Abstract
Mutation rate is one of the most fundamental parameters in genetics and evolutionary biology because mutation rate has major impacts on the incidence of disease, the amount of genetic variation, and the rate and trajectory of evolution. Based on estimates of synonymous nucleotide diversity in Escherichia coli, a recent study claimed that the per-nucleotide mutation rate in a gene decreases with the rise of its expression level or the intensity of purifying selection and that this trend reflects adaptive risk management. Here, we demonstrate that this argument is theoretically untenable, especially in the lack of mechanisms that simultaneously tune the mutabilities of multiple genes with similar fractions of deleterious mutations. Analyzing published genome sequences of E. coli mutation accumulation lines, we show that mutation rates are actually higher in more highly expressed genes, similar to previous genome-wide observations in Salmonella typhimurium, Saccharomyces cerevisiae, and the human germline. These general patterns likely arise from transcription-associated mutagenesis that exceeds transcription-coupled repair.
Keywords: mutation rate, expression level, natural selection, E. coli
Because mutation is the ultimate source of genetic variation and evolution, accurately measuring its rate is of central importance (Baer et al. 2007; Hodgkinson and Eyre-Walker 2011). Although eukaryotic mutation rates can be estimated relatively easily from either polymorphic or divergence levels of neutral genomic regions, this approach is problematic for prokaryotes because of the scarcity of neutral regions, due to their compact genomes that are subject to strong selection owing to their typically large effective population sizes (Lynch and Conery 2003). Using the genome sequences of 34 natural strains of Escherichia coli, Martincorena et al. (2012) estimated synonymous nucleotide diversity (θS) of individual genes and statistically converted it to θS′, such that it is uncorrelated with five factors known to affect θS. Using θS′ as a proxy for mutation rate, they reported lower mutation rates in genes that are more highly expressed or under stronger purifying selection, which led to the suggestion that mutation rates of different genes have been differentially reduced by natural selection to lessen the mutational harm (Martincorena et al. 2012). Here, we show that this conclusion is theoretically and empirically untenable.
In the lack of recombination such as in asexual organisms, the fitness advantage (k) conferred by a gene-specific antimutator approximates the reduction in deleterious mutation rate of the gene (Δµd) (Kimura 1967; Lynch 2011). Imagine two genes in which the fraction of deleterious mutations is f1 and f2, respectively. The fitness advantage of a given reduction in mutation rate (Δµ) will differ between the two genes by Δk = k1 − k2 = Δµd1 − Δµd2 = Δµf1 − Δµf2 = Δµ (f1 − f2) = ΔµΔf, where Δf = f1 − f2. To select for a mutation rate difference between the two genes, Δk has to exceed the inverse of the effective population size (Kimura 1983); otherwise, natural selection is dwarfed by genetic drift and is unexpected to result in a mutation rate difference between the two genes. The ratio of nonsynonymous to synonymous nucleotide diversity ranges from nearly 0 to approximately 0.3 among E. coli genes (Martincorena and Luscombe 2013). Assuming that Δf between genes is attributable predominantly to nonsynonymous mutations, the maximal Δf between two genes would be approximately 0.3 × 0.76 = 0.228, where 0.3 is the maximal among-gene difference in the fraction of nonsynonymous mutations that are deleterious and 0.76 is the expected proportion of mutations that are nonsynonymous in E. coli (Lee et al. 2012). The average mutation rate in E. coli is µ = 2.1 × 10−7 per gene per generation (see Materials and Methods), and θS′ is approximately 9.6% lower for the most conserved genes, compared with the least conserved genes (see Materials and Methods). If this θS′ difference reflects Δµ, Δk = (2.1 × 10−7 × 9.6%) × 0.228 = 4.6 × 10−9. Because of recombination, the actual Δk in E. coli is probably even smaller (Kimura 1967). There is no known mechanism of antimutation at the gene level that could explain the findings of Martincorena et al. (2012). The subsequently proposed mechanisms (Martincorena and Luscombe 2013), such as the folding of segments of single-stranded DNA during transcription, work at scales of approximately 10 nucleotides (Hoede et al. 2006). Thus, the above value of Δk per gene would be a combined effect of approximately 100 antimutators, each with an average fitness effect of 4.6 × 10−11. This tiny effect is much smaller than the inverse of the effective population size (Ne) of E. coli, despite previous estimates of Ne varying from 105 to 109 (Ochman and Wilson 1987; Bulmer 1991; Hartl et al. 1994; Charlesworth and Eyre-Walker 2006). Clearly, the observed θS′ differences among genes could not have been caused by gene-specific selective reduction of mutation rate. Although it remains theoretically possible for natural selection to act on a modifier that simultaneously tunes the mutation rates of tens to hundreds of genes based on their f values, no such molecular mechanism is known.
A plausible explanation of the results by Martincorena et al. is that θS′ does not accurately measure mutation rate because of the existence of uncontrolled confounding factors, such as the known selection against Shine–Dalgarno-like sequences in coding regions (Li et al. 2012) and selection for mRNA folding (Park et al. 2013), or unknown. We therefore re-estimated the mutation rate of individual E. coli genes from single-nucleotide substitutions observed in mutation accumulation (MA) lines of the wild-type (WT) and MutL− backgrounds (Lee et al. 2012). Because the MA lines went through repeated single-cell bottlenecks (Lee et al. 2012), there was little selection except on essential genes. We therefore focus our analysis on nonessential genes and then use essential genes to confirm the results. The lack of mismatch repair rendered the MutL− lines approximately 150 times more mutable (Lee et al. 2012) and therefore more powerful than the WT lines for detecting mutation rate differences among genes.
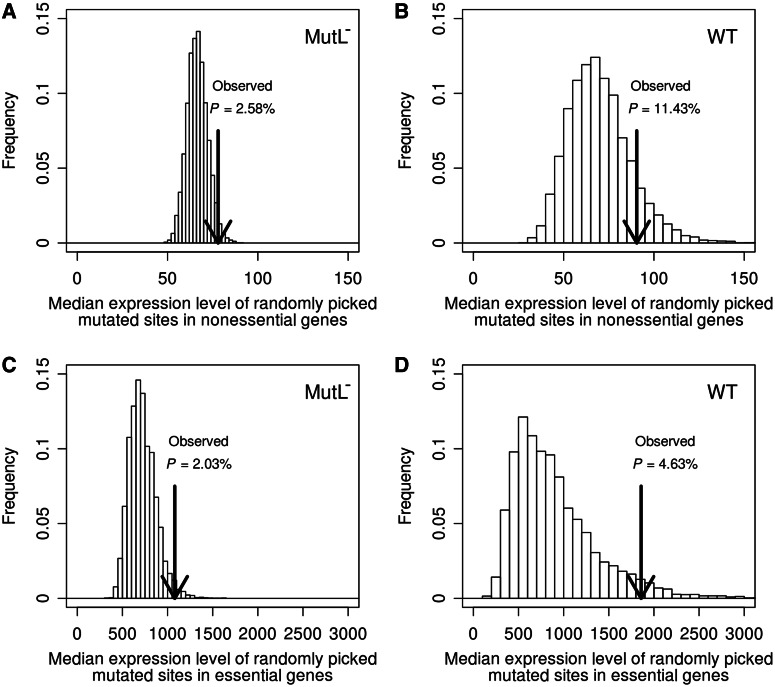
Contrary to the claim by Martincorena et al., we find a weak but significant positive correlation between the expression level of a nonessential gene and its per site mutation rate in the MutL− background (Spearman’s rank correlation ρ = 0.060, P < 0.0001). This correlation remains significant (ρ = 0.053, P < 0.0007) after the control of among-gene variation in percent guanine + cytosine nucleotides (GC%) that may influence gene mutability (Hodgkinson and Eyre-Walker 2011). Although the correlation is positive, it is not significant in WT before (ρ = 0.008, P > 0.5) or after (ρ = 0.007, P > 0.5) the control of GC%, likely due to the fact that the WT lines contain much fewer mutations (166), compared with the MutL− lines (1,346). We verified these results by comparing the median expression level of the observed mutated sites (ELobs) with the corresponding value from the same number of randomly picked sites (ELran). To control the mutability differences among the four bases, we required the leading strand nucleotide frequencies (Lee et al. 2012) among the randomly picked sites to equal their respective frequencies among the observed mutated sites. In the MutL− background, ELobs is 17% greater than the average ELran (fig. 1a). In 97.42% of 10,000 sets of randomly picked sites, ELobs exceeds ELran, confirming significantly higher mutation rates at more highly expressed sites (fig. 1a). In the WT background, although ELobs is 29% greater than the average ELran, their difference is not significant, again possibly due to the small sample size (fig. 1b).
Fig. 1.
Median expression levels of mutated sites of nonessential genes in (a) MutL− and (b) WT MA lines and of essential genes in (c) MutL− and (d) WT lines are higher than those of randomly picked sites. Expression levels are in arbitrary units but can be compared among the four panels. In each panel, the bars show the frequency distribution of the median expression level of the same number of randomly picked sites as the actual mutated sites, derived from 10,000 sets of random sites, whereas the arrowhead indicates the median expression level of the actual mutated sites. P value indicates the fraction of the 10,000 times in which the median expression level of the randomly picked sites exceeds that of the actual mutated sites. The leading strand nucleotide frequencies among the randomly picked sites are constrained to equal their respective frequencies among the actual mutated sites.
Only approximately 7% of E. coli genes are essential (Baba et al. 2006), but our analysis of these genes yielded similar results. For instance, there is a significant positive correlation between the expression level of an essential gene and its mutation rate in the MutL− background before (ρ = 0.151, P = 0.011) and after controlling for GC% (ρ = 0.157, P = 0.008). Although the corresponding correlation in WT is positive, it is not significant before (ρ = 0.047, P > 0.4) or after (ρ = 0.039, P > 0.5) the control of GC%. For essential genes, ELobs is 49% greater than the average ELran in the MutL− background (P = 0.0203; fig. 1c), and the corresponding number is 105% in the WT background (P = 0.0463; fig. 1d).
Note that, in their initial analysis of the MA lines, Lee et al. (2012) mentioned that mutation rate and gene expression level are uncorrelated, but they provided no information on how this result was obtained. Contrary to the MA lines, large laboratory populations of E. coli could be subject to natural selection even at synonymous sites and thus are unsuitable for verifying the θS′-based observations (Maddamsetti et al. 2012; Martincorena and Luscombe 2012).
It is improbable that our observations in the MutL− background are artifacts of the loss of mismatch repair, because mismatch repair fixes DNA replication errors in an expression-independent, nonsequence-specific manner (Schofield and Hsieh 2003). Furthermore, the observed trends in MutL− are also present in WT, and ELobs/ELran is not lower in WT than MutL−. Although the laboratory condition used in the MA experiment differs from E. coli’s natural environment, there is currently no evidence that such an environmental change would differentially alter mutation rate according to gene expression level. Note that in the analysis by Martincorena et al., gene expression levels were measured in a laboratory condition despite that θS′ was calculated using natural strains.
Our observations from the MA lines, coupled with the strong positive correlation between the expression level of a gene and its protein sequence conservation (Pal et al. 2001; Rocha and Danchin 2004), predict higher mutabilities of genes with more conserved protein sequences (i.e., subject to stronger purifying selection). This tendency, in contrast to the claim by Martincorena et al. (2012), is indeed observed in both MutL− (ρ = 0.055, P < 0.009; see Materials and Methods) and WT (ρ = 0.011, P > 0.5) for nonessential genes. The same trend is found for essential genes (ρ = 0.104, P = 0.139 for the MutL− lines; ρ = 0.091, P = 0.198 for the WT lines).
With the caveat that essential genes are subject to some purifying selection in the MA experiment, we directly compare the observed mutation frequencies between essential and nonessential genes. In the WT background, the mean mutation frequency per site in nonessential and essential genes is 5.46 × 10−5 and 7.37 × 10−5, respectively, and their difference is not significant (P = 0.39, Mann–Whitney U test). In the MutL− background, the corresponding numbers are 3.73 × 10−4 and 3.06 × 10−4, respectively, and their difference is again not significant (P = 0.41). Thus, the MA lines provide no significant evidence for lowered mutation rates in essential genes compared with nonessential genes.
In sum, Martincorena et al.’s conclusion of gene-specific selective optimization of mutation rates in E. coli is not supported theoretically or empirically. If anything, E. coli mutation rates appear higher in more highly expressed genes, at least in the MA lines. Previous results of the relationship between gene expression level and mutation rate in E. coli varied, because of the use of few genes or inappropriate mutation rate estimators (Mellon and Hanawalt 1989; Eyre-Walker and Bulmer 1995; Beletskii and Bhagwat 1996). Our genome-wide result in E. coli echoes the observations from the MA lines of the bacterium Salmonella typhimurium (Lind and Andersson 2008) and the yeast Saccharomyces cerevisiae (Park et al. 2012), as well as the comparative genomic evidence from the yeast and the human germline (Park et al. 2012). Mechanistically, gene expression is known to impact mutation rate by transcription-associated mutagenesis (Kim and Jinks-Robertson 2012) and transcription-coupled repair (Hanawalt and Spivak 2008). That mutation rate increases with gene expression level at the genomic scale suggests that the overall effect of transcription-associated mutagenesis on mutation rate surpasses that of transcription-coupled repair.
Materials and Methods
Martincorena et al. (2012) used an E. coli RNA-Seq expression data set generated in their laboratory (Kahramanoglou et al. 2011). We found a publicly avialable E. coli RNA-Seq data set (Giannoukos et al. 2012) that is approximately 50 times the size of their data set in terms of the total number of sequencing reads mapped to open reading frames. Although gene expression levels estimated from the two data sets are correlated (ρ = 0.71, P < 10−30), the larger data set is expected to offer more precise estimates and therefore was used in our analysis. Both the E. coli mutation and gene expression data were from strain MG1655 grown in LB at 37 °C and were downloaded from National Center for Biotechnology Information. The mutation data (Lee et al. 2012) have the accession numbers of SRA054030 and SRA054031, whereas the Ribo-Zero RNA-Seq expression data (Giannoukos et al. 2012) have the accession numbers of SRR441615, SRR441637, SRR441644, SRR441655, SRR441662, SRR441697, SRR442249, SRR442255, SRR442258, SRR442262, SRR442267, SRR442269, SRR442271, SRR442291, SRR442294, and SRR442307. The RNA-Seq paired-end reads (101 bases/read) were aligned to the MG1655 genome sequence using BWA v5.9, with parameters: -q 5 -l 32 -k 2 -t 4 -o 1 (Giannoukos et al. 2012). Only uniquely mapped read pairs with ≤2 mismatches in each read were considered. The expression level of a nucleotide position was defined as the number of reads covering the site. Positions encompassed by but not directly mapped to by a read pair were also treated as being covered by the read pair. The expression level of a gene was defined by the mean expression level of its nucleotide positions annotated by Ensembl. Overlapping regions of multiple genes were not considered. Classification of E. coli essential and nonessential genes followed Martincorena et al. (2012), which was based on an earlier study (Baba et al. 2006).
Escherichia coli and S. typhimurium one-to-one orthologs were identified by reciprocal best hits implemented in BLASTP with default parameters. Protein sequence conservation was defined by the protein sequence identity between one-to-one orthologs, calculated by ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default parameters. Proteins with identities less than 50% were removed to guard against the inclusion of xenologs arising from horizontal gene transfers. The median θS′ of the 10% most conserved proteins and that of the 10% least conserved proteins were compared. The per generation mutation rate in E. coli is on average 2.2 × 10−10 per nucleotide (Lee et al. 2012) or approximately 2.1 × 10−7 per gene (based on the mean coding sequence length of 951 nucleotides in E. coli [Zhang 2000]).
Acknowledgments
The authors thank Calum Maclean, Wenfeng Qian, the associate editor, and three anonymous reviewers for valuable comments. This work was supported, in part, by the research grant R01GM067030 from the U. S. National Institutes of Health to J.Z.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100050. 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- Beletskii A, Bhagwat AS. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. The rate of adaptive evolution in enteric bacteria. Mol Biol Evol. 2006;23:1348–1356. doi: 10.1093/molbev/msk025. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Bulmer M. Synonymous substitution rates in enterobacteria. Genetics. 1995;140:1407–1412. doi: 10.1093/genetics/140.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoukos G, Ciulla DM, Huang K, et al. (13 co-authors) Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012;13:R23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Moriyama EN, Sawyer SA. Selection intensity for codon bias. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 2011;12:756–766. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- Hoede C, Denamur E, Tenaillon O. Selection acts on DNA secondary structures to decrease transcriptional mutagenesis. PLoS Genet. 2006;2:e176. doi: 10.1371/journal.pgen.0020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. On the evolutionary adjustment of spontaneous mutation rates. Genet Res. 1967;9:23–34. [Google Scholar]
- Kimura M. Cambridge: Cambridge University Press; 1983. The neutral theory of molecular evolution. [Google Scholar]
- Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Andersson DI. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci U S A. 2008;105:17878–17883. doi: 10.1073/pnas.0804445105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The lower bound to the evolution of mutation rates. Genome Biol Evol. 2011;3:1107–1118. doi: 10.1093/gbe/evr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Maddamsetti R, Hatcher PJ, Cruveiller S, Médigue C, Barrick JE, Lenski RE. Horizontal gene transfer may explain variation in θs. 2012 arXiv:1210.0050. [Google Scholar]
- Martincorena I, Luscombe NM. Response to “horizontal gene transfer may explain variation in theta_s”. 2012 arXiv:1211.0928. [Google Scholar]
- Martincorena I, Luscombe NM. Non-random mutation: the evolution of targeted hypermutation and hypomutation. Bioessays. 2013;35:123–130. doi: 10.1002/bies.201200150. [DOI] [PubMed] [Google Scholar]
- Martincorena I, Seshasayee AS, Luscombe NM. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485:95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Ochman H, Wilson AC. Evolutionary history of enteric bacteria. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington (DC): ASM Press; 1987. pp. 1649–1654. [Google Scholar]
- Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Chen X, Yang JR, Zhang J. Differential requirements for mRNA folding partially explain why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2013;110:E678–E686. doi: 10.1073/pnas.1218066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Qian W, Zhang J. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 2012;13:1123–1129. doi: 10.1038/embor.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol Biol Evol. 2004;21:108–116. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- Zhang J. Protein-length distributions for the three domains of life. Trends Genet. 2000;16:107–109. doi: 10.1016/s0168-9525(99)01922-8. [DOI] [PubMed] [Google Scholar]