Abstract
The GacS/GacA two-component regulatory system activates the production of secondary metabolites including phenazines crucial for biological control activity in Pseudomonas chlororaphis 30-84. To better understand the role of the Gac system on phenazine regulation, transcriptomic analyses were conducted by comparing the wild-type strain to a gacA mutant. RNA-seq analysis identified 771 genes under GacA control, including many novel genes. Consistent with previous findings, phenazine biosynthetic genes were significantly downregulated in a gacA mutant. The transcript abundances of phenazine regulatory genes such as phzI, phzR, iopA, iopB, rpoS, and pip also were reduced. Moreover, the transcript abundance of three noncoding RNAs (ncRNAs) including rsmX, rsmY, and rsmZ was significantly decreased by gacA mutation consistent with the presence of consensus GacA-binding sites associated with their promoters. Our results also demonstrated that constitutive expression of rsmZ from a non-gac regulated promoter resulted in complete restoration of N-acyl-homoserine lactone (AHL) and phenazine production as well as the expression of other gac-dependent secondary metabolites in gac mutants. The role of RsmA and RsmE in phenazine production also was investigated. Overexpression of rsmE, but not rsmA, resulted in decreased AHL and phenazine production in P. chlororaphis, and only a mutation in rsmE bypassed the requirement for GacA in phenazine gene expression. In contrast, constitutive expression of the phzI/phzR quorum sensing system did not rescue phenazine production in the gacA mutant, indicating the direct posttranscriptional control by Gac on the phenazine biosynthetic genes. On the basis of these results, we propose a model to illustrate the hierarchic role of phenazine regulators modulated by Gac in the control of phenazine production. The transcriptomic analysis also was used to identify additional genes regulated by GacA that may contribute to the biological control capability of strain 30-84.
Keywords: Biological control, Gac, phenazine, posttranscriptional regulation, Pseudomonas, two-component signal transduction
Introduction
The GacS/GacA two-component signal transduction system (TCST) is highly conserved among Gram-negative bacteria, including many beneficial biological control and plant pathogenic bacteria (Heeb and Haas 2001). The Gac system was originally identified as a global activator of antibiotic and cyanide synthesis in Pseudomonas, hence the acronym Gac (Haas and Keel 2003). It is now known that Gac regulates the expression of secondary metabolites (including hydrogen cyanide [HCN], phenazines, 2,4-diacetylphloroglucinol [DAPG], pyoluteorin, and the phytohormone indole-3-acetic acid), extracellular enzymes, and several carbon storage compounds as well as an oxidative stress response and other functions that play roles in biological control or plant pathogenicity (Hassan et al. 2010).
Global regulation by Gac in many Pseudomonas species was shown to act through the small RNA-binding proteins RsmA and RsmE, members of the RsmA/CsrA family (Reimmann et al. 2005; Lapouge et al. 2008). According to the current model, these small RNA-binding proteins act as negative regulators of gene expression by binding to 5′-GGA-3′ motifs located in the 5′ leading sequence of target mRNAs (Blumer et al. 1999; Heeb and Haas 2001). The binding of these proteins reduces target protein levels either by a reduction in mRNA translation (by blocking the recruitment of the 30S ribosomal subunit), by a reduction in mRNA stability (by targeting messages for degradation), or both (Dubey et al. 2003). Gac also is required for the expression of the small regulatory noncoding RNAs (ncRNAs), rsmX, rsmY, and rsmZ (Lapouge et al. 2008; Humair et al. 2010). These ncRNAs have multiple stemloop structures that interact with RsmA and RsmE, sequestering them, and relieving posttranscriptional repression (Babitzke and Romeo 2007).
The rhizosphere-colonizing bacterium Pseudomonas chlororaphis 30-84 is a member of a group of phenazine-producing bacteria with beneficial agronomic applications (Pierson and Pierson 2010). Strain 30-84 is a biological control agent capable of suppressing take-all disease of wheat caused by the fungal pathogen Gaeumannomyces graminis var. tritici (Ggt) (Pierson and Thomashow 1992). Strain 30-84 produces phenazine-1-carboxylic acid (PCA), 2-hydroxy-phenazine-1-carboxylic acid (2OHPCA), and a small amount of 2-hydroxy-phenazine (2OHPZ) (Pierson and Thomashow 1992). Phenazines are responsible for the majority of strain 30-84's ability to inhibit fungal pathogens and have been shown to be important for 30-84 to persist in the rhizosphere (Mazzola et al. 1992) and to form biofilms in vitro (Maddula et al. 2006).
Phenazine biosynthesis in 30-84 is controlled by a complex regulatory network involving quorum sensing, Gac and other TCSTs, sigma factor rpoS, and other regulatory genes (Pierson and Pierson 2010). Phenazine biosynthesis is regulated directly by the PhzR/PhzI quorum sensing system (Pierson et al. 1994; Wood and Pierson 1996; Wood et al. 1997). PhzR is a LuxR family transcriptional regulator that activates the expression of the phenazine biosynthetic genes in response to the accumulation of N-acyl-homoserine lactone (AHL) signals. Mutation in phzR abolishes the expression of the phenazine biosynthetic genes and multiple copies of phzR in trans significantly increases phenazine production (Pierson et al. 1994). PhzI, a LuxI homolog, is an AHL synthase that produces the AHL signals to which PhzR is most responsive (Wood et al. 1997). In 30-84, mutations in Gac result in loss of phenazine, HCN, exoprotease, lipase, gelatinase, and AHL production as well as losses in the capacity for pathogen inhibition, biofilm formation, and rhizosphere competence (Chancey et al. 1999; Maddula et al. 2006). Other genes with demonstrated roles in phenazine production in P. chlororaphis include pip (phenazine inducing protein), the rpeA/rpeB TCST, and iopA/iopB (inducers of phenazine) (Girard et al. 2006a; van Rij 2006; Wang et al. 2012b). Pip, a homolog of the TetR family of transcriptional regulators (Ramos et al. 2005) was shown to promote phenazine production in 30-84 and P. chlororaphis PCL1391 by enhancing phzI and phzR expression (Girard et al. 2006a; Wang et al. 2012a,b). In both P. chlororaphis 30-84 and PCL1391, the expression of pip is regulated by the sigma factor rpoS. In 30-84, the RpeA/RpeB TCST also regulates pip (Wang et al. 2012a,b). In PCL1391, two additional genes, iopA and iopB, positively contribute to phenazine production through RpoS (van Rij 2006). Pseudomonas chlororaphis 30-84 also has a second quorum sensing system, CsaI/CsaR, that controls exoprotease production and cell aggregation, but plays a minor role in phenazine regulation under laboratory conditions (Zhang and Pierson 2001).
GacS/GacA also negatively regulates a spectrum of traits by mechanisms that are as yet largely uncharacterized. For example, inactivation of GacA resulted in the differential expression of 635/6,147 (10%) of the genes in P. protegens (fluorescens) strain Pf-5 (Hassan et al. 2010). Of these, 288 genes were upregulated in a gacA mutant. On solid medium colonies of gac mutants often appear hyper-fluorescent and larger than wild type (WT), suggesting that traits related to iron acquisition (e.g., siderophores) or motility may be among the traits negatively regulated by Gac (Chancey et al. 1999). Negative regulation by Gac is particularly interesting given that many Pseudomonas species identified for their biological control potential exhibit phenotypic variation resulting from spontaneous mutation in gacS or gacA (van den Broek et al. 2005). Spontaneous gac mutants are especially prominent in fermentation culture, where they often outgrow the WT population resulting in a deficiency of secondary metabolites essential for biological control activity (Duffy and Defago 2000). In comparison, the prevalence of gac mutants in natural environments (e.g., in the rhizosphere) suggests some benefit to the Gac− phenotype and thus selection at some level to maintain it. In contrast to liquid culture, in the rhizosphere both the WT (Gac+ phenotype) and the Gac mutants (Gac− phenotype) survive better in mixed populations than in uniform populations, demonstrating a benefit to maintaining phenotypic variation (Chancey et al. 2002). More recently, it was shown that the WT and gac mutants interact mutualistically in biofilms (Driscoll et al., 2011). The improved survival of both Gac+ and Gac− phenotypes in mixed populations suggests that mutants arising de novo or introduced within the inoculum contribute to the survival of 30-84 in the rhizosphere, however, the molecular mechanisms responsible remain unknown.
In this study, the functionality of major components of the Gac/Rsm signal transduction pathway in 30-84 was demonstrated. Transcriptomic analysis using RNA-seq was performed comparing WT and a gacA mutant to examine whether the expression of known phenazine regulatory genes including phzR/phzI, pip, rpoS, and rpeA/rpeB were controlled by GacA. We used mutation and complementation analysis of genes within the Gac/Rsm pathway as well as phenazine regulatory genes to determine their hierarchic role in the control of phenazine production. This study expands the current knowledge of the Gac/Rsm regulon, especially its role in phenazine regulation, and based on our results a model describing the regulatory network controlling phenazine gene expression in 30-84 is proposed. The transcriptomic analysis also facilitated the identification of additional genes regulated by GacA that may contribute to the biological control capability of 30-84. Of special interest were genes with increased expression in the gacA mutant as these may provide insights into the benefits of the Gac− phenotype observed in mixed populations.
Experimental Procedures
Bacterial stains and growth conditions
Bacterial strains and plasmids are listed in Table S1. Oligonucleotides and primers are listed in Table S2. Liquid Luria Broth (LB) medium or AB minimal medium supplemented with 2% casamino acids (AB + 2% CAA) (Difco, Becton Dickinson and Company, Franklin Lakes, NJ) were used for culturing P. chlororaphis as described previously (Wang et al. 2012b). The following antibiotics were added to the medium when necessary: ampicillin (Ap) 100 μg/mL, kanamycin (Km) 50 μg/mL, rifampin (Rif) 100 μg/mL, tetracycline (Tc) 50 μg/mL, and gentamicin 30 μg/mL.
DNA manipulation and sequence analysis
Standard procedures were used for plasmid isolation, cloning, restriction enzyme digestion, and T4 DNA ligation (Sambrook and Russell 2001). Polymerase chain reaction (PCR) was carried out using Invitrogen Taq DNA polymerase (Life Technologies, Carlsbad, CA) at 95°C for 5 min, followed by 30 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 90 sec, and a final elongation step of 70°C for 10 min. DNA sequencing was performed at the Genome Technology Lab within the Texas A&M University Institute for Plant Genomics and Biotechnology. Nucleotide and amino acid homology searches were conducted using the blast programs (BLASTN and BLASTP, respectively) on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST). The P. chlororaphis subspecies aureofaciens 30-84 genome sequence was recently deposited in GenBank (GenBank Accession #: PRJNA67533). The P. chlororaphis 30-84 phenazine biosynthetic operon contains seven conserved biosynthetic genes phzXYFABCD (Pierson et al. 1995; Mavrodi et al. 1998), which correspond to phzABCDEFG (according to the P. fluorescens nomenclature, Mavrodi et al. 1998). Here, we use the P. chlororaphis nomenclature to conform to the original literature.
RNA preparation for RNA-seq and qPCR analyses
Three biological replicates of every strain were started from single colonies located on three separate plates containing AB + 2% CAA and then transferred to 10 mL AB + 2% CAA broth. All cultures were grown at 37°C with shaking (150 rpm) to an approximate OD600 = 1.2. Cell cultures collected at OD600 = 1.2 were diluted to OD600 = 0.3 with AB + 2% CAA broth. RNA extraction was performed as described previously (Wang et al. 2012b) with one exception: contaminating genomic DNA was removed off-column with Turbo DNA-free DNAse (Life Technologies, Carlsbad, CA). Elimination of contaminating DNA was confirmed via qPCR amplification of the rpoD gene with SYBR green® dye on an ABI 9400HT PCR machine (Life Technologies, Carlsbad, CA). RNA samples were ethanol-precipitated and resuspended in 0.1% diethylpyrocarbonate (DEPC). Ribosomal RNA (rRNA) was depleted from ∼9 μg of total RNA using the RiboZero rRNA depletion kit (for Gram-negative bacteria, Epicentre Biotechnologies, Madison, WI). Two separate samples were prepared for each treatment using this protocol. At the elution step, the two samples were pooled and concentrated using RiboMinus concentration modules (Life Technologies, Carlsbad, CA). RNA quantification throughout was achieved using a GE NanoVue Plus spectrophotometer (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) and RNA quality was monitored during preparation for library construction and during library construction with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) at the Texas A&M GTL.
Library construction and sequencing
Strand-specific cDNA libraries were constructed using the SOLiD Total RNA-Seq kit according to the manufacturer's instructions with the following modifications: (1) for RNA fragmentation, 300 ng of rRNA-depleted RNA were incubated at 95°C for 5 min. Bioanalyzer traces were used to confirm that the largest proportion of RNA fragments were around 200 bp. (2) Bioanalyzer traces following reverse transcription were used to confirm that hybridization and ligation of RNA adapters and subsequent reverse transcription of the RNA to cDNA was successful. (3) Amplification of cDNA was carried out following reverse transcription, but preceding the size-selection step. Samples were barcoded by replacing the SOLiD 3′ primer with the appropriate barcoding primer as directed in the SOLiD instructions. Size selection of DNA amplicons ranging from 200 to 300 bp was performed using an E-Gel iBase system (SYBR Safe 2% SizeSelect gel) (Life Technologies, Carlsbad, CA). This step was repeated to ensure that adapter–adapter dimers were fully eliminated before sequencing. Paired-end sequencing was carried out by the University of Texas Genomic Sequencing and Analysis Facility on a Life Technologies SOLiD 5500xl sequencing system. Targeted sequencing depth was six-million paired-end reads per sample.
Read mapping and visualization and transcript quantification
Filtering and alignment of the sequencing data was performed at the UTGSAF using the AB SOLiD BioScope Whole Transcriptome pipeline (v1.3), for whole-transcriptome RNA-seq analysis. Based on gene annotation information, the pipeline produced genome alignment results as a compressed binary version of the Sequence Alignment Map (BAM files). Mapped reads were visualized using BamView in Artemis 13.2.0 (Rutherford et al. 2000).
RNA-seq data analysis
To determine the transcriptional abundance for each gene, the number of reads that mapped within each annotated sequence was determined. The number of reads per kb of transcript per million mapped reads (RPKM) was used to normalize the raw data (Mortazavi et al. 2008), and mean RPKM values were determined for both the WT and gacA mutant samples. The complete dataset from this study has been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) with the Accession No. GSE43083. A ratio of the mean RPKM values (gacA mutant/WT) was computed for each gene and displayed by gene order from the chromosome origin of replication (Fig. 1). Comparisons were performed using a modified t-test and genes with differences in gene expression based on the ratio were considered for further analysis when the P-value was less than 0.05 (except as otherwise mentioned) and the expression ratio was ≥2.0 or ≤0.5 (Table S3) (Wang et al. 2011).
Figure 1.
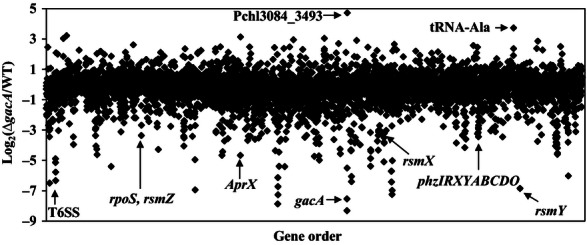
Differential gene expression between wild-type (WT) Pseudomonas chlororaphis 30-84 and a gacA mutant. Each point represents one of the 5980 annotated genes in the 30-84 genome, with the x-axis showing gene order (from the DNA replication origin), and the y-axis showing the log2 of transcript abundance for each gene in the gacA mutant relative to the WT strain. The arrows point to a few of the genes or gene clusters that are differentially expressed. Pchl3084_3493, conserved hypothetical protein; tRNA-Ala, alanine transfer RNA; T6SS, type VI secretion system; rpoS, stationary phase sigma factor; rsmX, rsmY, rsmZ: ncRNAs; AprX, metallopeptidase; AprA, metalloprotease.
qPCR methods and analysis
qPCR was performed at the Texas A&M GTL using a previously described method (Wang et al. 2012a,b). RNA was reverse transcribed using random primers (Invitrogen) and Superscript III (Invitrogen) at 50°C for 1 h and inactivated at 75°C for 15 min. SYBR Green reactions were performed using the ABI 7900 HT Fast System (Applied Biosystems, Foster City, CA) in 384 well optical reaction plates. Aliquots (1 μL) of cDNA (2 ng/reaction) or water (no-template control) were used as template for qPCR reactions with Fast SYBR Green PCR Master Mix (Applied Biosystems) and primers (500 nmol/L final concentration). Primer pairs gacSRT1-gacSRT2, gacART1-gacART2, rpeBRT1-rpeBRT2, pipRT1-pipRT2, phzYRT1-phzYRT2, phzRRT1-phzRRT2, phzIRT1-phzIRT2, hfqRT1-hfqRT2, rpoDRT1-rpoDRT2, iopBRT1-iopBRT2, 16SRT1-16SRT2, and rpoSRT1-rpoSRT2 were used to detect the expression of gacS, gacA, rpeB, pip, phzY, phzR, phzI, hfq, rpoD, iopB, 16S rRNA, and rpoS genes, respectively (Table S2). qPCR amplifications were carried out at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min, and a final dissociation curve analysis step from 65 to 95°C. Two technical replicates of each of three biological replicates were used for each experiment. Gene expression levels were analyzed using the relative quantification (ΔΔCt) method (Wang et al. 2011, 2012a,b). The housekeeping sigma factor rpoD was used as the reference gene to normalize our samples (ΔCt = Cttarget − CtrpoD). A relative quantification (RQ) value was calculated as 2 exp-(ΔΔCt = ΔCttarget − ΔCtreference) for each gene. A P-value was computed using a t-test to measure the significance associated with each RQ value. Variations were considered statistically significant when the P-value was <0.05. RQ values for the gacA mutant were then normalized to those of WT.
Cloning of rsmA, rsmE, rsmX, rsmY, rsmZ, and phzR genes
The rsmA and rsmE flanking sequences were used to design primers (rsmA1-rsmA2 and rsmE1-rsmE2) to amplify these genes and their promoter sequences. Following amplification, DNA fragments and vectors (pLAFR3) were digested by EcoRI and BamHI and ligated. The resulting plasmids were verified by sequencing and designated as pLAFR3-rsmA and pLAFR3-rsmE, respectively. Plasmids were introduced into P. chlororaphis strains by triparental mating as described previously (Wang et al. 2012b). Transformants were selected on LB plates supplemented with appropriate antibiotics.
Primers pairs rsmX1-rsmX2, rsmY1-rsmY2, rsmZ1-rsmZ2, and phzR1-phzR2 were used to amply rsmX, rsmY, rsmZ, and phzR genes. Primers rsmX1, rsmY1, rsmZ1, and phzR1 contain constitutive-hybrid Ptac promoters (De Boer et al. 1983) that ensure the correct transcription start site. The amplified DNA fragments and vectors (pLAFR3 for ncRNAs and pPROBE-GT2 for phzR) were digested by EcoRI and BamHI and ligated resulting in plasmids pLAFR3-Ptac-rsmX, pLAFR3-Ptac-rsmY, pLAFR3-Ptac-rsmZ, and pGT2-Ptac-phzR. The genotypes were confirmed by both enzymatic digestion and sequencing.
Construction of an rsmE mutant
A 30-84 genomic library was introduced into strain 30-84ZN (phzB::lacZ) via triparental mating. Exconjugants were screened for alterations in phenazine gene expression as indicated by differences in β-galactosidase activity on medium containing X-gal. Cosmid pLSP298 caused a visible reduction in phzB expression and was chosen for further analysis. Cosmid pLSP298 was mutated using EZ::TN <KAN-2> (Epicentre Biotechnologies, Madison, WI) and cosmids containing insertions were introduced into strain 30-84ZN via triparental mating and compared with strain 30-84ZN (pLSP298) for phzB::lacZ expression. One EZ::TN insertion (pLSP298EZ::TN5-11-2) that no longer reduced phzB expression was cloned as a BamHI fragment containing the EZ::TN KmR gene into pIC20H (Marsch et al. 1984). The regions adjacent to the cloned region of the EZ::TN were sequenced using the EZ::TN supplied primer and shown to be rsmE. Cosmid pLSP298EZ::TN5-11-2 was introduced into strain 30-84ZW, a ΔgacA β-galactosidase reporter (phzB::lacZ, gacA−) via triparental mating. As a result, the rsmE::EZ::TN insertion was introduced into the genome of 30-84ZW via marker exchange to create a ΔgacA β-galactosidase reporter with a mutation in rsmE, designated 30-84ZWE (phzB::lacZ, gacA−, rsmE−).
Quantification of rsmZ promoter activity
A 630 bp SalI fragment immediately upstream of rsmZ that contained the rsmZ promoter (PrsmZ) was cloned into the SalI site within pIC20H. A 681 bp EcoR1-HindIII fragment from the resulting plasmid was cloned into pPROBE-KT2 upstream of the promoter-less GFP gene to create pKT2-PrsmZ-gfp. A HindIII fragment containing a promoter-less lacZ gene was cloned from plasmid pKOK6.1 and inserted into the unique HindIII site between PrsmZ and gfp in pKT2-PrsmZ-gfp resulting in pKT2-PrsmZ-lacZ-gfp. Plasmid pKT2-PrsmZ-lacZ-gfp was introduced into the 30-84Ice strain and gac mutants via triparental mating. GFP expression for strains containing pPrsmZ-lacZ-gfp was determined visually and quantified by β-galactosidase activity (Miller 1972).
Quantification of phenazine production
Pseudomonas chlororaphis strains were grown with aeration at 28°C in LB for 24 h. Phenazines were extracted and quantified by UV-visible light spectroscopy as described previously (Wang et al. 2012a,b). Briefly, triplicate 10 mL cultures grown overnight at 28°C were centrifuged (5000g), and the supernatants were acidified to ca. pH 2 with concentrated HCl. Phenazines were extracted with an equal volume of benzene for 6 h. Following separation and evaporation of the benzene phase under a stream of air, phenazines were resuspended in 0.5 mL of 0.1 N NaOH, and serial dilutions were quantified via absorbance at 367 nm. The absorbance for each sample was normalized to the total absorbance of the 10 mL culture. Strain 30-84ZN, used as a negative control, carries a mutation in the phzB gene and is deficient in phenazine production.
AHL extraction and biological assays
Total AHL extractions were prepared from cell-free supernatants of strain 30-84, ΔgacS, or ΔgacA with plasmid pLAFR3 containing either no insert (NI) or rsmZ. The cell-free supernatants were made from overnight cultures grown in LB medium (OD600 ∼1.8) as described previously (Pierson et al. 1998). Briefly, 5 mL cultures were grown overnight at 28°C with shaking in LB broth to an OD600 of 1.8. The cultures were centrifuged and supernatants were mixed with an equal volume of acidified ethyl acetate. The ethyl acetate phase was evaporated and the dried extracts containing AHLs were suspended in a volume of LB equal to the original culture, which was subsequently filter-sterilized (0.45 μm). AHL production was quantified by inoculating the extracted AHLs with the AHL-specific reporter P. chlororaphis strain 30-84I/Z (phzI−, phzB::lacZ). Strain 30-84I/Z is deficient in AHL production, but responds to exogenously added AHL's by producing β-galactosidase. β-galactosidase activity was determined subsequently on cultures grown shaking at 28°C after 24 h (Wood and Pierson 1996). The assays were repeated at least three times.
Phenotypic analyses and fungal inhibition assays
The production of siderophore, exoprotease, gelatinase, amylase, lipase, and HCN was qualitatively measured (Chancey et al. 1999, 2002). Strains were spotted onto appropriate media: KMB plates + FeCl3·6H2O (final concentration 1 mmol/L) for fluorescence, skim milk plates for exoprotease, gelatin plates for gelatinase, starch plates for amylase, and tributyrin plates for lipase, respectively. For HCN production, a 1-inch piece of cyantesmo cyanide indicator paper (Macherey-Nagel, Germany) was attached to the inside lid of sealed plates. Cell growth and phenotypes were measured after 24 and 48 h. For pathogen inhibition, overnight cultures of 30-84 and derivatives were spotted onto triplicate LB + 0.5% potato dextrose agar plates. After 2 days of growth at 28°C, a 5 mm plug of G. graminis var. tritici was placed in the center of the plates. After 4 days, zones of inhibition, the distance between the edge of the bacterial colony and the fungal mycelium, were measured (Whistler and Pierson 2006). The assays were repeated twice.
Bacterial swimming motility assays
For P. chlororaphis WT, gac mutants, and 30-84Z strain, bacterial cell suspensions were grown overnight in LB broth. Five microliter of the bacterial suspension was plated onto the center of motility agar plates (10 g tryptone, 5 g NaCl, 2.5 g agar per liter distilled water) as previously described (Wang 2011). Diameters were determined following incubation at 28°C for 48 h. The experiments were repeated at least three times.
Results and Discussion
Transcriptomic analyses of phenazine biosynthetic and regulatory genes
The RNA-seq analysis identified 771 genes that were differentially expressed in the gacA mutant compared with the WT (Fig. 1; Table S3). Among these genes, the transcripts of 551 genes were underrepresented and 220 genes were overrepresented in the gacA mutant relative to WT. Genes encoding components of the Gac signal transduction pathway were among those differentially regulated (Fig. 1).
Transcript abundance patterns in P. chlororaphis 30-84 WT
To better understand the role of the Gac system on phenazine regulation, transcript abundance of the phenazine biosynthetic and regulatory genes in the WT were measured. The mean RPKM determined by RNA-seq for gacA and gacS in the P. chlororaphis 30-84 WT were 266.9 and 29.2 (Table 1), respectively, demonstrating that in the WT, gacA expression is about ninefold higher than gacS under these experimental conditions (e.g., grown in AB + 2% CAA to OD600 = 1.2). RPKM values for rsmA and rsmE in WT were similar to each other (151.8 and 292.7, respectively), and to gacA, whereas values for the three ncRNAs rsmX, rsmY, and rsmZ were significantly higher. In P. protegens Pf-5, GacA transcriptionally activates rsmX, rsmY, and rsmZ by binding to conserved motifs in their promoter regions (Humair et al. 2010). Analysis of rsmX, rsmY, and rsmZ in P. chlororaphis 30-84 revealed the conserved palindromic consensus sequence typical of GacA-controlled ncRNA genes and a conserved −10 hexamer (TAATCT) promoter sequence identical to that found in P. protegens Pf-5 (Humair et al. 2010) (Fig. 2A). Analogous to P. protegens Pf-5, the rsmX and rsmY promoter sequences are similar in length, but the rsmZ promoter sequence is longer (Fig. 2A). Consistent with rsmY having a promoter with the highest similarity to the conserved GacA binding site, rsmY was expressed at the highest level (RPKM = 38112.4), approximately 22-fold higher than rsmX (RPKM = 1486.5) or rsmZ (RPKM = 1679.6) in the WT (Table 1).
Table 1.
Mean transcript abundance and ratio of abundances (ΔgacA/WT) of the phenazine biosynthetic and regulatory genes in the gacA mutant compared with the WT strain
| Gene ID | Gene | Protein description | Mean RPKM WT | Mean RPKM ΔgacA | ΔgacA/WT | P-value |
|---|---|---|---|---|---|---|
| gacS/gacA | ||||||
| Pchl3084_3491 | gacA | Response regulator | 266.9 | 1.43 | 0.01 | 0.00 |
| Pchl3084_4333 | gacS | Sensor protein | 29.17 | 21.15 | 0.73 | 0.05 |
| phz genes | ||||||
| Pchl3084_4951 | phzX | Phenazine biosynthesis protein | 40.50 | 4.52 | 0.11 | 0.03 |
| Pchl3084_4952 | phzY | Phenazine biosynthesis protein | 37.87 | 5.28 | 0.14 | 0.03 |
| Pchl3084_4953 | phzF | Phenazine biosynthesis protein | 20.00 | 2.63 | 0.13 | 0.03 |
| Pchl3084_4954 | phzA | Phenazine biosynthesis protein | 8.52 | 0.78 | 0.09 | 0.15 |
| Pchl3084_4955 | phzB | Phenazine biosynthesis protein | 13.47 | 2.08 | 0.15 | 0.04 |
| Pchl3084_4956 | phzC | Phenazine biosynthesis protein | 8.24 | 1.90 | 0.23 | 0.00 |
| Pchl3084_4957 | phzD | Phenazine biosynthesis protein | 7.63 | 1.40 | 0.18 | 0.10 |
| Pchl3084_4958 | phzO | Phenazine biosynthesis protein | 44.23 | 21.70 | 0.49 | 0.06 |
| Regulatory genes | ||||||
| Pchl3084_4949 | phzI | Autoinducer synthase | 5.24 | 1.50 | 0.29 | 0.01 |
| Pchl3084_4950 | phzR | Transcriptional activator | 125.1 | 67.12 | 0.54 | 0.02 |
| Pchl3084_1189 | rpoS | Sigma factor | 1045.7 | 101.8 | 0.10 | 0.03 |
| Pchl3084_5155 | pip | Phenazine-inducing protein | 123.4 | 105.4 | 0.85 | 0.04 |
| Pchl3084_1659 | iopA | Inducer of phenazine A | 49.10 | 8.35 | 0.17 | 0.01 |
| Pchl3084_1660 | iopB | Inducer of phenazine B | 20.71 | 2.51 | 0.12 | 0.00 |
| Pchl3084_3224 | rpeA | Sensor histidine kinase | 46.78 | 49.28 | 1.05 | 0.67 |
| Pchl3084_3225 | rpeB | Response regulator | 45.37 | 55.20 | 1.22 | 0.07 |
| Pchl3084_2449 | csaI | Autoinducer synthase | 8.15 | 8.72 | 1.07 | 0.65 |
| Pchl3084_2451 | csaR | Transcriptional activator | 0.54 | 0.38 | 0.71 | 0.50 |
| Pchl3084_0554 | hfq | RNA chaperone | 1642.19 | 1408.2 | 0.84 | 0.28 |
| Pchl3084_4387 | rsmA | Translational regulator | 151.82 | 307.24 | 2.02 | 0.03 |
| Pchl3084_2024 | rsmE | Translational regulator | 292.7 | 204.83 | 0.70 | 0.06 |
| Pchl3084_3970 | rsmX | ncRNA | 1486.51 | 201.13 | 0.14 | 0.02 |
| Pchl3084_5419 | rsmY | ncRNA | 38112.4 | 329.2 | 0.01 | 0.02 |
| Pchl3084_1190 | rsmZ | ncRNA | 1679.64 | 261.26 | 0.16 | 0.05 |
Figure 2.

Promoter regions of ncRNA and phenazine biosynthetic genes. (A) Alignment of the rsmX, rsmY, and rsmZ promoter regions of Pseudomonas chlororaphis 30-84. The consensus Upstream Activation Sequence recognized by GacA (UAS) (Humair et al., 2010) is shown. The putative UAS for each promoter region is boxed and conserved nucleotides are in bold. The −10 promoter elements are also indicated. (B) RNA-seq profile showing sequence reads across the phz promoter region. RNA-seq results were visualized using the Artemis (http://www.sanger.ac.uk/resources/software/artemis/) genome browser. Transcription and translation start sites are shown below the profile. (C) Nucleotide sequence of the phz promoter. Boxed nucleotides indicate sequences involved in transcription initiation (PhzR Box, RNA polymerase binding site), whereas sequences involved in translation initiation (ribosome or RsmE binding sites) regulatory elements are underlined. Arrow indicates the direction of transcription from the start site.
We also looked at the relative transcript abundance of other regulatory genes associated with phenazine production or quorum sensing (Table 1). In the WT, hfq and rpoS were expressed at very high levels with RPKM values of 1642.2 and 1045.7 (the average RPKM value is 119.5), respectively. These two regulators were of interest because the RNA chaperone, Hfq, has been shown previously to control luxR-type regulators (Wilf et al. 2012) and the sigma factor, RpoS, to control phzR/phzI through pip (Girard et al. 2006a; Wang et al. 2012b). RPKM values for the two essential phenazine regulators pip and phzR were 123.4 and 125.1, around 10-fold lower than those of hfq and rpoS. The RPKM value for the other quorum sensing regulator csaR, which plays a minor role in phenazine regulation under laboratory conditions, was only 0.54 suggesting it was barely expressed under the assay conditions. The RPKM values for both quorum sensing signal synthases, phzI and csaI, were 5.2 and 8.2, respectively.
Similar to other phenazine-producing bacteria, the core phenazine, PCA, is encoded by a conserved set of biosynthetic genes (Mavrodi et al. 2006; Mentel et al. 2009); in 30-84 these are phzXYFABCD (Pierson et al. 1995). According to the RNA-seq analysis, the transcription start site of the phzX was predicted 115 nt upstream of the translation start site (ATG) (Fig. 2B). This was consistent with the expected structure of a quorum sensing-regulated promoter containing a −10 hexamer and a Lux-box located at the −35 site predicted for PhzR binding (Qin et al. 2007) (Fig. 2C). The transcript abundance of the phz genes showed a staircase-like pattern from phzX to phzD (Table 1), suggesting decreased transcription of genes with position along the phz operon. In many phenazine-producing Pseudomonas species, PCA is then modified into other derivatives by one or more accessory genes encoding modifying enzymes (Mavrodi et al. 2006). In 30-84, PCA is modified into the 2-hydroxy forms, 2OHPCA and 2OHPZ, by an enzyme encoded by phzO, adjacent to the phenazine biosynthetic operon (Maddula et al. 2008). It was shown previously that the expression of phzO was driven by its own promoter (Maddula et al. 2008) and this was supported by the RNA-seq analysis (data not shown). The expression of phzO was similar to that of phzX (RPKM values of 40.5 and 44.2, respectively, Table 1).
Phenazine biosynthetic and regulatory gene transcripts are in lower abundance in the gacA mutant relative to WT
In order to identify genes regulated by GacA in 30-84, mean RPKM values for the gacA mutant were compared with WT values. As expected, the RPKM value for gacA was very low in the gacA mutant compared with the WT (1.43 vs. 266.9, respectively) demonstrating that the gacA message was successfully disrupted, whereas expression of gacS was unchanged (Table 1). Mutation in gacA also resulted in lower abundance of the three ncRNAs; the greatest fold change was for rsmY (∼115-fold decrease). The RPKM values for rsmX, rsmY, and rsmZ were decreased to 201.1, 329.2, and 261.3, respectively (Table 1). These data indicate that GacA positively regulates the expression of rsmX, rsmY, and rsmZ in P. chlororaphis 30-84. Interestingly, the transcript abundance of rsmA was 2.2-fold higher in the gacA mutant compared with WT 30-84, whereas that of the closely related rsmE was relatively unchanged (Table 1).
Consistent with the deficiency in phenazine production observed in gac mutants, mutation in gacA resulted in 2- to 11-fold decreases in the transcript abundance of the phenazine biosynthetic genes (Table 1). Transcripts of six regulatory genes involved in phenazine production, iopA, iopB, rpoS, pip, phzR, and phzI, were underrepresented by one- to ninefold in the gacA mutant as compared with WT, whereas other regulatory genes such as hfq, csaI/R, and rpeA/rpeB were not appreciably altered (Table 1). The expression of these phenazine regulatory and biosynthetic genes was verified by qPCR (Fig. 3). The results suggest that GacA controls the expression of the phenazine biosynthesis genes in 30-84 partly by activating the expression of phzR, phzI, pip, rpoS, and iopA/iopB.
Figure 3.
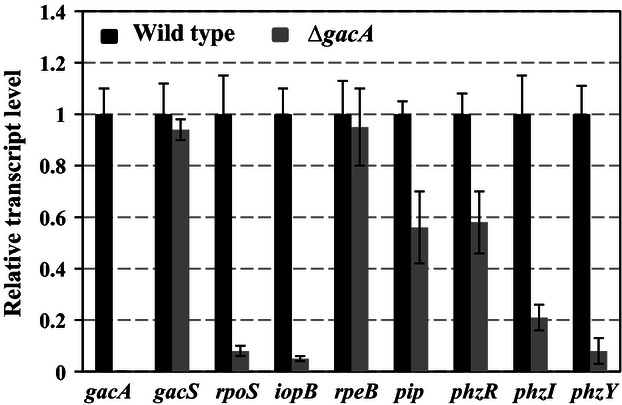
Verification of RNA-seq data by quantitative reverse transcription-polymerase chase reaction (qPCR). The relative fold change of gacA, gacS, rpoS, iopB, rpeB, pip, phzI, phzR, and phzB genes in gacA mutant compared towith the WT strain. The gene rpoD was used as the reference. Cells were grown in AB minimal medium + 2% casamino acid for 18 h with shaking to an OD600 of 1.2. Data points represent means of three replicates ± standard deviations. These experiments were repeated at least three times and similar results were obtained.
Genetic analyses of the phenazine biosynthetic and regulatory genes in 30-84
Expression of phzR/phzI does not rescue phenazine production in a gacA mutant
It was shown previously that the expression of phzI and AHL production are significantly reduced in 30-84 gacS and gacA mutants (Chancey et al. 1999). RNA-seq analysis indicated that both quorum sensing genes phzI and phzR were significantly decreased in the gacA mutant. One hypothesis is that this deficiency in quorum sensing is the reason for the lack of phenazine production by gacA mutants. However, if Gac operates by controlling phenazine production posttranscriptionally, constitutive expression of quorum sensing genes should not complement a gac mutant. To test this, the phzR gene was cloned into a medium copy vector driven by the Ptac promoter, and the plasmid with or without the phzR insertion was introduced into either WT, 30-84R (phzR−), or the gacA mutant. As expected, complementation of the phzR mutant by the pGT2-Ptac-phzR plasmid or the phzI mutant 30-84I by the addition of AHLs fully rescued phenazine production (Fig. 4A and B). In contrast, constitutive expression of phzR with the addition of AHLs to the growth medium did not rescue phenazine production in the gacA mutant (Fig. 4A and B). These results demonstrate that the underexpression of the PhzR/PhzI quorum sensing system in Gac mutants is not solely responsible for the phenazine deficiency observed.
Figure 4.
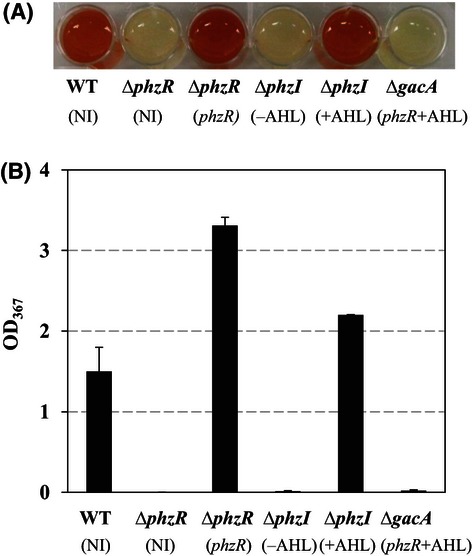
Phenazine production by WT and mutants with additional copies of phzR and supplemental AHLs. (A) Bacterial strains were grown in 2 mL LB medium in a 24-well plate at 28°C without shaking. The photo was taken 48 h after inoculation. (B) Bacterial strains were grown in LB medium for 24 h at 28°C with shaking at 200 rpm. Treatments included strains with the plasmid pGT-2 containing either no insert (NI) or the plasmid containing a Ptac-phzR insertion, AHLs were added to the growth medium as indicated. Phenazine was extracted and quantified at OD367. AHLs were extracted as in ethyl acetate as described previously and total AHL quantified by ability to rescue phenazine production in the phzI mutant 30-84I. Data points represent means of three replicates ± standard deviations.
RsmE, but not RsmA, regulates phenazine and AHL production
Pseudomonas chlororaphis 30-84 has both of the RNA-binding proteins RsmA and RsmE (this study) and they share 72% and 87% sequence identity and similarity, respectively. Amino acid identity of 30-84 RsmA and RsmE to their respective homologs in other Pseudomonas species was high (e.g., 100% and 93% identical to P. protegens Pf-5, P. fluorescens Pf01, P. syringae DC3000, P. aeruginosa PAO1; 94% and 91% identical Escherichia coli O157). The current model predicts that overexpression of RsmA or RsmE in the WT should provide phenotypes that are similar to gac mutants (Heeb and Haas 2001). To determine the roles of RsmA and RsmE in the regulation of phenazine production, multiple copies of rsmA and rsmE were introduced separately into the WT strain. Phenazine production decreased 7.1-fold in the WT strain harboring pLAFR3-rsmE (compared to WT with pLAFR3-NI) whereas overexpression of rsmA did not alter phenazine production in the WT strain (Fig. 5A). To confirm that rsmA was overexpressed in the WT, qPCR was performed and results showed that multiple copy of rsmA leads to 12.8 ± 0.6-fold transcript increase in the WT strain. To correlate phenazine production with phenazine gene expression, phzB::lacZ expression was measured in 30-84ZN harboring these plasmids. Consistent with phenazine production, multiple copies of rsmE reduced 30-84ZN β-galactosidase activity from 487.0 ± 78.6 to 48.1 ± 5.2 in 30-84ZN with pLAFR3-NI compared with pLAFR3-rsmE, respectively; the -galactosidase activity produced by strain 30-84ZN harboring pLAFR3-rsmA was similar to the 30-84ZN (pLAFR3-NI) control (511.0 ± 42.1 vs. 487.0 ± 78.6, respectively) (Fig. 5B). The result that RsmE, but not RsmA is involved in the regulation of phenazines is consistent with the observation in Pseudomonas sp. M18 that RsmA negatively controls pyoluteorin production but not phenazine production (Zhang et al. 2005). As pyoluteorin genes were not detected in P. chlororaphis 30-84, the target(s) of RsmA remains to be discovered.
Figure 5.

Role of RsmE in AHL and phenazine production. (A) Phenazine production by 30-84 with plasmid pLAFR3 containing either no insert (NI), rsmA, or rsmE. Bacterial strains were grown in LB medium for 24 h at 28°C with shaking. Phenazine was extracted as described previously and total phenazine was quantified at OD367. Data points represent means of three replicates ± standard deviations. (B) Repression of phzB expression in 30-84ZN by introduction of extra copies of rsmE but not by introduction of extra copies of rsmA. Treatments included 30-84ZN with plasmid pLAFR3 containing either NI, rsmE, or rsmA. 30-84ZN is a translational phzB::lacZ fusion. The β-galactosidase activities (Miller Units) were determined in triplicate (mean ± SE). (C) Suppression of AHL production by rsmE in trans. AHL accumulation was quantified from overnight cultures of WT with pLAFR3 containing either NI or rsmE using the AHL-specific reporter 30-84I/Z (phzI−, phzB::lacZ). The relative amount of AHL was determined by β-galactosidase assays. (D) Effect of inactivation of rsmE on phzB expression. Treatments included the phzB::lacZ reporter (30-84ZN), a phzB::lacZ/GacA− derivative (30-84ZW), and a phzB::lacZ/GacA−/RsmE− derivative (30-84ZWE). Bacterial cultures were grown in LB and phzB expression was measured as β-galactosidase activity after 24 h. Data are the means of two replicates from one representative experiment.
To determine whether RsmE also inhibited AHL production, the effect of overexpression of rsmE on AHL signal production was measured. Total AHL signal was extracted from 24 h bacterial cultures of WT (pLAFR3-NI) and WT (pLAFR3-rsmE). The relative amount of AHL signal was quantified by activation of β-galactosidase activity in strain 30-84IZ (Fig. 5C). β-galactosidase activity of strain 30-84IZ was approximately eightfold lower when the reporter strain was grown in the presence of AHLs prepared from overnight cultures of WT (pLAFR3-rsmE) compared with AHLs prepared from WT (pLAFR3-NI). These results indicate that RsmE overexpression also resulted in a significant decrease in AHL production.
To determine whether RsmE functions downstream of GacA in the regulation of phenazine gene expression, cosmid pLSP298EZ:TN5-11-2 containing a transposon insertion that inactivated rsmE was introduced into the genome of the gacA mutant 30-84ZW via marker exchange mutagenesis. Comparison of phzB::lacZ expression in strains 30-84ZN, 30-84ZW, and 30-84ZWE indicated that loss of rsmE restored β-galactosidase activity to the gacA mutant (Fig. 5D).
Constitutive expression of rsmZ rescues phenazine and AHL production in gac mutants
We further examined whether constitutive expression of ncRNAs would remove the need for a functional Gac system for phenazine production in strain 30-84. The rsmX, rsmY, and rsmZ genes were cloned under the control of a constitutive Ptac promoter such that the +1 site of the promoter was at the beginning of these genes. Although strains ΔgacS (pLAFR3-NI) and ΔgacA (pLAFR3-NI) failed to synthesize measurable amounts of phenazines, strain ΔgacS (pLAFR3-Ptac-rsmZ) and ΔgacA (pLAFR3-Ptac-rsmZ) produced approximately the same amount of phenazines as WT (pLAFR3-Ptac-rsmZ) (Fig. 6A). In contrast, constitutive expression of rsmX did not rescue phenazine production (data not shown). The expression rsmX was confirmed using qPCR, the gacA mutant containing rsmX plasmid showed 2.4-fold increase of rsmX expression than the WT strain. Constitutive expression of rsmY by the Ptac promoter appeared to be lethal in gac mutants, as no transformants were recovered with electroporation or triparental mating (data not shown).
Figure 6.
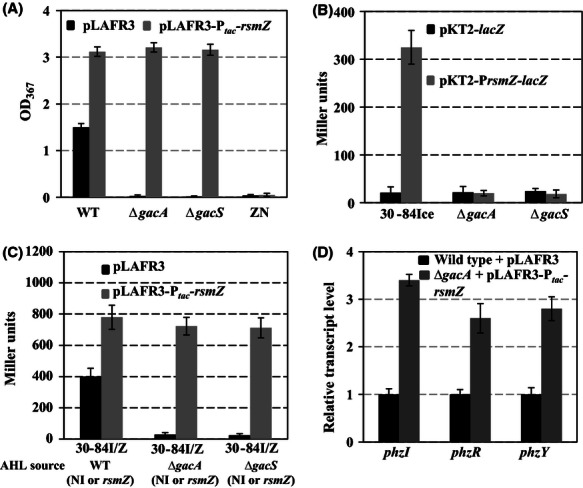
Impact of rsmZ on phenazine and AHL production. (A) Phenazine production by different derivatives in vitro. Treatments included strains with plasmid pLAFR3 containing either no insert (NI) or Ptac-rsmZ. Growth conditions were as in (Fig. 5A). Phenazine was extracted in benzene and the amount of phenazine was measured at OD367. Data points represent means of three replicates ± standard deviations. Similar results were obtained in at least three independent experiments. (B) Promoter activity of the rsmZ gene in WT, gacS, and gacA was determined by β-galactosidase activities. Treatments included strains with plasmid pKT-2 containing either a promoter-less lacZ insertion or the rsmZ promoter fused to the promoter-less lacZ gene (PrsmZ-lacZ). Insertions were made in front of the promoter-less gfp gene, already present on the plasmid. Bacterial strains were grown in LB medium for 24 h at 28°C with shaking to an OD600 of ∼1.8. Each value is the average from two different cultures ± standard deviations with two independent experiments. The experiment was repeated at least twice and similar results were obtained. (C) Effect of rsmZ on AHL production. Total AHL extractions were prepared from cell-free supernatants of WT, ΔgacS, or ΔgacA with plasmid pLAFR3 containing either NI or Ptac-rsmZ. Cell-free supernatants were made from overnight cultures grown in LB medium (OD600 ∼1.8). The relative amount of AHL in each extract was determined by β-galactosidase assays using the AHL-specific reporter 30-84I/Z (phzI−, phzB::lacZ). Bars represent β-galactosidase activity measured in Miller Units of 30-84I/Z grown with the AHL extracts indicated. Each value is the average from three different cultures ± standard deviations. (D) Relative expression of phzI, phzR, and phzY genes in the gacA mutant (harboring the pLAFR3-Ptac-rsmZ) compared with WT (harboring pLAFR3 empty vector) as determined by qPCR. rpoD was used as the reference gene. Cells were grown in AB minimal medium + 2% casamino acid for 18 h with shaking to an OD600 of 1.2. Data points represent means of three replicates ± standard deviations. These experiments were repeated at least three times and similar results were obtained.
We further explored the regulatory role of GacA on ncRNA expression by characterizing the effect of mutations in gacA on rsmZ promoter activity using the reporter plasmid pKT2-PrsmZ-lacZ-gfp (e.g., by measuring fluorescence or β-galactosidase activity). The reporter plasmid was introduced into a gacA and a gacS mutant as well as 30-84Ice (phzB::inaZ), which has a functional gacA, but does not produce phenazines (thus no interference with GFP fluorescence). Strain 30-84Ice (pKT2-PrsmZ-lacZ-gfp) was brightly fluorescent whereas the Gac mutants with the reporter plasmid were not (data not shown). Introduction of the plasmid into the gacA or gacS mutant resulted in an 11-fold reduction in expression of β-galactosidase relative to strain 30-84Ice (pKT2-PrsmZ-lacZ-gfp) (Fig. 6B). These results confirm that the Gac system positively regulates the expression of rsmZ.
To determine whether constitutive expression of rsmZ also regulated AHL signal production, the effect of constitutive expression of rsmZ on AHL signal production was measured. Total AHL signal was extracted from 24 h bacterial cultures and the relative amount of AHL signal present was quantified by activation of β-galactosidase activity in strain 30-84IZ (Fig. 6C). The β-galactosidase activity of the 30-84IZ reporter was significantly higher when grown with AHLs prepared from strains ΔgacS (pLAFR3-Ptac-rsmZ) and ΔgacA (pLAFR3-Ptac-rsmZ) compared with AHLs prepared from ΔgacS (pLAFR3-NI) and ΔgacA (pLAFR3-NI); β-galactosidase activities of the 30-84IZ reporter were 712.0 ± 77.1 and 723.1 ± 56.2 as compared with 28.9 ± 12.2 and 24.1 ± 10.2, respectively. This increase (27-fold) in phzB expression with constitutive rsmZ expression supports the hypothesis that the presence of the rsmZ ncRNA bypasses the requirement for a functional Gac system for both phenazine and AHL production. qPCR analyses confirmed that the transcript abundance of phzI, phzR, and phzY in the gacA mutant was two- to fourfold higher in the presence of rsmZ (Fig. 6D). Collectively, these results indicate that Gac regulates phenazine production and quorum sensing by activating the expression of rsmZ.
Constitutive expression of rsmZ also qualitatively restored HCN, exoprotease, gelatinase, and lipase production and reduced fluorescence to WT levels in strain 30-84W (data not shown). Consistent with restoration of these capabilities, constitutive expression of rsmZ also reinstated 30-84's ability to inhibit the mycelial growth of the fungal pathogen Ggt. Zones of Ggt mycelial inhibition by WT, ΔgacA, ΔgacA (pLAFR3-NI), and ΔgacA (pLAFR3-rsmZ) were 8.4 ± 0.9, 0.0 ± 0.3, 0.2 ± 0.4, and 9.4 ± 0.8 mm, respectively.
Model of Gac/Rsm regulation of phenazines in 30-84
In this study, the major components of the Gac/Rsm signal transduction pathway in P. chlororaphis 30-84 were identified and characterized (Fig. 7). For example, we discovered that the genome of 30-84 contains genes encoding both RsmE and RsmA. Only overexpression of rsmE in the WT reduced the gene expression and production of phenazine. Similarly only deletion of rsmE in a gacA mutant restored both, demonstrating that rsmE deletion bypassed the need for a functional GacA to produce phenazines. The genome of 30-84 contains the gene sequences for the three small ncRNAs, rsmX, rsmY, and rsmZ. A functional Gac system is required for rsmZ expression and that expression of rsmZ from a constitutive promoter restored phenazine, HCN, exoprotease, lipase, and gelatinase production and reduced fluorescence to WT levels in a gacA mutant. In contrast, constitutive expression of rsmX did not restore any of the phenotypes tested, and constitutive expression of nor rsmY could not be achieved. The importance of the rsmZ in Gac regulation of products essential for biological control is interesting given that rsmZ has the longest promoter region, suggesting the potential for multiple protein interactions in its regulation. The observation that rsmZ plays a different role than the other two ncRNAs in Gac-mediated regulation has not been reported previously. However, the observation that transcript abundances of both rsmX and particularly rsmY are significantly lower in a GacA mutant is consistent with the hypothesis that they are involved in Gac-mediated regulation of as yet unidentified targets. Consistent with Gac/Rsm exerting posttranscriptional control of phenazine production, previous work showed that the introduction of a cosmid containing the entire phenazine biosynthetic cluster and phzR and phzI in trans on a low copy plasmid failed to complement a gacA mutant for phenazine production (Chancey et al. 1999). In this study, we bypassed transcriptional regulation of quorum sensing by providing PhzR (from a constitutive promoter) and purified AHL signal, confirming the prediction that Gac/Rsm controls phenazine gene expression posttranscriptionally.
Figure 7.
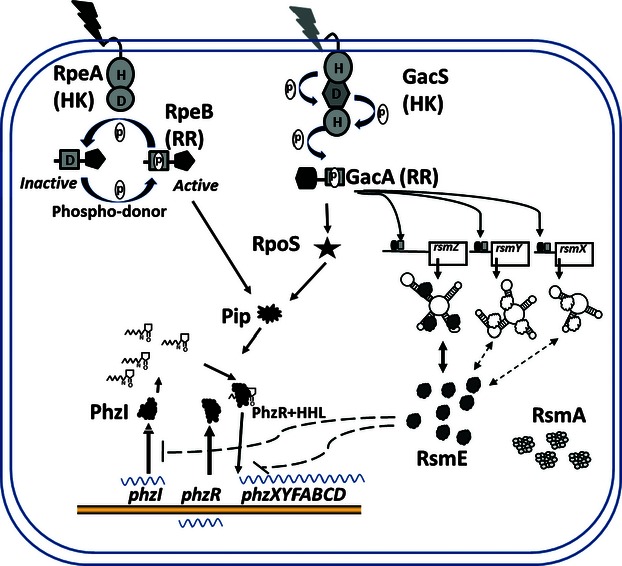
Proposed model for the regulation of phenazine biosynthesis by the Gac-Rsm system in Pseudomonas chlororaphis strain 30-84. The model shows the presumed position of the GacS/GacA system in relation to other known regulators of phenazine production. GacS is a transmembrane protein according to in silico predictions based on amino acid sequences. Amino acids H (histidine) and D (aspartate) involved in phosphorylation are indicated. The shaded area of GacA indicates the DNA-binding domain. Solid arrows and blunt lines point to genes (or processes) that are positively or negatively affected, respectively. Dashed arrows or lines indicate unknown or as yet uncharacterized regulatory pathways. GacA positively controls the expression of rsmX, rsmY, and rsmZ, which in turn activates phenazine production by titrating the translation suppressor RsmE. Although the transcript abundance of rsmA was 2.2-fold higher in the gacA mutant compared with WT 30-84, RsmA does not appear to play a role in the regulation of phenazines under the experimental conditions tested here. A functional GacA is also required for increased expression of other phenazine regulatory genes, including rpoS, pip, and phzR/phzI. HK, histidine kinase; RR, response regulator; P, phosphoryl group.
The transcriptomic data demonstrated that GacA controls the expression of the phenazine biosynthetic genes (phzXYFABCD and phzO), the quorum sensing genes phzR and phzI, and also several additional regulatory genes involved in phenazine gene expression including pip, rpoS, and iopA/iopB. In P. chlororaphis stains 30-84 and PCL1391, Pip promotes phenazine production by enhancing phzI and phzR expression (Girard et al. 2006a; Wang et al. 2012a,b). In both strains, the expression of pip is regulated by the sigma factor rpoS, which in turn is regulated by GacA. In 30-84 pip also is regulated by the RpeB/RpeA TCST system (Wang et al. 2012a,b). Complementation assays showed that constitutive expression of pip or phzR rescued phenazine production in rpeB mutants whereas multiple copies of rpeB or pip failed to rescue phenazine production in a phzR mutant; for both strains, phzR expression restored phenazine production in a pip mutant (Wang et al. 2012a,b). Furthermore, expression of gacS, gacA, and rpoS were unchanged in an rpeB mutant, and conversely the expression of rpeA and rpeB were unchanged in an rpoS mutant (Wang et al. 2012a,b). These results indicate that RpoS and the RpeB/RpeA independently regulate phenazine production using pip as a common regulatory intermediate. Consistent with these findings, data presented here showed that expression of rpeB and rpeA were not altered by mutation of gacA, indicating that RpeB/RpeA and GacS/GacA also do not regulate phenazines in a hierarchic manner. However, the effect of gacA mutation on rpoS and pip expression suggests that GacA influences the expression of additional phenotypes via RpoS and Pip.
Transcriptomic analyses of genes controlled by Gac: a preliminary survey
GacS/GacA functions as a positive regulator of secondary metabolism and exoenzyme production
In 30-84, gac mutants are not only deficient in phenazine production, but also HCN, exoprotease, and lipase (Chancey et al. 1999). Consistent with these observations, the transcript abundances of genes that encode for HCN, exoprotease/exopeptidase, and lipase activities were lower in the gacA mutant than in WT (Table 2). Additionally, the transcript abundances of 30-84 genes annotated to encode pyrrolnitrin biosynthesis (prnABCD) and four genes involved in chitin degradation were lower in the gacA mutant than in WT (Table 2). Some of these results were confirmed by qPCR analysis using the hcnA, aprA, and prnA genes (data not shown). These results are consistent with previous findings showing a reduction in the abundance of transcripts of the biosynthetic genes for HCN, pyrrolnitrin, and protease in a P. protegens Pf-5 gacA mutant (Hassan et al. 2010). In Pf-5, the products of these genes have been demonstrated to be important for biological control activity.
Table 2.
Mean transcript abundance and ratio of abundances (ΔgacA/WT) of genes involved in secondary metabolism and iron uptake in the gacA mutant compared with WT strain
| Gene ID | Gene | Protein description | Mean RPKM WT | Mean RPKM ΔgacA | ΔgacA/WT | P-value |
|---|---|---|---|---|---|---|
| Hydrogen cyanide | ||||||
| Pchl3084_2378 | hcnA | Hydrogen cyanide synthase | 746.54 | 243.92 | 0.33 | 0.01 |
| Pchl3084_2379 | hcnB | Hydrogen cyanide synthase | 474.43 | 70.53 | 0.15 | 0.00 |
| Pchl3084_2380 | hcnC | Hydrogen cyanide synthase | 705.07 | 66.39 | 0.09 | 0.00 |
| Protease | ||||||
| Pchl3084_3127 | aprA | Metalloprotease | 100.83 | 4.77 | 0.04 | 0.02 |
| Pchl3084_2293 | aprX | Metallopeptidase | 164.03 | 6.44 | 0.04 | 0.01 |
| Lipase | ||||||
| Pchl3084_3120 | Lipase | 9.4 | 3.4 | 0.37 | 0.00 | |
| Pchl3084_3439 | Lipase | 16.0 | 4.8 | 0.3 | 0.01 | |
| Pchl3084_3943 | Lipase | 25.9 | 11.4 | 0.44 | 0.01 | |
| Pchl3084_0883 | Phospholipase | 50.8 | 10.6 | 0.21 | 0.02 | |
| Pyrrolnitrin | ||||||
| Pchl3084_3143 | prnD | Aminopyrrolnitrin oxidase | 6.94 | 0.57 | 0.08 | 0.04 |
| Pchl3084_3144 | prnC | Halogenase | 12.53 | 0.85 | 0.07 | 0.02 |
| Pchl3084_3145 | prnB | Pyrrolnitrin biosynthesis | 6.88 | 1.23 | 0.18 | 0.00 |
| Pchl3084_3146 | prnA | Tryptophan halogenase | 16.45 | 1.83 | 0.11 | 0.01 |
| Chitinase | ||||||
| Pchl3084_2020 | Chitin-binding domain protein | 36.06 | 4.28 | 0.12 | 0.03 | |
| Pchl3084_2021 | Chitinase | 31.17 | 3.46 | 0.11 | 0.01 | |
| Pchl3084_3180 | Chitinase | 57.95 | 1.83 | 0.03 | 0.02 | |
| Pchl3084_3181 | Chitin-binding domain protein | 51.78 | 3.44 | 0.07 | 0.01 | |
| Iron uptake | ||||||
| Pchl3084_3935 | pvdA | L-ornithine 5-monooxygenase | 2.32 | 5.11 | 2.20 | 0.01 |
| Pchl3084_3936 | fpvI | Sigma-70 factor | 12.77 | 23.78 | 1.86 | 0.00 |
| Pchl3084_0320 | TonB-dependent outer membrane receptor | 1.71 | 6.19 | 3.61 | 0.02 | |
| Pchl3084_0855 | TonB-dependent outer membrane receptor | 23.45 | 85.19 | 3.63 | 0.00 | |
| Pchl3084_2518 | TonB-dependent outer membrane receptor | 2.71 | 7.17 | 2.65 | 0.00 | |
| Pchl3084_3174 | TonB-dependent outer membrane receptor | 8.26 | 16.77 | 2.03 | 0.02 | |
| Pchl3084_3659 | TonB-dependent outer membrane receptor | 3.05 | 6.43 | 2.11 | 0.00 | |
| Pchl3084_3848 | TonB-dependent outer membrane receptor | 7.11 | 15.08 | 2.12 | 0.00 | |
| Pchl3084_5801 | ExbB | TonB system transport protein | 55.52 | 116.65 | 2.10 | 0.04 |
Transcripts of genes potentially involved in type VI secretion system, signal transduction, and carbon storage are in lower abundance in the gacA mutant relative to WT
Genes annotated as encoding components of a type VI secretion system (T6SS) also were notably underexpressed (2- to ∼100-fold) in the gacA mutant relative to WT, suggesting positive control by GacA (Table S4). Again, this is similar to previous findings showing that the expression of Pf-5 genes homologous to the P. aeruginosa T6SS (HIS-I type) was reduced in a gacA mutant from early to late growth phase (Hassan et al. 2010). In contrast to Pf-5 where all 16 of the differentially expressed genes encoding the T6SS system were located in the same gene cluster, the 24 differentially expressed T6SS genes identified in this study were distributed among six different loci. Previous studies also demonstrated GacS/GacA control of the T6SS in P. aeruginosa and P. syringae (Silverman et al. 2012). In P. aeruginosa, the T6SS delivers bacteriolytic effectors to target cells and is associated with competition among bacterial species (Russell et al. 2011; Julie et al. 2012). Future studies will be required to determine whether 30-84 contains a functional T6SS and if so, its function.
Genes annotated as encoding enzymes involved in polyhydroxyalkanoate (PHA) synthesis and utilization also were underexpressed (2- to ∼100-fold) in the gacA mutant relative to WT, suggesting positive control by GacA (Table S5). PHAs are bacterial storage materials produced by many fluorescent pseudomonads including P. aeruginosa, P. putida, and P. chlororaphis (aureofaciens) under nutrient conditions in which carbon is in relatively high abundance, but other nutrients are limited (Nishikawa et al. 2002; Pham et al. 2004; Hervas et al. 2008). Genes included phaD, a TetR-like transcriptional regulator that in P. putida KT2442 activates the pha cluster (phaC1ZC2D) conferring the ability to degrade PHAs (de Eugenio et al. 2010). In P. oleovorans GPo1, phaD is transcribed as part of the pha operon and controls its own transcription as well as the transcription of the phaIF operon, which in turn is involved in the regulation of pha genes (Prieto et al. 1999). Consistently, in this study, the transcripts of phaC1, phaZ, phaC2, phaI, and phaF also were underrepresented (2- to 17-fold) in the gacA mutant relative to the WT (Table S5). Previously, it was shown that the PHA synthase phaC2 is underexpressed in an rpoS mutant of P. chlororaphis PCL1391 (Girard et al. 2006b). Work with P. aeruginosa PAO1 demonstrated that mutants deficient in PHA biosynthesis were less tolerant of heat shock both in liquid culture and in biofilms and were altered in attachment and biofilm architecture relative to WT (Pham et al. 2004). Moreover, comparison of alginate production by PHA-negative derivatives of WT PAO1 and the alginate-overproducing strain, FRD1, led the authors to suggest that the alginate and PHA pathways compete for acyl-CoA precursors (Pham et al. 2004). In 30-84, the transcript abundance of the alginate regulatory gene algK was fourfold higher in the gacA mutant compared with WT, however, the majority of genes annotated as being involved in alginate synthesis were unchanged. The roles of PHA or alginate production and utilization by 30-84 have yet to be determined, but it is interesting to consider given PHA's potential for influencing bacterial survival and biofilm formation.
Differential transcript abundance of 23 genes annotated as being involved in signal transduction or transcription regulation suggest that these genes also were regulated by GacA (Table S6). Among them, transcripts of 21 genes were underrepresented in the gacA mutant compared with WT. Most of these genes are functionally uncharacterized suggesting there remains a large gap in our understanding of P. chlororaphis gene regulation by the Gac system.
Transcripts of genes involved in iron acquisition are in higher abundance in the gac mutant relative to WT: potential contribution to the Gac− phenotype
Hyper-fluorescence is a consistent phenotype of gac mutants among fluorescent pseudomonads, including 30-84 (Chancey et al. 2002). It was shown previously that the Gac system negatively controls a large number of genes involved in iron acquisition (Hassan et al., 2010). Transcripts of nine genes annotated to be involved in iron acquisition were two- to threefold more abundant in the gacA mutant than the WT (Table 2). Six of them were annotated as TonB-dependent outer membrane receptors potentially involved in the uptake of ferric pyoverdine siderophores. According to current siderophore uptake models, when ferric pyoverdine is recognized by its cognate TonB outer membrane receptor, it is transported into the periplasmic space via a two-gate mechanism (Schalk 2008). TonB and proton motive force provide the energy for siderophore uptake and cause the release of the iron, which is subsequently transported into the cytoplasm through an ABC transporter (Schalk 2008). Consistent with increased iron uptake, transcripts of pvdA (involved in fluorescent pyoverdine biosynthesis), exbB (a cytoplasmic membrane protein), and fpvI (an RNA polymerase sigma-70 factor that recruits RNA polymerase for expression of iron transport genes) were overrepresented in the gacA mutant compared with WT. Previously it was shown that insertion of Tn5 into the pvdA locus of 30-84 resulted in the loss of fluorescence on low iron media (data not shown), linking fluorescence with pyoverdine synthesis. These results are consistent with overproduction of pyoverdine being partially responsible for the hyper-fluorescent phenotype typical of gac mutants of Pseudomonas and suggest a possible benefit to having Gac− phenotypes in mixed populations.
Transcripts of genes involved in preparation for exponential growth are in higher abundance in the gac mutant relative to WT: GacA regulation of sigma factors and genes involved in translation
Protein synthesis is a highly coordinated process that requires ribosomes composed of multiple proteins and ribosomal RNAs, mRNA, tRNAs, initiation and elongation factors, and many other genes. One of the most dramatic transcriptional consequences of loss of gacA in P. chlororaphis 30-84 was the overexpression (two- to fourfold) relative to the WT of 50 genes involved in translation, including those encoding most of the large and small subunit ribosomal proteins, the translation initiation factor IF-3, and the three translation elongation factors (Table S7). Because the ribosomal protein transcripts were increased in the gacA mutant, it is logical to speculate that rRNAs also were increased. However, due to the rRNA depletion steps, the RNA-seq only detected small amounts of rRNA transcripts. Therefore, we performed qPCR to quantify the relative abundance of the 16S rRNA in the gacA mutant compared with the WT. Relative to WT, the abundance of 16S rRNA was significantly higher (1.8 ± 0.5-fold) in the gacA mutant, suggesting higher expression of ribosomal RNA. In addition, tRNA genes for transferring aspartate, glutamine, lysine, isoleucine, and alanine were overexpressed in the gacA mutant compared with WT; whereas threonine and glycine tRNAs were underexpressed (Table S7). A unique consequence of these findings was that due to the consistency of rpoD expression relative to 16S rRNA abundance, rpoD was used as the reference for all qPCR analyses comparing gene expression in the gacA mutant and WT.
In previous study with 30-84 and other pseudomonads, it was found that mutations in gacA or gacS resulted in earlier transition of cells from lag to exponential phase growth, and this was suggested as an explanation for the competitive advantage of gac mutants in liquid broth cultures (Duffy and Defago 2000; Chancey et al. 2002; van den Broek et al. 2005; Driscoll et al., 2011). Although the precise mechanism(s) that facilitate gac mutants entering exponential growth phase earlier than the WT is unclear, we speculate that the reduction in rpoS expression in gac mutants relative to WT may contribute to this effect. Differential regulation of the stationary phase sigma factor RpoS, but not the housekeeping sigma factor RpoD in a gacA mutant is particularly interesting given that RpoS competes with RpoD for binding to RNA polymerase core enzymes (Farewell et al. 1998; Battesti et al. 2011). This competition, in part, determines overall patterns of gene expression in the cell. Most research on RpoS has focused on its role in the transition from exponential to stationary phase growth. However, recently Rolfe et al. (2012) showed that during the transition from lag to exponential phase growth rpoS expression increased transiently, presumably to enable the cell to respond to oxidative challenges due to the introduction of the stationary phase cells into oxygenated fresh medium. In contrast, levels of RpoD increased twofold during this transition period. A reduction in levels of RpoS in gacA mutants would result in higher levels of expression of RpoD-regulated genes, which are involved in primary growth. Increased levels of transcription of translational machinery, including ribosomal proteins, rRNAs, tRNAs, and accessory proteins in the gac mutant as reported here also would favor earlier entry into exponential growth.
The role of GacA in bacterial motility
The Gac system has been shown to differentially control motility in various bacteria. For example, it positively contributes to motility in E. coli K-12 (Wei et al. 2001), P. protegens Pf-5 (Hassan et al. 2010), and P. syringae pv. tabaci (Marutani et al. 2008), but negatively in P. fluorescens F113 (Martínez-Granero et al. 2012) and Erwinia amylovora Ea1189 (Zhao et al. 2009). In both Pf-5 and P. syringae pv tabaci, the Gac system positively influences the level of transcription of the flagellar genes (Marutani et al. 2008; Hassan et al. 2010). Consistent with the potential role of GacA in enhanced bacterial motility, colonies of 30-84 gac mutants typically appear larger on standard culture plates. Bacterial motility was assessed for strain WT, ΔgacS, ΔgacA, and 30-84ZN by inoculating bacterial cells onto motility plates (0.25% agar) and measuring the diameter of the area colonized by bacterial cells at 24 and 48 h as described previously (Wang et al. 2011). The Gac mutants exhibited enhanced motility compared with that of the WT strain 12 and 24 h following inoculation (Fig. S1A and B). In contrast, phenazine deficient derivative 30-84ZN (phzB::lacZ) exhibited a similar motility pattern to the WT suggesting that the increased motility in the gac mutant was not due to lack of phenazine production. Diameters of colonies for the WT, ΔgacS, ΔgacA, and 30-84ZN at 48 h were about 4.8 ± 0.1, 6.4 ± 0.3, 6.5 ± 0.4, and 4.8 ± 0.3 cm (longest dimension from the inoculation point), respectively. These results indicate that the Gac system negatively regulates bacterial motility in 30-84.
It was demonstrated previously that swimming motility in P. fluorescens F113 is under negative control by Gac and that this downregulation occurs through the repression of the flagella master regulatory gene fleQ (Martínez-Granero et al. 2012). One of the factors affecting motility is flagella production. Moreover, a P. chlororaphis 30-84 fliM mutant is nonmotile (L. S. Pierson et al., unpubl. data). However, RNA-seq analyses revealed that the transcript abundance of flagellar genes such as fliA, fliM, and fliC were not significantly altered in the gacA mutant (Fig. S1C), and these data were verified by qPCR (Fig. S1D). The results suggest that the impact of GacA on motility may be secondary to flagella structure, possibly related to flagellar rotation. Another possibility is that increased swimming motility is related to enhanced surfactant production, as observed in other bacterial species (Wilf et al., 2012; Daniels et al. 2004). However, known surfactant biosynthetic genes such as the rhlA or homologs were not detected in P. chlororaphis 30-84 genome.
Conclusions
The role of the ncRNAs rsmY and rsmZ in the Pseudomonas Gac/Rsm signal transduction pathway was first detected by their binding capacity to the regulatory protein RsmA and subsequently RsmE in P. fluorescens CHA0, where their interaction contributed to the regulation of HCN, exoprotease, and 2,4-diacetylphloroglucinol biosynthesis important for the biological control capability of the strain (Heeb et al. 2002; Reimmann et al. 2005). In our study, the functionality of the components of the Gac/Rsm signal transduction pathway in the regulation of phenazines and other genes important for biological control in strain 30-84 was examined. Only rsmZ and rsmE are linked to phenazine production, however, further RNA–protein binding experiments are needed to verify whether direct interactions between RsmE and the ncRNA rsmZ are involved in the regulation of phenazine biosynthesis. Interestingly, in P. fluorescens CHA0 rsmE expression was shown to be regulated negatively by RsmA (Reimmann et al. 2005). What role RsmA plays in Gac-mediated regulation in 30-84 has yet to be determined. In P. fluorescens CHA0 and in P. aeruginosa, the regulatory effect of gacA was attributed exclusively to the direct control of only two ncRNAs rsmY and rsmZ (Heeb et al. 2002; Brencic and Lory 2009). In P. aeruginosa, single rsmY and rsmZ mutants synthesize intermediate levels of phenazines, whereas the double rsmY/rsmZ mutant produces less (Kay et al. 2006). Although constitutive expression of rsmZ completely restored phenazine, HCN, and exoprotease production in a 30-84 gacA mutant, we cannot exclude the possibility that rsmY also partially contributes to the production of these key biological control compounds. Future study will be directed toward identifying the potential roles of the three ncRNAs in strain 30-84.
In most Pseudomonas species, phenazine production is controlled by combinations of conserved regulatory systems integrated into sensory networks potentially responsive to environmental, nutritional, population, and metabolic inputs (reviewed in Pierson and Pierson 2010). Transcriptomic analysis revealed that the expression of regulatory genes known to be involved in phenazine production (including phzR and phzI, pip, and rpoS) as well as other genes annotated as being involved in trascriptional regulation were affected by a gacA in mutation 30-84. Our genetic analysis demonstrated that whereas a gacA mutation could be bypassed by either mutation of rsmE or constitutive expression of rsmZ, it could not be rescued by expression of quorum sensing (e.g., constitutive expression of phzR with the addition of AHLs) or other regulatory genes. These data provide evidence that the phenazine biosynthesis genes are direct targets of Gac/Rsm regulation and highlight the strength of gene deletion/overexpression analysis in combination with transcriptomic profiling as an approach to study complex regulatory mechanisms. These results also illustrate the multiple-level regulatory role of the Gac/Rsm system in the control of phenazines and suggest that the far-reaching influences of GacA on the transcriptome are likely mediated through intermediate transcriptional regulatory genes.
Our transcriptional profiling revealed that GacA regulates a wide array of biological functions in P. chlororaphis 30-84. In this study, transcriptomic analysis identified 771 genes that were differentially expressed in the GacA mutant compared with WT 30-84, with 551 genes downregulated in the gacA mutant under our experimental conditions. Consistent with previous observations that gac mutants are phenazine, HCN, exoprotease, and lipase deficient, the transcript abundances of genes that code for these phenotypes were reduced in the gacA mutant compared with WT 30-84. Additionally, the transcript abundances of 30-84 genes annotated to encode pyrrolnitrin biosynthesis and chitin degradation also were reduced. These results are consistent with previous findings demonstrating that GacS/GacA functions as a positive regulator of secondary metabolites that contribute to biological control activity in Pseudomonas. Other genes positively regulated by GacA in strain 30-84 that could be of potential importance in rhizosphere survival, plant–microbe interaction, and biological control include genes annotated as coding for components of a T6SS. Beyond genes involved in well-defined functional processes, the gacA regulon included a gene cluster that is involved in the utilization of polyhydroxyalkanoates, important storage molecules in many pseudomonads. Further studies will be required to determine whether either strain contains a functional T6SS and whether strain 30-84 is capable of utilizing polyhydroxyalkanoates. Future studies also will be directed at how Gac-mediated regulation of these potentially beneficial genes is altered under rhizosphere conditions.
Many Pseudomonas species identified for biological control demonstrate phenotypic variation resulting from spontaneous mutation in gacS or gacA. However, phenotypic variation derived from conserved mutation of specific regulatory genes is not limited to Pseudomonas, and has been observed in many Gram− and Gram+ species resulting from mutations in gac or other regulatory genes (van den Broek et al. 2005). The prevalence of Pseudomonas gac mutants in laboratory and natural environments suggests that there is some benefit to the gac mutant phenotype and thus selection at some level to maintain phenotypic variation in the species. Previous study with strain 30-84 provides evidence that the presence of the gac mutants in WT-Gac mixed populations in biofilms and on plant roots can be beneficial (Chancey et al. 2002; Driscoll et al., 2011). Thus, of particular interest in this study was the identification of genes that are expressed at a higher level in 30-84 gacA mutants. Our transcriptomic analysis showed that 220 genes that were overexpressed in the gacA mutant under our experimental conditions. The most obvious group, genes involved in iron uptake, may increase iron availability especially in the biofilm context. Increased protein translation is another possible benefit, as the shorter transition to exponential growth for gac mutants allows them to quickly establish a population. It is worth noting that 52 upregulated genes in the gacA mutant are hypothetical or unknown genes, including the highest upregulated gene (Pchl30-84_3493, 26.2-fold increase) (Table S3). Functional characterizations of these highly expressed genes are currently underway.
Acknowledgments
This project was supported by the USDA NIFA NRI Competitive Grants Project no. 2008-35319-21879 and the NSF Integrated Organismal Biology Project no. IOS-1035157. We thank Scott Hunicke-Smith and members of the University of Texas Genomic Sequencing and Analysis Facility for assistance with many aspects of the RNA-Seq analysis. We also thank Joyce Loper and Ian Paulsen for leading the USDA/NSF Microbial Genome Initiative Project: Genomics Based Discovery of Novel Traits in Plant-Associated Pseudomonas species that resulted in the production of the high quality draft genomic sequence of P. chlororaphis 30-84.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. (A) Migration phenotypes of WT, gac mutants and 30-84ZN at 48 h after incubation. (B) Comparison of the movement distance of WT, gac mutants and 30-84ZN. Cells were inoculated on the surface of a motility plate containing 0.25% agar as described previously (Wang 2011). Plates were incubated at 28°C for 48 h, during which the displacement diameter to the outermost edge of the movement area was measured. The experiment was performed at least three times in triplicate. (C) Expression levels of fliA, fliC, and fliM in WT and a gacA mutant. RPKM values representing levels of gene expression were calculated from RNA-seq analysis and log transformed. Data points represent means of three replicates ± standard deviations. (D) Verification of RNA-seq data with qPCR. Bacterial strains were grown overnight in LB broth and reinoculated in 5 mL AB medium + 2% casamino acid. Relative expression of fliA, fliC, and fliM genes, normalized to the expression of rpoD gene, was determined by qPCR after ∼18 h growth (OD600 at 1.2).
Table S1. Bacterial strains and plasmids used in this study.
Table S2. Oligonucleotides used for gene cloning and qPCR.
Table S3. Differentially expressed genes in the gacA mutant compared with the WT strain.
Table S4. Mean transcript abundance and ratio of abundances (ΔgacA/WT) of type VI secretion system (T6SS) genes in the gacA mutant compared with the WT.
Table S5. Mean transcript abundance and ratio of abundances (ΔgacA/WT) of polyhydroxyalkanoate (PHA) biosynthetic genes in the gacA mutant compared with the WT.
Table S6. Mean transcript abundance and ratio of abundances (ΔgacA/WT) of regulatory genes in the gacA mutant compared with the WT.
Table S7. Mean transcript abundance and ratio of abundances (ΔgacA/WT) of genes involved in protein metabolism in the gacA mutant compared with the WT.
References
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer C, Heeb S, Pessi G, Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek D, Chin-A-Woeng TFC, Bloemberg GV, Lugtenberg BJJ. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J. Bacteriol. 2005;187:593–600. doi: 10.1128/JB.187.2.593-600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancey ST, Wood DW, Pierson LS., III Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 1999;65:2294–2299. doi: 10.1128/aem.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancey ST, Wood DW, Pierson EA, Pierson LS., III Survival of Gac mutants of the biological control bacterium Pseudomonas aureofaciens 30–84 in the wheat rhizosphere. Appl. Environ. Microbiol. 2002;68:3308–3314. doi: 10.1128/AEM.68.7.3308-3314.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J, Vanderleyden J, Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- De Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll WW, Pepper JW, Pierson LS, III, Pierson EA. Spontaneous Gac mutants of Pseudomonas biological control strains: cheaters or mutualists? Appl. Environ. Microbiol. 2011;77:7227–7235. doi: 10.1128/AEM.00679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, et al. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 2003;185:4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy BK, Defago G. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 2000;66:3142–3150. doi: 10.1128/aem.66.8.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Eugenio LI, Galán B, Escapa IF, Maestro B, Sanz JM, García JL, et al. The PhaD regulator controls the simultaneous expression of the pha genes involved in polyhydroxyalkanoate metabolism and turnover in Pseudomonas putida KT2442. Environ. Microbiol. 2010;12:1591–1603. doi: 10.1111/j.1462-2920.2010.02199.x. [DOI] [PubMed] [Google Scholar]
- Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- Girard G, Barends S, Rigali S, Lugtenberg ET, van Rij BJ, Bloemberg GV. Pip, a novel activator of phenazine biosynthesis in Pseudomonas chlororaphis PCL1391. J. Bacteriol. 2006a;188:8283–8293. doi: 10.1128/JB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard G, Lugtenberg ET, van Rij BJ, Bloemberg GV. Roles of psrA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology. 2006b;152:43–58. doi: 10.1099/mic.0.28284-0. [DOI] [PubMed] [Google Scholar]
- Haas D, Keel C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 2003;41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LDH, et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 2010;12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- Heeb S, Haas D. Regulatory roles of the Gac two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe Interact. 2001;14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 2002;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas AB, Canosa I, Santero E. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J. Bacteriol. 2008;190:416–420. doi: 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humair B, Wackwitz B, Haas D. GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 2010;76:1497–1506. doi: 10.1128/AEM.02014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julie M, Brunet YR, Cascale E, Mougous JD. Structure and Regulation of the T6SS System. Annu. Rev. Microbiol. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, et al. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Maddula VSRK, Zhang Z, Pierson EA, Pierson LSIII. Quorum sensing and phenazines are involved in biofilm formation by Pseudomonas chlororaphis strain 30–84. Microb. Ecol. 2006;52:289–301. doi: 10.1007/s00248-006-9064-6. [DOI] [PubMed] [Google Scholar]
- Maddula VSRK, Pierson EA, Pierson LSIII. Altering the ratio of phenazines in Pseudomonas chlororaphisaureofaceins) strain 30–84: effects on biofilm formation and pathogen inhibition. J. Bacteriol. 2008;190:2759–2766. doi: 10.1128/JB.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch JL, Erfle M, Wykes EJ. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- Martínez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, Rivilla R. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS ONE. 2012;7:e31765. doi: 10.1371/journal.pone.0031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutani M, Taguchi F, Ogawa Y, Hossain MM, Inagaski Y, Toyoda K, et al. Gac two-component system in Pseudomonas syringae pv. tabaci is required for virulence but not for hypersensitive reaction. Mol. Genet. Genomics. 2008;279:313–322. doi: 10.1007/s00438-007-0309-y. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Ksenzenko VN, Bonsall RF, Cook RJ, Boronin AM, Thomashow LS. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2–79. J. Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp biosynthesis and regulation. Annu. Rev. Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LSIII. Contribution of phenazine antibiotic biosynthesis to the ecological competance of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentel M, Ahuja EG, Mavrodi DV, Breinbauer R, Thomashow LS, Blankenfeldt W. Of two make one: the biosynthesis of phenazines. ChemBioChem. 2009;10:2295–2304. doi: 10.1002/cbic.200900323. [DOI] [PubMed] [Google Scholar]
- Miller JH. Assay of β-galactosidase. In: Miller JH, editor. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- Miller WG, Leveau JH, Lindow SE. Improved gfp and inaZ Broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Ogawa K, Miyasaka F. Cloning and molecular analysis of poly(3-hydroxyalkanoate) biosynthesis genes in Pseudomonas aureofaciens. Curr. Microbiol. 2002;44:132–135. doi: 10.1007/s00284-001-0063-z. [DOI] [PubMed] [Google Scholar]
- Pham TH, Webb JS, Rehm BHA. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiology. 2004;150:3405–3413. doi: 10.1099/mic.0.27357-0. [DOI] [PubMed] [Google Scholar]
- Pierson LS, III, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 2010;86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson LS, III, Thomashow LS. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30–84. Mol. Plant Microbe Interact. 1992;5:330–339. doi: 10.1094/mpmi-5-330. [DOI] [PubMed] [Google Scholar]
- Pierson LS, III, Keppenne VD, Wood DW. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens is regulated by PhzR in response to cell density. J. Bacteriol. 1994;176:3966–3974. doi: 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson LS, III, Gaffney T, Lam S, Gong F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens 30–84. FEMS Microbiol. Lett. 1995;134:299–307. doi: 10.1111/j.1574-6968.1995.tb07954.x. [DOI] [PubMed] [Google Scholar]
- Pierson EA, Wood DW, Cannon JA, Blachere FM, Pierson LS., III Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant Microbe Interact. 1998;11:1078–1084. [Google Scholar]
- Prieto MA, Bühler B, Jung K, Witholt B, Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J. Bacteriol. 1999;181:858–868. doi: 10.1128/jb.181.3.858-868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin N, Callahan SM, Dunlap PV, Stevens AM. Analysis of LuxR regulon gene expression during quorum sensing in Vibrio fischeri. J. Bacteriol. 2007;189:4127–4134. doi: 10.1128/JB.01779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, et al. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C, Valverde C, Kay E, Haas D. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 2005;187:276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij ET. Environmental and molecular regulation of phenazine-1-carboxamide biosynthesis in Pseudomonas chlororaphis strain PCL1391. Leiden University; 2006. Ph.D. [Thesis], ISBN 9789090205830. [Google Scholar]
- Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, Cameron ADS, et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J. Bacteriol. 2012;194:686–701. doi: 10.1128/JB.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. T6SS delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schalk IJ. Metal trafficking via siderophores in Gram-negative bacteria: specificities and characteristics of the pyoverdine pathway. J. Inorg. Biochem. 2008;102:1159–1169. doi: 10.1016/j.jinorgbio.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B, Dahlbreck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Urbana-Champaign: University of Illinois; 2011. Regulation of amylovoran biosynthesis in Erwinia amylovora. Available at http://hdl.handle.net/2142/24468 (accessed May 16, 2013) [Google Scholar]
- Wang D, Calla B, Vimolmangkang S, Wu X, Korban SS, Huber SC, et al. The orphan gene ybjN conveys pleiotropic effects on multicellular behavior and survival of Escherichia coli. PLoS ONE. 2011;6:e25293. doi: 10.1371/journal.pone.0025293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Qi M, Calla B, Korban SS, Clough SJ, Sundin GW, et al. Genome-wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora. Mol. Plant Microbe Interact. 2012a;25:6–17. doi: 10.1094/MPMI-08-11-0207. [DOI] [PubMed] [Google Scholar]
- Wang D, Yu J, Pierson LS, Pierson EA. Differential regulation of phenazine biosynthesis by RpeA and RpeB in Pseudomonas chlororaphis strain 30–84. Microbiology. 2012b;158:1745–1757. doi: 10.1099/mic.0.059352-0. [DOI] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüß BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Whistler CA, Pierson LS., III Repression of phenazine antibiotic production in Pseudomoas aureofaciens strain 30–84 by RpeA. J. Bacteriol. 2006;185:3718–3725. doi: 10.1128/JB.185.13.3718-3725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilf NM, Williamson NR, Ramsay JP, Poulter S, Bandyra KJ, Salmond GP. The RNA chaperone, Hfq, controls two luxR-type regulators and plays a key role in pathogenesis and production of antibiotics in Serratia sp. ATCC 39006. Environ. Microbiol. 2012;13:2649–2666. doi: 10.1111/j.1462-2920.2011.02532.x. [DOI] [PubMed] [Google Scholar]
- Wood DW, Pierson LS., III The phzI gene of Pseudomonas aureofaciens 30–84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene. 1996;168:49–53. doi: 10.1016/0378-1119(95)00754-7. [DOI] [PubMed] [Google Scholar]
- Wood DW, Gong F, Daykin M, Williams P, Pierson LS., III N-acyl-homoserine lactone-mediated regulation of phenazine gene expression in Pseudomonas aureofaciens 30–84 in the wheat rhizosphere. J. Bacteriol. 1997;179:7663–7670. doi: 10.1128/jb.179.24.7663-7670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pierson LS., III A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 2001;67:4305–4315. doi: 10.1128/AEM.67.9.4305-4315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Wang SL, Geng HF, Ge YH, Huang XQ, Hu HB, et al. Differential regulation of rsmA gene on biosynthesis of pyoluteorin and phenazine-1-carboxylic acid in Pseudomonas sp. M18. World J. Microbiol. Biotechnol. 2005;21:883–889. [Google Scholar]
- Zhao YF, Wang D, Nakka S, Sundin GW, Korban SS. Systems level analysis of two-component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics. 2009;10:245. doi: 10.1186/1471-2164-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


