Abstract
N-methyl-d-aspartate (NMDA) receptors are ligand-gated ion channels assembled from GluN1 and GluN2 subunits. We used a series of N-hydroxypyrazole-5-glycine (NHP5G) partial agonists at the GluN2 glutamate binding site as tools to study activation of GluN1/GluN2A and GluN1/GluN2D NMDA receptor subtypes. Using two-electrode voltage-clamp electrophysiology, fast-application patch-clamp, and single-channel recordings, we show that propyl- and ethyl-substituted NHP5G agonists have a broad range of agonist efficacies relative to the full agonist glutamate (<1–72%). Crystal structures of the agonist binding domains (ABDs) of GluN2A and GluN2D do not reveal any differences in the overall domain conformation induced by binding of the full agonist glutamate or the partial agonist propyl-NHP5G, which is strikingly different from ABD structures of 2-amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propanoate (AMPA) and kainate receptors bound to full and partial agonists. Subsequent evaluation of relative NHP5G agonist efficacy at GluN2A-GluN2D chimeric subunits implicates the amino-terminal domain (ATD) as a strong determinant of agonist efficacy, suggesting that interdomain interactions between the ABD and the ATD may be a central element in controlling the manner by which agonist binding leads to channel opening. We propose that variation in the overall receptor conformation, which is strongly influenced by the nature of interdomain interactions in resting and active states, mediates differences in agonist efficacy and partial agonism at the GluN2 subunits.
Introduction
N-methyl-d-aspartate (NMDA) receptors are ligand-gated ion channels that mediate glutamatergic neurotransmission. They are mainly assembled from two GluN1 and two GluN2 subunits, and are activated upon binding of glycine and glutamate to GluN1 and GluN2, respectively (Traynelis et al., 2010). The GluN2 subunits (GluN2A-D) have different temporal and spatial expression patterns in the brain (Akazawa et al., 1994; Monyer et al., 1994), and are responsible for the marked differences in function among the NMDA receptor subtypes (Vicini et al., 1998; Yuan et al., 2009). NMDA receptors are critically involved in many neuronal functions, including synaptic plasticity and neuronal development, but are also implicated in many pathologic conditions, such as ischemia and traumatic brain injury, as well as Parkinson's, Huntington's, and Alzheimer's diseases (Lau and Zukin, 2007; Traynelis et al., 2010).
The agonist binding domain (ABD) of ionotropic glutamate receptor subunits is formed by two protein segments, denoted S1 and S2, which form a kidney-shaped structure (Armstrong et al., 1998; Furukawa and Gouaux, 2003; Furukawa et al., 2005; Naur et al., 2005, 2007). The agonist binds in a pocket located in the cleft between the upper and lower lobes, D1 and D2, respectively. Agonist binding induces closure of the cleft formed between D1 and D2, which is the initial conformational change within a sequence that leads to channel opening (Armstrong and Gouaux, 2000) [see also Hansen et al. (2007) and Kumar and Mayer (2013)]. The mechanism by which agonist binding induces channel gating through these conformational changes has been intensively studied to understand how glutamate receptors function. In that regard, partial agonists are useful tools to study the mechanism of receptor activation, because, like full agonists, they induce the conformational changes that lead to channel gating, but with lower efficacy (Banke and Traynelis, 2003; Erreger et al., 2005; Kussius and Popescu, 2009; Kussius et al., 2010). Partial agonists can also be used to identify structural elements that are associated with channel activation and provide opportunities to test ideas about functional steps that represent conformational changes needed for channel opening.
Much of the variation in function between NMDA receptor subtypes has been attributed to the modulatory amino-terminal domain (ATD) of the GluN2 subunit (Gielen et al., 2009; Yuan et al., 2009) [see also Hansen et al. (2010)]. The amino acid sequences of the GluN2 ATDs are highly variable with only 19% identical residues among the four GluN2A-D subunits. Open probability, deactivation time, and pharmacology of the NMDA receptor subtypes are to a large extent influenced by the ATD (Gielen et al., 2009; Yuan et al., 2009). Residues that interact with the agonist in the binding pocket are fully conserved among the GluN2 subunits, and are presumably not responsible for differences in function between the NMDA receptor subtypes (Laube et al., 1997; Anson et al., 1998; Chen et al., 2005; Furukawa et al., 2005). However, the GluN2 ABD, which is highly conserved (63% identical residues), harbors some structural elements that can mediate differences in deactivation time course and pharmacology of the NMDA receptor subtypes (Kinarsky et al., 2005; Horak et al., 2006; Erreger et al., 2007; Chen et al., 2008; Costa et al., 2010; Acker et al., 2011; Hansen and Traynelis, 2011; Vance et al., 2011; Hansen et al., 2012). From studies on the modulatory role of the ATD (Gielen et al., 2008, 2009; Yuan et al., 2009; Zhu et al., 2013) [reviewed in Hansen et al. (2010) and Paoletti (2011)], it can be inferred that interactions exist between the ATD and the ABD that influence the energetics of conformational changes induced by agonist binding and the efficacy by which the agonist activates the receptor. To study this in more detail, partial agonists that show pronounced variation in activity at the different NMDA receptor subtypes could be useful tools to study subtype-specific receptor function.
Here, we have used a series of N-hydroxypyrazole-5-glycine (NHP5G) compounds that are partial agonists at the glutamate binding site of the GluN2 subunit to identify the molecular determinants that control agonist efficacy at the individual NMDA receptor subtypes. Ethyl- and propyl-substituted NHP5G agonists have been shown to exhibit a broader range of relative efficacies at GluN2 subunits (<1–70% relative to glutamate-activated currents) (Clausen et al., 2008) than previously known for partial NMDA receptor agonists (60–100%) (Erreger et al., 2007; Hansen et al., 2008). Thus, these compounds provide a unique opportunity to probe structural elements that mediate differences in efficacy between NMDA receptor subtypes.
Materials and Methods
DNA Constructs and Ligands.
Wild-type cDNAs for rat GluN1-1a (GenBank accession numbers U11418 and U08261; hereafter GluN1), rat GluN2A (D13211), and rat GluN2D (L31611) were provided by Drs. S. Heinemann (Salk Institute, La Jolla, CA), S. Nakanishi (Osaka Bioscience Institute, Osaka, Japan), and P. Seeburg (Max Planck Institute for Medical Research, Heidelberg, Germany). The chimera with ATD interchanged between GluN2A and GluN2D [2A-(2D ATD)] was previously described (Yuan et al., 2009). All other chimeras were generated using standard molecular biology methods. Detailed information about the GluN2A-GluN2D chimeric junctions is summarized in Supplemental Table 1. Site-directed mutagenesis was performed using the QuikChange method (Stratagene, Cedar Creek, TX). The amino acids are numbered according to the full-length protein, including the signal peptide. For expression in Xenopus oocytes, DNA constructs were linearized by restriction enzymes, purified using phenol/chloroform extraction followed by ethanol precipitation, and used as templates for in vitro transcription to produce cRNAs with the mMessage mMachine kit (Ambion, Austin, TX). Racemic (R,S)-ethyl-NHP5G (Et-NHP5G) and (R,S)-n-propyl-NHP5G (Pr-NHP5G) were synthesized and purified as previously described (Clausen et al., 2008). MK-801 [(+)-MK 801 maleate] was purchased from Tocris Bioscience (R&D Systems, Inc., Minneapolis, MN). All other ligands were purchased from Sigma-Aldrich (Saint Louis, MO).
Preparation and Injection of Xenopus Oocytes.
Mature female Xenopus laevis were anesthetized in a 0.3% MS-222 (3-aminobenzoic acid ethyl ester; Sigma-Aldrich) solution for 10–15 minutes before the oocytes were surgically removed. The follicle layer was subsequently removed by treating the oocytes with 0.5 mg/ml collagenase (type IA; Sigma-Aldrich) in OR-2 buffer (in mM: 82.5 NaCl, 2.0 KCl, 1.0 MgCl2, and 5.0 HEPES pH 7.6) at room temperature for 2–3 hours. The following day, healthy-looking oocytes (stage V–VI) were injected with cRNAs encoding GluN1 and GluN2 at a 1:2 ratio at a total volume of 50 nl (0.2–10 ng total cRNA). The cRNA was diluted with RNase-free water to give responses in the 300–1500 nA range. After cRNA injection, the oocytes were stored at 15–19°C in Barth’s solution (in mM: 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4, 5 Tris-HCl, pH 7.4 with NaOH) supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) as well as 100 µg/ml gentamicin (Fisher Scientific, Pittsburgh, PA).
Two-Electrode Voltage-Clamp Recordings.
Recordings were performed 2–5 days after cRNA injection at room temperature (23°C) using a two-electrode voltage-clamp amplifier (OC725; Warner Instruments, Hamden, CT). The signal was low-pass filtered at 20 Hz (4-pole, -3 dB Bessel; Frequency Devices, Haverhill, MD) and digitized using PCI-6025E or USB-6212 BNC data acquisition boards (National Instruments, Austin, TX). Oocytes were placed in a custom-made chamber and continuously perfused (approximately 5 ml/min) with oocyte recording solution containing (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2, 0.01 EDTA (pH 7.4 with NaOH). Solutions were applied by gravity, and solution exchange was controlled through a digital 8-modular valve positioner (Hamilton, Reno, NV). Recording electrodes were filled with 3.0 M KCl, and current responses were recorded at a holding potential of −40 mV. Data acquisition, voltage control, and application of solutions were controlled using custom-made software. All ligand stock solutions for two-electrode voltage-clamp recordings were prepared in oocyte recording solution adjusted to pH 7.4.
Whole-Cell Patch-Clamp and Single-Channel Recordings.
Approximately 48 hours before the experiments, human embryonic kidney (HEK) 293 cells (CRL 1573; American Type Culture Collection, Rockville, MD) were plated in 24-well plates on 5-mm glass coverslips (Warner Instruments) coated with 0.1 mg/ml poly(d-lysine). The culture medium was Dulbecco’s modified Eagle’s medium with GlutaMax-I (Invitrogen, Carlsbad, CA), 10% dialyzed fetal bovine serum, 10 U/ml penicillin, and 10 μg/ml streptomycin. Cells were transfected with plasmid cDNAs encoding GluN1 and GluN2 subunits, as well as green fluorescent protein, at a ratio of 1:1:1 using the calcium phosphate precipitation method (Chen and Okayama, 1987). To transfect four wells in a 24-well plate, 10 µl plasmid DNA (0.2 µg/µl; total 2 µg DNA) was mixed with 65 µl H2O and 25 µl 1 M CaCl2. This DNA-CaCl2 solution was then mixed with 100 µl 2 × BES solution composed of (in mM) 50 N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES), 280 NaCl, 1.5 Na2HPO4 (pH 6.95 with NaOH), incubated for 3–5 minutes, and 50 µl of this solution (which is 200 µl total) was then added dropwise to each of four 24-wells containing cells and 500 µl culture medium. The culture medium containing the transfection mix was replaced after 4–5 hours to minimize cytotoxicity. NMDA receptor antagonists d,l-2-amino-5-phosphonovalerate (DL-APV; 200 µM) and 7-chlorokynurenic acid (200 µM) were added to the culture medium immediately after transfection, and experiments were performed approximately 24 hours after transfection.
Single-channel recordings from outside-out patches were performed at −80 mV at room temperature (23°C) using an Axopatch 200B amplifier (Molecular Devices, Union City, CA) with the low-pass filter set at 10 kHz. The signal was then filtered using an 8 kHz 8-pole low-pass filter (-3 dB Bessel; Frequency Devices), digitized using Digidata 1322A or 1440A (Molecular Devices) at 40 kHz with pCLAMP software (Molecular Devices). Recording electrodes (8–11 ΜΩ) were made from thick-walled glass micropipettes (G150F-4; Warner Instruments), coated with Sylgard 184 (Warner Instruments), and filled with internal solution composed of (in mM) 110 d-gluconate, 110 CsOH, 30 CsCl, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 NaATP, and 0.3 NaGTP (pH 7.35 with CsOH). The extracellular recording solution was composed of (in mM) 150 NaCl, 10 HEPES, 3 KCl, 0.5 CaCl2, 0.01 EDTA (pH 8.0 with NaOH). Each outside-out patch was recorded in ligand solutions in the following sequence: 1) glutamate plus glycine (1 mM and 100 µM, respectively), 2) Pr-NHP5G plus glycine (2 mM and 100 µM, respectively), 3) Pr-NHP5G plus glycine plus DL-APV (2 mM, 100 µM, and 500 µM, respectively), and 4) 100 µM glycine alone (i.e., in the absence of any glutamate-site agonist). Each condition was recorded for several minutes and the patch was washed for at least 4 minutes with extracellular solution between each application.
Whole-cell voltage-clamp recordings were performed at −60 mV using an Axopatch 200B amplifier (Molecular Devices) at room temperature (23°C). Recording electrodes (3–4 ΜΩ) were made from thin-walled glass micropipettes (TW150F-4; World Precision Instruments, Sarasota, FL) pulled using a horizontal puller (P-1000; Sutter Instrument Company, Novato, CA). The electrodes were filled with internal solution (see above). The extracellular recording solution (as above, but supplemented with 20 mM d-mannitol) was adjusted to pH 7.4 with NaOH (or pH 8.0 for MK-801 block experiments). Rapid solution exchange was achieved using a two-barrel theta-glass pipette controlled by a piezoelectric translator (Burleigh Instruments, Fishers, NY). Junction currents between undiluted and diluted (1:2 in water) extracellular recording solution were used to estimate speed of open tip solution exchange after recordings and typically had 10–90% rise times of 0.4–0.8 milliseconds. All ligand stock solutions for whole-cell patch-clamp and single-channel recordings were prepared in extracellular recording solution. Macroscopic response time courses of NMDA receptor responses were analyzed using ChanneLab software (www.synaptosoft.com; Synaptsoft, Decatur, GA).
Single-Channel Analysis.
Outside-out patches contained between 2 and 4 channels judged by the detection of multiple openings occurring simultaneously. Only stretches of data with no double openings were analyzed. Recordings were digitally postfiltered (8-pole Bessel) at 4 kHz using Clampfit 9 software (Molecular Devices), and idealization of single-channel records was performed using the time course fitting method (SCAN; www.ucl.ac.uk/Pharmacology/dcpr95.html); open time and amplitude histograms were analyzed using maximum likelihood fitting (EKDIST; www.ucl.ac.uk/Pharmacology/dcpr95.html). Using the calculated effective cutoff frequency for the final, combined filtering of the recording (fc = 3.37 kHz), and the calculated filter rise time (Tr = 99 µs), the open and shut time resolutions were determined to be 0.54Tr = 53 µs and 0.31Tr = 31 µs, respectively. These resolutions were imposed on the idealized recordings as previously described before amplitude and open duration histograms were generated (Colquhoun, 1994). For calculation of open durations, subconductance levels were ignored and adjacent open periods with different amplitudes were combined to create a single open period. For calculation of unitary conductance, the holding potential was corrected for liquid junction potential in the single-channel experiments, which was measured to be +4.5 ± 0.5 mV (n = 5), and the reversal potential assumed to be 0 mV.
Simulations of MK-801 Inhibition.
The relationship between open probability and the rate of MK-801 inhibition for NMDA receptors was simulated in ChanneLab software (Synaptsoft) by adapting a previously described model that explains the mechanism of trapping blockers such as MK-801 (Blanpied et al., 1997). The parameters used in the model were determined experimentally as previously described; values for glutamate association k+ = 1.04 × 107 M−1 s−1, and glutamate dissociation k− = 73 s−1 are from Erreger et al. (2005), and values at pH 7.6 for α = 359 s−1, β = 488 s−1, α′ = 5600 s−1, β′ = 220 s−1, kd+ = 2.9 s−1, kd− = 8.7 s−1, kon = 3.2 × 107 M−1 s−1, and koff = 6.3 s−1 are from Dravid et al. (2007). This model is for wild-type GluN1/GluN2A activated by glutamate at pH 7.6 at a membrane potential of −60 mV. Open probability was then changed by varying the opening rate β between 25 and 1200 s−1; for each condition, the time course of inhibition by 1 µM MK-801 was simulated and the inhibition time constant (τMK-801) was determined using a mono-exponential fit. The inhibition time constant (τMK-801) was used to calculate the rate of inhibition (1/τMK-801).
Data Analysis.
Concentration–response data were analyzed using GraphPad Prism 5.2 (GraphPad Software, La Jolla, CA). Data for individual oocytes were fitted to the Hill equation using variable slope, I = Imax/(1 + 10^((logEC50 − log[A]) * nH)), where Imax is the maximum current in response to the agonist, nH is the Hill slope, [A] is the agonist concentration, and EC50 is the agonist concentration that produces half-maximum response. For graphical presentation, data points from individual oocytes were normalized to the current response to saturating glutamate plus glycine in the same recording and averaged.
The equilibrium constant (Kb) for inhibition of GluN1/GluN2A by Pr-NHP5G was estimated using the Schild method (Arunlakshana and Schild, 1959). Determination of dose ratios from individual oocytes was simplified by generating two-point agonist concentration-response curves in the absence and presence of increasing concentrations of antagonist [see Wyllie and Chen (2007)]. For each oocyte, the two-point concentration–response curves were plotted on a log-log scale and each two-point curve was then fitted with a straight line with the same slope. The parallel fitted lines were then used to calculate the dose ratio for each antagonist concentration, allowing generation of a Schild plot and determination of Kb for each oocyte. Thus, the mean and S.E.M. for Kb were calculated using values determined for multiple oocytes.
X-Ray Crystallography.
The GluN2A and GluN2D ABD proteins were recombinantly expressed and purified by previously published methods (Vance et al., 2011). Both GluN2A and GluN2D ABD proteins were expressed by the bacterial cell-line, Origami B (DE3), and purified in the presence of 1 mM glutamate. The purified samples were concentrated to 5 mg/ml and dialyzed against a buffer containing 10 mM Tris-HCl (pH 8), 50 mM NaCl, and 0.1 mM (R)-Pr-NHP5G. The purified proteins were crystallized by vapor diffusion in hanging drops containing 2 µl of proteins and 1 µl of reservoir solution at 18°C. The reservoir solution for crystallization of GluN2A ABD contained 100 mM HEPES-NaOH (pH 7.0), 10–18% polyethylene glycol 800, and 100 mM calcium acetate, whereas that of GluN2D ABD contained 100 mM 3-(cyclohexylamino)propanesulfonic acid, 3.5–4.5 M sodium formate, and 8–12% 1,4-butanediol. Crystals appeared 1–2 days after the setup and grew to a full size after 5–7 days. The crystals were cryoprotected by addition of 20% glycerol to the reservoir solution for GluN2A ABD and increasing 1,4-butanediol concentration to 14%. All of the X-ray diffraction datasets were collected at X25 at the National Synchrotron Light Source at Brookhaven National Laboratory. Structures of GluN2A and GluN2D ABDs in complex with (R)-Pr-NHP5G were solved by molecular replacement using the structural coordinates of GluN2A and GluN2D ABDs in complex with glutamate [Protein Data Bank (PDB) IDs 1FTJ and 3OEN, respectively]. The molecular replacement models were subjected to simulated annealing to reduce model bias first, and refined using the PHENIX program until convergence of Rwork and Rfree. Atomic coordinates and structure factors for the structures of GluN2A and GluN2D ABDs in complex with Pr-NHP5G were deposited in the PDB (IDs 4JWX and 4JWY, respectively).
Results
Activation of GluN1/GluN2A and GluN1/GluN2D NMDA Receptors by NHP5G Agonists.
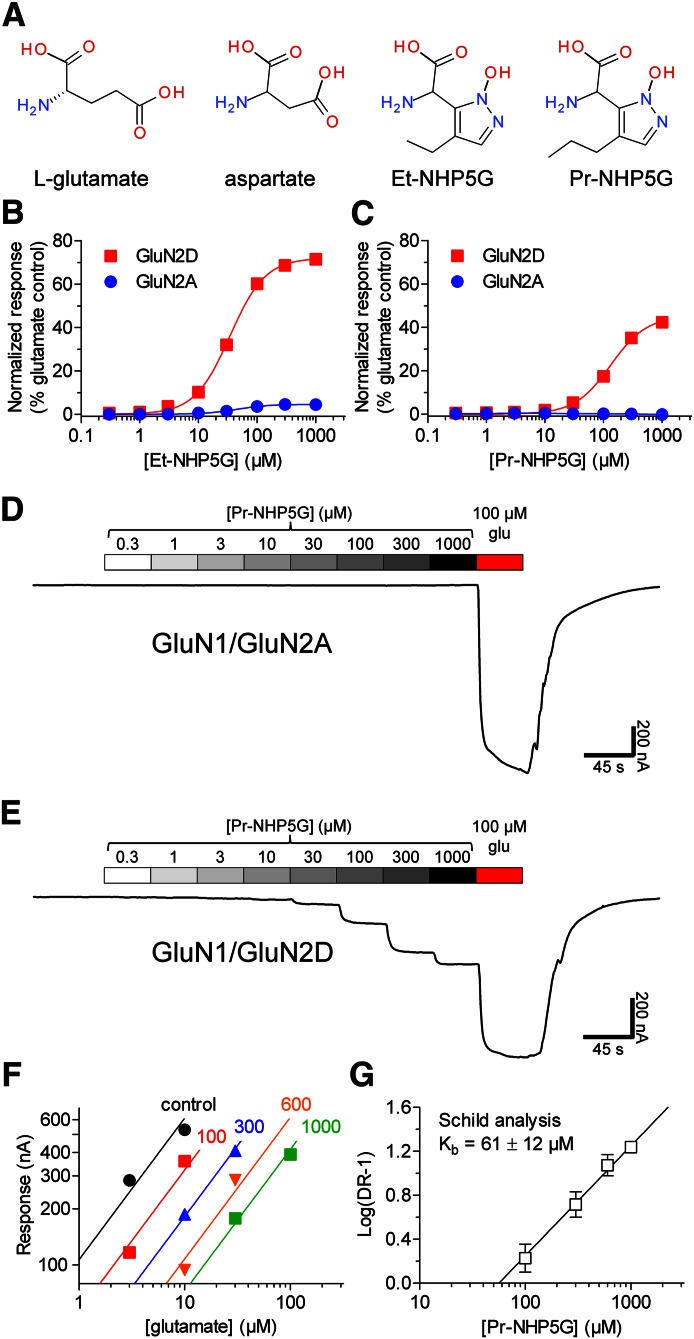
Partial agonists that show subunit selectivity can be used to identify specific structural elements that influence activation of receptor subtypes. The NHP5G series of agonists represents a particularly useful set of compounds with which to probe questions about activation of NMDA receptors because these agonists possess an unusually wide range of efficacies across different NMDA receptor subunits (Hansen et al., 2005; Clausen et al., 2008) [see also Risgaard et al. (2010)]. Ethyl- and propyl-substituted NHP5G (i.e., Et-NHP5G and Pr-NHP5G, respectively) interact with the glutamate binding site in the GluN2 NMDA receptor subunit (Fig. 1A). Et-NHP5G activates recombinant GluN1/GluN2A with an EC50 of 51 ± 5 µM (n = 6) and GluN1/GluN2D with an EC50 of 34 ± 1 µM (n = 6) (Fig. 1B). The maximal response to Et-NHP5G relative to the maximal response to glutamate (i.e., relative agonist efficacy) is strikingly different at GluN1/GluN2A (5% ± 1%; n = 6) and GluN1/GluN2D (72% ± 1%; n = 6). Pr-NHP5G shows even stronger subunit-selective relative efficacy in that it activates GluN1/GluN2D with an EC50 of 133 ± 6 µM and a relative agonist efficacy of 45% ± 1% (n = 7), but does not appear to activate detectable macroscopic responses from GluN1/GluN2A even at 1 mM (n = 8) (Fig. 1, C–E).
Fig. 1.
Relative efficacy of NHP5G agonists at GluN1/GluN2A and GluN1/GluN2D receptors. (A) Chemical structures of glutamate (l-glutamate), aspartate, Et-NHP5G, and Pr-NHP5G. (B and C) Concentration-response data measured using two-electrode voltage-clamp recordings for Et-NHP5G and Pr-NHP5G at GluN1/GluN2A and GluN1/GluN2D receptors expressed in Xenopus oocytes. The fitted maximal response to agonist is normalized to the maximal response to glutamate measured in the same recording. (D and E) Representative recordings of Pr-NHP5G concentration-response data for GluN1/GluN2A and GluN1/GluN2D receptors expressed in Xenopus oocytes. (F) Averaged two-point agonist concentration-response data from five oocytes expressing GluN1/GluN2A in the absence and presence of increasing concentrations of Pr-NHP5G as indicated above the linear fit (values are Pr-NHP5G in micromolar). The parallel fitted lines were used to calculate the dose ratio for each antagonist concentration (see Materials and Methods). (G) Determination of Kb for inhibition of GluN1/GluN2A by Pr-NHP5G using Schild analysis. Data are from five oocytes.
We determined the equilibrium binding constant (Kb) of Pr-NHP5G on GluN1/GluN2A by Schild analysis using the assumption that Pr-NHP5G is a competitive antagonist at the glutamate binding site. Schild plots for individual oocytes gave slopes that were not significantly different from unity (see Materials and Methods). The data for each oocyte were therefore refitted with a slope constrained to unity and Kb was determined for each oocyte. This analysis gave a mean Kb of 61 ± 12 µM (n = 5) for binding of Pr-NHP5G to GluN2A (Fig. 1, F and G). Thus, Pr-NHP5G appears to be a subunit-selective NMDA receptor ligand in that it is capable of activating one NMDA receptor subunit (GluN2D) and competitively inhibiting another subunit (GluN2A).
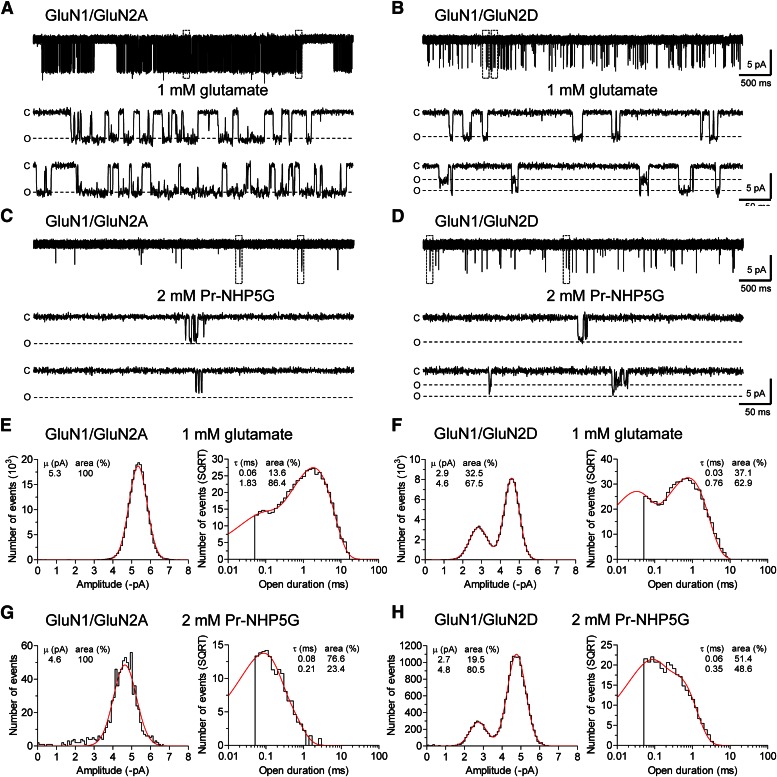
To determine whether Pr-NHP5G is capable of activating GluN1/GluN2A with low agonist efficacy, we evaluated the properties of individual channel openings in outside-out excised patches from HEK293 cells in response to application of Pr-NHP5G. As a control, openings of GluN1/GluN2A receptors recorded in response to 1 mM glutamate plus 100 µM glycine had a mean open time of 1.78 ± 0.06 milliseconds and a mean conductance of 64 ± 1 pS (n = 3) (Fig. 2A). No subconductance levels were detected. Interestingly, openings of GluN1/GluN2A receptors were detected in response to 2 mM Pr-NHP5G plus 100 µM glycine with a mean open time that was 10-fold lower (0.172 ± 0.004 milliseconds; n = 3) than glutamate-induced openings (Fig. 2C). The openings activated by Pr-NHP5G had a mean conductance of 53 ± 4 pS (n = 3). Whereas this value is significantly different (P > 0.05; unpaired, two-tailed t test) from that for channel openings activated by glutamate (64 ± 1 pS; n = 3), the openings activated by Pr-NHP5G were exceptionally brief, some of which may be at the limit of the imposed resolution (see Materials and Methods). Thus, the mean conductance for openings activated by Pr-NHP5G may be underestimated because the recordings will have openings that have not reached full amplitude before they begin to close due to the limitations of filtering and data acquisition. The patches contained between two and four channels judged by the detection of multiple openings occurring simultaneously in response to glutamate plus glycine; only stretches of data with no double openings were analyzed. For the same period of time, the relative number of openings in 2 mM Pr-NHP5G compared with the number of openings in glutamate was 11% ± 9% for GluN1/GluN2A. In the presence of 2 mM Pr-NHP5G plus 500 µM DL-APV, the relative number of openings was reduced approximately 300-fold to 0.04% ± 0.03%, confirming that Pr-NHP5G-induced channel openings were from activation of GluN1/GluN2A (n = 3). The GluN1/GluN2A openings observed in the presence of Pr-NHP5G were not spontaneous or activated by a contaminating glutamate-site agonist, since no openings were detected in the presence of 100 µM glycine alone (2–5 minutes of recordings; n = 3).
Fig. 2.
Glutamate- and Pr-NHP5G-activated channel openings from GluN1/GluN2A and GluN1/GluN2D receptors. (A and C) Representative recordings of GluN1/GluN2A channels activated by 1 mM glutamate or 2 mM Pr-NHP5G in outside-out patches from HEK293 cells. (B and D) Representative recordings of GluN1/GluN2D channels activated by 1 mM glutamate or 2 mM Pr-NHP5G. Glycine (100 µM) is present in all recordings; recordings for each receptor are from the same patch. The boxed openings in the top recordings are displayed on a faster time scale below. C and O indicate closed and open states. The open state level is shown as that determined in the recordings from the same patch in the presence of glutamate; the briefest openings may not reach full amplitude in the recorded traces due to the resolution imposed by the filtering. (E and F) Open point amplitude and open duration histograms for activation of GluN1/GluN2A and GluN1/GluN2D receptors by 1 mM glutamate (plus 100 µM glycine). The fitted probability density functions are shown in red. The number of events per bin in the open duration histogram is shown as the square root of the number events per bin and was best fitted to two exponential components. Data are pooled from three patches for GluN1/GluN2A and four patches for GluN1/GluN2D. (G and H) Open point amplitude and open duration histograms for activation of GluN1/GluN2A and GluN1/GluN2D receptors by 2 mM Pr-NHP5G (plus 100 µM glycine). Data are pooled from three patches for each receptor.
We also recorded and analyzed openings of GluN1/GluN2D receptors activated by 1 mM glutamate or 2 mM Pr-NHP5G (Fig. 2). Unitary current amplitudes were similar for GluN1/GluN2D activated by either glutamate or Pr-NHP5G, with a prominent subconductance level. The mean open time in response to Pr-NHP5G was about 2-fold briefer than that observed for glutamate. Table 1 summarizes single-channel properties of GluN1/GluN2A and GluN1/GluN2D receptors activated by glutamate or Pr-NHP5G.
TABLE 1.
Single-channel properties of NMDA receptors activated by glutamate or Pr-NHP5G
Data are from recordings of GluN1/GluN2A or GluN1/GluN2D channels in outside-out patches from HEK293 cells activated by 1 mM glutamate or 2 mM Pr-NHP5G. Glycine (100 µM) was present in all applications. A single conductance level was detected for GluN1/GluN2A activated by either glutamate or Pr-NHP5G. Only stretches of data with no double openings were analyzed. Outside-out patches used for generation of Pr-NHP5G data were also used for recordings in glutamate. Data are presented as the mean ± S.E.M., except for open durations, which were obtained by simultaneously fitting all analyzed openings (i.e., no double openings) from all patches (i.e., data from patches were pooled before analysis).
| GluN1/GluN2A |
GluN1/GluN2D |
|||
|---|---|---|---|---|
| Glutamate | Pr-NHP5G | Glutamate | Pr-NHP5G | |
| Mean open time, ms | 1.78 ± 0.06 | 0.172 ± 0.004 | 0.71 ± 0.06 | 0.34 ± 0.08 |
| Open duration, ms | ||||
| τ1 | 0.06 (14%) | 0.08 (77%) | 0.03 (37%) | 0.06 (51%) |
| τ2 | 1.83 (86%) | 0.21 (23%) | 0.76 (63%) | 0.35 (49%) |
| Conductance, pS | ||||
| γ1 | — | — | 33 ± 1 (25%) | 31 ± 1 (26%) |
| γ2 | 64 ± 1 (100%) | 53 ± 4 (100%) | 55 ± 1 (75%) | 52 ± 2 (74%) |
| Number of openings | 21,896 | 3050 | 30,542 | 11,136 |
| Number of patches | 3 | 3 | 4 | 3 |
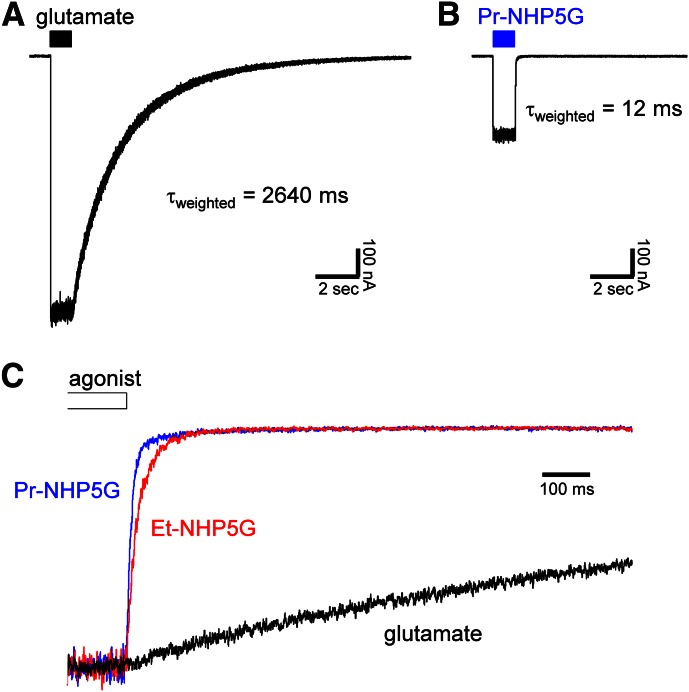
To evaluate whether differences exist in activation time course of GluN1/GluN2D receptors by glutamate or NHP5G agonists, we recorded macroscopic responses from receptors expressed in HEK293 cells using fast-application whole-cell patch-clamp. The parameters describing the macroscopic response time course are summarized in Table 2. Increased desensitization of responses to NHP5G agonists is not mediating the partial agonism of the NHP5G agonists, because desensitization was not observed for responses to either glutamate or NHP5G agonists (Fig. 3). Response rise times were slightly slower for activations by NHP5G agonists compared with glutamate, whereas there were marked differences in the deactivation time course of GluN1/GluN2D activated by these ligands. Responses to 1 mM Et-NHP5G and 2 mM Pr-NHP5G deactivated 69-fold and 220-fold faster, respectively, than glutamate (Fig. 3C; Table 2).
TABLE 2.
Time course of macroscopic responses from GluN1/GluN2D receptors
Data are from whole-cell patch-clamp recordings of GluN1/GluN2D receptors expressed in HEK293 cells. Ligands were applied using rapid solution exchange (see Materials and Methods). Concentrations used were 1 mM glutamate, 1 mM Et-NHP5G, and 2 mM Pr-NHP5G. Glycine (100 µM) was present in all applications. Desensitization was not observed for responses to either of the agonists. The deactivation time course for responses to glutamate were best described using dual-exponential fits and therefore two time constants are listed (τfast and τslow), whereas the deactivation time course for responses to NHP5G agonists were best described using a mono-exponential fit and therefore only one time constant is listed (τfast). Weighted time constants τweighted were calculated as previously described (Vance et al., 2012). Data are presented as the mean ± S.E.M.
| Agonist | Deactivation |
Cells | |||||
|---|---|---|---|---|---|---|---|
| Rise Time (10–90%) | τfast | τslow | % Fast | τweighted | Relative Imax (% Glutamate) | ||
| ms | ms | ms | ms | n | |||
| Glutamate | 7.6 ± 0.6 | 1030 ± 140 | 3360 ± 220 | 31 ± 4 | 2640 ± 160 | 100 | 7 |
| Et-NHP5G | 12 ± 3 | 38 ± 8 | 100 | 38 ± 8 | 61 ± 2 | 4 | |
| Pr-NHP5G | 16 ± 1 | 12 ± 1 | 100 | 12 ± 1 | 28 ± 1 | 3 | |
Fig. 3.
Macroscopic time course of responses to NHP5G agonists. (A and B) Representative whole-cell patch-clamp recordings of wild-type GluN1/GluN2D expressed in HEK293 cells and activated by (A) 1 mM glutamate or (B) 2 mM Pr-NHP5G. The cells were voltage-clamped at −60 mV, and ligands were applied using rapid solution exchange (see Materials and Methods). Glycine (100 µM) was present in all applications. (C) Overlay of normalized responses is shown for wild-type GluN1/GluN2D activated by 1 mM glutamate (black), 1 mM Et-NHP5G (red), or 2 mM Pr-NHP5G (blue). Only fragments of recordings showing the deactivation time course are shown. The parameters describing the macroscopic response time course are summarized in Table 2.
In summary, these results demonstrate that Et-NHP5G and Pr-NHP5G are GluN2 agonists with similar potencies at GluN1/GluN2A and GluN1/GluN2D receptors, but a wide range of relative agonist efficacies and marked differences in both single-channel properties and macroscopic response kinetics.
Binding of NHP5G to the GluN2A and GluN2D ABDs.
To directly assess whether the atomic contacts to Pr-NHP5G or the overall conformation of the ABDs of GluN2A and GluN2D might vary, we solved crystal structures of the isolated ABDs of both GluN2A and GluN2D in complex with Pr-NHP5G. The crystals for both GluN2A and GluN2D ABDs gave rise to high-quality X-ray diffraction data at 1.5 and 2.0 Å, respectively (Table 3). The crystal structures were solved by molecular replacement using structural coordinates of GluN2A ABD in complex with glutamate or GluN2D LBD in complex with glutamate as search probes. These high resolution crystal structures allow us to unambiguously visualize molecular determinants for recognition of Pr-NHP5G.
TABLE 3.
Data collection and refinement statistics
Values in parentheses are the low-resolution limits for the highest resolution shell of data.
| GluN2A Pr-NHP5G | GluN2D Pr-NHP5G | |
|---|---|---|
| Crystal space group | P41212 | C2221 |
| Crystal unit cell parameters | ||
| a, Å | 52.0 | 60.0 |
| b, Å | 52.0 | 113.9 |
| c, Å | 198.8 | 95.7 |
| α = β = γ, ° | 90 | 90 |
| Molecules per asymmetric unit | 1 | 1 |
| Data collection statistics | ||
| Beamline | NSLS ×25 | NSLS ×25 |
| Wavelength, Å | 1.10 | 1.10 |
| Resolution range, Å | 50.0–1.5 | 30.0–2.0 |
| Completeness, % | 98.4 (86.4) | 98.4 (99.9) |
| Redundancy | 12.6 (6.6) | 6.1 (5.6) |
| Rmerge, % | 9.6 (37.9) | 7.2 (45.6) |
| I/σ | 11.9 (4.0) | 17.4 (4.3) |
| No. of unique reflections | 44,489 | 22,209 |
| Refinement statistics | ||
| Resolution, Å | 19.2–1.6 | 19.5–2.0 |
| Rwork, % | 16.0 | 19.1 |
| Rfree, % | 19.5 | 23.3 |
| Root-mean-square distance | ||
| Bond lengths, Å | 0.007 | 0.008 |
| Bond angles, ° | 1.15 | 1.17 |
| Atoms, n | ||
| Protein | 2364 | 2126 |
| Ligand | 14 | 14 |
| Waters | 466 | 182 |
| Average B factor, Å2 | 17.5 | 39.1 |
| Ramachandran plot, % | ||
| Preferred | 96.7 | 94.5 |
| Allowed | 3.3 | 5.5 |
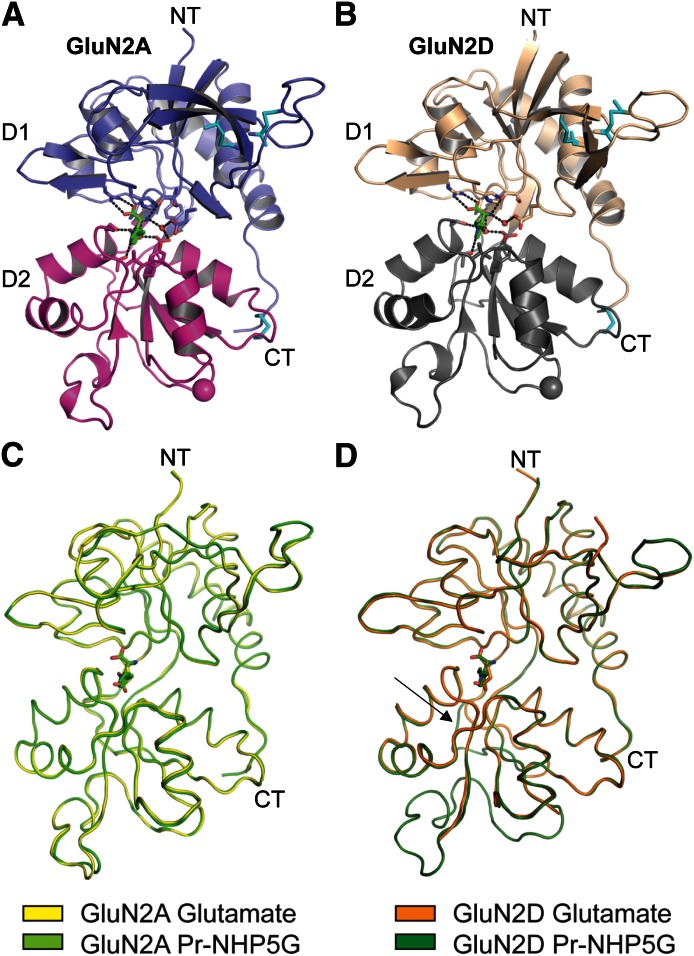
The crystal structures of GluN2A and GluN2D ABDs have the overall clamshell-like architectures composed of D1 and D2 as previously reported (Furukawa et al., 2005; Vance et al., 2011) (Fig. 4, A and B). In both GluN2A and GluN2D, Pr-NHP5G binds at the interdomain cleft between D1 and D2, where agonists such as glutamate bind. A series of crystallographic studies of ionotropic glutamate receptor ABDs have shown a pattern of conformational alteration of the ABD clamshells, where agonists and antagonists stabilize closed and open conformations, respectively. This conformational pattern is conserved in all of the ionotropic glutamate receptor classes, including the GluA2 2-amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propanoate (AMPA), GluK1 kainate, and GluN1 NMDA receptor subunits (Armstrong and Gouaux, 2000; Furukawa and Gouaux, 2003; Hogner et al., 2003; Mayer et al., 2006). In this crystallographic study, the structures of both GluN2A and GluN2D LBDs in complex with Pr-NHP5G are stabilized in the closed conformation that is virtually identical to the glutamate-bound structures. Moreover, there is no detectable difference in the degree of domain closure between the two lobes of the ABD. Indeed, the crystal structure of GluN2A ABD bound to glutamate can be superimposed to the one bound to Pr-NHP5G with 278 of 285 Cα positions showing a root-mean-square distance of 0.30 Å over both D1 and D2 of the bilobed structures (Fig. 4, C and D). Similarly, GluN2D ABD bound to glutamate can be superimposed to the Pr-NHP5G bound structure with root-mean-square distance of 0.34 Å for 242 of 285 Cα positions.
Fig. 4.
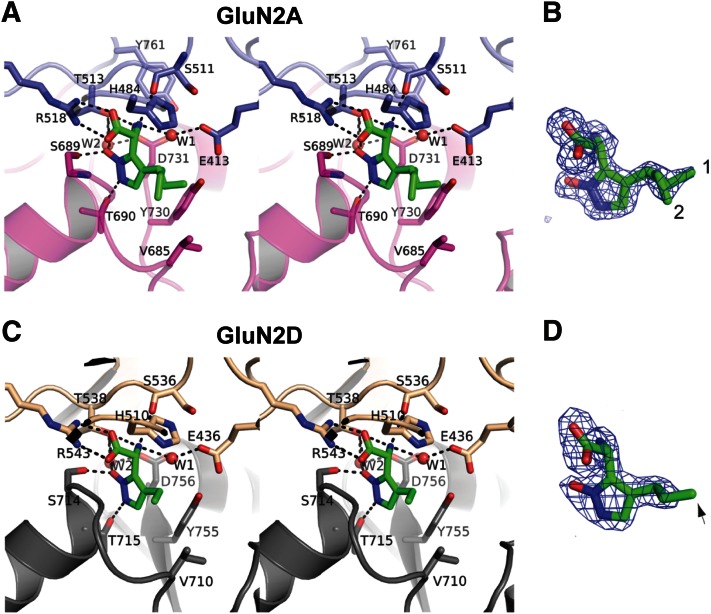
Crystal structures of GluN2A and GluN2D ABDs bound to Pr-NHP5G. Crystal structures of GluN2A ABD (A) and GluN2D ABD (B) with N-terminus (NT) and C-terminus (CT) on top and bottom, respectively. The GluN2A and GluN2D ABD structures are solved at 1.6 and 2.0 Å resolutions, respectively. Both GluN2A and GluN2D ABDs have bilobed clamshell-like architectures composed of D1 (blue for GluN2A and wheat for GluN2D) and D2 (magenta for GluN2A and gray for GluN2D) characteristic of ionotropic glutamate receptor ABDs. Pr-NHP5G (green stick) binds at the D1-D2 interface in both GluN2A and GluN2D. The GluN2A and GluN2D ABD structures contain three conserved disulfide bonds (cyan sticks). Spheres at the bottom of the LBD structures represent the α-carbons of glycine in the Gly and Thr residues replacing the transmembrane domains. (C) Structural comparison of GluN2A ABD bound to glutamate (yellow; PDB ID 1FTJ) and to Pr-NHP5G (green). The two structures are superimposed with root-mean-square distance of 0.30 Å over 278 Cα positions. (D) Structural comparison of GluN2D ABD bound to glutamate (orange; PDB ID 3OEN) and to Pr-NHP5G (dark green). The two structures are superimposed with root-mean-square distance of 0.34 Å for 242 Cα positions. The arrow indicates the conformational variation in the hinge region on the backside of the GluN2D ABD that is not observed in the GluN2A. This variation in GluN2D correlates with the deactivation time course of GluN1/GluN2D receptors and depends on the structure of the activating ligand (Vance et al., 2011).
Comparison of glutamate and Pr-NHP5G bound structures reveals a conformational variation in the hinge region on the backside of the GluN2D ABD that is not observed in the GluN2A ABD (Fig. 4, C and D). This variation in GluN2D has previously been shown to correlate with the deactivation time course of GluN1/GluN2D receptors and to depend on the structure of the activating ligand in that glutamate induced a markedly slower deactivation time course than other GluN2 agonists (Vance et al., 2011). The extraordinary fast deactivation time course observed for Pr-NHP5G compared with glutamate (Fig. 3; Table 2) is consistent with the structural variation observed in the hinge region of the GluN2D ABD structures (i.e., the hinge region in Pr-NHP5G-bound GluN2D ABD adopts conformation seen for ABD in complex with fast deactivating ligands) (Fig. 4D). However, structural variation in this hinge region of the receptor is unique to the GluN2D ABD and is not observed in structures of the GluN2A ABD. The variation in this region is therefore unable to explain the pronounced difference between agonist efficacies of Pr-NHP5G and glutamate at GluN1/GluN2A receptors.
Detailed inspection of the binding site reveals the strong similarity in the mode of Pr-NHP5G binding between GluN2A and GluN2D. In both GluN2A and GluN2D binding pockets, Pr-NHP5G binds to the receptor via a series of direct polar interactions between the α-carboxyl group of Pr-NHP5G and Arg518 (GluN2A) or Arg543 (GluN2D) and between α-amino group of Pr-NHP5G and Thr513 (GluN2A) or Thr538 (GluN2D). The N-hydroxypyrazole ring is oriented so that their polar groups form interaction with Ser689 and Thr690 (GluN2A) or Ser714 and Thr715 (GluN2D). Two water molecules, W1 and W2, form hydrogen bonds and further strengthen the binding (Fig. 5, A and C). One difference in the binding modes of Pr-NHP5G to GluN2A and GluN2D is in the orientation of the propyl group. In GluN2A, there is clear electron density for two conformations for the end carbon of the propyl group of Pr-NHP5G (Fig. 5B). The orientation of the propyl group is such that both conformers favor formation of Van der Waal contacts with GluN2A Val685. In GluN2D, there is no electron density for the end carbon atom of the propyl group, which implies random orientations of the end carbon atom and perhaps no interaction with Val710 (Fig. 5D). Thus, the electron density appears as if Et-NHP5G, instead of Pr-NHP5G, is bound to the receptor. Since Et-NHP5G is a partial agonist on both GluN2A and GluN2D, we speculate that formation of Van der Waal interaction between the propyl group and GluN2A Val685 or GluN2D Val710 work toward antagonism, whereas weaker Van der Waal interaction may increase agonist efficacy. This idea is consistent with a previous study, showing that relative agonist efficacy is reduced when the extent of steric clash (i.e., Van der Waal interaction) between agonists and the residue at this position in GluN2B (Val686) is increased (Hansen et al., 2005) [see also Risgaard et al. (2010)]. Furthermore, substitutions on the glutamate backbone [e.g., (2S,4R)-4-methyl glutamate (SYM2081)] that favor GluN1/GluN2D activation are predicted from docking studies to be directed into this same general vicinity within the GluN2 binding pocket (Erreger et al., 2007).
Fig. 5.
The Pr-NHP5G binding sites. Stereoviews of the Pr-NHP5G binding sites in GluN2A (A) and GluN2D (C). Binding of Pr-NHP5G (green sticks) involve residues from both D1 and D2 and water molecules (W1 and W2; red spheres). Dotted lines represent polar interactions. Shown on the right (B and D) are Fo-Fc omit difference Fourier maps for Pr-NHP5G contoured at 3.5 σ. In the GluN2A ABD structure, clear electron density for two conformations of Pr-NHP5G (1 and 2) is present. In the GluN2D LBD structure, there is no apparent electron density for the end carbon of the Pr-NHP5G (D; arrow), thus, it is omitted from the final structural coordinate file.
The agonist-bound conformations of glutamate- and NHP5G-bound GluN2A and glutamate- and NHP5G-bound GluN2D are strikingly similar structures. Moreover, binding of the full agonist glutamate or the partial agonist Pr-NHP5G to the isolated GluN2A and GluN2D ABDs induced the same degree of domain closure (see Fig. 4), which is similar to structures of full and partial agonists bound to the isolated GluN1 ABD (Furukawa and Gouaux, 2003; Inanobe et al., 2005). In addition, the agonist binding pocket of GluN2D appears capable of accommodating slightly more spacious agonist substituents protruding toward Val710 compared with GluN2A, which might explain some of the differences in relative NHP5G agonist efficacy at GluN1/GluN2A and GluN1/GluN2D receptors. However, the crystal structures did not reveal gross conformational variation between glutamate and Pr-NHP5G-bound structures that can account for the large differences in agonist efficacy between glutamate and Pr-NHP5G. Similarly, the structures of the GluN2A and GluN2D ABDs did not reveal differences in the overall domain conformation or closure that can explain the marked variation in relative NHP5G agonist efficacy between the NMDA receptor subtypes. The striking similarity between the glutamate- and Pr-NHP5G-bound GluN2A ABDs is particularly surprising, since glutamate can activate GluN1/GluN2A to an open probability of approximately 0.5 (Schorge et al., 2005; Yuan et al., 2009), whereas Pr-NHP5G is barely capable of activating GluN1/GluN2A receptors (see Fig. 2). We therefore speculate that agonist efficacy at GluN1 and GluN2 NMDA receptor subunits are predominantly controlled by interactions between the ABD and other domains in the full-length subunit. This would place some of the key structural determinants of open probability outside the ABD in NMDA receptor subunits.
Structural Determinants of Partial Agonism.
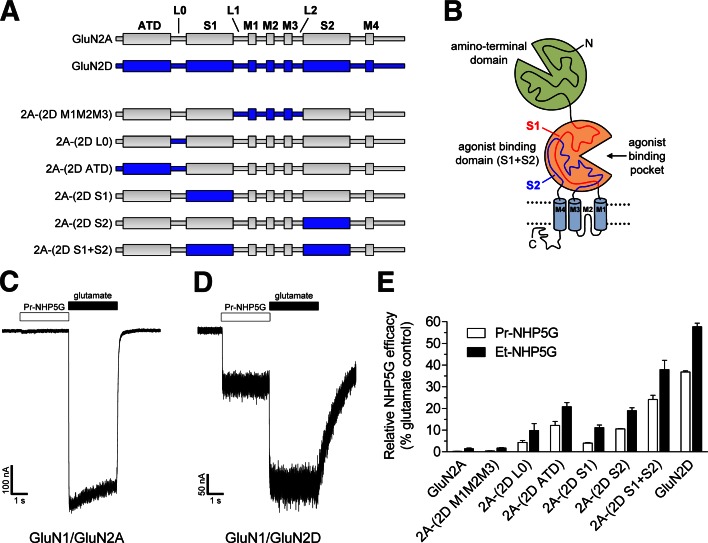
To identify structural elements within the full-length receptor that mediate the pronounced subtype-dependence of relative NHP5G agonist efficacies (i.e., partial agonism), we generated chimeric subunits between GluN2A and GluN2D (Fig. 6, A and B). The working hypothesis was that relative NHP5G agonist efficacies at GluN2A-containing receptors could be enhanced by replacing various segments in GluN2A with the corresponding segments from GluN2D. Whole-cell patch-clamp recordings of maximal responses to applications of NHP5G agonists and glutamate at wild-type and chimeric NMDA receptors expressed in HEK293 cells demonstrated that relative agonist efficacies of both Et-NHP5G and Pr-NHP5G could be increased in GluN2A by replacing segments of the extracellular regions with those of GluN2D (Fig. 6, C–E). Transfer from GluN2D into GluN2A of the ATD and the peptide linker (L0) connecting the ATD to the ABD resulted in marked increases in relative NHP5G agonist efficacies (Fig. 6E). Transfer of S1 alone [2A-(2D S1)] or S2 alone [2A-(2D S2)] from GluN2D into GluN2A also increased relative NHP5G agonist efficacies, but not to the extent observed with ATD. A more pronounced increase was observed when the entire ABD (S1+S2) was transferred. The relative agonist efficacies of the ligands, Et-NHP5G and Pr-NHP5G, increased in a similar fashion at the chimeras, suggesting that the activity of these two analogous ligands are controlled by the same structural determinants (Fig. 6E). Transfer of pore-forming elements in the GluN2D transmembrane domain and adjacent linkers (M1M2M3; M4 was not transferred) into GluN2A did not increase relative NHP5G agonist efficacies (Fig. 6E). This result suggests that structural elements comprising the channel pore, including the M3 transmembrane helix thought to form the gate and the pre-M1 region, do not contain key structural determinants that mediate the subtype-dependence of relative NHP5G agonist efficacies.
Fig. 6.
Structural determinants of partial agonism. (A) Linear representations of the polypeptide chains of GluN2A (gray), GluN2D (blue), and chimeric GluN2A-GluN2D subunits (see Supplemental Table 1 for chimeric junctions). (B) The illustration depicts the four semi-autonomous domains that comprise the NMDA receptor subunit, which are the ATD, the ABD, the transmembrane domain containing three transmembrane helices (M1, M3, and M4) and a membrane re-entrant loop (M2), and the intracellular carboxy-terminal domain. The ABD is formed by the two amino acid segments S1 and S2. (C and D) Representative whole-cell patch-clamp recordings from GluN1/GluN2A and GluN1/GluN2D receptors expressed in HEK293 cells. The receptors were activated by Pr-NHP5G (1.5 mM) or glutamate (1 mM) in the presence of glycine (100 µM). (E) Summary of maximal current responses to Et-NHP5G and Pr-NHP5G relative to maximal responses to glutamate (i.e., relative NHP5G efficacy) for wild-type and chimeric GluN2 subunits coexpressed with GluN1 in HEK293 cells.
The results also show unequivocally that elements outside of the ABD influence partial agonism of NHP5G agonists at the GluN2A subunit. This result is consistent with crystallographic data suggesting minimal differences between contact residues and overall architecture within the isolated glutamate- and Pr-NHP5G-bound ABDs of GluN2A and GluN2D. NMDA receptor subtype-dependent variation in partial agonism is therefore not exclusively controlled by the ABD and structural differences in this domain. These results strengthen the hypothesis that interactions exist between the ATD and the ABD that influence the energetics of conformational changes induced by agonist binding.
Estimated Open Probability in Glutamate for GluN2A-GluN2D Chimeras.
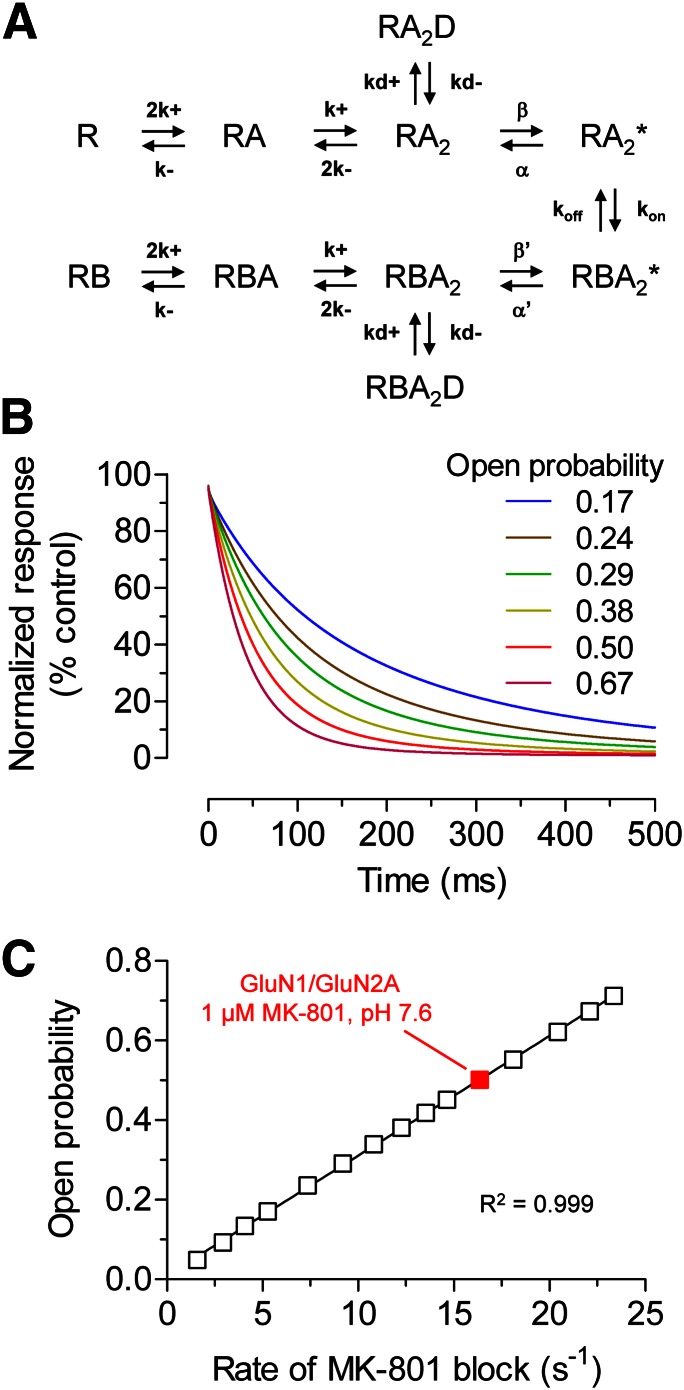
Variation in relative NHP5G efficacy (i.e., partial agonism) for GluN2A-GluN2D chimeras could be caused by changes in agonist efficacy of glutamate, NHP5G agonist, or both. To assess changes in glutamate efficacy, we estimated open probability of chimeric receptors maximally activated by glutamate using the rate of channel block by MK-801. Channel block by MK-801 can be considered irreversible on the time scale of these experiments and the rate of inhibition depends on open probability and the MK-801 association rate with the ion channel pore (Huettner and Bean, 1988; Jahr, 1992; Hessler et al., 1993; Rosenmund et al., 1993; Dzubay and Jahr, 1996) [see also Chen et al. (1999), Blanke and VanDongen (2008), and Vance et al. (2011)]. To substantiate the use of channel block by MK-801 as a method to estimate open probability, we initially evaluated the relationship between open probability and the rate of MK-801 inhibition using a previously described model that explains the mechanism of MK-801 block (Blanpied et al., 1997; Dravid et al., 2007). Using this model, the time courses of MK-801 inhibition were simulated at NMDA receptors with differences in open probability, revealing a strong linear correlation between the rate of MK-801 channel block and NMDA receptor open probability (Fig. 7). This result confirms the utility of MK-801 channel block as a method to estimate open probability of the GluN2A-GluN2D chimeras. We assume that MK-801 affinity is unchanged for chimeras, since they do not exchange elements of the ion channel pore.
Fig. 7.
Relationship between open probability and the rate of MK-801 inhibition for NMDA receptors. (A) The reaction scheme is adapted from a previously described model that can explain the mechanism of trapping blockers (e.g., MK-801) (Blanpied et al., 1997). R is receptor, A is agonist (glutamate), and B is channel blocker (MK-801). RBA2D and RA2D are desensitized states of receptors (R) with two bound agonists (A2) with and without bound blocker (B), respectively. Rate constants are described in the Materials and Methods. (B) The graph shows simulated time courses of inhibition by 1 µM MK-801 for GluN1/GluN2A receptors with variation in open probability. Open probability was changed by varying the opening rate β between 25 and 1200 s−1 (see Materials and Methods for details). The simulated inhibition time course of wild-type GluN1/GluN2A activated by glutamate at pH 7.6 at a membrane potential of −60 mV is shown as a red line (open probability is 0.50). The simulated responses are normalized to the steady-state response to a saturating concentration of glutamate in the absence of channel blocker. (C) The different open probabilities obtained by varying the opening rate β is plotted as a function of the rate of MK-801 inhibition. The inhibition time constants (τMK-801) were determined using mono-exponential fits to the simulated time courses, and the rates of inhibition (1/τMK-801) were calculated. There is a strong linear correlation (R2 = 0.999) between open probability and the rate of MK-801 inhibition. Simulated data for wild-type GluN1/GluN2A is shown in red.
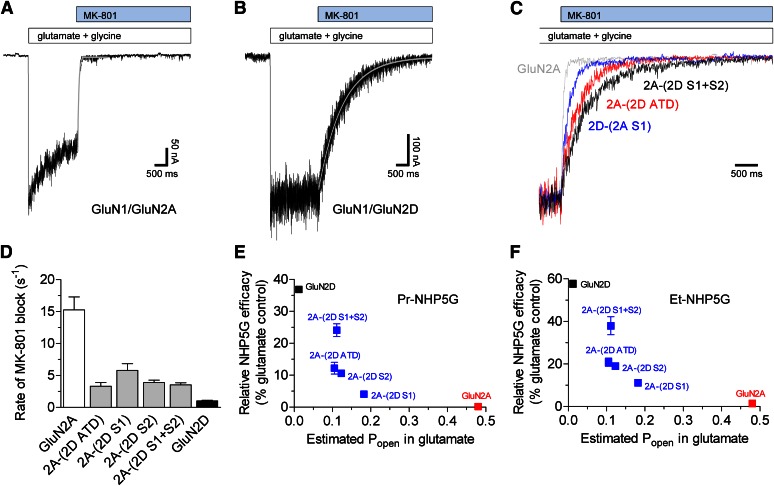
To determine the rate of MK-801 channel block, we recorded macroscopic responses from chimeric receptors expressed in HEK293 cells using rapid solution exchange and measured the rate of inhibition by 5 µM MK-801 in the continuous presence of saturating glutamate and glycine (Fig. 8; Table 4). All chimeras with regions of GluN2D inserted into GluN2A show marked reductions in estimated open probabilities in response to glutamate (2.6- to 4.5-fold lower than wild-type GluN1/GluN2A). Furthermore, the results also show that chimeras with increased relative NHP5G agonist efficacy also have reduced rate of MK-801 channel block when activated by glutamate (i.e., estimated glutamate open probability), suggesting that NHP5G agonists become more efficacious relative to glutamate as glutamate open probability decreases and the chimeric receptor gains elements of GluN2D (Fig. 8). The effects of the chimeras on partial agonism of NHP5G agonists are therefore not solely due to transfer of structural elements that specifically alter agonist efficacy of the NHP5G agonist without affecting agonist efficacy of glutamate. These results are consistent with our interpretation that interdomain interactions exist between the ATD and the ABD that control agonist efficacy, and also suggest that these interactions can have different effects on efficacy depending on the structure of the agonist.
Fig. 8.
Determination of rate of channel block by MK-801. (A and B) Representative whole-cell patch-clamp recordings of wild-type (A) GluN1/GluN2A and (B) GluN1/GluN2D receptors expressed in HEK293 cells. The cells were voltage-clamped at −60 mV, and ligands were applied using rapid solution exchange (see Materials and Methods). The responses to 1 mM glutamate plus 100 µM glycine at pH 8.0 were inhibited by 5 µM MK-801 and the inhibition time constants (τMK-801) were determined using a mono-exponential fit as indicated by the gray lines. (C) Overlay of normalized responses is shown for wild-type GluN1/GluN2A and GluN2A-GluN2D chimeras. Only fragments of recordings containing inhibition by MK-801 are shown. (D) Summary of the rates of MK-801 inhibition (1/τMK-801), which is linearly correlated to open probability (Popen), for wild-type receptors and GluN2A-GluN2D chimeric receptors activated by maximally effective glutamate and glycine. See Table 4 for values and estimated open probabilities. Data are mean ± S.E.M. from 5–7 cells. (E and F) Relationships between relative agonist efficacies of (E) Pr-NHP5G and (F) Et-NHP5G and estimated Popen in glutamate for GluN2A-GluN2D chimeras (shown in blue) as well as experimentally determined Popen for wild-type GluN1/GluN2A (red) and GluN1/GluN2D (black) [values from Yuan et al. (2009)]. Data for relative NHP5G agonist efficacies are from Fig. 5. Data are mean ± S.E.M.
TABLE 4.
Estimation of open probability using rate of MK-801 channel block
Data are from whole-cell patch-clamp recordings of GluN2 subunits coexpressed with GluN1 in HEK293 cells. Cells were voltage-clamped at −60 mV, and ligands were applied using rapid solution exchange. Steady-state responses to 1 mM glutamate plus 100 µM glycine at pH 8.0 were inhibited by 5 µM MK-801 and the inhibition time course was determined using a mono-exponential fit. The inhibition time constant (τMK-801) was then used to calculate the rate of inhibition (1/τMK-801), which is linearly correlated to open probability (Popen) assuming MK-801 affinity is constant, using Popen(chimera) = Popen(WT) * (τMK-801(WT)/τMK-801(chimera)). Chimera data were compared with Popen for wild-type GluN1/GluN2A. Data are presented as the mean ± S.E.M.
| GluN2 Subunit | Inhibition τMK-801 | Rate of Block 1/τMK-801 | Cells | Estimated Popen | Fold Change in Popen (Relative to Wild Type) |
|---|---|---|---|---|---|
| ms | s−1 | n | |||
| GluN2A | 71 ± 9 | 15 ± 2 | 6 | 0.48a | — |
| 2A-(2D ATD) | 374 ± 98 | 3.3 ± 0.6 | 6 | 0.10 | 0.22 |
| 2A-(2D S1) | 199 ± 37 | 5.8 ± 1.0 | 5 | 0.18 | 0.38 |
| 2A-(2D S2) | 266 ± 25 | 3.2 ± 0.4 | 5 | 0.12 | 0.26 |
| 2A-(2D S1+S2) | 308 ± 46 | 3.5 ± 0.3 | 7 | 0.11 | 0.23 |
| GluN2D | 1043 ± 98 | 1.0 ± 0.1 | 6 | 0.012a | — |
Indicates previously published Popen values for wild-type receptors determined using single-channel recordings (Yuan et al., 2009). To allow comparison with these values, the inhibition data were generated using the same compositions of intracellular and extracellular solutions as the previous study (i.e., pH 8.0 and 0.5 mM Ca2+ in the extracellular solution). The rate of MK-801 inhibition at pH 8.0 was lower than previously described rates at pH 7.6, consistent with MK-801 having a pKa of 8.37 (i.e., less protonated MK-801 at higher pH) (Dravid et al., 2007). All chimeras were blocked > 95% by 5 µM MK-801.
Discussion
We have used NHP5G-derived partial agonists at the glutamate binding site of NMDA receptors as tools to explore the molecular mechanism underlying partial agonism at NMDA receptors. We show through patch-clamp recordings that propyl-NHP5G is an exceptionally weak agonist at GluN1/GluN2A, capable only of generating brief and rare APV-sensitive channel openings. Surprisingly, X-ray crystallographic data show that the isolated ABD of GluN2A binds both Pr-NHP5G and the full agonist glutamate with similar overall configuration and atomic contacts. Furthermore, there is no detectable difference in the degree of domain closure at the GluN2A ABD for these two agonists that have pronounced differences in agonist efficacy. Analogous structural data for GluN2D show that partial agonists also show remarkably similar structural arrangements of the ABD without changes in domain closure, with the only divergent region being that observed in the hinge region in a comparison of glutamate to other agonists that display fast unbinding (Vance et al., 2011). Binding of partial agonists, such as 1-aminocyclopropane-1-carboxylic acid and 1-aminocyclobutane-1-carboxylic acid, to the glycine-binding GluN1 subunit has been reported to result in a conformational alteration in the hinge region of the ABD with similar degree of domain closure to the glycine-bound structure (Inanobe et al., 2005). Thus, the mechanism of partial agonist action in NMDA receptors appears to be different from that of AMPA and kainate receptors, where binding of partial agonist induces less domain closure than full agonist in crystal structures of the isolated ABDs (Armstrong et al., 1998; Armstrong and Gouaux, 2000; Hogner et al., 2002; Jin et al., 2003; Mayer, 2005; Hald et al., 2007; Venskutonytė et al., 2012).
Both structural and functional data suggest that structural elements outside the ABD control agonist efficacy at GluN2 subunits and that these elements influence the conformational changes induced by agonist binding in manner that is sensitive to the structure of the agonist. We propose that the overall receptor conformation, which is strongly influenced by the nature of interdomain interactions (e.g., between ABDs and ATDs) in resting and active states, controls agonist efficacy and partial agonism for the GluN2 subunits. However, our results do not rule out the possibility that the GluN2 ABD in solution or in full-length receptors with intact interdomain interactions may adopt conformations with different domain closure for partial and full agonists from the ones observed in crystal structures. For example, molecular dynamics simulations suggest that the GluN1 ABD can adopt an energetically stable conformation with intermediate domain closure not observed in the crystal structure upon binding of partial agonists (Ylilauri and Pentikäinen, 2012). In addition, these simulations indicated that partial agonists are unable to induce full domain closure of the GluN1 ABD if small forces are applied in a manner designed to mimic forces exerted by the ion channel in the full-length subunit. Despite the pronounced differences in agonist efficacy for Pr-NHP5G and glutamate, our structural data did not reveal a detectable difference in the degree of domain closure in the isolated, soluble ABDs of GluN2A and GluN2D bound to these agonists. The plausible explanation for this dichotomy between structural and functional data might be that the agonist-induced conformations are different for the isolated GluN2 ABDs and the ABDs in full-length GluN2 subunits.
We have used a chimeric strategy to search for elements that control agonist efficacy. Our chimera data implicate the ATD as a strong determinant of agonist efficacy, suggesting that interdomain interactions between the ABD and the ATD may be central elements in controlling the manner by which agonist binding leads to channel opening. The chimera data are supported by indirect measurement of open probability from MK-801 blocking rate. These functional data evaluate whether chimeric receptors have changed agonist efficacy and strengthen the overall conclusions. To our knowledge, this is the first demonstration that the ATD influences the relative efficacy of NMDA receptor agonists. Previous reports have demonstrated a role for the highly divergent GluN2 ATD in mediating a large portion of the functional differences among the NMDA receptor subtypes, including variation in single-channel open probability (Gielen et al., 2009; Yuan et al., 2009). Recently, it has been shown that perturbing the conformation of the GluN1 ATD affects functional and pharmacological receptor properties (Zhu et al., 2013). This study also suggested that the GluN1 ATD, like the GluN2 ATD, can control NMDA receptor function through intra- and inter-subunit allosteric interactions (Zhu et al., 2013). The modulatory role of the GluN1 ATD on NMDA receptor function is also supported by previous studies showing strong effects of alternative exon splicing that results in the insertion of 21 amino acids into the GluN1 ATD on agonist potency, deactivation time course, open probability, proton inhibition, Zn2+ inhibition, and polyamine potentiation (Durand et al., 1993; Hollmann et al., 1993; Williams et al., 1994; Traynelis et al., 1995, 1998; Rumbaugh et al., 2000; Vance et al., 2012). Furthermore, GluN2B-selective noncompetitive antagonists are known to bind to a site located at the interface between ATDs of GluN1 and GluN2B (Karakas et al., 2011).
The mechanism by which the ATD of NMDA receptor subunits influences conformational changes induced by agonist binding that trigger channel gating must involve long-range interdomain interactions. The ABD is likely a key structural determinant that transmits these interdomain interactions (Gielen et al., 2008), since the ABD is located between the modulatory ATD and the ion channel gate in the transmembrane domain. The ability of the ATD to influence agonist efficacy appears to be a distinct feature of NMDA receptor subunits, since deletion of the ATD in the GluA4 AMPA receptor subunit does not change receptor function (Pasternack et al., 2002). This indicates a different arrangement of interactions between the ATD and ABD in NMDA receptors compared with AMPA and kainate receptors.
The variation among the GluN2 ATDs suggests that differences exist in interdomain interactions among the GluN2 subunits that could mediate some of the functional variation among the NMDA receptor subtypes (Gielen et al., 2009; Yuan et al., 2009). We speculate that variation in resting conformations of the receptor subtypes and/or the energetically permissible agonist-induced conformational changes can be influenced by interactions between the ATD and the ABD, thereby allowing different conformational changes upon agonist binding among the GluN2 subunits. Future studies aimed at mapping these interactions could identify the intra- or inter-subunit domain interfaces that are responsible for the functional variation among NMDA receptor subtypes. Such interfaces might be useful targets for therapeutically relevant modulators that either potentiate or reduce receptor function under certain neuropathological conditions.
Supplementary Material
Acknowledgments
The authors thank Phoung Le, Jing Zhang, and Anel Tankovic for excellent technical assistance. They also thank the staff at X25 at the National Synchrotron Light Source at Brookhaven National Laboratory for beamline support.
Abbreviations
- ABD
agonist binding domain
- AMPA
2-amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propanoate
- ATD
amino-terminal domain
- BES
50 N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid
- DL-APV
D,L-2-amino-5-phosphonovalerate
- Et-NHP5G
(R,S)-ethyl-NHP5G
- HEK
human embryonic kidney
- MK-801
5H-dibenzo[a,d]cyclohepten-5,10-imine (dizocilpine maleate)
- MS-222
3-aminobenzoic acid ethyl ester
- NHP5G
N-hydroxypyrazole-5-glycine
- NMDA
N-methyl-d-aspartate
- PDB
Protein Data Bank
- Pr-NHP5G
(R,S)-n-propyl-NHP5G
- SYM2081
(2S,4R)-4-methyl glutamate
Authorship Contributions
Participated in research design: Hansen, Tajima, Perszyk, Vance, Ogden, Furukawa, Traynelis.
Conducted experiments: Hansen, Tajima, Perszyk, Vance, Ogden, Furukawa.
Contributed new reagents or analytic tools: Risgaard, Jørgensen, Clausen.
Performed data analysis: Hansen, Tajima, Perszyk, Vance, Ogden, Furukawa.
Wrote or contributed to the writing of the manuscript: Hansen, Tajima, Risgaard, Perszyk, Jørgensen, Vance, Ogden, Clausen, Furukawa, Traynelis.
Footnotes
This work was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grants NS036654 and NS065371]; the National Institutes of Health National Institute of Mental Health [Grant MH085926]; and the National Institutes of Health National Institute of Environmental Health Sciences [Grant T32-ES012870]; as well as the Robertson Research Fund of Cold Spring Harbor Laboratory; the Villum Kann Rasmussen Foundation; the Lundbeck Foundation; and the Japan Society for the Promotion of Science.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Acker TM, Yuan H, Hansen KB, Vance KM, Ogden KK, Jensen HS, Burger PB, Mullasseril P, Snyder JP, Liotta DC, et al. (2011) Mechanism for noncompetitive inhibition by novel GluN2C/D N-methyl-D-aspartate receptor subunit-selective modulators. Mol Pharmacol 80:782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. (1994) Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol 347:150–160 [DOI] [PubMed] [Google Scholar]
- Anson LC, Chen PE, Wyllie DJ, Colquhoun D, Schoepfer R. (1998) Identification of amino acid residues of the NR2A subunit that control glutamate potency in recombinant NR1/NR2A NMDA receptors. J Neurosci 18:581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28:165–181 [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E. (1998) Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395:913–917 [DOI] [PubMed] [Google Scholar]
- Arunlakshana O, Schild HO. (1959) Some quantitative uses of drug antagonists. Br Pharmacol Chemother 14:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke TG, Traynelis SF. (2003) Activation of NR1/NR2B NMDA receptors. Nat Neurosci 6:144–152 [DOI] [PubMed] [Google Scholar]
- Blanke ML, VanDongen AM. (2008) Constitutive activation of the N-methyl-D-aspartate receptor via cleft-spanning disulfide bonds. J Biol Chem 283:21519–21529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Boeckman FA, Aizenman E, Johnson JW. (1997) Trapping channel block of NMDA-activated responses by amantadine and memantine. J Neurophysiol 77:309–323 [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7:2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Luo T, Raymond LA. (1999) Subtype-dependence of NMDA receptor channel open probability. J Neurosci 19:6844–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PE, Geballe MT, Katz E, Erreger K, Livesey MR, O’Toole KK, Le P, Lee CJ, Snyder JP, Traynelis SF, et al. (2008) Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J Physiol 586:227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PE, Geballe MT, Stansfeld PJ, Johnston AR, Yuan H, Jacob AL, Snyder JP, Traynelis SF, Wyllie DJ. (2005) Structural features of the glutamate binding site in recombinant NR1/NR2A N-methyl-D-aspartate receptors determined by site-directed mutagenesis and molecular modeling. Mol Pharmacol 67:1470–1484 [DOI] [PubMed] [Google Scholar]
- Clausen RP, Christensen C, Hansen KB, Greenwood JR, Jørgensen L, Micale N, Madsen JC, Nielsen B, Egebjerg J, Bräuner-Osborne H, et al. (2008) N-Hydroxypyrazolyl glycine derivatives as selective N-methyl-D-aspartic acid receptor ligands. J Med Chem 51:4179–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D (1994) Practical analysis of single channel records, in Microelectrode Techniques: The Plymouth Workshop Handbook (Ogden D ed), pp 101–139, Company of Biologists, Cambridge, UK. [Google Scholar]
- Costa BM, Irvine MW, Fang G, Eaves RJ, Mayo-Martin MB, Skifter DA, Jane DE, Monaghan DT. (2010) A novel family of negative and positive allosteric modulators of NMDA receptors. J Pharmacol Exp Ther 335:614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid SM, Erreger K, Yuan H, Nicholson K, Le P, Lyuboslavsky P, Almonte A, Murray E, Mosely C, Barber J, et al. (2007) Subunit-specific mechanisms and proton sensitivity of NMDA receptor channel block. J Physiol 581:107–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand GM, Bennett MV, Zukin RS. (1993) Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc Natl Acad Sci USA 90:6731–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzubay JA, Jahr CE. (1996) Kinetics of NMDA channel opening. J Neurosci 16:4129–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Dravid SM, Snyder JP, Wyllie DJ, Traynelis SF. (2005) Mechanism of partial agonism at NMDA receptors for a conformationally restricted glutamate analog. J Neurosci 25:7858–7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Kristensen A, Chen PE, Hansen KB, Lee CJ, Yuan H, Le P, Lyuboslavsky PN, Micale N, et al. (2007) Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol Pharmacol 72:907–920 [DOI] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. (2003) Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J 22:2873–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. (2005) Subunit arrangement and function in NMDA receptors. Nature 438:185–192 [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. (2008) Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron 57:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. (2009) Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 459:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald H, Naur P, Pickering DS, Sprogøe D, Madsen U, Timmermann DB, Ahring PK, Liljefors T, Schousboe A, Egebjerg J, et al. (2007) Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid. J Biol Chem 282:25726–25736 [DOI] [PubMed] [Google Scholar]
- Hansen KB, Bräuner-Osborne H, Egebjerg J. (2008) Pharmacological characterization of ligands at recombinant NMDA receptor subtypes by electrophysiological recordings and intracellular calcium measurements. Comb Chem High Throughput Screen 11:304–315 [DOI] [PubMed] [Google Scholar]
- Hansen KB, Clausen RP, Bjerrum EJ, Bechmann C, Greenwood JR, Christensen C, Kristensen JL, Egebjerg J, Bräuner-Osborne H. (2005) Tweaking agonist efficacy at N-methyl-D-aspartate receptors by site-directed mutagenesis. Mol Pharmacol 68:1510–1523 [DOI] [PubMed] [Google Scholar]
- Hansen KB, Furukawa H, Traynelis SF. (2010) Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol 78:535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Ogden KK, Traynelis SF. (2012) Subunit-selective allosteric inhibition of glycine binding to NMDA receptors. J Neurosci 32:6197–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Traynelis SF. (2011) Structural and mechanistic determinants of a novel site for noncompetitive inhibition of GluN2D-containing NMDA receptors. J Neurosci 31:3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Yuan H, Traynelis SF. (2007) Structural aspects of AMPA receptor activation, desensitization and deactivation. Curr Opin Neurobiol 17:281–288 [DOI] [PubMed] [Google Scholar]
- Hessler NA, Shirke AM, Malinow R. (1993) The probability of transmitter release at a mammalian central synapse. Nature 366:569–572 [DOI] [PubMed] [Google Scholar]
- Hogner A, Greenwood JR, Liljefors T, Lunn ML, Egebjerg J, Larsen IK, Gouaux E, Kastrup JS. (2003) Competitive antagonism of AMPA receptors by ligands of different classes: crystal structure of ATPO bound to the GluR2 ligand-binding core, in comparison with DNQX. J Med Chem 46:214–221 [DOI] [PubMed] [Google Scholar]
- Hogner A, Kastrup JS, Jin R, Liljefors T, Mayer ML, Egebjerg J, Larsen IK, Gouaux E. (2002) Structural basis for AMPA receptor activation and ligand selectivity: crystal structures of five agonist complexes with the GluR2 ligand-binding core. J Mol Biol 322:93–109 [DOI] [PubMed] [Google Scholar]
- Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann S. (1993) Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron 10:943–954 [DOI] [PubMed] [Google Scholar]
- Horak M, Vlcek K, Chodounska H, Vyklicky L., Jr (2006) Subtype-dependence of N-methyl-D-aspartate receptor modulation by pregnenolone sulfate. Neuroscience 137:93–102 [DOI] [PubMed] [Google Scholar]
- Huettner JE, Bean BP. (1988) Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: selective binding to open channels. Proc Natl Acad Sci USA 85:1307–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E. (2005) Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron 47:71–84 [DOI] [PubMed] [Google Scholar]
- Jahr CE. (1992) High probability opening of NMDA receptor channels by L-glutamate. Science 255:470–472 [DOI] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. (2003) Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci 6:803–810 [DOI] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. (2011) Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinarsky L, Feng B, Skifter DA, Morley RM, Sherman S, Jane DE, Monaghan DT. (2005) Identification of subunit- and antagonist-specific amino acid residues in the N-Methyl-D-aspartate receptor glutamate-binding pocket. J Pharmacol Exp Ther 313:1066–1074 [DOI] [PubMed] [Google Scholar]
- Kumar J, Mayer ML. (2013) Functional insights from glutamate receptor ion channel structures. Annu Rev Physiol 75:313–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius CL, Popescu AM, Popescu GK. (2010) Agonist-specific gating of NMDA receptors. Channels (Austin) 4:78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius CL, Popescu GK. (2009) Kinetic basis of partial agonism at NMDA receptors. Nat Neurosci 12:1114–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. (2007) NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci 8:413–426 [DOI] [PubMed] [Google Scholar]
- Laube B, Hirai H, Sturgess M, Betz H, Kuhse J. (1997) Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron 18:493–503 [DOI] [PubMed] [Google Scholar]
- Mayer ML. (2005) Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron 45:539–552 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. (2006) Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci 26:2852–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. (1994) Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12:529–540 [DOI] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L, Vestergaard B, Egebjerg J, Gajhede M, Traynelis SF, et al. (2007) Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci USA 104:14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, Vestergaard B, Skov LK, Egebjerg J, Gajhede M, Kastrup JS. (2005) Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate. FEBS Lett 579:1154–1160 [DOI] [PubMed] [Google Scholar]
- Paoletti P. (2011) Molecular basis of NMDA receptor functional diversity. Eur J Neurosci 33:1351–1365 [DOI] [PubMed] [Google Scholar]
- Pasternack A, Coleman SK, Jouppila A, Mottershead DG, Lindfors M, Pasternack M, Keinänen K. (2002) Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor channels lacking the N-terminal domain. J Biol Chem 277:49662–49667 [DOI] [PubMed] [Google Scholar]
- Risgaard R, Hansen KB, Clausen RP. (2010) Partial agonists and subunit selectivity at NMDA receptors. Chemistry 16:13910–13918 [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Clements JD, Westbrook GL. (1993) Nonuniform probability of glutamate release at a hippocampal synapse. Science 262:754–757 [DOI] [PubMed] [Google Scholar]
- Rumbaugh G, Prybylowski K, Wang JF, Vicini S. (2000) Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J Neurophysiol 83:1300–1306 [DOI] [PubMed] [Google Scholar]
- Schorge S, Elenes S, Colquhoun D. (2005) Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J Physiol 569:395–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. (1998) Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci 18:6163–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF. (1995) Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science 268:873–876 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62:405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Hansen KB, Traynelis SF. (2012) GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J Physiol 590:3857–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Simorowski N, Traynelis SF, Furukawa H. (2011) Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun 2:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venskutonytė R, Frydenvang K, Hald H, Rabassa AC, Gajhede M, Ahring PK, Kastrup JS. (2012) Kainate induces various domain closures in AMPA and kainate receptors. Neurochem Int 61:536–545 [DOI] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. (1998) Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol 79:555–566 [DOI] [PubMed] [Google Scholar]
- Williams K, Zappia AM, Pritchett DB, Shen YM, Molinoff PB. (1994) Sensitivity of the N-methyl-D-aspartate receptor to polyamines is controlled by NR2 subunits. Mol Pharmacol 45:803–809 [PubMed] [Google Scholar]
- Wyllie DJ, Chen PE. (2007) Taking the time to study competitive antagonism. Br J Pharmacol 150:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylilauri M, Pentikäinen OT. (2012) Structural mechanism of N-methyl-D-aspartate receptor type 1 partial agonism. PLoS ONE 7:e47604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. (2009) Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci 29:12045–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. (2013) Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat Struct Mol Biol 20:477–485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.