranopyrrole core unit, as well as a suite of tailoring enzymes (e.g., four halogenases, an O-methyltransferase, and an N-glycosyltransferase) necessary for further modifications of the core structure. The N-glycosyltransferase is predicted to transfer either glucose or a pseudosugar (cyclitol) to the aglycone. A gene cassette encoding C7-cyclitol biosynthetic enzymes was identified upstream of the benzopyranopyrrole-specific ORFs. Targeted disruption of the gene encoding the N-glycosyltransferase, prlH, abolished pyralomicin production and recombinant expression of PrlA confirms the activity of this enzyme as a sugar phosphate cyclase (SPC) involved in the formation of the C7-cyclitol moiety.
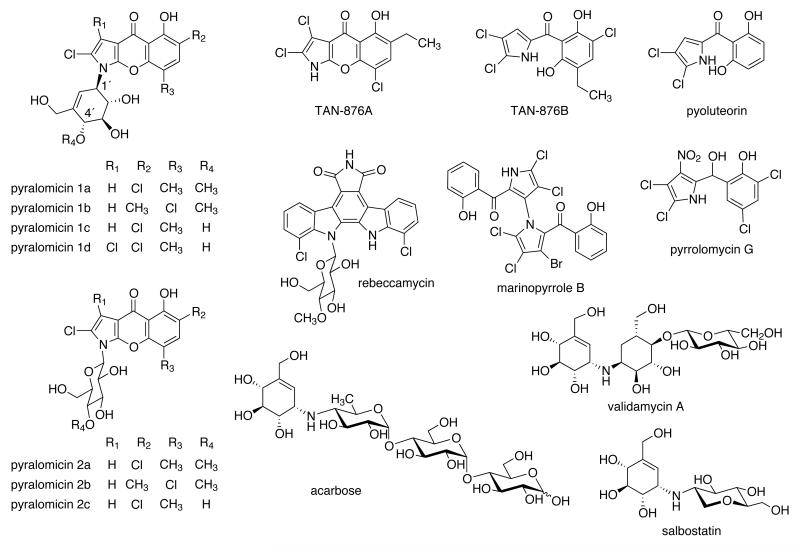
The pyralomicins are a group of antibiotics isolated from the soil bacterium Nonomuraea spiralis IMC A-0156 that contain a unique benzopyranopyrrole chromophore linked to a C7-cyclitol (pyralomicins 1a-1d) or glycosylated by glucose (pyralomicins 2a-2c) (Figure 1).1, 2 The aglycone region of the molecule resembles pyoluteorin, the pyrrolomycins, the marinopyrroles, TAN-876A and TAN-876B (Figure 1) and is predicted to house homologous enzymes within the biosynthetic pathway.3-7 The pyralomicins are active against various bacteria, particularly strains of Micrococcus luteus, an opportunistic Gram-positive bacterium that can cause endocarditis, intracranial suppuration, bacteraemia, and hepatic abscess.1 The antibacterial activity of the pyralomicins appears to be dependent upon the number and the position of chlorine atoms within the molecules, and the nature and methylation of the glycone.1, 2 Pyralomicin 1c, which contains the unmethylated cyclitol as the glycone, exhibits more potent antibacterial activity than its glucosyl analogue, pyralomicin 2c, and is also more active than its 4′-methylated congener, pyralomicin 1a, suggesting a role for the cyclitol moiety in the antimicrobial activity.
Figure 1.
Structures of the pyralomicins and related compounds.
Previous isotope-labeled feeding experiments have demonstrated that the pyralomicins contain a polyketide-peptide-derived core structure that is assembled from proline, two acetates and one propionate, and the cyclitol moiety is derived from 2-epi-5-epi-valiolone, implicating a sugar phosphate cyclase (SPC) enzyme in their biosynthetic pathway.8, 9 SPCs represent a superfamily of enzymes involved in the biosynthesis of a wide array of primary and secondary natural products, including well-known antibiotics such as rifamycin, neomycin, kanamycin, and gentamicin, the antifungal agent validamycin, the anti-diabetic agent acarbose, the antitumor agent mitomycin C, as well as the sunscreen compounds mycosporine-like amino acids.10-12 Detailed BLAST and maximum likelihood analysis of protein sequences from the superfamily of SPCs has revealed that while they uniformly share a number of highly conserved motifs, each family of SPC has a unique signature of altered binding-pocket residues.12 Thus, the primary sequence can be used as a tool to predict the enzymatic function and class of natural products that will be produced by any given SPC or used to develop degenerate primers to amplify specific SPC subclasses.
In this study, we have utilized the signature sequences within the SPC superfamily to design degenerate primers specific for the 2-epi-5-epi-valiolone synthase family of enzymes. The degenerate primers were then used to PCR amplify a 2-epi-5-epi-valiolone synthase homologue from the pyralomicin producer, N. spiralis IMC A-0156. Recombinant expression and biochemical characterization of the homologue, PrlA, have confirmed the activity of the enzyme as a 2-epi-5-epi-valiolone synthase. The prlA gene sequence was subsequently used as a homologous probe to screen a genomic DNA library from N. spiralis, identifying a 41-kb DNA sequence containing the gene cluster required for pyralomicin biosynthesis. Disruption of the gene encoding the N-glycosyltransferase PrlH abrogated pyralomicin production, confirming the critical function of PrlH in pyralomicin biosynthesis.
RESULTS AND DISCUSSION
Isolation of the pyralomicin biosynthetic gene cluster
To identify the biosynthetic gene cluster of pyralomicin, high molecular weight genomic DNA was isolated from N. spiralis, end-filled and ligated into pCC1Fos, Copy Control Fosmid (Epicentre), to yield a library of ~5,000 colonies. Degenerate primers were designed to homologous regions within the 2-epi-5-epi-valiolone synthases, AcbC and ValA, from the acarbose and validamycin pathways.13, 14 Subsequent primers were used to PCR amplify a fragment of a 2-epi-5-epi-valiolone synthase gene from the pyralomicin producer. This sequence served as a probe for screening the library. In addition, the pltA halogenase gene from the pyoluteorin cluster was used as a second heterologous probe.3 Results from the dual screening yielded eight overlapping fosmids.
Production and characterization of recombinant PrlA
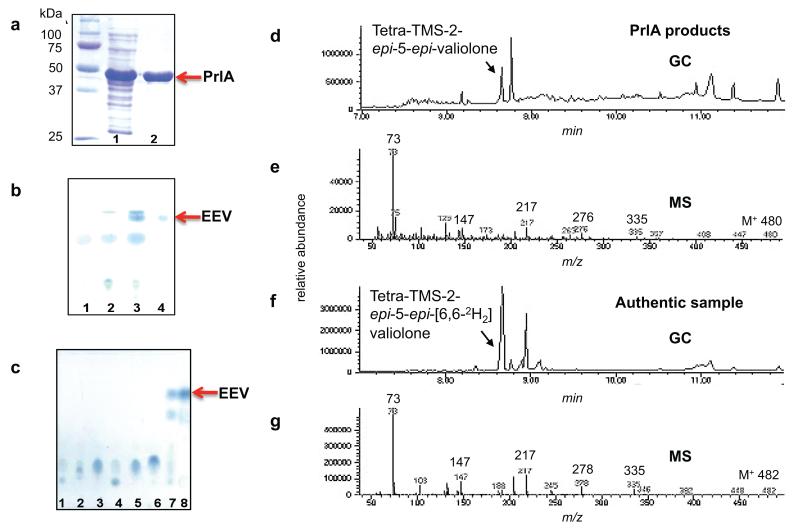
Chromosome walking within the region of the 2-epi-5-epi-valiolone synthase gene revealed the complete ORF of prlA. The PrlA homologue contains 57% and 54% identity with the ValA and SalQ, 2-epi-5-epi-valiolone synthases from the validamycin (Streptomyces hygroscopicus subsp. jinggangensis) and salbostatin (Streptomyces albus) gene clusters, respectively, whereas PrlA demonstrated only 22% identity with the AroB dehydroquinate synthase from Emericella nidulans, suggesting that sedoheptulose 7-phosphate is the native substrate for the enzyme.13-15 To test this hypothesis, the prlA gene was cloned into pRSET B expression plasmid (Invitrogen). Expression of the gene yielded a soluble 6XHis-PrlA fusion protein that was purified using nickel agarose chromatography (Figure 2a). Biochemical assays of the enzyme demonstrate that it is capable of catalyzing the cyclization of sedoheptulose 7-phosphate to 2-epi-5-epi-valiolone; however, it cannot effectively cyclize other sugar phosphate intermediates including DAHP, amino-DAHP, glucose 6-phosphate, fructose 6-phosphate, mannose 6-phosphate, and ribose 5-phosphate (Figures 2b – 2g). An isotopically labeled authentic sample, 2-epi-5-epi-[6-2H2]valiolone,16 was used for comparison (Fig. 2f – 2g). The enzyme product was detected as tetra-trimethylsilyl derivative of 2-epi-5-epi-valiolone [m/z 480 (M+)] with major fragment ions at m/z 335, 276, 217, 147 and 73 (Fig. 2e). This fragmentation pattern is consistent with that of the authentic sample, which showed m/z 482 (M+), 335, 278, 217, 147, 73 (Fig. 2g). The high specificity of PrlA as a 2-epi-5-epi-valiolone synthase is strong evidence that this enzyme is part of the pyralomicin biosynthetic gene cluster and is involved in the formation of the C7-cyclitol functional group incorporated into pyralomicins 1a-1d.
Figure 2.
Recombinant expression and characterization of PrlA. (a) Heterologously expressed and purified PrlA. lane 1, soluble fraction of PrlA; lane 2, Ni-NTA agarose-purified PrlA. (b) TLC analysis of PrlA enzyme activity. lane 1, boiled PrlA with sedoheptulose 7-phosphate; lane 2, cell free extract of E. coli harboring empty vector with sedoheptulose 7-phosphate; lane 3, PrlA with sedoheptulose 7-phosphate; lane 4, 2-epi-5-epi-valiolone standard. (c) TLC analysis of the substrate specificity of PrlA. Enzyme reaction mixtures contained 5 mM of the following substrates: lane 1, aminoDAHP; lane 2, DAHP; lane 3, glucose 6-phosphate; lane 4, fructose 6-phosphate; lane 5, mannose 6-phosphate; lane 6, ribose 5-phosphate; lane 7, sedoheptulose 7-phosphate; and lane 8, 2-epi-5-epi-valiolone standard. (d-g) GC-MS profiles of silylated PrlA reaction product and silylated 2-epi-5-epi-[6-2H2]valiolone standard. (d) GC trace of silylated PrlA reaction product. (e) MS fragmentation pattern of the major peak eluting at 8.65 – 8.75 minutes from the silylated PrlA reaction product [m/z 480 (M+)]. (f) GC trace of silylated 2-epi-5-epi-[6-2H2]valiolone standard and (g) MS fragmentation pattern of the major peak eluting at 8.65-8.75 minutes from the silylated 2-epi-5-epi-[6-2H2]valiolone standard [m/z 482 (M+)].
Sequencing and characterization of the prl gene cluster
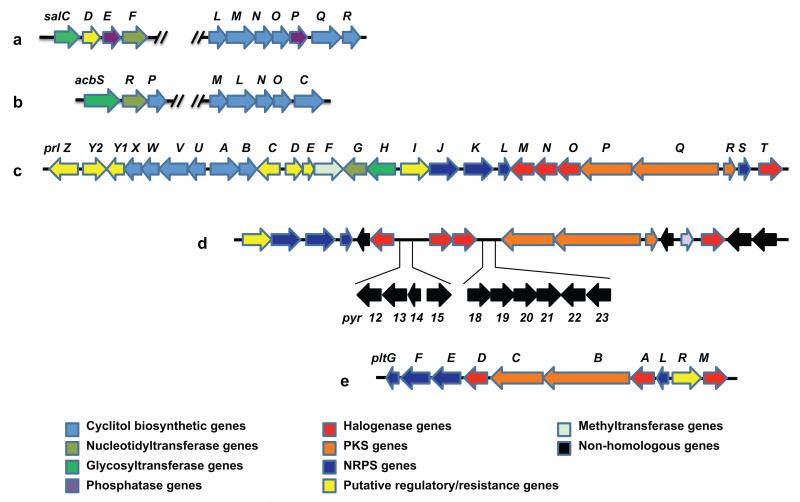
With the function of PrlA confirmed, three of the overlapping cosmids isolated in the screening process were sequenced using a combination of pyrosequencing and chromosome walking. A region of ~41 kb of DNA revealed 27 ORFs predicted to be involved in the biosynthesis of the pyralomicin aglycone and the cyclitol unit, as well as ORFs predicted to function in the regulation and transport (Figure 3 and Table S1). The upstream sequence ends with prlZ, whose gene product may be involved in the regulation of pyralomicin biosynthesis,17 whereas the downstream sequence ends with prlT, which encodes a halogenase protein. Additional sequencing of 5 kb of DNA on either side of the putative cluster did not reveal any additional open reading frames related to secondary metabolite biosynthesis. Thus, we believe that all of the genes required for pyralomicin biosynthesis are housed within the gene cassette provided in Figure 3.
Figure 3.
Genetic organization of the pyralomicin gene cluster (prl) compared with partial gene clusters of salbostatin (sal), acarbose (acb), pyrrolomycin (pyr), and pyoluteorin (plt). (a) salbostatin cluster. (b) acarbose cluster. (c) pyralomicin cluster. (d) pyrrolomycin cluster. (e) pyoluteorin cluster.
Formation of the C7-cyclitol unit
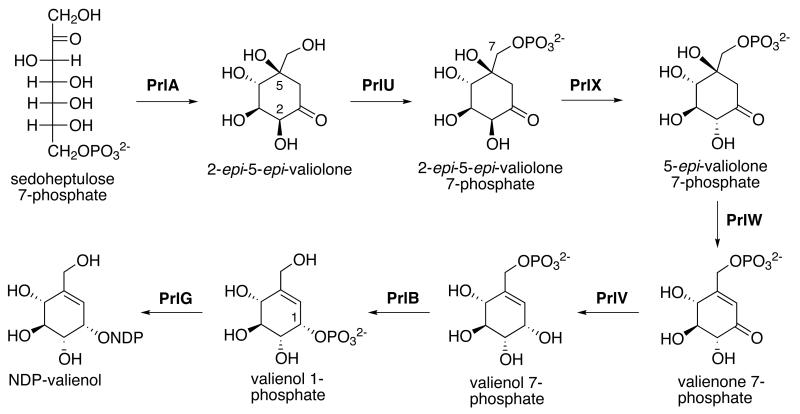
Production of the C7-cyclitol is proposed to be mediated through the actions of several enzymes within the pathway (Scheme 1). These include the 2-epi-5-epi-valiolone synthase PrlA, a putative phosphomutase (PrlB), a cyclitol kinase (PrlU), two cyclitol dehydrogenases (PrlV and PrlW), and a 2-epi-5-epi-valiolone phosphate epimerase (PrlX). This cassette of genes shares high homology with the cyclitol biosynthetic genes from the salbostatin and acarbose gene clusters (Figure 3, Table S1).15, 18 The gene prlF encodes an enzyme homologous to RebM from the rebeccamycin biosynthetic gene cluster.19 RebM is predicted to function as a glucose O-methyltransferase during rebeccamycin biosynthesis.20, 21 Similarly, the cyclitol moiety in pyralomicin 1a and 1b contain O-methyl groups at the C-4 position suggesting a role for PrlF. The PrlG protein has high similarity with galactose/hexose 1-phosphate uridylyltransferases including GalT from the milbemycin-producing S. bingchenggensis (63% identity/76% similarity), suggesting a role for PrlG in the activation of the cyclitol moiety to the nucleotidyl derivative.22, 23
Scheme 1.
Proposed mode of formation of the cyclitol moiety.
Formation of the core unit benzopyranopyrrole
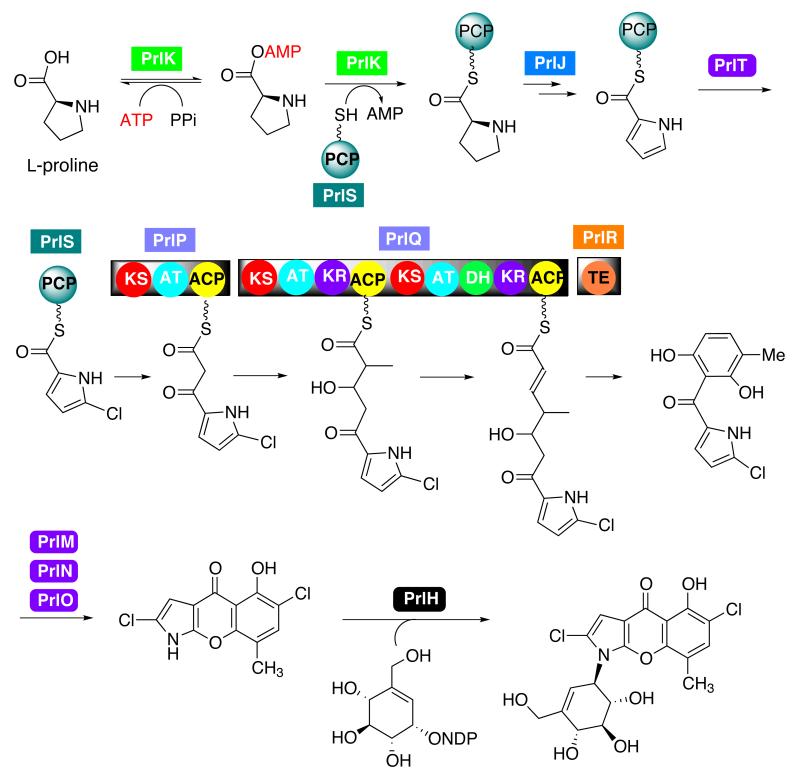
The biosynthesis of the aglycone portion of pyralomicin is proposed to take place via a suite of genes that share high identity with both the pyrrolomycin and pyoluteorin biosynthetic genes (Figure 3, Table S1).3, 24 These include genes encoding a putative nonribosomal peptide synthetase (NRPS) (PrlK), an acyl-CoA dehydrogenase (PrlJ), four halogenases (PrlM, PrlN, PrlO, and PrlT), two polyketide synthases (PrlP and PrlQ), an FAD reductase (PrlL), a stand-alone PCP domain (PrlS) and a stand-alone type II thioesterase (PrlR).
Biosynthesis of the aglycone is thought to be initiated through the activation of a proline residue via PrlK and covalent linkage to the small stand-alone PCP domain, PrlS (Scheme 2). Once tethered to the PCP domain through a thioester linkage, the proline residue is proposed to be modified by the PrlJ dehydrogenase and halogenated by PrlT to yield a chlorinated pyrrole intermediate. This is similar and consistent with the mechanism of proline activation and halogenation shown by Walsh and co-workers for pyoluteorin biosynthesis.25 PrlK shares high identity with other known NRPS adenylation domains including the Pyr8 (64% identity) and PltF (51% identity) acyl-CoA ligases from the pyrrolomycin and pyoluteorin pathways, respectively, and with the acyl-CoA ligase, CouN4 from the coumermycin pathway.3, 24, 26
Scheme 2.
Proposed mode of formation of the NRPS-PKS-derived backbone and attachment of the C7-cyclitol unit.
PrlT shares higher identity with Pyr17 (73% identity; from the pyrrolomycin pathway) and PltA (59% identity; from the pyoluteorin cluster) than with any of the other halogenases within these clusters, suggesting that this enzyme will function in the halogenation of the PCP-bound pyrrolyl intermediate. Extension of the halogenated pyrrolyl-intermediate is predicted to occur via the action of three PKS modules housed in PrlP and PrlQ. The nascent aglycone would then be released from the enzyme bound intermediate via the type II thioesterase, PrlR (Scheme 2). The remaining prl halogenases, PrlM, PrlN, and PrlO, demonstrate high homology with the pyrrolomycin halogenases Pyr11 (53% identity/67% similarity), Pyr17 (53% identity/71% similarity), and Pyr16 (47% identity/65% similarity) respectively, and are proposed to mediate the halogenation of the PKS-derived phenyl ring (Scheme 2).
Although the core structure of pyralomicin is similar to pyoluteorin and pyrrolomycin, it differs from the latter compounds in that a unique cyclization reaction involving the rearrangement of the proline residue occurs to yield a benzopyranopyrrole core. Interestingly, comparison of the pyralomicin biosynthetic gene cluster with the pyoluteorin and pyrrolomycin clusters has not revealed a novel enzyme candidate to mediate the proline rearrangement and ring cyclization. However, the pyralomicin cluster contains a total of four ORFs encoding putative halogenase enzymes, despite the pyralomicins described to date containing only two or three halogen atoms. Therefore, it is possible that one of those putative halogenase enzymes is involved in the formation of the benzopyranopyrrole core structure. There is precedence in the literature for cryptic halogenation pathways that lead to the formation of novel biosynthetic features. For example, in coronamic acid biosynthesis the CmaB halogenase mediates the chlorination of allo-isoleucine which is further modified by an intramolecular gamma-elimination of the chlorine atom to yield cyclopropyl ring formation.27 A similar reaction has been shown to occur during the biosynthesis of the kutzneride family of hexapeptidolactones.28 Within this pathway, KtzD mediates a cryptic halogenation that is further modified to a cyclopropyl intermediate by KtzA. Thus, during pyralomicin biosynthesis we predict that a FADH2-dependent halogenase is involved in proline ring rearrangement and the cyclization leading to the formation of the benzopyranopyrrole core structure (Scheme S1).
The hypothetical mechanism of these halogenases implies that the chlorination involves formation of hypochlorous acid (HOCl) or a more stable Lys-eNH-Cl (lysine chloramine) from reaction of HOCl with the active site residue lysine. The intramolecular migration of the carbonyl group of the pyrrole ring and the benzopyranopyrrole ring formation in pyralomicin and TAN-876A biosyntheses could well be initiated by the same type of reaction catalyzed by the halogenases (Scheme S1). Alternatively, the rearrangement reaction may be initiated by epoxidation of the pyrrole ring. However, no putative tailoring genes for such a reaction can be found within the pyralomicin gene cluster, suggesting that the latter mechanism is less likely. Overall, the ring cyclization event represents an unprecedented reaction mechanism that has not previously been described in other known biosynthetic pathways.
Attachment of the C7-cyclitol unit to the benzopyranopyrrole core
The pyralomicins are unique in that they are the only known natural products containing a pseudosugar (C7-cyclitol) moiety together with a PKS-derived aglycone. Many C7N-aminocyclitol-containing compounds, including acarbose and validamycin A, resemble oligosaccharides. Thus, the biosynthetic mechanism utilized by N. spiralis to incorporate this unique pseudosugar into an aglycone core structure is of interest.
The co-occurrence of another benzopyranopyrrole antibiotic TAN-876A and closely related TAN-876B (both contains no sugar or cyclitol) in Streptomyces sp. C-70899 suggests that the cyclitol attachment takes place at a late stage in pyralomicin biosynthesis.7 This reaction may be catalyzed by the putative N-glycosyltransferase enzyme, PrlH. PrlH has high homology (63% identity/73% similarity) with RebG, the N-glycosyltransferase from the rebeccamycin pathway.19 RebG is responsible for transferring D-glucose to an indolocarbazole aglycone core, which retains structural features that is similar to the benzopyranopyrrole core of pyralomicin (Figure 1).
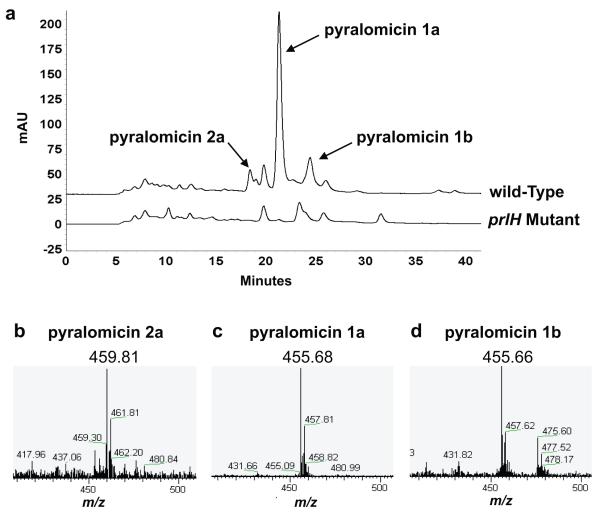
To test the involvement of prlH in pyralomicin biosynthesis, we inactivated the gene in N. spiralis. A 697 bp BamHI-ApaI fragment of the prlH gene was cloned into the pBsk vector. An apramycin resistance cassette and the oriT loci from pOJ446 were subsequently cloned into the pBsk vector backbone to give pKO3A4. Single crossover recombination of the prlH deletion plasmid pKO3A4 was achieved using conjugation of N. spiralis with E. coli ET12567 transformed with pKO3A4 and the pUB307 conjugation helper plasmid. The mutant was genotypically confirmed by PCR (Figure S1) and was grown side-by-side with wild-type N. spiralis in the pyralomicin production medium. Analysis of extracts from the prlH mutant strain using LC-MS shows that the mutant no longer produces the suite of pyralomicin analogues present in the wild-type strain (Figure 4). This provides evidence that prlH is essential for the biosynthesis of pyralomicin and that a single glycosyltransferase is required for the addition of either the glucose or cyclitol moiety into the pyralomicin aglycone core structure. However, neither the aglycone nor the cyclitol moiety could be detected in the culture broths of the mutant. This lack of aglycone accumulation may not be due to a polar effect, as the nucleotidyltransferase gene prlG is the only downstream gene present within the same operon as prlH. Rather, it may be due to either negative feedback inhibition of the intermediate(s) production or the lack of a suitable transport system for the aglycone. Nevertheless, the results confirm the critical function of PrlH in pyralomicin biosynthesis.
Figure 4.
Analysis of prlH deletion mutant. (a) HPLC elution peaks were monitored at 355 nm. (b-d) Mass spectral analysis of pyralomicin 1a (m/z 455.68 [M+H]+), 1b (m/z 455.68 [M+H]+), and 2a (m/z 459.81 [M+H]+), respectively.
In conclusion, we have cloned, sequenced, and identified the pyralomicin biosynthetic gene cluster from N. spiralis IMC A-0156. The cluster contains 27 ORFs predicted to encode all of the functions for pyralomicin biosynthesis. Those include the NRPS and PKS enzymes, which are required for the formation of the benzopyranopyrrole core unit, a suite of tailoring enzymes, as well as regulatory and transport proteins. Targeted disruption of the gene encoding the N-glycosyltransferase, prlH, abolished pyralomicin production and recombinant expression of PrlA confirms the activity of this enzyme as a sugar phosphate cyclase (SPC) involved in the formation of the C7-cyclitol moiety. Further studies are necessary to dissect the biosynthetic mechanisms of the unique ring cyclization and proline rearrangement and the attachment of the cyclitol unit to the benzopyranopyrrole core moiety.
MATERIALS AND METHODS
General Experimental Procedures
Low-resolution electrospray ionization (ESI) mass spectra were recorded on a ThermoFinnigan Liquid Chromatograph-Ion Trap Mass Spectrometer. Gas Chromatography-Mass Spectrometry was performed on a Hewlett Packard 5890 SERIES II Gas Chromatograph and a Hewlett Packard 5971 SERIES Mass Selective Detector. High Performance Liquid Chromatography was carried out on a C18 column (C18 Symmetry 10 × 150 mm, Waters) with the detector set at 355 nm. Thin layer chromatography was performed on aluminum sheets with silica gel 60 F254 (EMD chemicals Inc.).
Bacterial strains and culture conditions
E. coli strains DH10B, EPI300, ET12567, and BL21(DE3)pLysS were grown in LB Miller broth with appropriate supplements as indicated. Gene expression was conducted using LBBS (LB Miller broth containing 2.5 mM betaine and 1M sorbitol) supplemented with appropriate antibiotics.
Nonomuraea spiralis IMC A-0156 was kindly provided by Drs. Hiroshi Naganawa and Yoshikazu Takahashi at the Institute of Microbial Chemistry, Japan. N. spiralis was maintained on BTT agar (1% D-glucose, 0.1% yeast extract, 0.1% beef extract, 0.2% casitone, and 1.5% Bacto-agar, pH 7.4 prior to sterilization) at 30 °C. Spore formation in N. spiralis was induced by growth on modified minimal medium agar (0.05% L-asparagine, 0.05% K2HPO4, 0.02% MgSO4·7H2O, 1% D-glucose, 1.5% Bacto-agar, pH 7.4 prior to sterilization) at 30 °C for 1 to 2 weeks. For the production of pyralomicins, N. spiralis was grown in seed medium (2% D-galactose, 2% Dextrin, 1% Bacto-soytone, 0.5% corn steep liquor, 0.2% (NH4)2SO4, 0.2% CaCO3, a drop of silicone oil, pH 7.4 prior to sterilization) at 30 °C for 5-7 days. The seed culture (10%) was then innoculated into production medium (3% potato starch, 1.5% soy flour, 0.5% corn steep liquor, 0.2% yeast extract, 0.05% MgSO4·7H2O, 0.3% NaCl, 0.001% CoCl2, 0.3% CaCO3, 1-2 drops silicone oil, pH 7.4 prior to sterilization) and the culture was shook 200 rpm at 30 °C for an additional 5-7 days. Production of the pyralomicins corresponded to a light pink color change in the bacterial mycelium and a purple to wine coloration within the production medium. Growth of N. spiralis in RARE3 medium (1% D-glucose, 0.4% yeast extract, 1% malt extract, 0.2% Bacto-peptone, 0.2% MgCl2, 0.5% glycerol, pH 7.4 prior to sterilization) was used for conjugation experiments.
PCR amplification of 2-epi-5-epi-valiolone synthase homologues from N. spiralis
Degenerate primers, ValA-1S, ValA-2S, and ValA-2R (Table S2), were designed from sequence alignments of sugar phosphate cyclases, including the 2-epi-5-epi-valiolone synthases ValA from the validamycin pathway and AcbC from the acarbose pathway. Amplification using ValA-2S and ValA-1R were successful in PCR amplifying a homologue sequence from N. spiralis. The Promega Wizard Kit was used to isolate genomic DNA from N. spiralis according to manufacturer’s specifications. Promega GoTaq was used for PCR amplification reactions according to manufacturer’s specifications. 100 ng of template DNA was heated at 70 °C for 10 min prior to addition to the PCR reaction mixtures. PCR amplifications were conducted with 1 cycle at 95 °C for 7 min followed by 5 cycles of 95 °C for 30 sec, 42 °C for 30 sec, and 72 °C for 1 min, followed by 30 cycles of 95 °C for 30 sec, 48 °C for 30 sec, and 72 °C for 1 min, followed by 1 cycle of 72 °C for 5 min. Samples were held at 4 °C until they were analyzed by gel electrophoresis. PCR products of approximately 400 base pairs were gel purified, subcloned into pBsk (Stratagene), and submitted for DNA sequencing at the Center for Genome Research and Biocomputing (CGRB) at Oregon State University.
Construction and screening of the N. spiralis genomic library
N. spiralis was grown for 5 days in seed medium. Ten milliliters of culture were centrifuged at 1,400 × g and the cell pellet was utilized to isolate genomic DNA using a modified cetyl trimethylammonium bromide (CTAB) method. The cell pellet was washed with 10 mL of 10.3% sucrose, repelleted and the supernatant discarded. The pellet was washed twice with 10 mM Tris, 1.0 mM EDTA buffer, pH 7.4, repelleted by centrifugation and the supernatant discarded. The cell pellet was resuspended in 6 mL of lysis buffer (10 mM Tris pH 8.0, 25% sucrose, 0.08 mM EDTA, and 50 mg of lysozyme). The sample was incubated at 37 °C for 90 min. 100 mL of proteinase K (20 mg mL−1) and 3.6 mL of 10% SDS was added and the solution was incubated for an additional 90 min at 37 °C. 1.6 mL of CTAB (10%) was added to the solution and the solution was incubated at 65 °C for 10 min. An equal volume of 1:1 phenol-chloroform pH 8.0 was added to the solution and mixed by inversion until a homogenous emulsion was created. The solution was centrifuged and the aqueous layer removed to a fresh tube where it was extracted in phenol-chloroform for a second time. The aqueous solution was removed to a fresh tube and extracted using an equal volume of chloroform. The aqueous layer was removed, 1/10 volume of 3 M sodium acetate was added and the DNA was precipitated by adding one volume of room temperature isopropanol. The resulting DNA was spooled from the mixture and resuspended in 500 μl of sterile H2O.
The copy control fosmid library kit from Epicentre was utilized to prepare a genomic library using the N. spiralis DNA. Briefly, 20 μg of isolated DNA was end-filled and gel purified to recover 40 kb DNA fragments. N. spiralis DNA fragments were ligated into the fosmid vector, pCC1Fos, overnight at 16 °C. The ligation extract was packaged into lambda phage using the Gigapack III gold packaging extract from Stratagene according to manufacturer’s specifications. Phage extracts were used to infect competent E. coli EPI300 cells grown in LB Miller containing 10 mM MgCl2.
The library of approximately 5000 colonies was replica plated into liquid culture using the Genetix QPix2 colony picker into sets containing two 96-well plate replicates. All of the plates were grown overnight at 37 °C. One set of plates was used to make glycerol stock cultures (10% glycerol) that were stored at −80 °C. The second set of liquid plates was used to replica plate the library onto 150 mm agar plates containing 100 μl of copy control induction solution (Epicentre). Agar plates were grown at 37 °C overnight and were lifted onto N-hybond nitrocellulose membrane (Amersham). Briefly, the colony lifts were performed on prechilled agar plates. The nitrocellulose membrane was placed on the agar for 1 min and marked using a needle to retain orientation. The membrane was then floated in succession in three solutions for 15 min each: Denaturation solution (0.5 M NaOH, 1.5 M NaCl); Neutralization solution (1.5 M NaCl, 1.0 M Tris-HCl, pH 7.4); and 2×SSC (0.3 M NaCl, 0.03 M Na-citrate, pH 7.0). After the last incubation, the blots were crosslinked using UV light in a UV crosslinker at 1200 watts. After crosslinking the blots were incubated for 1 h at 37 °C with proteinase K solution (2.5 mg mL−1). The blots were then washed liberally with water, dried on Whatman paper overnight and stored at 4 °C until used for hybridization.
The lifts were hybridized in succession with the 2-epi-5-epi-valiolone homologue, prlA, and the pyoluteorin halogenase gene, pltA (kindly provided by Dr. Joyce Loper) in DIG-Easy Hyb Solution (Roche) containing DIG-labeled DNA probe (15 ng mL−1) overnight at 50 °C. After each round of hybridization the blots were washed twice with 2×SSC/0.1% SDS at room temperature for 5 min each and then subsequently washed twice with 0.5×SSC/0.1% SDS at 50 °C for 15 min each wash. The blots were then incubated in blocking solution [0.1 M maleic acid, 0.15 M NaCl, pH 7.5, 1× blocking reagent (Roche) and 5% non-fat dry milk] for 1 h at room temperature. The blots were incubated in blocking solution containing 1:10,000 dilution anti-dig monoclonal Ab for 30-45 min at room temperature. The blots were then washed three times for 15 min in wash buffer (0.1 M maleic acid, 0.15 M NaCl, and 0.3% Tween® 20, pH 7.5). The blots were briefly rinsed in detection buffer (0.1 M Tris, 0.1 M NaCl, pH 9.5) and incubated for 10 min in detection buffer containing 0.025 mM CSPD (Roche) at 37 °C. The blots were then put down to film for 30 min.
Positive colonies were inoculated into 20 mL LB cultures with chloramphenicol (12.5 μg mL−1) and grown overnight at 37 °C. Following overnight incubation, 20 μl of copy control induction solution, was added to the remaining culture. The cultures were then incubated for 5 h at 37 °C before they were harvested using the Bio-Rad miniprep kit. The resulting DNA was digested with BamHI and used to run a Southern analysis. Briefly, digested DNA samples were separated using agarose gel electrophoresis. DNA was transferred to N-hybond nitrocellulose membrane using capillary transfer. Southern membranes were hybridized and analyzed in the same manner as colony lifts from the library. Both DNA strands from three overlapping library clones housing either one or both of the screening probes were sequenced by a combination of chromosome walking and shotgun pyrosequencing. The 41-kb putative pyralomicin biosynthetic gene cluster was reconstituted using BioEdit software.
Expression of prlA and characterization of the recombinant enzyme
PCR was used to clone prlA into the pRSET B expression plasmid (Invitrogen) in frame with the 6XHis-tag at the BamHI/KpnI restriction sites. Primers PrlA-F2 and PrlA-R2 (Table S2) were used during the PCR amplification step. Resulting clones were confirmed by DNA sequencing and were subsequently transferred into E. coli BL21(DE3)pLysS. Transformed bacteria were grown in LBBS containing ampicillin (100 μg mL−1) and chloramphenicol (25 μg mL−1) at 37 °C until the O.D.600 reached 1.0 to 1.2. At this point the culture temperature was reduced to 25 °C and the expression of prlA was induced by the addition of 0.3 mM IPTG. Cultures were incubated at 25 °C for 15 h and bacterial cell pellets collected by centrifugation at 1,400 × g. Bacterial cell pellets were lysed in K2HPO4/KH2PO4 (25 mM), NaCl (300 mM), pH 7.4, containing 0.2 mM CoCl2 and the protease inhibitor 4-(2-aminoethyl)-benzenesulfonylfluoride (AEBSF) (200 μg mL−1). The cell lysate was sonicated for two 30 sec bursts on a Microson Ultrasonic Cell Disruptor XL on setting 6, and clarified by centrifugation to yield the cell free extract. This mixture was used directly in the cell-free protein assay or the enzyme was further purified using Ni-NTA agarose beads according to manufacturer’s specifications (Qiagen). The purified protein was dialyzed in 2 L dialysis buffer [K2HPO4/KH2PO4 pH 7.5 (25 mM), CoCl2 (0.2 mM)] and visualized using 10% SDS-PAGE followed by Coomassie staining.
Enzyme activity was analyzed by the addition of cell-free extract or purified protein to the reaction mixture, as described below. The enzyme assay (100 μl volume) was performed at 30 °C for 2 h in 25 mM phosphate buffer (pH 7.5) containing 50 μl of protein (0.3 mg mL−1), 5 mM substrate, 0.05 mM CoCl2, 1 mM NAD+ and 2 mM KF. The substrate, aminoDAHP was a gift from Prof. Heinz G. Floss. The remaining substrates, including sedoheptulose 7-phosphate, DAHP, glucose 6-phosphate, fructose 6-phosphate, mannose 6-phosphate, and ribose 5-phosphate, were purchased from Sigma. After incubation, samples were visualized using silica gel thin layer chromatography (TLC) (butanol-ethanol-water 9:7:4), and the substrate and reaction product were detected as blue spots with a cerium sulfate/phosphomolybdate-containing reagent. The reaction mixture was lyophilized and the reaction products were extracted with methanol. After drying, the extracts were mixed with drops of SIGMA SIL-A and dried under argon. The products were reextracted with n-hexane and analyzed by GC-MS.
Construction of the prlH deletion mutant
Construction of a prlH suicide vector to use in single crossover recombination experiments was designed using the pBsk cloning vector (Stratagene). A 697 base pair BamHI-ApaI fragment of the prlH gene was cloned into the pBsk vector for single crossover recombination into N. spiralis. To identify mutant colonies, an apramycin resistance cassette from pOJ446 was cloned downstream of the partial prlH gene into the pBsk vector backbone at the KpnI restriction site. In addition, the 800 base pair oriT loci from pOJ446 was also cloned into the knockout vector at the NotI restriction site to enable conjugal transfer of the vector using the helper plasmid pUB307. The completed prlH knockout vector is referred to as pKO3A4.
Single crossover recombination of the prlH deletion plasmid pKO3A4 was achieved using conjugation of N. spiralis with E. coli ET12567 transformed with pKO3A4 and the pUB307 conjugation helper plasmid. N. spiralis was inoculated and grown in RARE3 medium at 30 °C for 5-7 days. The night before conjugation, N. spiralis cultures were re-inoculated at a 1:10 dilution into RARE3 medium and grown overnight at 30 °C. An overnight culture of E. coli ET12567 transformed with pKO3A4 and pUB307 was inoculated into LB Miller broth supplemented with thiamine hydrochloride (25 μg mL−1), 0.1% D-glucose, 25 mM MOPS, chloramphenicol (25 μg mL−1), kanamycin (50 μg mL−1), and apramycin (50 μg mL−1) and incubated at 37 °C. On the day of conjugation, cultures of N. spiralis and E. coli cultures were centrifuged at 1,400 × g and the supernatant removed. The N. spiralis cells were resuspended directly in 1/10 volume of RARE3. The E. coli cells were washed 3 times in RARE3 medium to remove antibiotics and traces of LB Miller broth and were then resuspended in 1/10 volume of RARE3 medium. Aliquots of 500 μl of E. coli and 500 μl of N. spiralis were mixed and incubated without shaking for 16, 24, 48, or 72 h at 30 °C. Following incubation, 200 μl aliquots of 1-fold, 10-fold, and 100-fold dilutions were plated onto BTT agar containing apramycin (50 μg mL−1) and nalidixic acid (25 μg mL−1). Growth of mutant strains was evaluated after 4-6 weeks incubation at 30 °C. Mutant colonies were restreaked onto BTT agar containing apramycin (50 μg mL−1) and nalidixic acid (25 μg mL−1). Growth of mutant strains in RARE3 liquid broth was used to make 10% glycerol stocks that were stored at −80 °C.
Analysis of prlH deletion mutant
Wild-type and prlH deletion strains of N. spiralis were cultured for 5-7 days in seed medium at 28 °C. Seed cultures were subsequently diluted 1:10 into production medium and grown for an additional 5-7 days. Upon harvesting, cultures of wild-type and prlH deletion strains of N. spiralis were acidified to pH 3 using HCl. Bacterial mycelia and other precipitates were removed from the culture medium via centrifugation at 1,400 × g. The acidified culture medium was extracted three times with an equal volume of butyl acetate. Butyl acetate fractions were dried by the addition of sodium sulfate, clarified by filtration, and reduced using rotary evaporation. Resulting extracts were analyzed using LC-MS on a Waters C18 Symmetry Prep column with a 25% to 100% MeOH gradient. Peaks were monitored at 355 nm. Peaks corresponding to pyralomicin metabolites were identified using electrospray ionization mass spectrometry.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Drs. Hiroshi Naganawa and Yoshikazu Takahashi at the Institute of Microbial Chemistry for providing N. spiralis IMC A-0156, and Micah Walters for technical assistance. The project described was supported by grant R01AI061528 from the National Institute of Allergy and Infectious Diseases and by the Herman-Frasch Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health (NIH).
Footnotes
ASSOCIATED CONTENT
Tables S1 and S2, Figure S1, and Scheme S1. This material is available free of charge via the Internet at http://pubs.acs.org.
The GenBank accession number for the sequence of the pyralomicin gene cluster is JX424761.
REFERENCES AND NOTES
- (1).Kawamura N, Kinoshita N, Sawa R, Takahashi Y, Sawa T, Naganawa H, Hamada M, Takeuchi T. J. Antibiot. 1996;49:706–709. doi: 10.7164/antibiotics.49.706. [DOI] [PubMed] [Google Scholar]
- (2).Kawamura N, Sawa R, Takahashi Y, Isshiki K, Sawa T, Naganawa H, Takeuchi T. J. Antibiot. 1996;49:651–656. doi: 10.7164/antibiotics.49.651. [DOI] [PubMed] [Google Scholar]
- (3).Nowak-Thompson B, Chaney N, Wing JS, Gould SJ, Loper JE. J. Bacteriol. 1999;181:2166–2174. doi: 10.1128/jb.181.7.2166-2174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ezaki N, Shomura T, Koyama M, Niwa T, Kojima M, Inouye S, Ito T, Niida T. J. Antibiot. 1981;34:1363–1365. doi: 10.7164/antibiotics.34.1363. [DOI] [PubMed] [Google Scholar]
- (5).Kaneda M, Nakamura S, Ezaki N, Iitaka Y. J. Antibiot. 1981;34:1366–1368. doi: 10.7164/antibiotics.34.1366. [DOI] [PubMed] [Google Scholar]
- (6).Hughes CC, Prieto-Davo A, Jensen PR, Fenical W. Org. Lett. 2008;10:629–631. doi: 10.1021/ol702952n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Funabashi Y, Takizawa M, Tsubotani S, Tanida S, Harada S, Kamiya K. J. Takeda Res. Lab. 1992;51:73–89. [Google Scholar]
- (8).Kawamura N, Sawa R, Takahashi Y, Sawa T, Naganawa H, Takeuchi T. J. Antibiot. 1996;49:657–660. doi: 10.7164/antibiotics.49.657. [DOI] [PubMed] [Google Scholar]
- (9).Naganawa H, Hashizume H, Kubota Y, Sawa R, Takahashi Y, Arakawa K, Bowers SG, Mahmud T. J. Antibiot. 2002;55:578–584. doi: 10.7164/antibiotics.55.578. [DOI] [PubMed] [Google Scholar]
- (10).Asamizu S, Xie P, Brumsted CJ, Flatt PM, Mahmud T. J. Am. Chem. Soc. 2012;134:12219–12229. doi: 10.1021/ja3041866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Balskus EP, Walsh CT. Science. 2010;329:1653–1656. doi: 10.1126/science.1193637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wu X, Flatt PM, Schlorke O, Zeeck A, Dairi T, Mahmud T. ChemBioChem. 2007;8:239–248. doi: 10.1002/cbic.200600446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Stratmann A, Mahmud T, Lee S, Distler J, Floss HG, Piepersberg W. J. Biol. Chem. 1999;274:10889–10896. doi: 10.1074/jbc.274.16.10889. [DOI] [PubMed] [Google Scholar]
- (14).Yu Y, Bai L, Minagawa K, Jian X, Li L, Li J, Chen S, Cao E, Mahmud T, Floss HG, Zhou X, Deng Z. Appl. Environ. Microbiol. 2005;71:5066–5076. doi: 10.1128/AEM.71.9.5066-5076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Choi WS, Wu X, Choeng YH, Mahmud T, Jeong BC, Lee SH, Chang YK, Kim CJ, Hong SK. Appl. Microbiol. Biotechnol. 2008;80:637–645. doi: 10.1007/s00253-008-1591-2. [DOI] [PubMed] [Google Scholar]
- (16).Mahmud T, Tornus I, Egelkrout E, Wolf E, Uy C, Floss HG, Lee S. J. Am. Chem. Soc. 1999;121:6973–6983. [Google Scholar]
- (17).Nolan M, Sikorski J, Jando M, Lucas S, Lapidus A, Glavina Del Rio T, Chen F, Tice H, Pitluck S, Cheng JF, Chertkov O, Sims D, Meincke L, Brettin T, Han C, Detter JC, Bruce D, Goodwin L, Land M, Hauser L, Chang YJ, Jeffries CD, Ivanova N, Mavromatis K, Mikhailova N, Chen A, Palaniappan K, Chain P, Rohde M, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. Stand. Genomic Sci. 2010;2:29–37. doi: 10.4056/sigs.631049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wehmeier UF, Piepersberg W. Appl. Microbiol. Biotechnol. 2004;63:613–625. doi: 10.1007/s00253-003-1477-2. [DOI] [PubMed] [Google Scholar]
- (19).Sanchez C, Butovich IA, Brana AF, Rohr J, Mendez C, Salas JA. Chem. Biol. 2002;9:519–531. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- (20).Singh S, McCoy JG, Zhang C, Bingman CA, Phillips GN, Jr., Thorson JS. J. Biol. Chem. 2008;283:22628–22636. doi: 10.1074/jbc.M800503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS. ChemBioChem. 2006;7:795–804. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]
- (22).Gras SL, Mahmud T, Rosengarten G, Mitchell A, Kalantar-Zadeh K. ChemPhysChem. 2007;8:2036–2050. doi: 10.1002/cphc.200700222. [DOI] [PubMed] [Google Scholar]
- (23).Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, Zhang J, Gao AL, Liu CX, Zhu ZX, Xiang WS. J. Bacteriol. 192:4526–4527. doi: 10.1128/JB.00596-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhang X, Parry RJ. Antimicrob. Agents Chemother. 2007;51:946–957. doi: 10.1128/AAC.01214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Thomas MG, Burkart MD, Walsh CT. Chem. Biol. 2002;9:171–184. doi: 10.1016/s1074-5521(02)00100-x. [DOI] [PubMed] [Google Scholar]
- (26).Xu H, Wang ZX, Schmidt J, Heide L, Li SM. Mol. Genet. Genomics. 2002;268:387–396. doi: 10.1007/s00438-002-0759-1. [DOI] [PubMed] [Google Scholar]
- (27).Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- (28).Neumann CS, Walsh CT. J. Am. Chem. Soc. 2008;130:14022–14023. doi: 10.1021/ja8064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.