Abstract
The periodontal pathogen Porphyromonas gingivalis has two different lipopolysaccharides (LPSs) designated O-LPS and A-LPS, which are a conventional O-antigen polysaccharide and an anionic polysaccharide that are both linked to lipid A-cores, respectively. However, the precise mechanisms of LPS biosynthesis remain to be determined. In this study, we isolated a pigment-less mutant by transposon mutagenesis and identified that the transposon was inserted into the coding sequence PGN_2005, which encodes a hypothetical protein of P. gingivalis ATCC 33277. We found that (i) LPSs purified from the PGN_2005 mutant were shorter than those of the wild type; (ii) the PGN_2005 protein was located in the inner membrane fraction; and (iii) the PGN_2005 gene conferred Wzz activity upon an Escherichia coli wzz mutant. These results indicate that the PGN_2005 protein, which was designated WzzP, is a functional homolog of the Wzz protein in P. gingivalis. Comparison of amino acid sequences among WzzP and conventional Wzz proteins indicated that WzzP had an additional fragment at the C-terminal region. In addition, we determined that the PGN_1896 and PGN_1233 proteins and the PGN_1033 protein appear to be WbaP homolog proteins and a Wzx homolog protein involved in LPS biosynthesis, respectively.
Keywords: LPS biosynthesis, periodontal pathogen, WbaP protein, Wzx protein, Wzz protein
Introduction
Porphyromonas gingivalis is a Gram-negative anaerobic bacterium considered a major etiological agent in chronic periodontitis (Haffajee and Socransky 1994) and may be associated with systemic conditions, such as cardiovascular diseases (Pussinen and Mattila 2004), preterm low birth weight (Madianos et al. 2001), and rheumatoid arthritis (Lundberg et al. 2010). The surface components of P. gingivalis, such as the major cell surface macromolecules including exopolysaccharide (EPS), lipopolysaccharide (LPS), and capsular polysaccharide (CPS) (or K-antigen) and the extracellular/surface cysteine proteases including Arg-gingipain (Rgp) and Lys-gingipain (Kgp), are considered important virulence factors in the pathogenesis of periodontal diseases (Lamont and Jenkinson 1998).
For many bacteria, surface polysaccharides play critical roles in immune modulation and evasion (Comstock and Kasper 2006). As surface polysaccharides constitute the outer layer of the outer membrane and form a defensive barrier against the host's immune system, their highly antigenic variations can lead to strain-specific properties. Porphyromonas gingivalis has two distinct polysaccharides on the cell surface: LPSs and CPSs (Arndt and Davey 2010). LPS consists of three general components: O-antigen polysaccharide, core oligosaccharide, and lipid A. Paramonov et al. (2001) demonstrated that the O-antigen of P. gingivalis strain W50 consists of a tetrasaccharide repeating unit composed of (6)-α-d-Glcp-(1-4)-α-l-Rhap-(1-3)-β-d-GalNAc-(1-3)-α-d-Galp-(1). In addition, it was recently shown that P. gingivalis synthesizes another surface polysaccharide, which is distinct from O-LPS and capsular polysaccharide (Paramonov et al. 2005; Aduse-Opoku et al. 2006; Rangarajan et al. 2008). Initially, the anionic polysaccharide (APS) was thought to be associated with the cell envelope through an unknown mechanism. As the APS was found to be anchored to the cell surface by lipid A, it was categorized as an LPS molecule and designated A-LPS (Rangarajan et al. 2008). Curtis et al. (1999) obtained a monoclonal antibody (mAb 1B5) that was originally raised against the catalytic domain of RgpA protease. The mAb 1B5 cross-reacts with A-LPS and recognizes a phosphorylated branched mannan in the APS repeating unit (Paramonov et al. 2005).
Our previous study indicated that a porR gene encoding a putative aminotransferase plays a role in colonial pigmentation on blood agar plates. A porR mutant presented a decrease of cell-associated Rgp and Kgp activities and no reduction of secreted Rgp or Kgp activity, and the mAb 1B5 did not recognize any products of the porR mutant (Shoji et al. 2002). Furthermore, mutants of vimA, vimE, vimF (Vanterpool et al. 2005a,b), wbpB (Slaney et al. 2006), rfa (Sato et al. 2009), waaL (Rangarajan et al. 2008), wzy (Paramonov et al. 2009), gtfB (Yamaguchi et al. 2010), PGN_0242 and PGN_0663 (Shoji et al. 2011) also exhibited no immunoreaction to mAb 1B5, indicating that these genes as well as porR are involved in the A-LPS biosynthesis. porR and wbpB are predicted to be involved in the initial synthesis of structural sugar(s) within APS. The rfa gene is thought to be involved in the synthesis of the core oligosaccharide of LPS (Sato et al. 2009). The vimA, vimE, and vimF genes play a role in the regulation of gingipain activities, but their precise roles in A-LPS biosynthesis are still unknown. Further study is therefore needed to identify the other factors that are required for the biosynthesis of both A-LPS and O-LPS.
The study of porR indicated that gingipains and hemagglutinin proteins are linked to the A-LPS, suggesting that A-LPS plays a critical role in the anchorage of cell surface virulence factors (Shoji et al. 2002). The gingipains and hemagglutinin proteins possess a conserved C-terminal domain (CTD) in their primary sequences. We recently demonstrated that CTD-containing proteins are secreted onto the cell surface via the Por secretion system (PorSS)/Type IX secretion system (T9SS) (Sato et al. 2010; Shoji et al. 2011; Sato et al. 2013; McBride and Zhu 2013). Among the CTD proteins, RgpB (Nguyen et al. 2007), TapA (Kondo et al. 2010), HBP35 (Shoji et al. 2010, 2011), and CPG70 (Chen et al. 2011) have been shown to form diffuse bands on an SDS-PAGE (sodium dodecyl sulfate polyacrylamid gel electrophoresis) gel, suggesting that they are linked to A-LPS. To understand the pathogenesis of P. gingivalis, it is important to analyze not only A-LPS biosynthesis but also the glycosylated form of CTD proteins.
In LPS biosynthesis pathways, the lipid A-core and the O-antigen are independently assembled on the cytoplasmic side of the inner membrane. Subsequently, these molecules are separately translocated to the periplasmic side of the inner membrane. Next, the lipid A-core and the O-antigen are joined covalently by the O-antigen ligase, WaaL. The mature LPS is transported onto the cell surface by the LPS transport proteins (Sperandeo et al. 2009). Three steps that are important for LPS biosynthesis are completed by the inner membrane proteins. First, the initiating enzyme, such as WbaP or WecA, links a sugar residue with one phosphate onto an undecaprenyl monophosphate (UndP) at the cytoplasmic side of the inner membrane. Then, further sugar residues are sequentially added by glycosyl transferases using nucleotide-activated sugars as substrates. After UndPP-glycans are assembled, these are transported either via the Wzx-like flippases or the adenosine triphosphate (ATP)-binding cassette transporters (ABC transporters), such as the Wzt and Wzm proteins. Finally, polysaccharides from the UndPP carrier, which are polymerized by Wzy and Wzz proteins, are transferred on terminal sugar residues of the lipid A-core by an O-antigen ligase, WaaL. It has been shown that PGN_1242 and PGN_1302 correspond to the Wzy and WaaL proteins, respectively (Rangarajan et al. 2008; Paramonov et al. 2009; Haurat et al. 2011). However, other responsible genes encoding initiation enzymes for UndP, UndPP glycans transporters, or a Wzz protein that is critical for LPS biosynthesis have yet to be identified.
In this study, we found that the P. gingivalis PGN_2005 protein is a functional homolog of the Wzz protein. Our results also suggest that the PGN_1896 and PGN_1233 proteins and the PGN_1033 protein play roles as, respectively, the WbaP homolog proteins and the Wzx homolog protein involved in P. gingivalis LPS biosynthesis.
Experimental Procedures
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Tables S2 and S3, respectively.
Media and conditions for bacterial growth
Porphyromona gingivalis strains were grown anaerobically (80% N2, 10% CO2, 10% H2) in enriched brain–heart infusion (BHI) broth (Becton Dickinson, Franklin Lakes, NJ) or on enriched tryptic soy (TS) agar plates (Nissui, Tokyo, Japan) supplemented with 5 μg/mL hemin (Sigma, St. Louis, MO) and 0.5 μg/mL menadione (Sigma). For blood agar plates, defibrinated laked sheep blood was added to enriched tryptic soy agar at 5%. Luria-Bertani (LB) broth and LB agar plates were used for growth of Escherichia coli strains. Antibiotics were used at the following concentrations: ampicillin (Ap; 100 μg/mL for E. coli, 10 μg/mL for P. gingivalis), erythromycin (Em; 10 μg/mL for P. gingivalis), gentamycin (Gm; 50 μg/mL for P. gingivalis), kanamycin (Km; 30 μg/mL for E. coli), spectinomycin (Sp; 80 μg/mL for E. coli), and tetracycline (Tc; 0.7 μg/mL for P. gingivalis).
Chemicals
The proteinase inhibitors Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) and iodoacetamide were purchased from Wako (Japan), and leupeptin was obtained from the Peptide Institute (Japan).
Sequence analysis
The genome sequence of P. gingivalis ATCC 33277 (GenBank: AP009380; Naito et al. 2008) was examined for the presence of the target gene with the transposon. Homology analysis was performed with BLAST. Secondary structure prediction of membrane proteins was performed using the SOSUI program (http://bp.nuap.nagoya-u.ac.jp/sosui/).
Transposon mutagenesis
Tn4400' transposon mutagenesis of P. gingivalis ATCC 33277 was performed as described previously (Yamaguchi et al. 2010).
Construction of P. gingivalis strains
The oligonucleotides used in this study are listed in Table S4. The general manipulation of DNA, restriction and mapping of plasmids, and transformation of E. coli and P. gingivalis were described in detail elsewhere (Shoji et al. 2010). The chromosomal DNA from P. gingivalis ATCC 33277 was used as the template for cloning purposes. The construction of various mutants from P. gingivalis ATCC 33277 or complemented strains from the mutants are described in Data S1 (Feldman et al. 1999; Nagano et al. 2007; Shi et al. 1999; Simon et al. 1983).
Enzymatic assay
Kgp and Rgp activities were determined using the synthetic substrates benzyloxycarbonyl-l-histidyl-l-glutamyl-l-lysine-4-methyl-7-coumarylamide (Z-His-Glu-Lys-MCA) and carbobenzoxy-l-phenyl-l-arginine-4-methyl-7-coumarylamide (Z-Phe-Arg-MCA). The released 7-amino-4-methyl-coumarin was measured at 460 nm (excitation at 380 nm).
Hemagglutination
Overnight cultures of P. gingivalis strains in enriched BHI medium were centrifuged, washed once with phosphate buffered saline (PBS), and suspended in PBS at an optical density of 0.5 at 595 nm. The bacterial suspensions were then diluted in a twofold series with PBS. A 100-μL aliquot of each suspension was mixed with an equal volume of defibrinated sheep erythrocyte suspension (1% in PBS) and incubated in a round-bottom microtiter plate at room temperature for 3 h.
Gel electrophoresis and immunoblot analysis
SDS-PAGE and immunoblot analysis were performed as described previously (Shoji et al. 2010, 2011).
Preparation of P. gingivalis LPS
Purification of the P. gingivalis LPS from the wild type, porK, porT, porU, porV, porW, sov, and porR mutants was performed by using an LPS purification reagent (Intron, Korea). As the amount of LPS from the PGN_2005 mutant was relatively small, a separate purification of LPS from the wild type or the PGN_2005 mutant was performed using the hot phenol method as described previously (Shoji et al. 2002). LPS was visualized by silver staining.
Preparation of antiserum
Preparation of the anti-HBP35 used as anti-HBP35 rabbit polyclonal antibody (Abiko et al. 1990), mAb 1B5 used as anti-A-LPS (Curtis et al. 1999), and mAb TDC-5-2-1 used as anti-O-LPS (Maruyama et al. 2009) has been described previously. Preparation of mAb TDC-5-2-1 is described in detail in Data S1. To prepare the mouse antiserum against the peptides, one peptide corresponding to the amino acid region (E361-L375) within the catalytic domain of RgpB (PGN_1466), in which a cysteine residue was synthesized at the N-terminus of the peptide, was constructed and conjugated to keyhole limpet hemocyanin (Sigma Genosys, Tokyo, Japan). To raise antiserum against RgpB, mice were immunized by EveBioscience Co., Ltd. (Wakayama, Japan). To prepare mouse antiserum against the peptides corresponding to the C-terminal amino acid region of PGN_2005 (L395-Y560), the region of PGN_2005 was amplified with PGN_2005expFw/PGN_2005expBw and was cloned into the pET30 Ek/LIC vector (Novagen, Darmstadt, Germany), yielding pKD889. Then, pKD889 was transformed into E. coli BL21(DE3). The recombinant PGN_2005 (L395-Y560) protein expressed in E. coli BL21(DE3) was purified by His-tag affinity purification and injected into mice. The antiserum against PGN_2005 (L395-Y560) was collected from the immunized mice at Biomedical Research Centre, Centre for Frontier Life Sciences in Nagasaki University. Animal care and experimental procedures were performed in accordance with the Guidelines for Animal Experimentation of Nagasaki University with approval from the Institutional Animal Care and Use Committee.
Cell fractionation analysis
Sample preparation and all procedures were followed as described previously (Murakami et al. 2002).
Sucrose density gradient centrifugation
A whole cell envelope suspension (1.5 mL) from the wild-type fully grown culture (200 mL) was applied to the discontinuous sucrose density gradient (1.6 mL of 2.02 mol/L, 5.6 mL of 1.44 mol/L, and 4.0 mL of 0.77 mol/L sucrose in HEPES buffer) as described previously (Murakami et al. 2002). The gradients were centrifuged at 100,000g for 48 h at 4°C in a Beckman Coulter (Brea, CA) SW41 rotor. Fractions (0.5 mL) were collected from the top of the centrifuge tube. NADH-dependent ferricyanide reductase activity was measured as an indicator for the inner membrane by the method described by Futai (1974).
In vivo complementation analysis
Strain EVV16 (Escherichia coli W3110 ΔwzzB::Km [Vinés et al. 2005]) was used for in vivo complementation analysis. Bacteria were cultured at 37°C in LB medium supplemented with Ap (100 μg/mL), Km (30 μg/mL), Sp (80 μg/mL), and 0.2% (wt/vol) arabinose as appropriate. The cloning of the PGN_2005 gene was performed by amplifying its coding region using the forward primer (5′-CCATGGCCATGACTGAGAAATCATTTCGAA-3′) carrying a NcoI site (underlined) located 8 bases upstream of the PGN_2005 start codon (double underlined) and the reverse primer (5′-AAGCTTTTAATACCTATCCAACCATAGCAC-3′), which included a HindIII site (underlined) and a stop codon (double underlined). The PCR product was cloned into pUC118, yielding pKD890. The PGN_2005 gene region was digested with NcoI and HindIII from the pKD890 and was cloned into the same sites of pBAD/Myc-His A (Invitrogen, Carlsbad, CA), yielding pKD891. pMF19-containing rhamnosyltransferase was introduced into E. coli W3110 and EVV16. Then, each complementation plasmid was introduced into E. coli cells. Escherichia coli LPSs were extracted using an LPS purification reagent (Intron, Korea). LPS was resolved by electrophoresis in a 10% SDS-polyacrylamide gel and visualized by silver staining.
Phylogenetic tree analysis of the Wzz proteins
The phylogenetic tree of the Wzz proteins was constructed by using the concatenated or whole amino acid sequences of Wzz from various bacterial strains. The amino acid sequences of the Wzz family were aligned, and ambiguous portions of the alignment were removed using BioEdit Sequence Alignment Editor software. MEGA5.05 software was used to generate the phylogenetic tree using the neighbor-joining method.
Results
Identification of a gene disrupted by Tn4400' transposon insertion
A collection of P. gingivalis ATCC 33277 random-transposon mutants was screened on blood agar plates for colonial pigmentation. The chromosomal DNA of a less-pigmented mutant (KDP205) was digested with HindIII, self-ligated and cloned into E. coli by the marker rescue method with selection for the bla gene in Tn4400' DNA. The sequencing of the cloned DNA fragment revealed that the insertion site of the transposon was located 246 bp downstream from the first nucleotide residue of the initiation codon of PGN_2005 (Fig. 1A). Complementation analysis revealed that the complemented strain was more pigmented than the PGN_2005 mutant (Fig. 1B).
Figure 1.
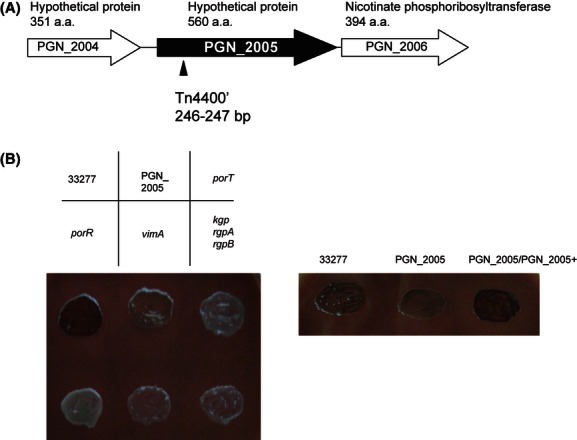
Physical map of the area around the PGN_2005 gene and pigmentation of PGN_2005 mutant. Physical map of the PGN_2005 gene region (A). A triangle indicates the Tn4400' insertion site of the PGN_2005 insertion mutant. Colony pigmentation (B). Porphyromonas gingivalis cells were anaerobically grown on blood agar plates at 35°C for 2 days.
We next examined Rgp and Kgp protease activities (Fig. 2A). The cell-associated Rgp and Kgp activities of the PGN_2005 mutant decreased by nearly 80% compared with that of the wild type. In the complemented strain, the partial recovery of the cell-associated gingipain activities relative to the PGN_2005 mutant was achieved. We did not observe significant differences in the Rgp and Kgp activities from the culture supernatants between the wild type and the PGN_2005 mutant (Fig. 2A). The Kgp activity from the culture supernatant of the complemented strain was twofold more than that of the wild type (Fig. 2A). In addition, we examined the hemagglutination activity of the P. gingivalis strains (Fig. 2B). The hemagglutination titer of the PGN_2005 mutant was twofold less than that of the wild type.
Figure 2.
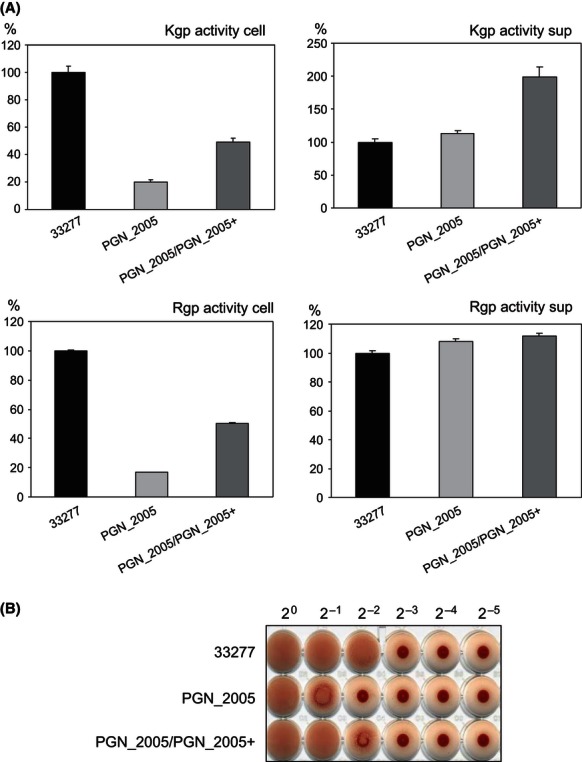
Gingipain and hemagglutination activities of Porphyromonas gingivalis. Porphyromonas gingivalis cells were anaerobically grown in enriched BHI medium at 35°C. The Kgp and Rgp activities of the cell lysates (cell) and vesicle-containing culture supernatants (sup) of ATCC 33277 (wild type), PGN_2005, or PGN_2005/PGN2005+ were measured (A). The hemagglutination activities of various P. gingivalis strains were measured (B). Twofold serial dilutions of various P. gingivalis cells were mixed with 1% sheep red blood cells and stored for 3 h at room temperature.
The PGN_2005 gene product has been annotated as a hypothetical protein of P. gingivalis ATCC 33277 (Naito et al. 2008). Homology searches revealed that genes orthologous to PGN_2005 are present in Porphyromonas asaccharolytica (Poras_0604 in strain DSM20707), Porphyromonas uenosis (PORUE0001_0693 in strain 60-3), and Porphyromonas endodontalis (POREN0001_1785 in strain ATCC 35406; Fig. S1). An analysis with the SOSUI program predicted that the PGN_2005 protein is located in the inner membrane and has two transmembrane regions (Fig. S1).
Profiles of two LPSs of the PGN_2005 mutant display lower bands than those of the wild type
The decrease of cell-associated Rgp and Kgp activities and the lack of decrease of secreted Rgp or Kgp activity in the PGN_2005 mutant suggested that the PGN_2005 mutant should be categorized as a porR-type mutant. We then examined the PGN_2005 mutant for LPS. MAb 1B5, which recognizes a glycan epitope of anionic polysaccharide bound to lipid A (called A-LPS), has been successfully used in the past as an anti-A-LPS antibody. In addition to mAb 1B5, we used mAb TDC-5-2-1 in this study, which recognizes LPS containing a normal polysaccharide O-side chain bound to lipid A (now referred as O-LPS). This was indicated by the following results: (i) mAb TDC-5-2-1, which was obtained by inoculation of cell extracts from P. gingivalis TDC60 to mice, reacted to LPS purified from the strain with a ladder-like structure pattern (Maruyama et al. 2009); (ii) mAb TDC-5-2-1 strongly reacted to cell lysates of P. gingivalis ATCC 33277, W83, TDC60, GAI7802, and HG66 with a ladder-like structure pattern and weakly reacted to lysates of strains TDC117, TDC275, and SU63 (Fig. 3A); (iii) cell lysates of P. gingivalis strain HG66, which displayed no reactivity against mAb 1B5, were recognized by mAb TDC-5-2-1 (Fig. 3A); and (iv) mAb TDC-5-2-1 recognized the low-molecular-mass product(s) of the cell lysates of the PGN_1242 (wzy) mutant but not those of the PGN_1251 (gtfB) and PGN_1302 (waaL) mutants (Fig. 3B). LPS from the wzy mutant possesses one repeat unit of O-antigen attached to a lipid A-core, whereas LPSs from the waaL and gtfB mutants possess a lipid A-core only and a truncated one-repeat unit of O-antigen attached to a lipid A-core, respectively. As the difference between the wzy and waaL mutants is one repeat unit of O-antigen only, mAb TDC-5-2-1 appears to recognize a glycan epitope within one repeat unit of O-antigen in O-LPS. To determine whether the PGN_2005 mutant has any defects in LPS biosynthesis or glycosylated CTD proteins, such as HBP35 and RgpB, we performed immunoblot analyses with mAb 1B5, mAb TDC-5-2-1, anti-HBP35, and anti-Rgp antibodies, respectively (Fig. 4). Both mAb 1B5 and mAb TDC-5-2-1 recognized low-molecular-mass products in the PGN_2005 mutant cell lysate compared with the wild type. We have recently shown that the HBP35 is secreted via the PorSS and is glycosylated with A-LPS, which is observed as diffuse bands on an SDS-PAGE gel (Shoji et al. 2010, 2011). Therefore, we examined whether the HBP35 in the PGN_2005 mutant is glycosylated. Only discrete 41-, 40-, and faint 39-kDa HBP35 bands were observed, but not the diffuse bands of HBP35 in the PGN_2005 mutant (Fig. 4). The diffuse bands of RgpB were also not present in the PGN_2005 mutant, as revealed by immunoblot analysis with anti-Rgp antibody (Fig. 4). The complemented strain displayed similar immunoreactive bands when mAb 1B5 or mAb TDC-5-2-1 was used to test against the wild-type cell lysate (Fig. 4). Furthermore, we also confirmed that LPS purified from the PGN_2005 mutant by a hot phenol method is shorter than that of the wild type and forms less than 10 repeat units of O-antigen linked to lipid A-cores (Fig. 5). With these results taken into consideration, the PGN_2005 protein is likely to be a homolog of Wzz protein, which belongs to the polysaccharide copolymerase family. These proteins play a role in determining the chain length of O-antigen.
Figure 3.
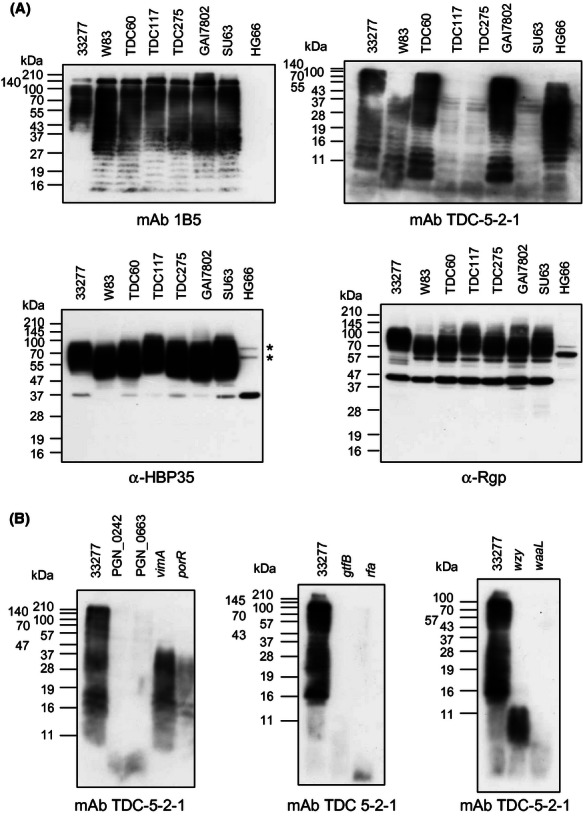
Immunoblot analyses of various Porphyromonas gingivalis strains. The cell lysates of various P. gingivalis strains were subjected to SDS-PAGE, and immunoblot analyses were performed with anti-HBP35, anti-Rgp, mAb1B5, or mAb TDC-5-2-1. The asterisks indicate nonspecific cross-reactive bands (A). Immunoblot analyses of various P. gingivalis A-LPS-deficient mutants. Cell lysates of various P. gingivalis A-LPS-deficient mutants were subjected to SDS-PAGE, and immunoblot analysis was performed with mAb TDC-5-2-1 (B).
Figure 4.
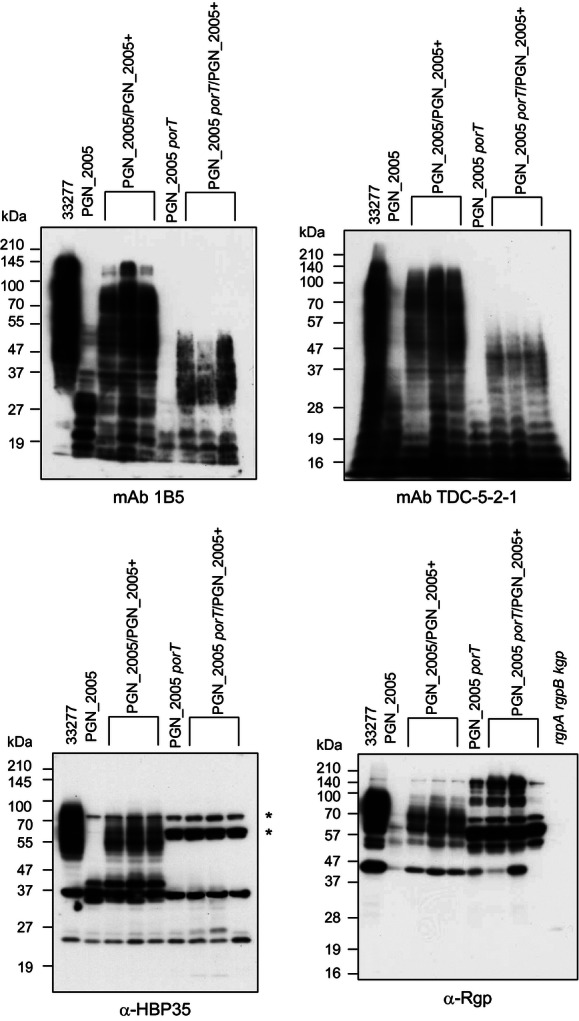
Immunoblot analyses of various Porphyromonas gingivalis strains. Immunoblot analyses of cell lysates of various P. gingivalis strains were performed with mAb 1B5, mAb TDC-5-2-1, anti-HBP35, or anti-Rgp. Three sets of PGN_2005/PGN_2005+ or PGN_2005 porT/PGN_2005+ strains were obtained from each single clone. The asterisks indicate nonspecific cross-reactive bands.
Figure 5.
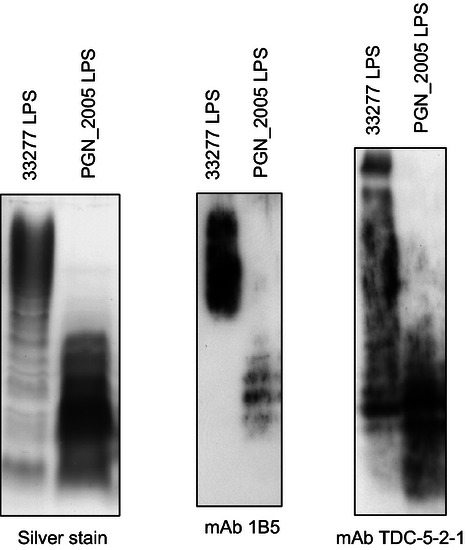
Purified LPS from the wild type or the PGN_2005 mutant. The LPS fraction was purified by the hot phenol method and subjected to SDS-PAGE followed by silver staining. Immunoblot analyses were also performed with mAb 1B5 and mAb TDC-5-2-1.
The PGN_2005 protein localizes in the inner membrane
As the PGN_2005 protein is predicted to possess two transmembrane regions but not a typical signal sequence in the N-terminus as the Wzz proteins do and to localize in the inner membrane, we examined whether the PGN_2005 protein localized in the inner membrane. The C-terminal cytoplasmic region of the PGN_2005 protein is longer than that of other Wzz proteins. We obtained an anti-PGN_2005 antibody against the C-terminal cytoplasmic region of the PGN_2005 protein. Immunoblot analysis revealed that the anti-PGN_2005 antibody recognized the 55-kDa protein in the wild type, but not in the PGN_2005 mutant (Fig. 6A). Although the apparent molecular mass of 55 kDa for the monomer is considerably lower than the predicted value of 63.2 kDa, such aberrations in molecular masses have frequently been encountered in SDS-PAGE analyses of integral membrane proteins (Abeyrathne and Lam 2007). Cell fractionation analysis revealed that the 55-kDa protein was observed in the total membrane fraction (Fig. 6B). Next, we separated the total membrane fraction of the wild type into inner and outer membrane fractions by sucrose density gradient centrifugation. We collected 20 fractions starting from the top of the centrifuge tube. Then, we measured the NADH-dependent ferricyanide reductase activity, which is characteristic of the inner membrane fraction. Fraction number 9, which displayed the highest NADH-dependent ferricyanide reductase activity, was used as the inner membrane fraction, and fraction number 20 was used as the outer membrane fraction (justified below). As shown in Figure 6C, the 55-kDa PGN_2005 protein of the inner membrane fraction was observed more than that of the outer membrane fraction. As we reported previously (Shoji et al. 2010), the outer membrane fraction, as separated by sucrose density gradient centrifugation, contained diffuse bands of HBP35 and RgpB and also displayed the presence of A-LPS and O-LPS. These results indicated that the PGN_2005 protein was located in the inner membrane, which is consistent with the localization of Wzz proteins.
Figure 6.
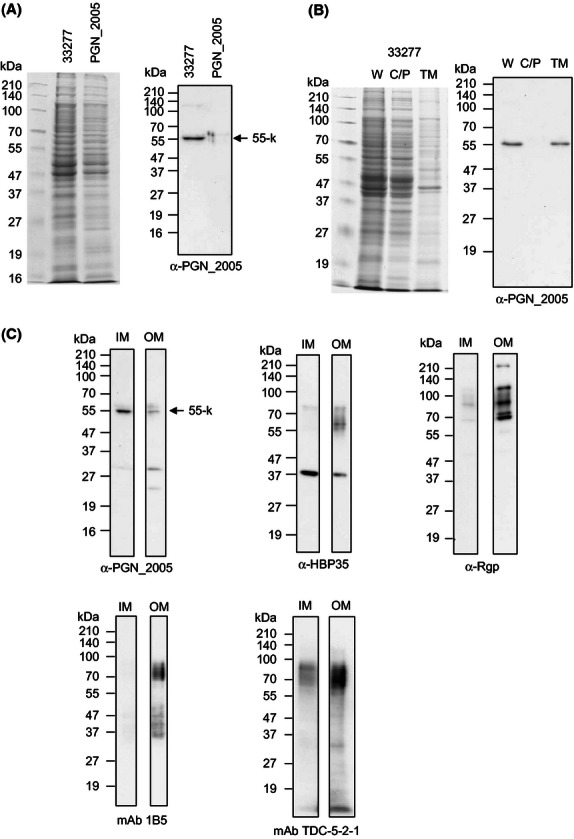
Localization of the PGN_2005 protein in Porphyromonas gingivalis. The cell lysates of the wild type and the PGN_2005 mutant were subjected to immunodetection with anti-PGN_2005 (A). Cell fractionation analysis from the wild type. W, C/P, and TM indicate the whole cell lysate, cytoplasm/periplasm, and total membrane fraction, respectively (B). Five micrograms of protein from the inner membrane (IM) or outer membrane (OM) fractions that were separated by sucrose density gradient centrifugation from the membrane fraction of the wild type were subjected to immunodetection with anti-PGN_2005, anti-HBP35, anti-Rgp, mAb 1B5, and mAb TDC-5-2-1 (C).
PGN_2005 protein confers Wzz activity upon an E. coli wzz mutant
To determine whether the PGN_2005 protein has Wzz activity, we conducted a heterologous complementation analysis using an E. coli wzzB mutant. As shown in Figure 7, the PGN_2005 protein, which is expressed in E. coli, plays a role in determining the chain length of the E. coli O-antigen. The Wzz activity of PGN_2005 is between that of WzzB-ST, which is from Salmonella typhimurium, and that of WzzB-SF, which is from Shigella flexneri. We designated the PGN_2005 protein WzzP.
Figure 7.
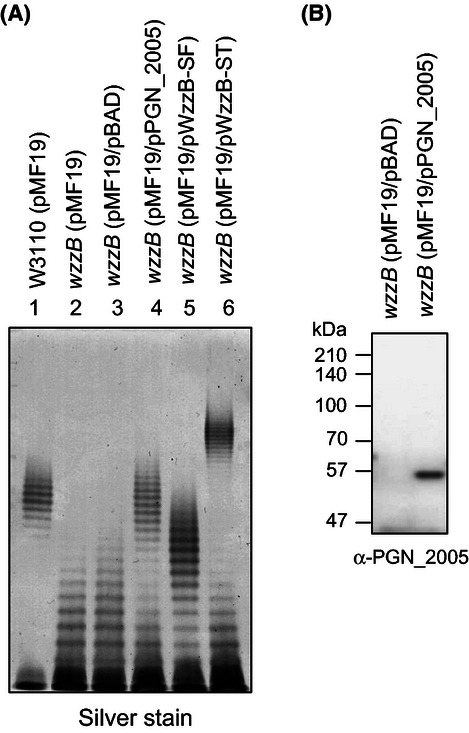
Analysis of LPS to assess Wzz activity by heterologous complementation. Silver-stained polyacrylamide gel displaying the O-antigen LPS profiles of Escherichia coli W3110 (lane 1), EVV16 (wzzB) containing pMF19 (lane 2), EVV16/pMF19 containing pBAD vector control (lane 3), EVV16/pMF19 containing PGN_2005-expressing plasmid from Porphyromonas gingivalis (lane 4), EVV16/pMF19 containing WzzB-expressing plasmid (pWzzB-SF) from Shigella flexneri (lane 5), and EVV16/pMF19 containing WzzB-expressing plasmid (pWzzB-ST) from Salmonella typhimurium (lane 6) (A). Immunoblot analysis of the cell lysates was performed with anti-PGN_2005 mouse polyclonal antiserum to confirm the expression of the PGN_2005 protein in the E. coli EVV16 (pMF19) strain (B).
Comparison of the amino acid sequence of P. gingivalis WzzP with those of Wzz proteins of other bacteria
Wzz proteins of other bacteria such as E. coli and Bacteroides fragilis consist of 300–400 amino acids, whereas P. gingivalis WzzP consists of 560 amino acids (Fig. 8). A comparison of the complete amino acid sequences of those Wzz proteins revealed that WzzP shared weak similarity with the Wzz proteins in other bacteria and that the Wzz protein had a long C-terminal cytoplasmic region consisting of approximately 160 amino acid residues, while other Wzz proteins contained short C-terminal cytoplasmic regions consisting of approximately 10 amino acid residues. These differences were conserved among WzzP homologs in the genus Porphyromonas. The C-terminal cytoplasmic region of WzzP has no sequence similarity to any proteins other than the WzzP-like proteins. We then compared these proteins in the forms lacking the nonhomologous C-terminal regions. The results were similar to that of the whole length comparison (Fig. S2).
Figure 8.
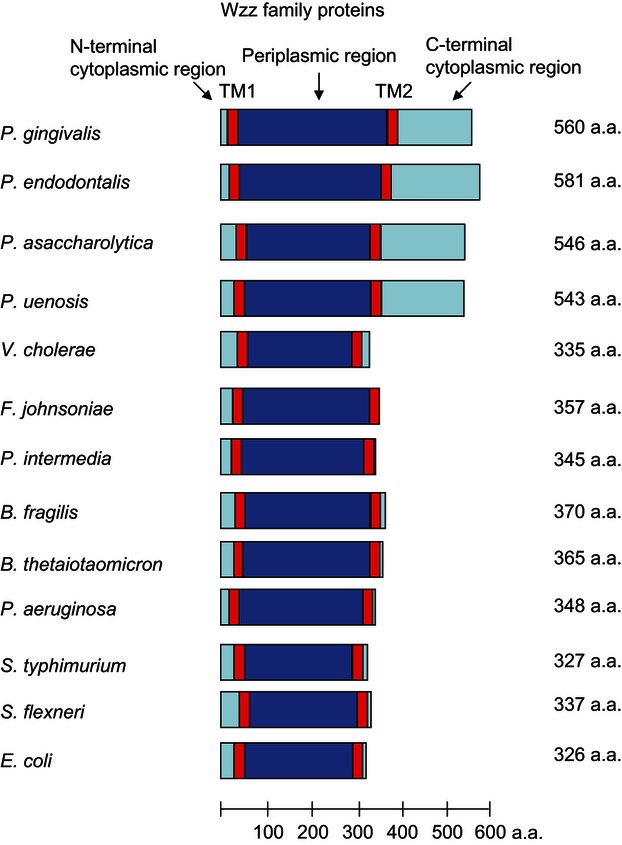
Comparison of Wzz family proteins. The transmembrane regions of various Wzz family proteins were analyzed using the SOSUI program.
WbaP and Wzx proteins are involved in A-LPS biosynthesis
A homology search revealed that PGN_1233 and PGN_1896 were putative WbaP homolog proteins and that PGN_0223 was a putative WecA homolog protein. To determine which genes were involved in the biosynthesis of the two LPSs, we constructed the PGN_1233 (putative wbaP paralog), PGN_1896 (putative wbaP), and PGN_0223-0227 (putative wecA- UDP-N-acetyl-d-mannosaminuronic acid dehydrogenase-glycosyl transferase-hypothetical protein-glycosyl transferase) mutants. As shown in Figure 9, the PGN_1896 mutant presented no immunoreaction to mAb 1B5 and no diffuse bands of HBP35 and RgpB. Interestingly, the molecular masses of the immunoreactive products of the PGN_1896 mutant probed with mAb TDC-5-2-1 decreased, similar to the phenotype of the porR mutant. This result suggests that the PGN_1896 gene product contributes to the biosynthesis of the glycan epitope of mAb 1B5. The immunoreactive products of the PGN_1233 mutant were similar to those of the wild type. To reveal the significances of the PGN_1233 and PGN_1896 genes, we constructed a PGN_1233 PGN_1896 double mutant. The immunoreactive products of the PGN_1233 PGN_1896 double mutant probed with mAb TDC-5-2-1 were significantly less than was observed with PGN_1233 and PGN_1896 single mutants, suggesting that both the PGN_1233 and PGN_1896 genes play a role in the biosynthesis of the glycan epitope of mAb TDC-5-2-1. However, the phenotype of the PGN_0223-0227 deletion mutant including the PGN_0223 region was similar to that of the wild type, suggesting that the PGN_0223 gene product does not play a role in the biosynthesis of the glycan epitopes of mAbs 1B5 and TDC-5-2-1, respectively.
Figure 9.
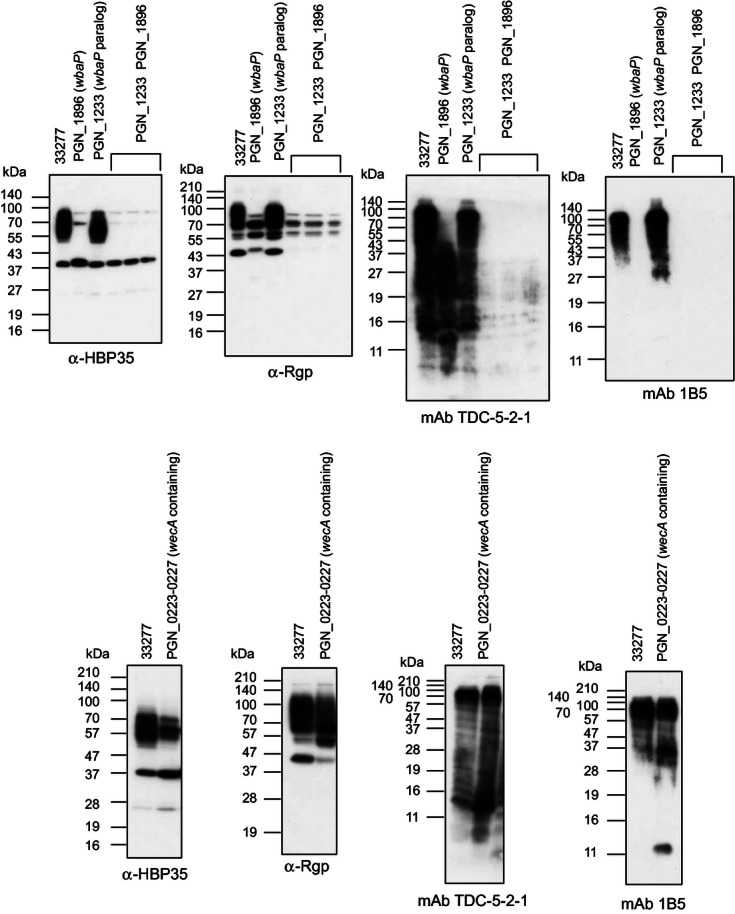
Immunoblot analyses of various Porphyromonas gingivalis mutants related to the first initiation enzyme of UndPP-glycan with anti-HBP35, anti-Rgp, mAb 1B5, and mAb TDC-5-2-1. Cell lysates of various P. gingivalis mutants were subjected to SDS-PAGE and immunoblot analysis with anti-HBP35, anti-Rgp, mAb 1B5, and mAb TDC-5-2-1.
As PGN_1242 and PGN_1302 have been identified as Wzy and WaaL, respectively, and PGN_2005 was identified as a Wzz protein in this study, we focused on PGN_1033 (putative wzx), PGN_1917-1916 (putative ABC transporter–ABC transporter), PGN_2066 (putative wzt), PGN_2072 (putative wzt), PGN_1523-1525 (putative wza-wzc-wzb), and PGN_1362-1363 (putative exported transglycosylase protein-wzt) genes and constructed the relevant mutants to determine which genes were involved in LPS biosynthesis. We found that neither of the monoclonal antibodies recognized any products of the PGN_1033 mutant (Fig. 10). The diffuse bands of HBP35 and RgpB were also not observed with the PGN_1033 mutant (Fig. 10). These results suggest that the PGN_1033 gene product plays a part in the biosynthesis of the two LPSs and belongs to the Wzx protein family, and acts as an UndPP-glycans flippase, although we have no experimental evidence of the enzymatic activity.
Figure 10.
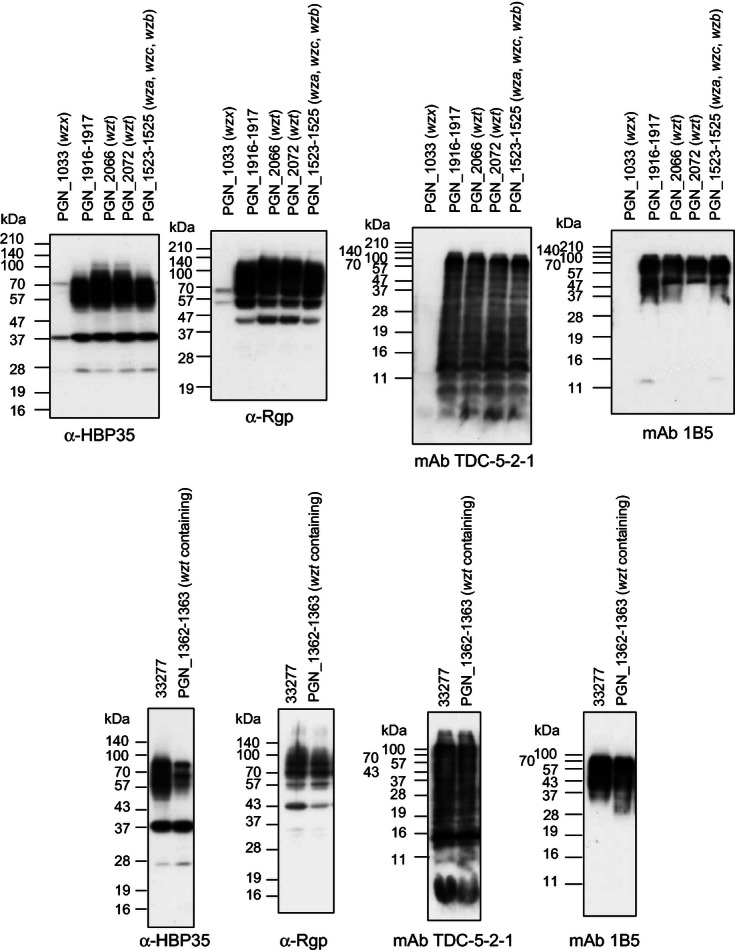
Immunoblot analysis of various Porphyromonas gingivalis mutants related to the O-antigen flippase with anti-HBP35, anti-Rgp, mAb 1B5, and mAb TDC-5-2-1. Cell lysates of various P. gingivalis mutants were subjected to SDS-PAGE and immunoblot analysis with anti-HBP35, anti-Rgp, mAb 1B5, and mAb TDC-5-2-1.
Discussion
LPS is composed of O-antigen, core polysaccharide, and lipid A. Biosynthesis of the O-antigen in P. gingivalis is assumed to occur by Wzy- and WaaL-dependent pathway (Paramonov et al. 2009). Briefly, the initiating reaction to form UndPP-glycan occurs via WbaP or WecA, which functions as a galactose-1-phosphate transferase and a GlcNAc-1-phosphate transferase, respectively. Then, UndPP-glycans are achieved on the cytoplasmic side of the inner membrane by specific glycosyltransferases, and are transported onto the periplasmic side of the inner membrane via a Wzx flippase located in the inner membrane. Polymerization of O-antigen occurs in the periplasm and is dependent on the inner membrane polymerase Wzy. The resulting O-antigen chains are typically clustered around a certain number of O-antigen repeat units determined by Wzz, the chain length regulator, also located in the inner membrane (Hug and Feldman 2011). After polymerization, the O-antigen chains are ligated to the preformed lipid A-core by the inner membrane ligase, WaaL, and the mature LPS is transported to the outer membrane by LPS transport proteins (Sperandeo et al. 2009). On the other hand, it has been shown that polymeric O-antigen assembly can also occur by an ATP-binding cassette (ABC) transporter-dependent pathway. Generally, the ABC transporter formed by Wzm and Wzt, which are the permease and ABC transporter, respectively, or a single protein, such as Wzk, is required for the transfer of the undecaprenyl-linked polymer to the periplasmic side of the inner membrane, where it is ligated to the lipid A-core and translocated to the outer membrane (Hug and Feldman 2011).
In this study, we isolated a novel pigment-less mutant by transposon mutagenesis. The transposon-inserted gene was identified as the PGN_2005 gene, the product of which is annotated as a hypothetical protein of P. gingivalis ATCC 33277 (Naito et al. 2008). The PGN_2005 protein appears to be a functional homolog of Wzz proteins for the following reasons: (i) the PGN_2005 protein was located in the inner membrane (Fig. 6); (ii) the secondary structure prediction of the amino acid sequence of PGN_2005 indicated that the protein has two transmembrane regions and one extended periplasmic loop region (Figs. 8 and S1); (iii) the PGN_2005 protein shared weak amino acid sequence homology with conventional Wzz proteins of other bacteria (Fig. S2); (iv) cell lysate products or purified LPS from the PGN_2005 mutant reacted with the LPS-specific antibodies mAb 1B5 and mAb TDC-5-2-1, resulting in shorter bands than those observed in the wild type (Figs. 4 and 5); and (v) the PGN_2005 gene conferred Wzz activity upon an E. coli wzz mutant (Fig. 7). We designated the PGN_2005 gene wzzP. The WzzP-like proteins can be found in P. asaccharolytica (Poras_0604), P. uenosis (PORUE0001_0693), and P. endodontalis (POREN0001_1785). Interestingly, all the bacteria belong to the genus Porphyromonas. The flanking genes of the wzzP homologs are also identical among P. gingivalis, P. asaccharolytica, P. endodontalis, and P. uenosis; however, they are unlikely to be involved in LPS biosynthesis. Most of the genes involved in LPS biosynthesis are scattered in the chromosomes of bacteria belonging to the phylum Bacteroidetes (Table S1).
It has been shown that Wzz proteins determine the chain length of O-antigen polysaccharides synthesized by Wzy. In the absence of Wzz, modality, that is, the nonrandom chain length of O-antigen, is lost, and predominantly short or long polysaccharides are produced (Bastin et al. 1993; Morona et al. 1995). A successive in vitro bacterial polysaccharide biosynthesis assay using Wzy and Wzz proteins has been reported (Woodward et al. 2010). And very recently, Kalynych et al. (2012) proposed that Wzz proteins bind and stabilize the growing O-antigen intermediates and prevent the premature release of the growing O-antigen, and once the O-antigen contains the appropriate number of the repeat units that can no longer be bound by Wzz, it is released. Most of Wzz proteins are 36- to 40-kDa inner membrane proteins with variable sequence identity (approximately 15–80%), whereas the WzzP protein of P. gingivalis consists of 560 amino acids and produces a 55-kDa protein on an SDS-PAGE gel (Figs. 6 and 7). The main difference between the WzzP protein and Wzz proteins is that the WzzP protein has a long C-terminal cytoplasmic region consisting of approximately 160 amino acid residues, while Wzz proteins contain a short C-terminal cytoplasmic region consisting of approximately 10 amino acid residues (Fig. 8). BLAST searches revealed that the C-terminal cytoplasmic region of WzzP has no sequence similarity to any proteins other than the WzzP-like proteins. The C-terminal cytoplasmic region of the WzzP protein may not play an important role in Wzz activity. It has been shown that the oligomerization state of Wzz proteins is critical for their function (Tang et al. 2007; Papadopoulos and Morona 2010) and that several periplasmic regions of the proteins are important for the activity (Kalynych et al. 2011). The WzzP protein may form the oligomerization state and function with Wzy; however, the periplasmic loop region of WzzP has a weak sequence similarity to those of Wzz proteins.
It has been reported that the Wzz proteins in Vibrio cholera and B. fragilis play roles in the biosynthetic pathways of the O-antigen capsule and the high-molecular-mass polysaccharides, respectively (Attridge et al. 2001; Patrick et al. 2009). To determine the effect of P. gingivalis WzzP on capsular polysaccharides, we constructed a wzzP (PG0056) mutant from strain W83 possessing K1, the capsular antigen, as strain ATCC 33277 lacks the K1 antigen. Similar to the ATCC 33277 wzzP mutant, the W83 wzzP mutant presented shorter bands that were immunoreactive to mAbs 1B5 and TDC-5-2-1. The W83 wzzP mutant was shown to be encapsulated (data not shown), suggesting that WzzP plays a role in the biosynthesis of LPS, but not in that of capsular polysaccharide. Wzc homolog protein (PGN_1524) has weak similarity to WzzP. Wzc proteins possess two transmembrane regions and a long C-terminal cytoplasmic domain and undergo the transphosphorylation of several C-terminal tyrosine residues to translocate the capsular polysaccharides (Whitfield 2006). In contrast, WzzP proteins do not possess such a tyrosine-rich region. Several bacteria, such as S. typhimurium, S. flexneri, and Pseudomonas aeruginosa, possess two types of Wzz proteins, and the Wzz proteins differentially regulate the length of LPS (Morona et al. 2003; Murray et al. 2003; Kintz et al. 2008). In P. gingivalis, no wzzP paralogs were identified in the genome. LPSs with various O-antigen chain lengths produced by Wzz proteins are important for resistance to complement (Murray et al. 2006). WzzP may be crucial for the resistance of P. gingivalis to complement, as PorR is responsible for complement resistance (Slaney et al. 2006).
Nakao et al. (2006) constructed wbaP homolog (PGN_1896), wecA homolog (PGN_0223), and wzt homolog (PGN_1363) mutants from strain ATCC 33277 by insertion of the ermF-ermAM cassette into the target genes and found that the distribution of the O-antigen ladder in the mutants did not change compared with that of the wild type. We have sometimes found that the distribution of the O-antigen ladder is different between an insertion mutant and a deletion mutant; this difference is presumably due to residual activity of the insertion mutant. To exclude this residual activity, we constructed deletion mutants of PGN_1896 (wbaP homolog), PGN_1233 (wbaP homolog), PGN_1033 (wzx homolog), PGN_1362-1363 (exported transglycosylase-wzt homolog), PGN_2066 (wzt homolog), PGN_2072 (wzt homolog), PGN_1916-1917 (putative ABC transporter–putative ABC transporter), and PGN_1523-1525 (wza-wzc-wzb homologs encoding capsular polysaccharide transporter). We found that WbaP homolog (PGN_1896) is critical for A-LPS biosynthesis, and Wzx homolog (PGN_1033) is critical for both biosynthesis of A-LPS and O-LPS (Figs. 1). As mAb TDC-5-2-1-immunoreactive products significantly decrease in PGN_1896 and PGN_1233 (wbaP homologs) double-mutant cell lysates, both PGN_1896 and PGN_1233 genes are important for the formation of the glycan epitope recognized by mAb TDC-5-2-1. However, the double mutant had faint ladder bands with mAb TDC-5-2-1, suggesting that other initiating enzyme(s) that can form UndPP-glycan are present (Fig. 9). It has recently been shown that PorS (PGN_1235) possesses Wzx flippase activity (Haurat et al. 2011). As cell lysates of the porS mutant were still immunoreactive with mAb 1B5, albeit at a much reduced level, other gene(s) responsible for the Wzx activity may exist. We found that PGN_1033 is critical for the biosynthesis of both A-LPS and O-LPS, suggesting that it acts as an O-antigen flippase, as it has high similarity to other Wzx proteins (Fig. 11).
Figure 11.

Transport model of LPS and CTD proteins. The first initiation enzymes of UndPP-glycan for two LPSs in Porphyromonas gingivalis are WbaP-like proteins (PGN_1896 and PGN_1233). Assembly of UndPP-glycans is achieved at cytoplasmic side of the inner membrane, and the block is then transported onto the periplasmic side of the inner membrane by Wzx (PGN_1033). The nonrandom (modal) chain length of O-antigen is dictated by Wzy and Wzz proteins, which correspond to an O-antigen polymerase (PGN_1242) and O-antigen chain length regulator (PGN_2005), respectively. Then, O-antigen is ligated to preformed lipid A-cores by O-antigen ligase (PGN_1302), resulting in LPS. LPS is transported to the outer membrane by LPS transport proteins, which are poorly characterized in P. gingivalis. The C-terminal domain proteins are transported to the outer membrane by Sec and the Por secretion system/Type IX secretion system (PorSS/T9SS). Currently, the precise glycosylation mechanism of the CTD proteins remains uncertain.
Recently, it has been shown that mature LPS is transported to the outer membrane by the “Lpt machinery.” LPS transport proteins involved in the process are located at the inner membrane (LptB, LptC, LptF, and LptG), in the periplasm (LptA), and at the outer membrane (LptD and LptE) (Sperandeo et al. 2009; Chng et al. 2012; Okuda et al. 2012). We found that PGN_1553 contains LptA-, LptC-, or LptD-like domains, PGN_0669, PGN_1512, PGN_0884, and PGN_0260 are the best-matched P. gingivalis equivalents of LptB, LptC, LptD, and LptE, respectively, and PGN_0642 contains LptF- or LptG-like domains. Genes encoding Lpt proteins are essential in E. coli, but not in Neisseria meningitides (Bos and Tommassen 2011). As we have failed to construct deletion mutants for PGN_1553 and PGN_0884 thus far, it is possible that these genes may encode essential Lpt proteins (data not shown).
Porphyromonas gingivalis displays black pigmentation on blood agar plates. The black pigmentation is the result of storage of the μ-oxo-dimeric form of heme (iron protoporphyrin IX) on the cell surface (Smalley et al. 1998). It has been shown that Lys-gingipain can degrade hemoglobin protein, which holds heme molecules; this correlates with the inability to pigment, as a mutant with the kgp gene encoding a Lys-gingipain displayed a pigment-less phenotype on blood agar plates (Okamoto et al. 1998; Curtis et al. 2002). Our previous study demonstrated that the PorT protein is critical for the secretion of the gingipain-related proteins, which consist of the Arg-gingipains, Lys-gingipain, and HagA (Sato et al. 2005). These proteins have a conserved CTD in their primary sequences (Seers et al. 2006). Via genome analysis, we revealed that P. gingivalis strains have 34 and 33 proteins, including a conserved C-terminal domain, in strains W83 and strain ATCC 33277, respectively. For the CTD family proteins, the CTD region is required for proper secretion and attachment onto the cell surface. We recently have shown that P. gingivalis has a novel secretion apparatus, termed the PorSS (T9SS) (Sato et al. 2010). This system consists of 11 proteins, including PorK, PorL, PorM, PorN, PorP, PorQ, PorT, PorU (Glew et al. 2012), PorV/Pg27/LptO (Ishiguro et al. 2009; Chen et al. 2011), PorW, and Sov (Saiki and Konishi 2007), and plays a role in secreting the CTD proteins. It has been shown that RgpB, TapA, HBP35, and CPG70, which belong to the CTD family proteins, present diffuse bands on SDS-PAGE gels (Nguyen et al. 2007; Kondo et al. 2010; Shoji et al. 2010; Chen et al. 2011). Through GFP-fusion analysis, we recently demonstrated that 22 C-terminal amino acids of HBP35 are required to secrete via the PorSS (T9SS) and form the diffuse bands in the wild type (Shoji et al. 2011). In the PGN_2005 mutant, we found that Arg-gingipain and the HBP35 protein are secreted via the PorSS (T9SS), and relatively small amounts of the proteins are associated with the cell (Fig. 4). porR and vimA mutants and strain HG66, which possess O-LPS but lack A-LPS, produce no diffuse HBP35 and RgpB bands on a gel (Fig. 3A and B; Shoji et al. 2011). This result indicates that A-LPS is critical for forming the diffuse bands of HBP35 and RgpB. We have recently shown that the mAb 1B5-immunoreactive products of the PorSS (T9SS)-related mutant cell lysates had lower in molecular masses than those of the wild type (Fig. S3), suggesting that the P. gingivalis wild type has abundant glycosylated CTD proteins bound to A-LPS (Shoji et al. 2011). Interestingly, the mAb TDC-5-2-1-immunoreactive products of the PorSS-related mutant cell lysates were also lower in molecular masses than those of the wild type (Fig. S3). Why are mAb TDC-5-2-1-immunoreactive products of the PorSS (T9SS)-related mutants lower in molecular masses than those of the wild type? As it is unlikely that CTD proteins bind to O-LPS, A-LPS may include a glycan epitope that is recognized by mAb TDC-5-2-1. However, saccharides composing APS recognized by mAb 1B5 are not involved in the conventional O-antigen (Paramonov et al. 2001, 2005). To elucidate the source of this contradiction, further analysis is needed to identify the glycan epitope of mAb TDC-5-2-1.
It has been reported that not only CTD proteins but also OMP85 (Nakao et al. 2008) and Mfa1 (Zeituni et al. 2010) proteins are glycosylated in P. gingivalis. As the latter two proteins present discrete bands, the glycosylation mechanism of those proteins may be different from that of CTD proteins. In other bacteria, N-linked or O-linked protein glycosylation mechanisms are reported, and these protein glycosylation mechanisms are mediated by specific oligosaccharyltransferases (Hug and Feldman 2011). It has been shown that Campylobacter jejuni PglB has dual functions as an N-linked oligosaccharyl transferase and an O-antigen ligase (WaaL) (Feldman et al. 2005), and Burkholderia thailandensis PglLBt and V. cholera PglLVc have an O-linked oligosaccharyl transferase and contain a Wzy_C domain that is also present in WaaL (Gebhart et al. 2012). In P. gingivalis, the best-matched Wzy_C domain was found in PGN_1302 (WaaL) (Rangarajan et al. 2008). Further analysis is needed to determine whether PGN_1302 has protein glycosylation activity. PorSS-related genes are conserved in the Bacteroidetes phylum but are lacking in gut bacteria such as B. fragilis and Bacteroides thetaiotaomicron (Sato et al. 2010). We found that other bacteria within the Bacteroidetes phylum lack some A-LPS biosynthesis-related genes present in P. gingivalis (Table S1). MAb 1B5 does not recognize the cell lysates of Prevotella intermedia, Tannerella forsythia, and Flavobacterium johnsoniae (data not shown). Therefore, the mechanism of A-LPS biosynthesis and the glycosylation mechanism of CTD proteins may be specific to P. gingivalis among members of the Bacteroidetes phylum. As the glycosylation mechanism provides the robust pigmentation, gingipain activities, and hemagglutinin activity (Figs. 1B and 2A and B), it is important to identify this unknown mechanism. As shown in Figure 1, the biosynthesis pathway of the two LPSs is undertaken by WbaP homologs (PGN_1896 and PGN_1233), Wzx homolog (PGN_1033), Wzy homolog (PGN_1242), WzzP homolog (PGN_2005), and WaaL homolog (PGN_1302). LPS biosynthetic pathways are classified into four types: Salmonella enterica LPS, P. aeruginosa LPS, E. coli LPS, and Helicobacter pylori LPS (Hug and Feldman 2011). The LPS biosynthetic pathway of P. gingivalis is similar to that of S. enterica. A-LPS and O-LPS are present on the cell surface in the PorSS (T9SS)-related mutants (Shoji et al. 2011; data not shown), and the waaL mutant is PorSS (T9SS)-proficient because it demonstrated hemolysis activity on a blood agar plate (Rangarajan et al. 2008), suggesting that the LPS transport pathway is independent from PorSS (T9SS) (Fig. 11).
In conclusion, we found a novel P. gingivalis pigment-less mutant by transposon mutagenesis: the gene responsible was the PGN_2005 gene of strain ATCC 33277. On the basis of analysis of the LPS pattern of the PGN_2005 mutant, the location and structure of the PGN_2005 protein and complementation of an E. coli wzz mutant by the PGN_2005 gene, we believe that the PGN_2005 protein is Wzz of P. gingivalis, designated WzzP. In addition, in this study, we determined that the PGN_1896 and PGN_1233 proteins and the PGN_1033 protein appear to be WbaP homolog proteins and a Wzx homolog protein, respectively.
Acknowledgments
This study was supported by Grants-in-Aid (20249073 and 23792110 to K. N. and M. S., respectively) for scientific research from the Ministry of Education, Science, Sports, Culture, and Technology, Japan; by the Global COE Program at Nagasaki University to K. N.; and, in part, by the president's discretionary fund of Nagasaki University, Japan to M. S.. We thank to K. Nurse and M. A. Valvano for giving us the generous gifts of E. coli W3110, EVV16 strains, and pMF19. We also thank to S. Kalynych and M. Cygler for giving us the generous gifts of pBAD24 containing wzzB-SF and wzzB-ST.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Multiple sequence alignments of PGN_2005 homolog proteins. Amino acid sequences of PGN_2005 from Porphyromonas gingivalis, Poras_0604 from P. asaccharolytica, PORUE0001_0693 from P. uenosis, and POREN0001_1785 from P. endodontalis were compared. Red- or green-colored amino acids indicate putative transmembrane regions shown as TM1 and TM2, respectively (as judged by the SOSUI program).
Figure S2. Phylogenetic tree of the amino acid sequences of the Wzz family. The phylogenetic tree was constructed using concatenated amino acid sequences (A) or whole amino acid sequences (B). The scale bar indicates amino acid substitutions per site. Each number of amino acid sequences used for analysis is depicted.
Figure S3. Immunoblot analyses of various Porphyromonas gingivalis PorSS-related deficient mutants or of various purified LPSs. The cell lysates of various P. gingivalis PorSS-related deficient mutants were subjected to SDS-PAGE, and immunoblot analyses were performed with mAb 1B5 and mAb TDC-5-2-1 (A). The purified LPS fractions (using the Intron kit) from various strains were detected with mAb 1B5 or mAb TDC-5-2-1, as appropriate (B).
Data S1. Experimental procedures.
Table S1. Comparative genome analyses of A-LPS biosynthesis related genes.
Table S2. Bacterial strains used in this study.
Table S3. Plasmids used in this study.
Table S4. Oligonucleotides used in this study.
References
- Abeyrathne PD, Lam JS. WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O-antigen onto lipid A-core. Mol. Microbiol. 2007;65:1345–1359. doi: 10.1111/j.1365-2958.2007.05875.x. [DOI] [PubMed] [Google Scholar]
- Abiko Y, Hayakawa M, Aoki H, Kikuchi T, Shimatake H, Takiguchi H. Cloning of a Bacteroides gingivalis outer membrane protein gene in Escherichia coli. Arch. Oral Biol. 1990;35:689–695. doi: 10.1016/0003-9969(90)90091-n. [DOI] [PubMed] [Google Scholar]
- Aduse-Opoku J, Slaney JM, Hashim A, Gallagher A, Gallagher RP, Rangarajan M, et al. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 2006;74:449–460. doi: 10.1128/IAI.74.1.449-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A, Davey ME. Porphyromonas gingivalis: surface polysaccharides as virulence determinants. Interface Oral Health Sci. 2010;2009(Pt 9):382–387. [Google Scholar]
- Attridge SR, Fazeli A, Manning PA, Stroeher UH. Isolation and characterization of bacteriophage-resistant mutants of Vibrio cholerae O139. Microb. Pathog. 2001;30:237–246. doi: 10.1006/mpat.2000.0426. [DOI] [PubMed] [Google Scholar]
- Bastin DA, Stevenson G, Brown PK, Haase A, Reeves PR. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol. Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- Bos MP, Tommassen J. The LptD chaperone LptE is not directly involved in lipopolysaccharide transport in Neisseria meningitidis. J. Biol. Chem. 2011;286:28688–28696. doi: 10.1074/jbc.M111.239673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, et al. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol. Microbiol. 2011;79:1380–1401. doi: 10.1111/j.1365-2958.2010.07530.x. [DOI] [PubMed] [Google Scholar]
- Chng SS, Xue M, Garner RA, Kadokura H, Boyd D, Beckwith J, et al. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science. 2012;337:1665–1668. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Kasper DL. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–850. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Curtis MA, Thickett A, Slaney JM, Rangarajan M, Aduse-Opoku J, Shepherd P, et al. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 1999;67:3816–3823. doi: 10.1128/iai.67.8.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Aduse-Opoku J, Rangarajan M, Gallagher A, Sterne JA, Reid CR, et al. Attenuation of the virulence of Porphyromonas gingivalis by using a specific synthetic Kgp protease inhibitor. Infect. Immun. 2002;70:6968–6975. doi: 10.1128/IAI.70.12.6968-6975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Marolda CL, Monteiro MA, Perry MB, Parodi AJ, Valvano MA. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 1999;274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, et al. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai M. Orientation of membrane vesicles from Escherichia coli prepared by different procedures. J. Membr. Biol. 1974;15:15–28. doi: 10.1007/BF01870079. [DOI] [PubMed] [Google Scholar]
- Gebhart C, Ielmini MV, Reiz B, Price NL, Aas FE, Koomey M, et al. Characterization of exogenous bacterial oligosaccharyltransferases in Escherichia coli reveals the potential for O-linked protein glycosylation in Vibrio cholerae and Burkholderia thailandensis. Glycobiology. 2012;22:962–974. doi: 10.1093/glycob/cws059. [DOI] [PubMed] [Google Scholar]
- Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, et al. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J. Biol. Chem. 2012;287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 2011;286:1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug I, Feldman MF. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 2011;21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- Ishiguro I, Saiki K, Konishi K. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol. Lett. 2009;292:261–267. doi: 10.1111/j.1574-6968.2009.01489.x. [DOI] [PubMed] [Google Scholar]
- Kalynych S, Ruan X, Valvano MA, Cygler M. Structure-guided investigation of lipopolysaccharide O-antigen chain length regulators reveals regions critical for modal length control. J. Bacteriol. 2011;193:3710–3721. doi: 10.1128/JB.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalynych S, Valvano MA, Cygler M. Polysaccharide co-polymerases: the enigmatic conductors of the O-antigen assembly orchestra. Protein Eng. Des. Sel. 2012;25:797–802. doi: 10.1093/protein/gzs075. [DOI] [PubMed] [Google Scholar]
- Kintz E, Scarff JM, DiGiandomenico A, Goldberg JB. Lipopolysaccharide O-antigen chain length regulation in Pseudomonas aeruginosa serogroup O11 strain PA103. J. Bacteriol. 2008;190:2709–2716. doi: 10.1128/JB.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Ohara N, Sato K, Yoshimura M, Yukitake H, Naito M, et al. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect. Immun. 2010;78:2846–2856. doi: 10.1128/IAI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA-the citrullinated enolase connection. Nat. Rev. Rheumatol. 2010;6:727–730. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- Madianos PN, Lieff S, Murtha AP, Boggess KA, Auten RL, Jr, Beck JD, et al. Maternal periodontitis and prematurity. Part II: Maternal infection and fetal exposure. Ann. Periodontol. 2001;6:175–182. doi: 10.1902/annals.2001.6.1.175. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Hayakawa M, Zhang L, Shibata Y, Abiko Y. Monoclonal antibodies produced against lipopolysaccharide from fimA Type II Porphyromonas gingivalis. Hybridoma. 2009;28:431–434. doi: 10.1089/hyb.2009.0055. [DOI] [PubMed] [Google Scholar]
- McBride MJ, Zhu Y. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J. Bacteriol. 2013;195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R, Manning L, van den Bosch PA. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J. Bacteriol. 1995;177:1059–1068. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R, Daniels C, van den Bosch L. Genetic modulation of Shigella flexneri 2a lipopolysaccharide O antigen modal chain length reveals that it has been optimized for virulence. Microbiology. 2003;149:925–939. doi: 10.1099/mic.0.26141-0. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Imai M, Nakamura H, Yoshimura F. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 2002;110:157–162. doi: 10.1034/j.1600-0722.2002.11171.x. [DOI] [PubMed] [Google Scholar]
- Murray GL, Attridge SR, Morona R. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 2003;47:1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- Murray GL, Attridge SR, Morona R. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 2006;188:2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 2007;56:1536–1548. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R, Senpuku H, Watanabe H. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect. Immun. 2006;74:6145–6153. doi: 10.1128/IAI.00261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R, Tashiro Y, Nomura N, Kosono S, Ochiai K, Yonezawa H, et al. Glycosylation of the OMP85 homolog of Porphyromonas gingivalis and its involvement in biofilm formation. Biochem. Biophys. Res. Commun. 2008;365:784–789. doi: 10.1016/j.bbrc.2007.11.035. [DOI] [PubMed] [Google Scholar]
- Nguyen KA, Travis J, Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J. Bacteriol. 2007;189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- Okuda S, Freinkman E, Kahne D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science. 2012;338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos M, Morona R. Mutagenesis and chemical cross-linking suggest that Wzz dimer stability and oligomerization affect lipopolysaccharide O-antigen modal chain length control. J. Bacteriol. 2010;192:3385–3393. doi: 10.1128/JB.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramonov N, Bailey D, Rangarajan M, Hashim A, Kelly G, Curtis MA, et al. Structural analysis of the polysaccharide from the lipopolysaccharide of Porphyromonas gingivalis strain W50. Eur. J. Biochem. 2001;268:4698–4707. doi: 10.1046/j.1432-1327.2001.02397.x. [DOI] [PubMed] [Google Scholar]
- Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, et al. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 2005;58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- Paramonov N, Aduse-Opoku J, Hashim A, Rangarajan M, Curtis MA. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J. Bacteriol. 2009;191:5272–5282. doi: 10.1128/JB.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick S, Houston S, Thacker Z, Blakely GW. Mutational analysis of genes implicated in LPS and capsular polysaccharide biosynthesis in the opportunistic pathogen Bacteroides fragilis. Microbiology. 2009;155:1039–1049. doi: 10.1099/mic.0.025361-0. [DOI] [PubMed] [Google Scholar]
- Pussinen PJ, Mattila K. Periodontal infections and atherosclerosis: mere associations? Curr. Opin. Lipidol. 2004;15:583–588. doi: 10.1097/00041433-200410000-00013. [DOI] [PubMed] [Google Scholar]
- Rangarajan M, Aduse-Opoku J, Paramonov N, Hashim A, Bostanci N, Fraser OP, et al. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 2008;190:2920–2932. doi: 10.1128/JB.01868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki K, Konishi K. Identification of Porphymonalis gingivalis novel protein Sov required for the secretion of gingipains. Microbiol. Immunol. 2007;51:483–491. doi: 10.1111/j.1348-0421.2007.tb03936.x. [DOI] [PubMed] [Google Scholar]
- Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, et al. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 2005;280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- Sato K, Kido N, Murakami Y, Hoover CI, Nakayama K, Yoshimura F. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology. 2009;155:1282–1293. doi: 10.1099/mic.0.025163-0. [DOI] [PubMed] [Google Scholar]
- Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol. Lett. 2013;338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, et al. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 2006;188:6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglogin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA rgpB kgp, and hagA. J. Biol. Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, et al. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology. 2002;148:1183–1191. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- Shoji M, Shibata Y, Shiroza T, Yukitake H, Peng B, Chen YY, et al. Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 2010;10:e152. doi: 10.1186/1471-2180-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, et al. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS ONE. 2011;6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect. Immun. 2006;74:5352–5361. doi: 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley JW, Silver J, Marsh PD, Birss AJ. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the m-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem. J. 1998;331:681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Tang KH, Guo H, Yi W, Tsai MD, Wang PG. Investigation of the conformational states of Wzz and the Wzz.O-antigen complex under near-physiological conditions. Biochemistry. 2007;46:11744–11752. doi: 10.1021/bi701181r. [DOI] [PubMed] [Google Scholar]
- Vanterpool E, Roy F, Sandberg L, Fletcher HM. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect. Immun. 2005a;73:1357–1366. doi: 10.1128/IAI.73.3.1357-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanterpool E, Roy F, Fletcher HM. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect. Immun. 2005b;73:3971–3982. doi: 10.1128/IAI.73.7.3971-3982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinés E, Marolda CL, Balachandran A, Valvano MA. Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J. Bacteriol. 2005;187:3359–3368. doi: 10.1128/JB.187.10.3359-3368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Woodward R, Yi W, Li L, Zhao G, Eguchi H, Sridhar PR, et al. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 2010;6:418–423. doi: 10.1038/nchembio.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Sato K, Yukitake H, Noiri Y, Ebisu S, Nakayama K. A Porphyromonas gingivalis mutant defective in a putative glycosyltransferase exhibits defective biosynthesis of the polysaccharide portions of lipopolysaccharide, decreased gingipain activities, strong autoaggregation, and increased biofilm formation. Infect. Immun. 2010;78:3801–3812. doi: 10.1128/IAI.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeituni AE, McCaig W, Scisci E, Thanassi DG, Cutler CW. The native 67-kilodalton minor fimbria of Porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J. Bacteriol. 2010;192:4103–4110. doi: 10.1128/JB.00275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


