ABSTRACT
Enteropathogenic Escherichia coli (EPEC) is an attaching and effacing (A/E) human pathogen that causes diarrhea during acute infection, and it can also sustain asymptomatic colonization. A/E E. coli depletes host cell DNA mismatch repair (MMR) proteins in colonic cell lines and has been detected in colorectal cancer (CRC) patients. However, until now, a direct link between infection and host mutagenesis has not been fully demonstrated. Here we show that the EPEC-secreted effector protein EspF is critical for complete EPEC-induced depletion of MMR proteins. The mechanism of EspF activity on MMR protein was posttranscriptional and dependent on EspF mitochondrial targeting. EPEC infection also induced EspF-independent elevation of host reactive oxygen species levels. Moreover, EPEC infection significantly increased spontaneous mutation frequency in host cells, and this effect was dependent on mitochondrially targeted EspF. Taken together, these results support the hypothesis that A/E E. coli can promote colorectal carcinogenesis in humans.
IMPORTANCE
There is mounting evidence linking the gut microbiota with the induction of colorectal tumorigenesis. We previously described the downregulation of host cell mismatch repair (MMR) protein levels upon enteropathogenic Escherichia coli (EPEC) infection and speculated that this depletion may lead to an ablated DNA repair system. In this work, we identify EspF, a translocated EPEC effector protein, as one of the factors required for this phenotype and show that this effector protein must be targeted to the mitochondria in order to exert its effect. Furthermore, we found that the impaired mismatch repair system resulting from EPEC infection led to the generation of spontaneous mutations within host DNA at a site of microsatellite instability, a trait typical of colorectal tumors. Thus, this work provides a novel means by which enteric bacteria may promote colorectal carcinogenesis.
Introduction
Enteropathogenic Escherichia coli (EPEC) attaches intimately to intestinal epithelial cells and uses a type 3 secretion system (T3SS) to translocate multiple effector proteins into the host cell cytoplasm. Effectors induce cytoskeletal remodeling at the cell surface, forming actin pedestals on which the bacteria sit (1). EPEC effectors also induce a plethora of other host cellular changes. Multiple studies analyzing colonic mucosa samples from colorectal cancer (CRC) patients have demonstrated an association between adherent E. coli strains and CRC (2–4). In addition, evidence of a link between attaching and effacing (A/E) bacteria and cancer has been provided by the murine pathogen Citrobacter rodentium, which promotes tumorigenesis in Apcmin/+ mice (5) and facilitates chemically induced tumorigenesis (6). Whether the association between A/E bacteria and cancer is causal remains to be fully determined. The ability to modulate host protein expression raises the possibility that EPEC promotes oncogenic pathways in colonic epithelial cells. In support of this hypothesis, we recently described the ability of EPEC to deplete mismatch repair (MMR) proteins MSH2 and MLH1 in cultured colonic cells in a T3SS-dependent manner (2).
The MMR system corrects DNA base pair mismatches and insertion/deletion loops (IDL) caused by replication errors or DNA damaging agents. In mammalian cells, this system is orchestrated by protein heterodimers, termed the MutS and MutL complexes. MutSα is composed of the MSH2 and MSH6 proteins and represents the most abundant mismatch binding factor, while MutSβ is composed of MSH2 and MSH3. The MutL complexes contain MLH1 heterodimerized with either PMS2 (MutLα), PMS3 (MutLβ), or MLH3 (MutLγ). It is hypothesized that the MutS complexes recognize DNA base pair mismatches or IDL and then recruit MutL. A MutS/MutL conformation switch then results in the activation of exonuclease DNA degradation and eventual repair by DNA polymerase δ (reviewed in reference 7). Hence, MMR is ablated by loss of either one of the critical proteins (MSH2 or MLH1); this is exemplified by the rapid accumulation of spontaneous somatic mutations throughout the genome, particularly in long repeated sequences of 1 to 4 nucleotides, termed microsatellites, in the absence of either protein (8, 9). In addition to causing microsatellite instability (MSI), MMR disruption enhances somatic mutation of tumor suppressor genes such as Apc and Tp53, which are mutated in the majority of CRC (10). Heritable MMR gene mutations are the cause of hereditary nonpolyposis colorectal cancer (HNPCC) (Lynch syndrome) (11).
MMR gene silencing, either by somatic mutation or promoter hypermethylation, also contributes to sporadic CRC development (12, 13). However, a significant proportion of tumors display MSI without accompanying MMR gene mutation or hypermethylation (14), suggesting that alternative causes of MMR disruption exist. In the present study, we investigated the mechanism by which EPEC depletes MMR proteins and sought to establish whether this effect increases host mutagenesis.
RESULTS
EPEC depletes MMR proteins posttranscriptionally.
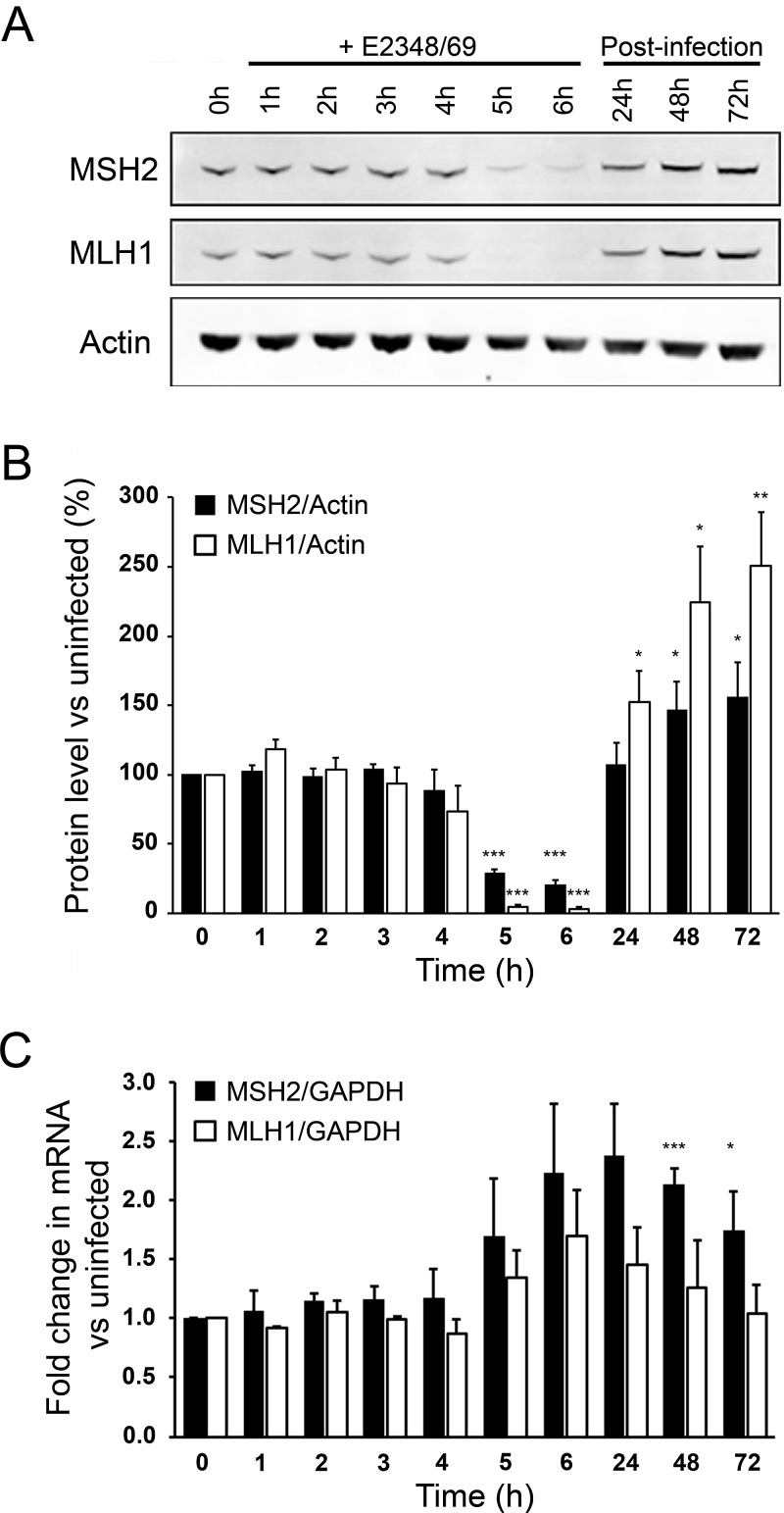
We previously reported that infection with wild-type EPEC strain E2348/69 induced a marked depletion of MSH2 and MLH1 protein levels in HT29 and SW480 colon cells. Given that promoter hypermethylation causes transcriptional silencing of MLH1, we speculated that MMR protein downregulation resulted from decreased transcription (2). Here we confirm that wild-type EPEC causes a dramatic depletion of MSH2 and MLH1 in HT29 cells after 4 to 5 h (Fig. 1A and B). Analysis of MSH6 levels also confirmed decreased expression of this protein at 6 h postinfection (see Fig. S1 in the supplemental material). As MSH6 stability is dependent on MSH2 expression (15), depletion of MSH6 may be a direct effect of infection or secondary to MSH2 depletion. Because of the importance of MSH2 and MLH1 for overall MMR competence, we focused our analysis on these two proteins. Treatment with gentamicin to kill EPEC after 6 h of infection fully restored MSH2 and MLH1 expression (Fig. 1A and B). Despite the dramatic drop in protein levels, quantitative real-time PCR (qRT-PCR) revealed that transcription of MSH2 and MLH1 was in fact elevated in response to infection (Fig. 1C). Elevated transcription corresponded with increased amounts of MSH2 and MLH1 protein in the postinfection period (Fig. 1B). We therefore conclude that loss of MMR proteins does not occur due to lower transcription. In contrast, the host cell appears to respond to EPEC infection by a compensatory increase in MMR gene transcription.
FIG 1 .
EPEC strain E2348/69 induces MSH2 and MLH1 protein depletion that is not due to transcriptional silencing. (A) Western blot showing MSH2 and MLH1 expression in HT29 cells cocultured with E2348/69 (wild-type EPEC) for up to 6 h. HT29 cells that were allowed to recover from infection were initially cocultured with strain E2348/69 for 6 h, then washed, and treated with antibiotics for up to 72 h postinfection. (B) MSH2 and MLH1 expression were quantitatively analyzed by secondary antibody infrared absorption analysis (Li-Cor Odyssey system) and corrected for loading using actin staining intensity. (C) Quantitative RT-PCR for MSH2 and MLH1 expression was performed on total mRNA extracts from the same cell populations used for protein analysis. Data are from three independent experiments, and values are means plus standard errors of the means (SEM) (error bars). The statistical significance of values compared to the value for control uninfected cells was assessed by Student’s t test and indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Adherence does not correlate with MMR protein depletion.
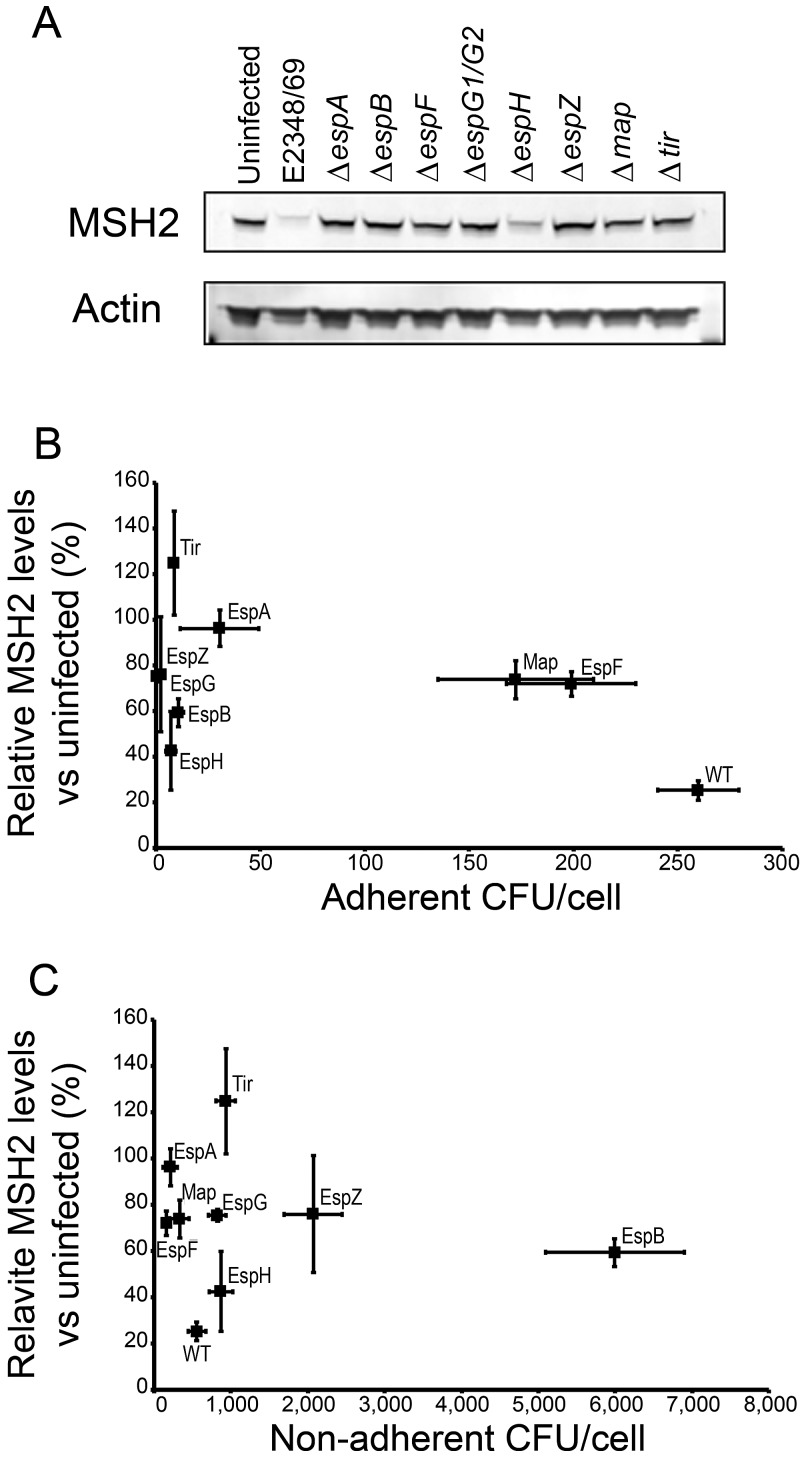
We showed previously that a mutant of EPEC E2348/69 lacking a crucial component of the T3SS (ΔespB mutant) was unable to fully deplete MSH2 and MLH1, suggesting that MMR protein depletion is dependent either on intimate attachment itself or on an effector protein(s) translocated during intimate attachment (2). To explore this hypothesis, we infected HT29 cells with a panel of E2348/69 T3SS effector and related mutants.
Western blots revealed that effector mutants of EPEC E2348/69 had different effects on MSH2 (Fig. 2A) and MLH1 protein levels (data not shown). However, microscopic examination revealed that the mutants also had different adhesion properties. This made it difficult to discern whether specific effector proteins were causing the changes in host MMR protein. For this reason, we quantified the adherent and nonadherent bacteria in cocultures after 5 h and quantified MSH2 and MLH1 expression in the same cultures to determine the relationship between the numbers of adherent bacteria and the levels of MSH2 and MLH1 protein. Interestingly, for MSH2, we observed a nonlinear relationship, indicating a role for effector proteins in modulating the levels of this protein (Fig. 2B). Wild-type EPEC had the highest adherence levels. The ΔespF and Δmap strains were the only mutants that adhered at levels comparable to, albeit lower than, the levels of the wild type, with adherence of the other mutants being markedly lower (Fig. 2B; see Fig. S2 in the supplemental material). It was clear that the number of adherent bacteria per cell did not predict the effect of infection on MMR protein expression. Despite having low adherence, the ΔespH mutant strain for example caused approximately 60% depletion of MSH2 levels (Fig. 2B), excluding this effector as a major contributor to MMR protein reduction. In contrast, the Δtir mutant strain, which showed a level of adherence similar to that of the ΔespH mutant, actually caused increased MSH2 expression. Furthermore, the mutant strains with the highest levels of adherence, the ΔespF and Δmap mutants, induced only a modest (~25%) decrease in MSH2 expression (Fig. 2B). MLH1 was more sensitive to depletion than MSH2, with every strain inducing a large depletion in MLH1, ranging from 60 to 100% (Fig. S2).
FIG 2 .
EPEC LEE effector mutants have differing abilities to adhere to HT29 cells and differing effects on MSH2 and MLH1 expression. (A) Western blot showing MSH2 expression in HT29 cells cocultured with EPEC strain E2348/69, effector protein mutants of E2348/69, and the ΔespA mutant (which is incapable of effector protein translocation) for 5 h. (B) HT29 cells were cocultured with EPEC E2348/69 (the wild-type [WT] strain), effector protein mutants of E2348/69, and the ΔespA mutant for 5 h, and bacterial attachment and MSH2 levels were quantified and plotted against one another. (C) Aliquots of the infection supernatant medium were diluted and spread onto agar plates to count nonadherent bacteria. The protein levels of MSH2 were plotted against the CFU of bacteria in the supernatant. Each experiment was repeated at least three times, and values are means (indicated by the markers) ± SEM (error bars).
Predictably, the mutant strains with low numbers of adherent bacteria (ΔespB, ΔespZ, ΔespH, ΔespG, and Δtir mutants) displayed high levels of nonadherent bacteria (Fig. 2C). Consistent with this trend, the wild-type strain and the Δmap and ΔespF mutant strains had the lowest numbers of nonadherent bacteria. There was no obvious correlation between the levels of nonadherent bacteria and MMR protein expression. Interestingly, while the ΔespF strain had the highest adherence levels of all the mutants; the amounts of MSH2 were relatively preserved compared with wild-type infection (71% ± 5.4% versus 25% ± 4.1% [Fig. 2B]). This result suggested that the locus of enterocyte effacement (LEE) effector EspF might play a major role in the EPEC infection-induced depletion of MSH2.
Complete host MMR protein depletion requires translocation and mitochondrial targeting of EspF.
To further establish the importance of EspF in depleting host MMR protein, we investigated the pathway involved. T3SS effectors influence host cell biology by targeting specific organelles and by regulating the level and activity of host cell proteins. EspF has an N-terminal sequence that targets the mitochondria and the nucleolus, and EspF also localizes to the host plasma membrane (16). Here, we examined the role of EspF mitochondrial targeting using a version of the espF gene that encodes a Leu-to-Glu amino acid substitution at position 16 (espFL16E); the resulting protein is inhibited in its ability to localize to mitochondria (17).
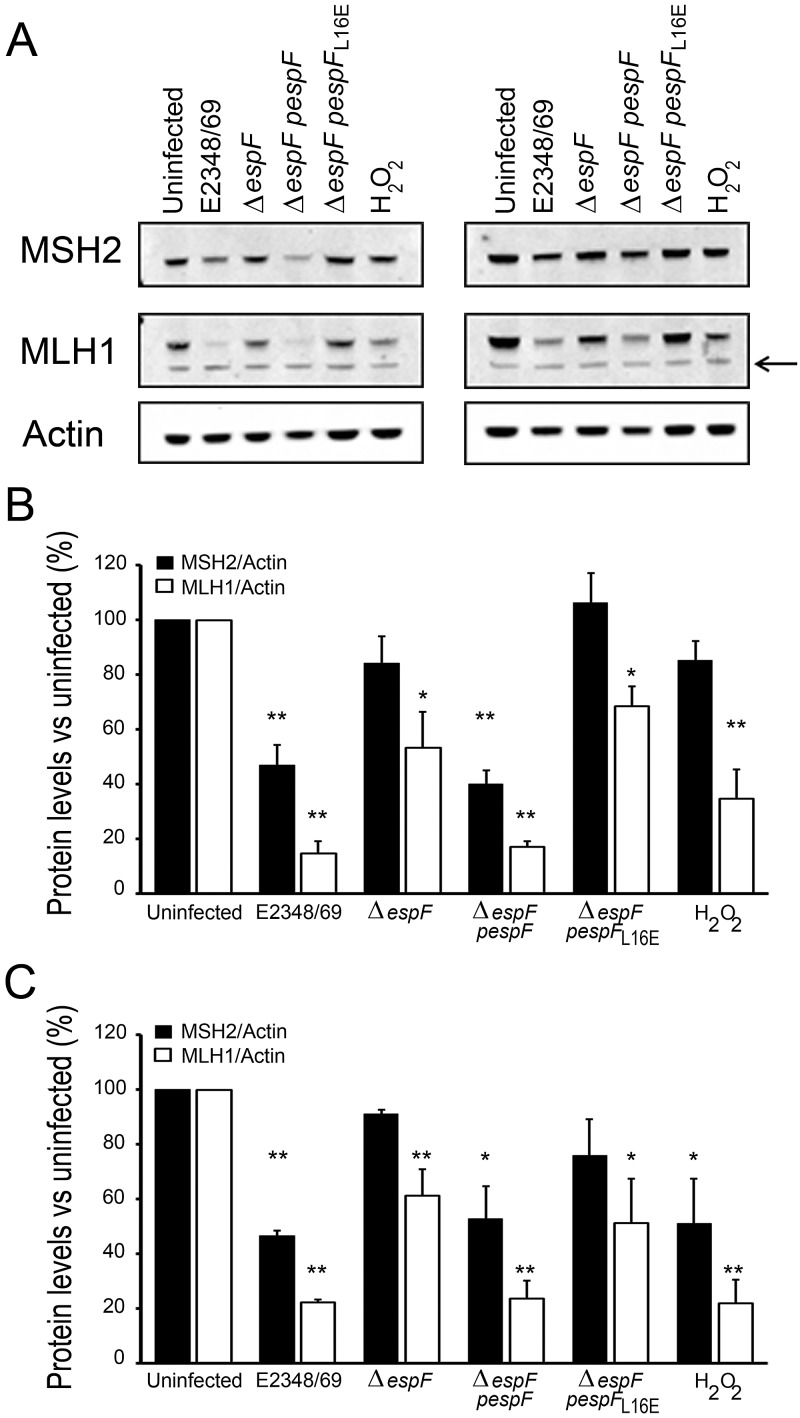
HT29 and SW480 cells were cocultured with wild-type EPEC (E2348/69), ΔespF mutant, ΔespF mutant complemented with plasmid-encoded espF (ΔespF pespF), and ΔespF mutant complemented with plasmid-encoded espFL16E (ΔespF pespFL16E). As expected, ΔespF induced only a modest decrease in MSH2 and MLH1 compared to EPEC E2348/69 (Fig. 3A to C). Complementation of the ΔespF mutant with wild-type EspF restored its ability to deplete MMR proteins, further validating the role of EspF in depleting MMR protein. However, complementation with EspFL16E that translocates into the host cytoplasm but is deficient in mitochondrial targeting did not restore the ability to deplete host MMR proteins (P < 0.001 compared with the ΔespF pespF strain for both MSH2 and MLH1 from infected HT29 cells). EspF is rapidly targeted to the mitochondria (within 30 min), causing a change in mitochondrial membrane potential (ΔΨm) as early as 1 h postinfection (18, 19). To determine whether MMR protein depletion was solely due to mitochondrial disruption by the effector, we assessed the levels of MMR proteins following treatment with an uncoupling compound that causes mitochondrial membrane depolarization, carbonyl cyanide m-chlorophenylhydrazone (CCCP). Cells were incubated for 7 h with increasing concentrations of CCCP from 1 µM to 50 µM, concentrations known to cause total loss of ΔΨm within 1 h (20). Alterations to the mitochondria caused by CCCP failed to decrease the levels of MSH2 or MLH1 at any of the concentrations tested (see Fig. S3 in the supplemental material), demonstrating that loss of ΔΨm is unlikely to be a mechanism by which EspF influences MMR protein expression.
FIG 3 .
MMR depletion was dependent on mitochondrial targeting of EspF. (A) EspF-dependent depletion of MSH2 and MLH1 proteins in HT29 cells (left) and SW480 cells (right) was assessed by Western blotting following 5 h infection with EPEC strain E2348/69, ΔespF mutant, ΔespF mutant complemented with plasmid-encoded espF (ΔespF pespF), or ΔespF mutant complemented with plasmid-encoded espFL16E (ΔespF pespFL16E). Cells were also treated with hydrogen peroxide (0.03% [vol/vol] H2O2) to induce oxidative stress. The black arrow indicates a cross-reactive band. (B and C) The expression of MSH2 and MLH1 in HT29 (B) and SW480 (C) cells were quantified using the Odyssey infrared imaging system and corrected for actin loading. Each experiment was repeated at least three times, and values are means (indicated by the markers) plus SEM (error bars). The statistical significance of values compared to the value for control uninfected cells was assessed by t test and indicated as follows: *, P < 0.01; **, P < 0.001.
Mitochondria are a potential source of cellular reactive oxygen species (ROS) (21). On the basis of evidence that oxidative stress depletes MMR protein levels in mammalian cells (22), we hypothesized that mitochondrial targeting of EspF might trigger elevated ROS levels, causing MMR protein depletion. To test whether the time scale and magnitude of MMR depletion caused by oxidative stress were comparable to EPEC infection, we treated cells with hydrogen peroxide (H2O2). H2O2 caused MSH2 and MLH1 protein depletion, which was similar in magnitude to that caused by E2348/69 (particularly in SW480 cells) over the same time course (Fig. 3A to C).
EPEC infection causes increased ROS levels in host cells independently of EspF.
To determine whether EPEC infection induced a significant increase in ROS, cells were subjected to an infection time course and analyzed for ROS levels by flow cytometry (Fig. 4A). To assess the dependence of ROS elevation on EspF, the panel of EspF mutants was tested. Increased ROS levels were detected after 3 h of infection for all strains tested (Fig. 4B). EPEC infection induced a 4.1-fold increase in ROS at 5 h compared with uninfected cells. While the highest ROS levels were observed for the ΔespF pespF strain (4.9-fold increase after 5 h), this was followed by the ΔespF strain (4.7-fold increase after 5 h), indicating that ROS levels were not dependent on EspF. In addition, there was no significant difference between the ΔespF pespF and ΔespF pespFL16E strains, indicating that EspF mitochondrial targeting was not involved in ROS induction. Cells infected with the ΔescN mutant (which cannot assemble a functional T3SS) displayed the lowest levels of ROS (3.6-fold increase after 5 h), but this value was not significantly different from the value for wild-type EPEC (Fig. 4B). Together, these results indicate that EPEC infection induced a significant increase in host cell ROS. However, this ROS increase was independent of T3S, and therefore does not explain the role of EspF in depleting MMR proteins.
FIG 4 .
EPEC infection induces an increase in ROS. (A) HT29 cells were infected with EPEC strain E2348/69, and the levels of ROS were determined by flow cytometry over time. (B) Infection of HT29 cells with EPEC strain E2348/69, ΔespF mutant, ΔespF pespF strain, ΔespF pespFL16E strain, or ΔescN mutant all resulted in elevated ROS levels after 3 h of infection. Data are from three independent experiments. The data points in panel B are means ± SEM (error bars).
EPEC infection increases mutation frequency at a site of microsatellite instability.
The failure to express functional DNA MMR proteins causes increased somatic mutation rate in vivo and strongly promotes colorectal carcinogenesis. Microsatellite DNA sequences are particularly susceptible to mutations normally corrected by the MMR system (9). MMR dysfunction therefore leads to accumulation of mutations in microsatellite sequences (and elsewhere in the genome), a phenomenon termed microsatellite instability (MSI). Interestingly, oxidative stress has been shown to enhance mutations in an MSI sequence within colonic cells deficient in MMR protein expression (23). With the knowledge that EPEC infection depletes MMR protein concomitant with elevated ROS levels, we sought to determine the functional consequences of these effects on host mutation frequency. To achieve this goal, we used a method that allows direct selection of cultured cells that have failed to repair spontaneously generated DNA mutations (24).
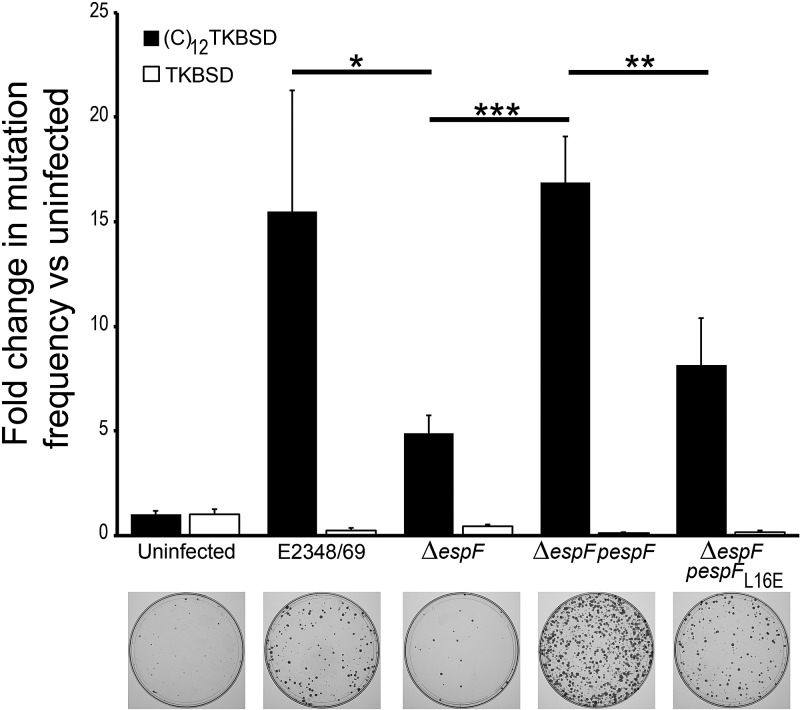
SW480 cells were stably transfected with a fusion gene containing a microsatellite sequence with 12 consecutive cytosine residues [(C)12 TKBSD] or a control fusion gene without the microsatellite sequence (thymidine kinase-blasticidin deaminase [TKBSD]). The (C)12 sequence is in frame with respect to the TK gene conferring ganciclovir sensitivity. Ganciclovir resistance results when a frameshift mutation occurs and is not repaired. The cells were infected for 6 h with EPEC strain E2348/69, ΔespF mutant, ΔespF pespF strain, or ΔespF pespFL16E mutant and subjected to ganciclovir selection. As expected, a low level of cell survival was observed in the TKBSD cells (Fig. 5) and in the uninfected (C)12 TKBSD cells, indicating competent DNA repair in these cells (Fig. 5). In marked contrast, EPEC infection induced a significant increase in mutation at the (C)12 site. Furthermore, this increase was EspF dependent, as determined by the significant decrease in mutation observed with the ΔespF strain versus the wild type. Restoration of EspF expression in the ΔespF strain restored the wild-type phenotype; however, complementation of ΔespF with EspFL16E did not, demonstrating that the effect was dependent on mitochondrial targeting.
FIG 5 .
The EPEC effector EspF increases mutation frequency at a microsatellite DNA site. SW480 cells transfected with a reporter fusion gene containing a (C)12 microsatellite sequence were subjected to infection with EPEC strain E2348/69, ΔespF mutant, ΔespF pespF strain, or ΔespF pespFL16E strain (black bars). Cells transfected with the fusion gene without the microsatellite sequence acted as a negative control (white bars). Examples of each of the (C)12TKBSD cell plates are displayed below their respective bars in the graph. Data are from six experiments, and values are means plus SEM. The statistical significance of values was assessed by t test and indicated as follows: *, P < 0.05; **, P < 0.005; ***, P < 0.001.
DISCUSSION
In the present study, we confirmed the prior observation that EPEC depletes MMR protein in colonic epithelial cells. We established that this effect did not occur via transcriptional silencing, as host cells responded to EPEC infection with increased MSH2 and MLH1 transcription, suggesting a possible compensatory response. This result demonstrates that MMR protein depletion is an EPEC-directed effect that the host cells attempt to counteract. Given these results and the rapidity of the effect, it seems likely that infection modulates MMR protein expression via a posttranslation mechanism. We demonstrated that the EPEC-secreted effector EspF was required for complete EPEC-induced MMR protein depletion and that the mitochondrial targeting motif of EspF was essential for this effect.
Oxidative stress is reported to cause MMR protein depletion (22), and we successfully validated this mechanism in our model using H2O2. We hypothesized that mitochondrial targeting of EspF might specifically enhance ROS generation in host cells and that elevated ROS levels would explain the enhanced ability of EspF-expressing strains to deplete MMR proteins. Contrary to our expectations, EspF-competent strains did not have an enhanced ability to promote host cell ROS generation. All of the strains we tested caused elevated ROS levels, suggesting that this effect does not require the T3SS. While ROS are not the mechanism by which EspF specifically depletes MMR protein, they may explain the residual MMR protein depletion caused by almost all strains tested. Intracellular ROS can cause oxidative modifications to both proteins and DNA (25, 26). The increased sensitivity to EPEC infection of MLH1 compared to MSH2 was a consistent trend in our experiments. Analysis of MLH1 reveals that it contains an oxidation susceptible motif (27) not present in MSH2. Furthermore, H2O2 caused a more marked depletion of MLH1 than MSH2. On the basis of this evidence, we speculate that infection-induced ROS contributes to MLH1 depletion by EPEC but that EspF is required for maximal depletion of MSH2, which is more resistant to ROS than MLH1 is.
While the exact mechanism by which EspF depletes MMR protein remains to be fully elucidated, the functional consequences of this effect were dramatic: we are the first to report that EPEC infection causes increased mutation frequency in host cells and that these mutations are substantially dependent on translocation and mitochondrial targeting of EspF. Interestingly, the only other EPEC effector mutant that displayed properties similar to those of the ΔespF strain was the Δmap mutant. Like the ΔespF mutant, the Δmap mutant showed high adherence and inability to induce complete MMR protein depletion. Map is also targeted to host mitochondria where it too causes a loss of ΔΨm (28). However, we found no evidence that loss of ΔΨm causes depletion of MMR proteins. In the future, it would be interesting to investigate the properties of a ΔespF Δmap double mutant or to ectopically express single EPEC effectors in host cells to help delineate the specific contributions of each effector.
Several other previously described activities of EPEC effectors also have the potential to promote cancer; in particular, infection can modulate host cell survival. EPEC effectors NleH1 and NleH2 can block apoptosis (29). EPEC-induced epidermal growth factor receptor (EGFR) activation also enhances the survival of infected cells; activated EGFR signaling is a feature of many cancers (30). Inhibition of host cell death also provides a rationale for EPEC effector-induced MMR disruption. Oxidative stress causes a variety of DNA lesions that are detected and repaired by the MMR pathway. However, when MMR proteins encounter high levels of DNA damage, they invoke a proapoptotic response via p53 to eliminate the damaged cell (31). By causing MMR protein depletion, EPEC could prevent the apoptotic response that may otherwise be triggered by the effects of EPEC-induced ROS on host DNA.
EPEC infection is a leading cause of infantile diarrhea in the developing world. In developed countries, EPEC is no longer considered a serious public health problem and is not routinely sought in clinical laboratories. Contemporary epidemiological data for EPEC in adults is therefore lacking. However, studies in Europe and Australia reveal that EPEC is carried by ~2.5 to 10% of healthy children (32, 33). Independent studies using tissue samples from adults with CRC demonstrate an association between mucosally adherent E. coli and CRC (2–4). Immunohistochemical analysis of human colonic mucosa cocultured with EPEC demonstrates that EPEC can enter colonic crypts and attach to epithelial cells in the proliferative compartment (2). The ability to asymptomatically colonize humans and to interact with proliferative cells in the crypt niche provides the biological context for EPEC to influence tumorigenesis. That EPEC can elevate ROS, suppress DNA repair, and increase mutation frequency in host cells provides a compelling mechanism by which these bacteria could promote CRC development.
CRC is a leading cause of cancer-related death worldwide. The majority of cases occur due to somatic mutations rather than inherited mutations, but the exact causes of the somatic mutations that initiate and drive sporadic CRC remain poorly defined. The importance of gene-environment interactions in causing CRC has been highlighted; however, conclusive research in this area is lacking (34). Mounting evidence suggests a causal link between bacterial infection and CRC and the potential contribution of pathogenic bacteria to CRC development has been noted (5, 6, 35).
Our data suggest that EPEC infection could promote somatic mutations (particularly in microsatellite DNA sequences) in colonic epithelium that does not feature genetic or epigenetic MMR silencing. Tumors developing in this way would be likely to show signs of MSI and critically, mutations in MSI sequences within oncogenes/tumor suppressor genes. Frameshift mutations in the tumor suppressor APC have been detected in tumors that do not display MMR gene mutation or hypermethylation (36). Many colorectal tumors display MSI that is not accompanied by MMR gene mutations or hypermethylation. The concept that alternative mechanisms account for MMR inactivation in a significant proportion of colorectal tumors is therefore well established (22, 23, 36).
Our findings provide a strong rationale for further investigation of A/E E. coli, particularly EPEC, in the context of colorectal carcinogenesis. Epidemiological studies in humans will be critical to establish a causal relationship. However, standard stool sample testing may not be an effective means to identify A/E bacteria due to their probable low levels in asymptomatic carriers. For this reason, we suggest analysis of tissue samples, e.g., biopsy samples or tissue samples removed during colorectal surgery.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Prior to infection, EPEC bacteria were cultured overnight in LB broth and appropriate antibiotics at 37°C. Overnight cultures were then diluted 1:50 in Dulbecco modified Eagle medium (DMEM) with nutrient mixture F-12 (DMEM/F12) with 5% fetal bovine serum (FBS) and maintained at 37°C for 1 h without agitation to activate the bacteria.
TABLE 1 .
Bacterial strains and plasmids used in this study
| Strain or plasmid | Mutation(s) | Gene carried on plasmid | Reference |
|---|---|---|---|
| Bacterial strainsa | |||
| E2348/69 (EPEC 0127:H7) | 37 | ||
| CVD452 | ΔescN | 38 | |
| UMD872 | ΔespA | 39 | |
| UMD864 | ΔespB | 40 | |
| UMD874 | ΔespF | 41 | |
| ΔespG Δorf3 mutant | ΔespG (ΔespG1 ΔespG2) | 42 | |
| ΔespH mutant | ΔespH | 43 | |
| MK34 | ΔespZ | 44 | |
| JAC719 | Δtir | 45 | |
| SE882 | Δmap | 46 | |
| Plasmids | |||
| pJN61 | espF | 19 | |
| pJN61L16E | espFL16E | This study | |
| pcDNA3-TKBSD | 24 | ||
| pcDNA3-(C)12TKBSD | 24 |
All bacterial strains were derived from EPEC strain E2348/69.
Tissue culture.
Human colorectal cancer cell lines HT29 and SW480 were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Cells were maintained at 37°C with 5% CO2 in DMEM/F12 medium (Gibco Invitrogen, NY) supplemented with 10% fetal bovine serum (Benchmark FBS; Gemini Bio-Products, CA). As the FBS brand/lot can have a considerable effect on bacterial growth rate (2), a single lot number of FBS was used for each set of experiments to provide consistency. For in vitro infections, cells were seeded in 12-well or 24-well plates (Corning, NY) and grown to a confluent monolayer.
Isolation of stable transfectants.
pcDNA3-TKBSD and pcDNA3-(C)12TKBSD were prepared as previously described (24) and used to transfect SW480 cells using the Xfect transfection reagent (Clontech, Palo Alto, CA). Two days posttransfection, selection was initiated by the addition of 10 µg/ml blasticidin S (EMD Millipore Corp., Billerica, MA). After 2 to 3 weeks, stable clones were isolated and additionally propagated with 100 µg/ml blasticidin S. To ensure thymidine kinase expression, clones were also subjected to ganciclovir (Sigma-Aldrich, St. Louis, MO) treatment (30 µM), and sensitivity was confirmed prior to experimentation.
In vitro coculture.
Activated bacterial cultures were added to confluent cell monolayers at a multiplicity of infection (MOI) of 30:1. After variable periods, cells were grown with 200 µg/ml gentamicin, 200 IU/ml penicillin, and 200 µg/ml streptomycin to kill bacteria and allow the cells to recover after infection. Whole-cell protein extracts were prepared from cell pellets after lysis in 1% Triton X-100 (in phosphate-buffered saline [PBS]) or radioimmunoprecipitation assay (RIPA) buffer, supplemented with Complete protease inhibitor cocktail (Roche Diagnostics, Germany). For qRT-PCR analysis, RNA was extracted from cell pellets using an RNeasy mini kit (Qiagen) according to the manufacturer’s instructions.
Quantification of mutation frequency.
To determine the frequency of mutation within the (C)12 microsatellite instability site of the TKBSD fusion gene, transfected cells we re infected for 6 h in DMEM/F12 medium containing 5% FBS. After infection, the cells were cultured for 24 h in DMEM/F12 medium containing 10% FBS, 100 IU/ml penicillin, and 100 µg/ml streptomycin (complete DMEM [cDMEM]) and 200 µg/ml gentamicin (Invitrogen) and cultured an additional 24 h with cDMEM supplemented with 100 µg/ml gentamicin. Forty-eight hours postinfection, infected and control uninfected cells were cultured in cDMEM containing 10 µg/ml gentamicin with or without 30 µM ganciclovir. After 7 days, total cell survival was calculated for the cells that did not receive ganciclovir selection. The ganciclovir selection medium was replaced every second day, and selection was continued for 14 days. After selection, the cells in petri dishes were fixed with 4% formaldehyde and stained with 0.1% crystal violet (Sigma-Aldrich), and the numbers of colonies were counted. The numbers of colonies were determined as a ratio of the cell survival. Cells transfected with TKBSD were subjected to the same treatment and acted as negative controls for the acquisition of mutations.
Western blotting.
Whole-cell protein extracts were resolved through precast 4 to 12% gradient Novex-NuPAGE gels (Invitrogen) and transferred to Immobilon-FL membranes (Millipore, MA). Blots were probed with mouse anti-MSH2 (Ab-2) (clone FE11) (diluted 1:500) (EMD Biosciences), mouse anti-MLH1 (diluted 1:1,000) (BD Biosciences, CA), and rabbit anti-beta-actin AC-15 (diluted 1:5,000) (Sigma-Aldrich) primary antibodies. IRDye 680- and IRDye 800-conjugated secondary antibodies diluted 1:15,000 (Li-Cor Biosciences, NE) were then applied. Infrared signals were detected and quantified using the Odyssey imaging system (Li-Cor Biosciences).
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) reactions were performed using Stratagene brilliant II SYBR green QRT-PCR (quantitative reverse transcription-PCR) master mix with low ROX dye (Agilent Technologies, CA) in a Stratagene Mx3005P instrument (Agilent Technologies, CA). The following primers were used: MSH2 forward, 5′-CAGTATATTGGAGAATCGCA; MSH2 reverse, 5′-AGGGCATTTGTTTCACC; MLH1 forward, 5′-GATTACCCCTTCTGATTGACA; MLH1 reverse, 5′-ACTGAGGCTTTCAAAACA; GAPDH forward (GAPDH stands for glyceraldehyde-3-phosphate dehydrogenase), 5′-CGGAGTCAACGGATTGGTCGTAT; and GAPDH reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC.
Flow cytometry.
After infection, the cells were resuspended in PBS containing the cell-permeant ROS indicator dye CM-H2DCFDA [5- (and 6-)-chloromethyl-2',7'-dichlorodihydrofluorescein diacetate, acetyl ester] (10 µM) (Invitrogen) and incubated with the probe for 30 min at 37°C in a 5% CO2 incubator. Oxidation of the probe resulted in a deacetylated fluorescent product that was detected using a BD LSR II flow cytometer (BD Biosciences). The relative amounts of ROS were determined by geometric mean fluorescence intensity (MFI).
SUPPLEMENTAL MATERIAL
EPEC infection of HT29 cells results in a depletion of MSH6 protein. Cells were infected with EPEC strain E2348/69 for the times indicated and assessed for MSH6 protein levels by Western blotting using a mouse anti-MSH6 antibody diluted 1:250 (BD Transduction Laboratories). Data are from three independent experiments; values are means plus SEM (error bars). Download
MLH1 depletion by EPEC is independent of bacterial adherence and T3SS. HT29 cells were cocultured with EPEC strain E2348/69, effector protein mutants of E2348/69, and the ΔespA mutant for 5 h, and bacterial attachment and MLH1 levels were quantified and plotted against one another. The experiment was repeated at least three times. Values are means (indicated by the markers) ± SEM (error bars). Download
HT29 cells demonstrate no depletion in MMR protein levels as a result of mitochondrial membrane depolarization. Cells were incubated with CCCP for 7 h at the concentrations indicated, and lysates were probed for MSH6, MSH2, and MLH1 protein levels. Data are from three independent experiments. Values are means plus SEM. Download
ACKNOWLEDGMENTS
We thank Greg Foster for providing pJN61L16E and Josef Jiricny for kindly donating pcDNA3-TKBDS and pcDNA3-(C)12TKBSD. We thank David Harrison and Scott Bader for the original inspiration for this work.
This work was supported by awards R21 CA141038 and R01 AI32074 from the National Institutes of Health. O.D.K.M. was supported by a United Kingdom Fulbright-AstraZeneca Research Scholarship.
Footnotes
Citation Maddocks ODK, Scanlon KM, Donnenberg MS. 2013. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio 4(3):00152-13. doi:10.1128/mBio.00152-13.
REFERENCES
- 1. Rosenshine I, Ruschkowski S, Stein M, Reinscheid DJ, Mills SD, Finlay BB. 1996. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 15:2613–2624 [PMC free article] [PubMed] [Google Scholar]
- 2. Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. 2009. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One 4:e5517. 10.1371/journal.pone.0005517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. 2004. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127:80–93 [DOI] [PubMed] [Google Scholar]
- 4. Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, Lochs H. 1998. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 115:281–286 [DOI] [PubMed] [Google Scholar]
- 5. Newman JV, Kosaka T, Sheppard BJ, Fox JG, Schauer DB. 2001. Bacterial infection promotes colon tumorigenesis in Apc(Min/+) mice. J. Infect. Dis. 184:227–230 [DOI] [PubMed] [Google Scholar]
- 6. Barthold SW, Jonas AM. 1977. Morphogenesis of early 1,2-dimethylhydrazine-induced lesions and latent period reduction of colon carcinogenesis in mice by a variant of Citrobacter freundii. Cancer Res. 37:4352–4360 [PubMed] [Google Scholar]
- 7. Jiricny J. 2006. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7:335–346 [DOI] [PubMed] [Google Scholar]
- 8. Jiricny J. 2000. Mediating mismatch repair. Nat. Genet. 24:6–8 [DOI] [PubMed] [Google Scholar]
- 9. Thibodeau SN, Bren G, Schaid D. 1993. Microsatellite instability in cancer of the proximal colon. Science 260:816–819 [DOI] [PubMed] [Google Scholar]
- 10. Sohn KJ, Choi M, Song J, Chan S, Medline A, Gallinger S, Kim YI. 2003. Msh2 deficiency enhances somatic Apc and p53 mutations in Apc+/-Msh2-/- mice. Carcinogenesis 24:217–224 [DOI] [PubMed] [Google Scholar]
- 11. Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapell A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. 2004. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 96:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. 1997. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 57:808–811 [PubMed] [Google Scholar]
- 13. Liu B, Nicolaides NC, Markowitz S, Willson JK, Parsons RE, Jen J, Papadopolous N, Peltomäki P, de la Chapelle A, Hamilton SR. 1995. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat. Genet. 9:48–55 [DOI] [PubMed] [Google Scholar]
- 14. Manders P, Spruijt L, Kets CM, Willems HW, Bodmer D, Hebeda KM, Nagtegaal ID, van Krieken JH, Ligtenberg MJ, Hoogerbrugge N. 2011. Young age and a positive family history of colorectal cancer are complementary selection criteria for the identification of Lynch syndrome. Eur. J. Cancer 47:1407–1413 [DOI] [PubMed] [Google Scholar]
- 15. Chang DK, Ricciardiello L, Goel A, Chang CL, Boland CR. 2000. Steady-state regulation of the human DNA mismatch repair system. J. Biol. Chem. 275:18424–18431 [DOI] [PubMed] [Google Scholar]
- 16. Holmes A, Mühlen S, Roe AJ, Dean P. 2010. The EspF effector, a bacterial pathogen’s Swiss army knife. Infect. Immun. 78:4445–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagai T, Abe A, Sasakawa C. 2005. Targeting of enteropathogenic Escherichia coli EspF to host mitochondria is essential for bacterial pathogenesis: critical role of the 16th leucine residue in EspF. J. Biol. Chem. 280:2998–3011 [DOI] [PubMed] [Google Scholar]
- 18. Dean P, Scott JA, Knox AA, Quitard S, Watkins NJ, Kenny B. 2010. The enteropathogenic E. coli effector EspF targets and disrupts the nucleolus by a process regulated by mitochondrial dysfunction. PLoS Pathog. 6:e1000961. 10.1371/journal.ppat.1000961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nougayrède JP, Donnenberg MS. 2004. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell. Microbiol. 6:1097–1111 [DOI] [PubMed] [Google Scholar]
- 20. Frieden M, James D, Castelbou C, Danckaert A, Martinou JC, Demaurex N. 2004. Ca(2+) homeostasis during mitochondrial fragmentation and perinuclear clustering induced by hFis1. J. Biol. Chem. 279:22704–22714 [DOI] [PubMed] [Google Scholar]
- 21. Kowaltowski AJ, de Souza-Pinto NC, Castilho RF, Vercesi AE. 2009. Mitochondria and reactive oxygen species. Free Radic. Biol. Med. 47:333–343 [DOI] [PubMed] [Google Scholar]
- 22. Chang CL, Marra G, Chauhan DP, Ha HT, Chang DK, Ricciardiello L, Randolph A, Carethers JM, Boland CR. 2002. Oxidative stress inactivates the human DNA mismatch repair system. Am. J. Physiol. Cell Physiol. 283:C148–C154 [DOI] [PubMed] [Google Scholar]
- 23. Gasche C, Chang CL, Rhees J, Goel A, Boland CR. 2001. Oxidative stress increases frameshift mutations in human colorectal cancer cells. Cancer Res. 61:7444–7448 [PubMed] [Google Scholar]
- 24. Cejka P, Marra G, Hemmerle C, Cannavó E, Storchova Z, Jiricny J. 2003. Differential killing of mismatch repair-deficient and -proficient cells: towards the therapy of tumors with microsatellite instability. Cancer Res. 63:8113–8117 [PubMed] [Google Scholar]
- 25. Freeman TA, Parvizi J, Della Valle CJ, Steinbeck MJ. 2009. Reactive oxygen and nitrogen species induce protein and DNA modifications driving arthrofibrosis following total knee arthroplasty. Fibrogenesis Tissue Repair 2:5. 10.1186/1755-1536-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohshima H, Tatemichi M, Sawa T. 2003. Chemical basis of inflammation-induced carcinogenesis. Arch. Biochem. Biophys. 417:3–11 [DOI] [PubMed] [Google Scholar]
- 27. Maisonneuve E, Ducret A, Khoueiry P, Lignon S, Longhi S, Talla E, Dukan S. 2009. Rules governing selective protein carbonylation. PLoS One 4:e7269. 10.1371/journal.pone.0007269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kenny B, Jepson M. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579–590 [DOI] [PubMed] [Google Scholar]
- 29. Hemrajani C, Berger CN, Robinson KS, Marchès O, Mousnier A, Frankel G. 2010. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc. Natl. Acad. Sci. U. S. A. 107:3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roxas JL, Koutsouris A, Viswanathan VK. 2007. Enteropathogenic Escherichia coli-induced epidermal growth factor receptor activation contributes to physiological alterations in intestinal epithelial cells. Infect. Immun. 75:2316–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin X, Ramamurthi K, Mishima M, Kondo A, Howell SB. 2000. p53 interacts with the DNA mismatch repair system to modulate the cytotoxicity and mutagenicity of hydrogen peroxide. Mol. Pharmacol. 58:1222–1229 [DOI] [PubMed] [Google Scholar]
- 32. Beutin L, Marchés O, Bettelheim KA, Gleier K, Zimmermann S, Schmidt H, Oswald E. 2003. HEp-2 cell adherence, actin aggregation, and intimin types of attaching and effacing Escherichia coli strains isolated from healthy infants in Germany and Australia. Infect. Immun. 71:3995–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pabst WL, Altwegg M, Kind C, Mirjanic S, Hardegger D, Nadal D. 2003. Prevalence of enteroaggregative Escherichia coli among children with and without diarrhea in Switzerland. J. Clin. Microbiol. 41:2289–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de la Chapelle A. 2004. Genetic predisposition to colorectal cancer. Nat. Rev. Cancer 4:769–780 [DOI] [PubMed] [Google Scholar]
- 35. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. 2010. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 107:11537–11542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Samowitz WS, Slattery ML, Sweeney C, Herrick J, Wolff RK, Albertsen H. 2007. APC mutations and other genetic and epigenetic changes in colon cancer. Mol. Cancer Res. 5:165–170 [DOI] [PubMed] [Google Scholar]
- 37. Levine MM, Nataro JP, Karch H, Baldini MM, Kaper JB, Black RE, Clements ML, O’Brien AD. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550–559 [DOI] [PubMed] [Google Scholar]
- 38. Jarvis KG, Girón JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U. S. A. 92:7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kenny B, Lai LC, Finlay BB, Donnenberg MS. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313–323 [DOI] [PubMed] [Google Scholar]
- 40. Donnenberg MS, Yu J, Kaper JB. 1993. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol. 175:4670–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McNamara BP, Koutsouris A, O’Connell CB, Nougayréde JP, Donnenberg MS, Hecht G. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Invest. 107:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elliott SJ, Krejany EO, Mellies JL, Robins-Browne RM, Sasakawa C, Kaper JB. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tu X, Nisan I, Yona C, Hanski E, Rosenshine I. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595–606 [DOI] [PubMed] [Google Scholar]
- 44. Kanack KJ, Crawford JA, Tatsuno I, Karmali MA, Kaper JB. 2005. SepZ/EspZ is secreted and translocated into HeLa cells by the enteropathogenic Escherichia coli type III secretion system. Infect. Immun. 73:4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- 46. Tomson FL, Viswanathan VK, Kanack KJ, Kanteti RP, Straub KV, Menet M, Kaper JB, Hecht G. 2005. Enteropathogenic Escherichia coli EspG disrupts microtubules and in conjunction with Orf3 enhances perturbation of the tight junction barrier. Mol. Microbiol. 56:447–464 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EPEC infection of HT29 cells results in a depletion of MSH6 protein. Cells were infected with EPEC strain E2348/69 for the times indicated and assessed for MSH6 protein levels by Western blotting using a mouse anti-MSH6 antibody diluted 1:250 (BD Transduction Laboratories). Data are from three independent experiments; values are means plus SEM (error bars). Download
MLH1 depletion by EPEC is independent of bacterial adherence and T3SS. HT29 cells were cocultured with EPEC strain E2348/69, effector protein mutants of E2348/69, and the ΔespA mutant for 5 h, and bacterial attachment and MLH1 levels were quantified and plotted against one another. The experiment was repeated at least three times. Values are means (indicated by the markers) ± SEM (error bars). Download
HT29 cells demonstrate no depletion in MMR protein levels as a result of mitochondrial membrane depolarization. Cells were incubated with CCCP for 7 h at the concentrations indicated, and lysates were probed for MSH6, MSH2, and MLH1 protein levels. Data are from three independent experiments. Values are means plus SEM. Download