Abstract
Bioinformatics and functional screens identified a group of Family A-type DNA Polymerase (polA) genes encoded by viruses inhabiting circumneutral and alkaline hot springs in Yellowstone National Park and the US Great Basin. The proteins encoded by these viral polA genes (PolAs) shared no significant sequence similarity with any known viral proteins but were remarkably similar to PolAs encoded by two of three families of the bacterial phylum Aquificae and by several apicoplast-targeted PolA-like proteins found in the eukaryotic phylum Apicomplexa, which includes the obligate parasites Plasmodium, Babesia, and Toxoplasma. The viral gene products share signature elements previously associated only with Aquificae and Apicomplexa PolA-like proteins and were similar to proteins encoded by prophage elements of a variety of otherwise unrelated Bacteria, each of which additionally encoded a prototypical bacterial PolA. Unique among known viral DNA polymerases, the viral PolA proteins of this study share with the Apicomplexa proteins large amino-terminal domains with putative helicase/primase elements but low primary sequence similarity. The genomic context and distribution, phylogeny, and biochemistry of these PolA proteins suggest that thermophilic viruses transferred polA genes to the Apicomplexa, likely through secondary endosymbiosis of a virus-infected proto-apicoplast, and to the common ancestor of two of three Aquificae families, where they displaced the orthologous cellular polA gene. On the basis of biochemical activity, gene structure, and sequence similarity, we speculate that the xenologous viral-type polA genes may have functions associated with diversity-generating recombination in both Bacteria and Apicomplexa.
Keywords: viral metagenomics, horizontal gene transfer, replication, DNA polymerase, Apicomplexa, Aquificae
Introduction
Viruses are increasingly recognized as highly abundant, diverse, and ecologically significant components of all ecosystems (Suttle 2007; Srinivasiah 2008; Clokie et al. 2011) and sources of molecular diversity in cellular genomes (Villarreal and DeFilippis 2000; Canchaya et al. 2003; Filee et al. 2003; Daubin and Ochman 2004; Chan et al. 2011). Viral genes also may have been catalysts of major evolutionary transitions (Forterre 2006). In this report, we combined functional and informatics approaches to identify a clade of viral polA-type DNA polymerase genes in viral metagenomes from three thermal springs in the western United States. We show that highly similar polymerases are found in the bacterial phylum Aquificae and the eukaryotic phylum Apicomplexa and invoke a key role of thermophilic viruses in lateral transfer of these polymerase genes. We suggest that these genes may be associated with dispersal of diversity-generating mechanisms between geothermal and moderate-temperature biomes.
The genes involved in replicating DNA, especially those encoding DNA polymerases, are ubiquitous and fundamental to life. The capacity to replicate the informational content of genomes was a critical step in the transformation from an abiotic to a biotic world (Martin and Russell 2003; Glansdorff et al. 2008). The basic mechanism of replication has been conserved throughout all life forms; however, it appears that the main replicative polymerases (PolA, PolB, and PolC) diverged relatively early (Koonin 2006). PolBs evolved as the main leading-strand replicative polymerases in the archaeal/eukaryal lineage, and PolCs evolved as the main replicative polymerases in Bacteria. The PolA lineage is confined to Bacteria, certain eukaryotic subcellular organelles of bacterial origin, and some bacteriophages. In Bacteria, PolAs function mainly in lagging-strand synthesis and DNA repair (Baker and Kornberg 1992). Bacterial PolAs usually comprise two domains, a functionally indivisible 3′-5′ proofreading exonuclease and DNA polymerase (3′exo/pol) domain and a separate 5′-3′ exonuclease (5′exo) domain, which functions in removal of RNA primers during lagging-strand synthesis and nick translation during DNA repair (Klenow and Overgaard-Hansen 1970; Setlow and Kornberg 1972; Beese and Steitz 1991). From a practical perspective, DNA polymerases, particularly thermostable DNA polymerases, are key elements in all major DNA and RNA amplification and sequencing methods, and polymerases described in this study have proven useful as molecular biology reagents (Schoenfeld et al. 2010; Moser et al. 2012; Perez et al. 2013).
Viruses have adopted a diverse range of replication strategies, some of which are almost completely dependent on host gene products, whereas others use mainly virus-encoded factors (Baker and Kornberg 1992; Weigel and Seitz 2006). Almost all large double-stranded DNA viruses encode DNA polymerases and often accessory proteins such as processivity factors (Kazlauskas and Venclovas 2011). As of August 2012, there were 1,031 bacteriophage genomes and 51 archaeal virus genomes in the EMBL-EBI sequence database (http://www.ebi.ac.uk/genomes; last accessed May 11, 2013). Only one archaeal virus (2% of total) has an annotated DNA polymerase gene (Peng et al. 2007), whereas 31% of the bacteriophage genomes in the database (316) encode apparent polymerases (204 polA genes [19.8%], 90 polB genes [8.7%], 18 polC genes [1.7%], 4 dnaE genes [0.4%], and 1 dinB gene [0.1%]).
Geothermal springs are especially well suited to studies of early evolution and biogeography and are windows into the subsurface biosphere. These natural formations are the sources of virtually all known thermophiles, the core genomes of which branch deeply within the tree of life, suggesting a thermophilic origin of life and a key role for certain essential genes, for example, those encoding thermostable polymerases, in early stages of evolution (Di Giulio 2003; Schwartzman and Lineweaver 2004). Unlike moderate-temperature environments that were sources of most viral genomes and metagenomic data sets, terrestrial hot springs may be more “island-like” because geothermally active regions are usually separated by large distances that limit transmission of thermophiles. Of particular interest for this study, no models accounting for significant subsurface connectivity between the Yellowstone Caldera and the western Great Basin have been proposed, although a limited extent of interbasin hydrologic flow through deep, fractured carbonates within the US Great Basin is a topic of debate (Anderson et al. 2006). Nevertheless, the continental subsurface represents a significant portion of the Earth’s habitable volume (Gold 1992) and extant biomass (Whitman et al. 1998) and is the least explored biome in the terrestrial critical zone (Richter and Mobley 2009).
Cultivation-based studies of thermophilic viruses have revealed six new morphological families of dsDNA viruses infecting the thermoacidophilic Archaea Sulfolobus and Acidianus (Prangishvili et al. 2006; Happonen et al. 2010; Pina et al. 2011), as well as additional viruses infecting thermophilic Bacteria, especially Thermus (Yu et al. 2006). However, thermophilic viruses have proven difficult to cultivate. Especially problematic is the bias resulting from the limited number of hosts that have been used to cultivate viruses. Metagenomics offers a powerful tool to study evolution and molecular diversity of natural populations. Most metagenomic studies of thermophilic viruses have focused on acidic springs dominated by Sulfolobus and Acidianus (Garrett et al. 2010; Snyder and Young 2011; Bolduc et al. 2012), with only one focused on circumneutral or alkaline springs (Schoenfeld et al. 2008). There is a fundamental difference between the two types of hot springs. Acidic hot springs are typically sourced by condensed water vapor and lack outflow channels, whereas circumneutral and alkaline springs are typically sourced by liquid water and often have high outflow into nearby meadows or streams, providing an obvious conduit for molecular communication with moderate-temperature environments (Nordstorm et al. 2005).
In this study, we addressed a fundamental shortcoming of most metagenomic studies, that is the reliance on primary sequence similarity to infer function, by combining functional and bioinformatic approaches to discover DNA polymerases in viral metagenomes from three geothermal springs in the western United States. The springs described in this study, Octopus Spring (OCT) in the Yellowstone Caldera, Great Boiling Spring (GBS) in the northwest Great Basin, and Little Hot Creek (LHC) in the Long Valley Caldera of the western Great Basin, have each been surveyed biologically and geochemically (Reysenbach et al. 1994; Huber et al. 1998; Costa et al. 2009; Vick et al. 2010). All have substantial populations of the genus Thermocrinis, which is a member of the bacterial phylum Aquificae. Aquificae diverged very near the separation of Archaea and Bacteria (Oshima et al. 2012), yet their genomes have been greatly impacted by lateral gene transfer (Boussau et al. 2008). Despite their high abundance in most hot springs (Spear et al. 2005), Aquificae have proven difficult to cultivate, and no cultivated viruses infecting members of this phylum have been described. Consequently, the contribution of viruses to the biology of the Aquificae remains unexplored. The viral PolAs described here showed unexpected similarity to PolAs of the Aquificae and the apicoplast-targeted PolA homolog, Pfprex of Plasmodium falciparum, and related PolAs in other pathogenic Apicomplexa. The sequence similarity between Aquificales PolA and Apicomplexa Pfprex proteins has been previously noted (Seow et al. 2005; Griffiths and Gupta 2006), although their relationship remained unexplained. The discovery of apparently xenologous PolAs in thermophilic viruses and prophage elements of a variety of unrelated mesophilic bacteria provides an obvious mechanism for lateral gene transfer across large phylogenetic distance and ecological space and between biomes of very different temperatures and chemistry. We speculate that the exchanged genes are associated with diversity-generating mechanisms.
Results and Discussion
Bioinformatic Discovery of polA Genes in Hot Springs Viral Metagenomes
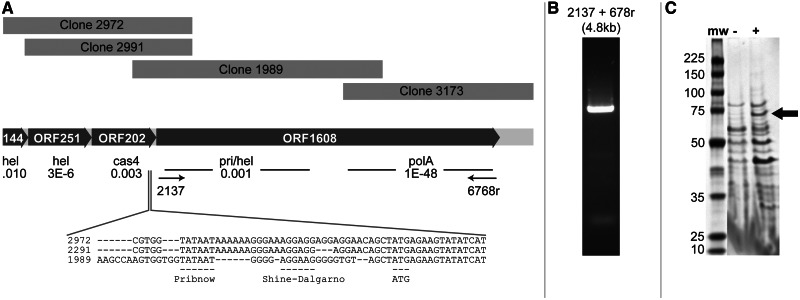
Viral polA genes were initially discovered by bioinformatic analysis of viral metagenomes. Viruses were collected and concentrated by tangential-flow filtration of bulk water from OCT in 2003 (OCT03) and from LHC in 2001 (supplementary fig. S1, Supplementary Material online). DNA isolated from these samples was used to create 3–5 kb insert clone libraries, which were Sanger sequenced by the Joint Genome Institute and analyzed by BLASTX as described (Schoenfeld et al. 2008), leading to the discovery of 256 clones with significant similarity to DNA polymerases (Schoenfeld et al. 2008; Moser et al. 2012). Of these, three clones were highly similar to each other and to various Aquificae polA genes and were designated OCT-3173, OCT-967, and LHC-488 (accession numbers for all the newly discovered PolAs are shown in fig. 2). All three PolAs contained conserved motifs associated with DNA polymerase (Pfam 00476) and 3′-5′ exonuclease (Pfam 01612) and demonstrated thermostable polymerase and exonuclease activity. Clone OCT-3173 was selected for large-scale expression, purification, and detailed biochemical characterization that showed the OCT-3173 PolA encodes the first known thermostable DNA polymerase with native reverse transcriptase, strand displacement, and proofreading activities (Moser et al. 2012). Although each of the three clones encoded functional polymerases when expressed in Escherichia coli, sequence analysis failed to identify transcriptional or translational starts and none appeared to contain a full open reading frame (ORF). Previous work had shown that, given the unusually high molecular diversity within viral populations, lower assembly stringencies can be useful in associating sequence reads that are otherwise be too divergent to join (Schoenfeld et al. 2008). These assemblies can then be validated by polymerase chain reaction (PCR) amplification across the junctions. Correspondingly, attempts to coassemble clones OCT-3173, OCT-967, or LHC-488 with other reads in the OCT03 metagenomic library at 95% nucleic acid identity (NAID) failed; however, when the assembly stringency was lowered to 75% NAID, three additional cloned sequences assembled with OCT-3173, producing a consensus ORF predicted to encode a 1,608-amino acid product, designated ORF1608 (fig. 1A). The assembly included a consensus Pribnow box (Pribnow 1975) and Shine–Dalgarno sequence (Shine and Dalgarno 1975) upstream of the apparent ORF1608 ATG start codon suggesting transcriptional and translational start sites (fig. 1A). Additional ORFs in the contig upstream of ORF1608 shared sequence similarity with cas4, a recB homolog with single-strand DNA nuclease activity associated with recombination (Zhang et al. 2012), and a SNF2-type, DEAD/DEAH box helicase-like protein (Fairman-Williams et al. 2010).
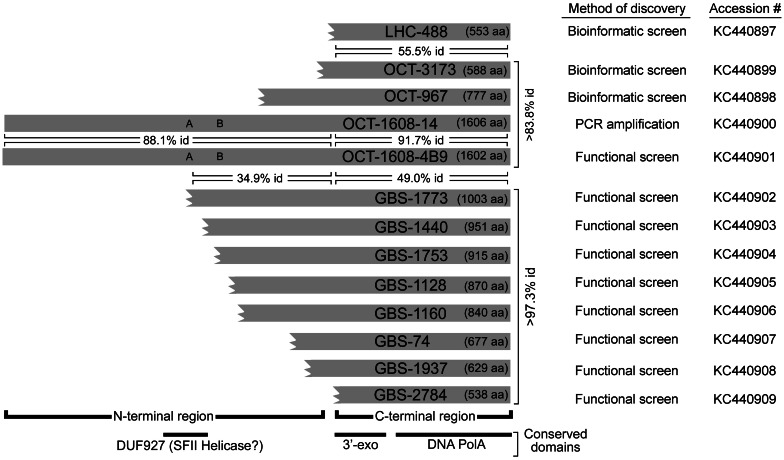
Fig. 2.
Schematic alignment of polA gene products of the indicated clones showing length in amino acids (aa) of conserved domains, method of discovery. Three letter codes specify the source hot spring: LHC (CA), OCT (YNP, WY), and GBS (NV). Truncated ORFs are shown with broken amino termini. Percent amino acid identities for the carboxy- and amino-terminal domains are indicated in horizontal brackets for selected pairs. Minimum percent identities for all pairwise comparisons within each biogeographic cluster are indicated in vertical text to the right of the sequences. A and B denote Walker A (P-loop) and B box motifs proposed to be involved in NTP binding and hydrolysis. GenBank accession numbers corresponding to the polA genes are shown.
Fig. 1.
Assembly, cloning, and expression of polA genes from OCT viral metagenomes. Four reads from the OCT-03 library (panel A, top) were assembled at 75% NAID to form a 7,410-nucleotide consensus contig (panel A, middle) that included three complete and one partial ORF. ORF251 was similar to hel (ACB83959.1), ORF202 to cas4 (ABN70290.1), and ORF ORF1608 had an amino-terminal domain similar to hel (EKQ55894.1) and a carboxy-terminal domain similar to polA (ADC89878.1). Expect values are shown. The sequence immediately adjacent to the 5′-terminus of ORF1608 included consensus transcriptional and translational start elements (panel A, bottom). Primers 2137 and 6768r, shown in panel A, were used to amplify the full-length OCT-1608 ORF from the OCT-07 DNA preparation (panel B). This PCR product (OCT-1608-14) was cloned and expressed to generate a thermostable DNA polymerase and migrates as a 70 kD protein (panel C) seen by SDS-PAGE in the induced (+) but not the uninduced (−) clone. SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
PCR primers were designed based on the composite ORF1608 sequence to amplify a full-length, native ORF. A viral DNA preparation from OCT sampled in 2007 (OCT07) was used because the entire OCT03 viral DNA preparation was consumed during library construction. PCR resulted in an appropriate-sized amplicon of 4,824 bp suggesting correct assembly (fig. 1B). Amplicons were expressed in E. coli, and heat-treated lysates were tested for thermostable polymerase activity. All clones showed activity when assayed at 70 °C (Schoenfeld T, unpublished observation). A clone designated OCT-1608-14 encoded a marginally more thermostable polymerase and was used for further analysis. This clone encoded a 3′exo/pol domain with 92% amino acid identity (AAID) with the entire 588-amino acid OCT-3173 ORF fragment (fig. 2).
Functional Screening of Viral Metagenome Expression Libraries and Mapping of Polymerase Activity
Functional screens were used to identify additional viral polA genes in clones based on expression of thermostable DNA polymerase activity. Screening of an 8–12-kb insert clone library from the OCT07 DNA preparation identified six additional polA clones with thermostable polymerase activity (supplementary table S1, Supplementary Material online) as well as large stretches of adjacent sequence. Although five of the clones were truncated, the DNA sequences within the regions of overlap were nearly identical to OCT-1608-14 (Schoenfeld T, unpublished observation). In contrast, OCT-1608-4B9 contained a complete ORF but shared approximately 90% AAID with the other full-length ORF from clone OCT-1608-14 (fig. 2).
Additional polA genes were identified by functional screening of a 3–5-kb insert plasmid clone library derived from a virus preparation from GBS, Gerlach, NV, collected in 2007 (supplementary fig. S1, Supplementary Material online). Screening of 2,800 clones identified 12 clones expressing DNA polymerase activity. DNA sequencing indicated that 11 of these were highly similar to one another and also related to the viral polA genes from OCT and LHC (fig. 2; supplementary fig. S2, Supplementary Material online). The 12th clone, GBS-347, was highly divergent and encoded a protein similar to a presumed primase/polymerase identified in a viral metagenome from a different hot spring (Garrett et al. 2010) and will be described separately.
The alignment of the full collection of the predicted PolAs enabled functional mapping of the domains. All clones shared conserved 3′-termini of the PolA ORFs (fig. 2). The sizes of the ORFs and ORF fragments ranged from 538 to 1,606 amino acids. Because all clones expressed similar thermostable DNA polymerase activity, no more than 538 amino acids, comprising the carboxy-terminal regions, are necessary and sufficient for polymerase activity. This 538 amino acid carboxy terminus corresponds to essentially the entirety of the sequence similarity to known polA genes and also included the conserved 3′-5′ proofreading exonuclease motif (Moser et al. 1997). Correspondingly, all the viral PolAs tested exhibited exonuclease activity (Moser et al. 2012). The identity of the 3′exo/pol domains of viruses was much higher within each hot spring (>83.8% AAID for OCT, >97.3% for GBS) than between springs (45–55% identity) (fig. 2), despite the intervening 4 years between collection of the two Octopus samples, OCT03 and OCT07.
Conservation and Possible Functions of Amino-Terminal Sequences
The amino-terminal regions of the viral PolAs from OCT07 and GBS samples were noticeably more divergent than the carboxy-terminal regions but still shared 34.9% AAID over the extent of the overlap between the largest GBS clone, GBS1773, and the OCT-1608-14 ORFs (fig. 2). The two full-length ORFs, OCT-1608-14 and OCT-1608-4B9, had sequence motifs that imply primase and/or helicase function. These include similarity to DUF927 (conserved domain with carboxy terminal P-loop NTPase; E value 4.8e-7) and COG5519 (Superfamily II helicases associated with DNA replication, recombination, and repair [Marchler-Bauer et al. 2011]), covering roughly amino acids 515–650 of the amino terminal regions. A consensus Walker A or P-loop motif (GxxxxGK[S/T]) and a potential Walker B motif (SIVLD in comparison to the consensus hhhhD, where h indicates a hydrophobic amino acid) suggest NTP binding and hydrolysis likely associated with helicase activity (Walker et al. 1982).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels of E. coli expressing the ORFs and ORF fragments from GBS all gave rise to a similar size product (∼55 kDa) when expressed in E. coli (supplementary fig. S2, Supplementary Material online), suggesting that the genes are expressed as larger polyproteins and cleaved within the cloned region, with amino-terminal regions being unstable or insoluble in the expression host. Similarly, expression of the full-length Oct-1608-14, which would be predicted to encode a protein of 189 kDa, resulted in a thermostable protein of about 75 kD (fig. 1C).
Phylogenetic Analysis of polA-Like Genes
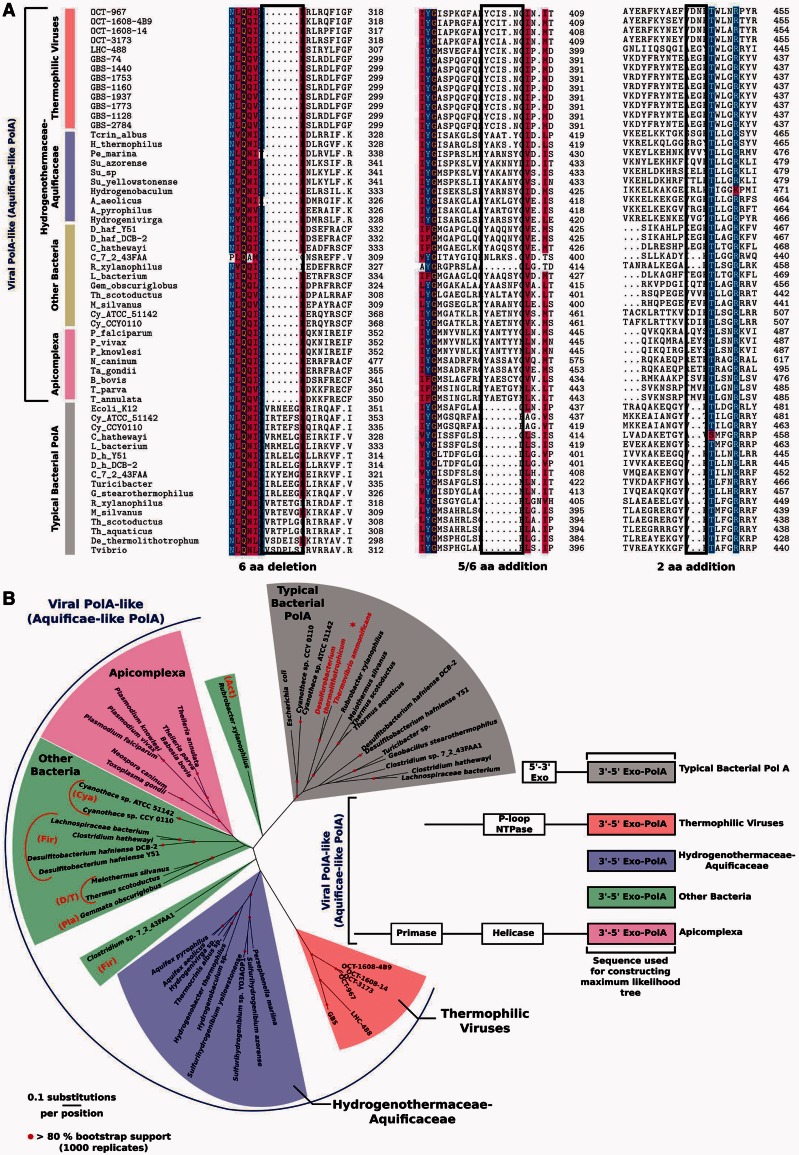
BLASTP analysis showed that the carboxy-terminal 3′exo/pol domains of the viral gene products were similar to those of the PolAs of several Aquificae species, typified by the Aquifex aeolicus PolA (Chang et al. 2001), and to 3′exo/pol domains of the nuclear-encoded, apicoplast-targeted DNA polymerases of several Apicomplexa species, typified by the Pfprex protein of P. falciparum (Seow et al. 2005). BLASTP also revealed closely related PolAs encoded by genomes of a few unrelated Bacteria belonging to the phyla Firmicutes, Actinobacteria, Deinococcus-Thermus, Cyanobacteria, and Planctomycetes. Alignment of these PolAs demonstrated that three indels that distinguish PolA-type polymerases of the Aquificaceae and Hydrogenothermaceae families from other bacterial PolAs (Griffiths and Gupta 2006) are conserved in the viral PolAs, the apicoplast-targeted PolAs of Apicomplexa, and the similar PolAs identified in various Bacteria by the BLASTP search (fig. 3). Phylogenetic analysis of the conserved 3′exo/pol domains of the viral and cellular PolA proteins confirmed these relationships and showed that PolAs discussed here comprise four major lineages related to, but distinct from, prototypical bacterial PolAs. These four groups include PolAs from: 1) thermophilic viruses; 2) Aquificaceae and Hydrogenothermaceae; 3) Apicomplexa; and 4) various Bacteria (fig. 3, supplementary fig. S3, Supplementary Material online). The viral PolAs diverged into clades from Yellowstone National Park (YNP) and the Great Basin. This biogeographic distribution is similar to that reported for a number of cellular thermophiles (Whitaker et al. 2003; Miller-Coleman et al. 2012) and prophages within genomes of Sulfolobus (Held and Whitaker 2009) and is evidence of a dispersal limitation. Notably, our own viral metagenomic studies from a variety of other apparently similar geothermal springs in YNP and the Great Basin showed related viral polA genes to be absent or extremely rare, demonstrating the disjointed nature of optimal habitat for this group of viruses.
Fig. 3.
Alignment of conserved indels of viral, Aquificae, and Apicoplast polA genes based on signature motifs identified in (Griffiths and Gupta 2006) (panel A). Representative prototypical bacterial PolAs are shown at the bottom for comparison. Maximum-likelihood phylogeny of carboxy-terminal 3′exo/pol domains of viral and cellular polA-like genes (panel B). Representative prototypical bacterial PolA proteins are shown for comparison (gray) with the Aquificae family Desulfurobacteriaceae emphasized (asterisk, red text). The region used for the analysis consisted of the 588 C-term amino acids corresponding to clone OCT-3173 (fig. 1). The schematic alignment (right) highlights the carboxy terminal regions used for analysis and shows divergence of amino-terminal PolA proteins. Branches with more than 80% bootstrap values (1,000 replicates) are indicated. Phylum-level designations for non-Aquificae bacterial hosts of viral polA genes are indicated by abbreviations as follows: Act, Actinobacteria; Cya, Cyanobacteria; Fir, Firmicutes; D/T, Deinococcus-Thermus; Pla, Planctomycetes. An analysis with the same alignment and a 50% mask shows similar results (supplementary fig. S3, Supplementary Material online).
Likely Orthologous Replacement of Bacterial polA Genes with Viral polA Genes in Two Families of Aquificae and Likely Virus–Host Relationship
The genomic distribution and phylogeny of the PolA xenologs in the viral and cellular genomes offer clues to their evolution (fig. 3). Two out of three families of the Aquificae, Aquificaceae, and Hydrogenothermaceae contain the viral-type PolA; however, the more deeply branching third family, Desulfurobacteriaceae, has a prototypical bacterial PolA. This suggests that a viral-type polA gene replaced its xenolog and became fixed in the genome of a common ancestor of two of three families of Aquificae following their divergence from the Desulfurobacteriaceae (L'Haridon et al. 2006; Hugler et al. 2007). Once fixed in the Aquificae, this polA gene probably descended vertically, because it mirrors the 16S rRNA gene phylogeny.
The 5′-3′ exonuclease (5′exo) domains provide additional support to this model. The 5′-3′ exonuclease domains of bacterial polA gene products, in contrast to 3′-5′ exonuclease domain, are quite independent from the rest of the polA gene product. As such, separation of the 5′exo domain from the 3′exo/pol domain, either by independent expression or proteolytic digestion, results in two functional polypeptides, a 5′-3′ exonuclease and a polymerase (Klenow and Overgaard-Hansen 1970; Setlow and Kornberg 1972). Consistent with the lateral gene transfer model, the Aquificaceae and Hydrogenothermaceae PolAs, along with all viral PolAs described here, lack a 5′exo domain and instead have an independent gene predicted to encode the 5′-3′ exonuclease, suggesting that this region of the prototypical bacterial polymerase gene was retained in the Aquificaceae and Hydrogenothermaceae after orthologous replacement of the 3′exo/pol domains by the viral genes.
In contrast, the extensive amino terminal region of the viral polA gene products, corresponding to the first approximately 1,000 amino acids of OCT-1608-14 and containing the putative helicase domain (DUF927), is absent from these Aquificae PolAs, implying that the replacement of the prototypical bacterial polA gene in the Aquificaceae and Hydrogenothermaceae must have substantially restructured the information processing within these organisms. The fact that these two families are the most abundant microorganisms in many >75 °C hot springs in YNP (Reysenbach et al. 1994; Spear et al. 2005) and probably worldwide suggests that this was one of probably many successful adaptations to the environment.
Coding bias reflects the shared use of host translational machinery by the host and virus, and correspondingly, tetranucleotide frequency tends to be conserved between host and virus (Pride et al. 2006; Pride and Schoenfeld 2008; Deschavanne et al. 2010). An analysis of tetranucleotide word frequency of a shotgun metagenome from the GBS planktonic microbial community along with contigs from the viral metagenome showed that contigs containing viral polA genes clustered closely with genomic sequences from the dominant Thermocrinis species of the GBS planktonic community (supplementary fig. S4, Supplementary Material online) (Dodsworth et al. 2011). Consistent with this observation, cultivation-independent censuses suggest Thermocrinis also dominate the pink streamer communities in the outflow channel of OCT (Reysenbach et al. 1994) and sediment/precipitate communities in the small source pool of LHC (Vick et al. 2010). The similar tetranucleotide word frequency and the abundance of Thermocrinis in all three hot springs studied imply that this genus may be the natural host for these viruses. If these viruses infect Thermocrinis, it is highly unusual that the viral replicases are so similar to those of the host because most virus-induced replicases are highly divergent from those of the host.
Evidence for Transient Presence of Viral-Type polA Genes in Other Bacterial Genomes
Xenologs of the 3′exo/pol domains of viral-type PolA proteins were also found encoded in the genomes of 11 bacteria representing five different phyla, each of which contained a second prototypical bacterial polA gene (fig. 3). Examination of the context of the viral PolA xenologs in these genomes revealed prophage-like elements, for example, integrases, glycosyltransferases, and transposases (Dodsworth J, Murugapiran S, unpublished observation), suggesting lateral gene transfer following lysogeny. These data suggest an evolutionary sequence in which viruses bearing polA genes infected bacteria, leading to incorporation of their polA genes into the genome by nonhomologous or site-specific recombination, without disruption of the native, bacterial polA genes. A similar pathway has been invoked to explain the presence of T3/T7-like RNA polymerase genes within cryptic prophage in genomes of a variety of unrelated bacteria that are not hosts for T3/T7 viruses (Filee and Forterre 2005). There is no evidence in any of the bacterial lineages of 1) the 5′exo coding regions of the polA genes, 2) preferences for insertion sites, or 3) any sequence that appeared to a remnant of the 5′-region.
The distribution of virus-like polA genes in bacterial genomes does not follow an obvious phylogenetic pattern, suggesting they are normally purged. However, it is notable that one pair of closely related strains of Desulfitobacterium hafniense (strains DCB-2 and Y51) and one other pair of closely related strains of Cyanothece spp. (strains ATCC 51141 and CCY 0110) each contain nearly identical polA genes coding proteins similar to the viral polA genes, so it appears that these genes were maintained over evolutionary timescales necessary for strain-level divergence. It is also interesting that coding regions corresponding to the amino-terminal putative primase/helicase region of the viral PolA are not observed in any of the bacterial genomes, which implies that this region is less useful or even detrimental. Alternatively, the proximal source of these polA xenologs may be a yet undiscovered group of related viruses encoding only the carboxy-terminal 3′exo/pol domain of the PolA protein although no evidence of such an intermediate viral gene was seen in our small sample of viral diversity. Interestingly, the PolA proteins of Rubrobacter xylanophilus DSM 9941 and Clostridium sp. 7_2_43FAA1 appear to be evolutionary chimeras of the viral-type and the prototypical bacterial PolAs based on the patterns of indels and their phylogenetic positions (fig. 3).
Possible Viral Origin for the Thermophilic, Nuclear-Encoded, Apicoplast-Targeted PolA of Apicomplexa
In addition to the Aquificae sequence similarity, BLASTP results also pointed to strong sequence similarity between the viral polA gene products and the nuclear-encoded, apicoplast-targeted DNA polymerases of several members of the phylum Apicomplexa. The Apicomplexa comprise a large group of unicellular, spore-forming eukaryotes that exist as obligate intracellular parasites. Examples include several very important human pathogens such as Plasmodium, the causative agent of malaria, Babesia, an emerging zoonosis transmitted by ticks and blood transfusions, and Toxoplasma, another pathogen. Common to most Apicomplexa species are apicoplasts, subcellular organelles believed to have been acquired by secondary endosymbiosis of an alga containing two plastids, one of which evolved into the apicoplast (Waller and McFadden 2005; Lim and McFadden 2010; McFadden 2011). Pfprex, the apicoplast-targeted DNA polymerase of P. falciparum, is the best studied of the apicoplast-targeted polymerases (Seow et al. 2005) and is clearly highly similar to the Aquificae PolA and to the viral PolA protein 3′exo/pol domains of this study (fig. 3).
In addition to sequence similarities in the 3′exo/pol domains, apicoplast DNA polymerases have remarkable structural similarities to the full-length viral PolAs, exemplified by OCT-1608-14 and OCT-1608-4B9 gene products. Similar to the OCT-1608 PolAs, the Apicomplexa PolA-like proteins are significantly larger than other PolAs (e.g., 1,613 and 2,016 predicted aa for the Babesia bovis and P. falciparum PolA-like ORFs, respectively). In fact, the 1,606-amino acid ORF of OCT-1608-14 is nearly identical in size to the Babesia protein. Interestingly, approximately 88 kDa portion of the P. falciparum PolA, including the polymerase domain, is cleaved from the amino-terminal region in vivo (Lindner et al. 2011), similar to the apparent cleavage of the viral PolAs when expressed in E. coli. Although it is unknown whether this apparent processing of the heterologously expressed viral PolA proteins is biologically relevant, it suggests that they may share a polyprotein structure with the P. falciparum Pfprex protein. Amino-terminal Walker A and B ATPase motifs associated with helicase activity in the Pfprex (Lindner et al. 2011) are conserved in apicoplast polymerases, as well as viral PolA proteins, although the overall sequence similarity between amino-terminal region of the apicoplast and viral polymerases is low (BLASTP e value > 0.1). This suggests that either the amino-terminal regions of these proteins shared a common ancestor but evolved much more rapidly than the carboxy-terminal 3′exo/pol domains or that the amino-terminal domains are a result of convergent evolution and are not xenologous.
Another previously unexplained property of the Pfprex protein points to an evolutionary connection. The Pfprex polymerase is optimally active at 75 °C (Huber et al. 1998; Seow et al. 2005), much higher than would be encountered during the Plasmodium life cycle and much higher than other Plasmodium proteins, but remarkably similar to the optimal growth temperature of Thermocrinis and to the geothermal springs sampled in this study (Huber et al. 1998).
An Evolutionary Model of Lateral Gene Transfer between Viruses, Bacteria, and Apicocomplexa Parasites
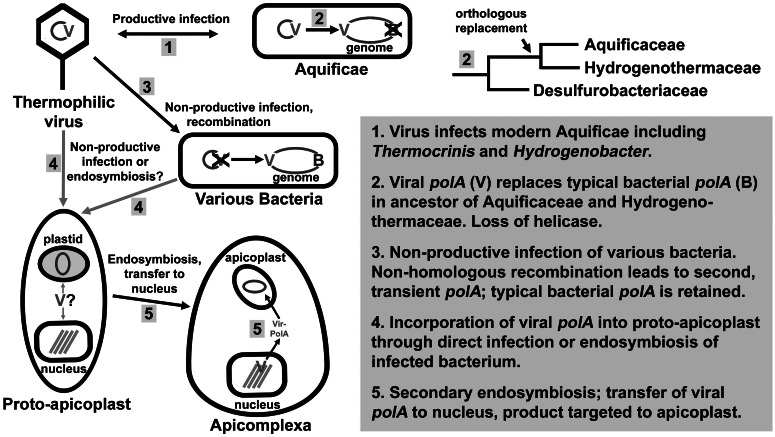
Considered together, the genomic context and distribution, phylogeny, and biochemistry of this clade of polA gene xenologs suggest that thermophilic viruses infecting Aquificae were key agents in the evolution of information processing systems of both Aquificae and Apicomplexa (fig. 4). We propose that a polA gene was transferred laterally between the viral population and a common progenitor of the families Aquificaceae and Hydrogenothermaceae. The 3′exo/pol coding region replaced the functionally equivalent 3′exo/pol domain but not the 5′exo domain of the previously existing bacterial polA gene. The amino-terminal region was either not transferred or was lost in the lineage that evolved into modern Aquificae, leaving only the 3′exo/pol domains. Additionally, we propose that a proto-apicoplast also obtained a viral-type polA xenolog, likely through endosymbiosis of a bacterium (proto-plastid) that had previously acquired a polA xenolog from a thermophilic virus. Following secondary endosymbiosis by the Apicomplexa lineage, this polA gene was transferred to the nucleus and targeted to the apicoplast, along with the majority of apicoplast-targeted proteins (Waller and McFadden 2005; Sato 2011). On the basis of this model, we propose that all the polA gene products with signature indels described in (Griffiths and Gupta 2006) were acquired and are therefore of viral origin. Given this ancestry and phylogenetic distribution, it is more appropriate to refer to these genes as virus-like and not Aquificae-like, especially given that only two families of Aquificae contain this distinctive polA gene.
Fig. 4.
Evolutionary model explaining the similarity of viral polA genes and those of two families of the Aquificae, some unrelated Bacteria, and the Apicomplexa.
A Possible Nonreplicative Role for the Viral DNA Polymerase
One limitation of viral metagenomics is that, lacking a host, biological function is difficult to test. The absence of a 5′exo domain in the viral, Aquificae, and Apicomplexa PolAs argues against a role in lagging strand synthesis or DNA repair. More compelling are certain anomalies unique to the polA genes of the thermophilic viruses and Apicomplexa that point to a possible role other than DNA replication. The putative polyprotein structure is unprecedented for DNA polymerases outside of this clade but very common in RNA replicases and reverse transcriptases (Eickbush and Jamburuthugoda 2008). Moreover, the reverse transcriptase activity of the OCT-3173 PolA (Moser et al. 2012) has no clear role in DNA replication but bears similarity to activities implicated in maintenance of telomeres (Bao and Cohen 2004) and in tropism switching mechanisms that prevent reinfection by phage (Liu et al. 2004; Medhekar and Miller 2007; Wang et al. 2011). Although we cannot exclude telomere processing as a function for the PolA, it seems more likely that this is an example of a bacterial tropism-switching mechanism. These mechanisms are known to use RNA intermediates copied by reverse transcriptases, although known examples use enzymes similar to retroviral/retroelement reverse transcriptases and not PolA-like proteins. The association of polA genes with likely diversity generating retroelements has been shown, but in these cases, similar to the others, reverse transcription appears to be performed by a separate retroviral-type reverse transcriptase (Kojima and Kanehisa 2008). Supporting a role in recombination is the adjacent recB homolog (fig. 1) that suggests a role of the operon in recombination rather than replication.
The role of the apicoplast is not clear although it is known to be indispensable for survival (Lim and McFadden 2010; McFadden 2011). Also unclear is the function of the apicoplast-targeted PolAs, which are also necessary for survival (Lindner et al. 2011) and have apparently been maintained over millions of years in the Apicomplexa (Mukhopadhyay et al. 2009). It has been presumed that Pfprex proteins function in replication of apicoplast DNA (Seow et al. 2005; Lindner et al. 2011), but strong evidence for this is lacking. If the role of this gene in thermophilic viruses is tropism switching then there may be a parallel role in Apicomplexa. Apicomplexa use genetic recombination to generate antigenic diversity as a means of evading host immune systems and for other virulence-related functions (Homer et al. 2000; Dzikowski et al. 2006; Scherf et al. 2008; Recker et al. 2011; Rovira-Graells et al. 2012; Witmer et al. 2012). It is possible that the viral-type polA-like genes function in Apicocomplexa as part of one of these systems or some other similar system. Supporting this model, the reported high in vivo mutation frequency and unusual mutation spectrum of Pfprex (Kennedy et al. 2011) would seem more consistent with a role in diversity generation than accurate replication.
Materials and Methods
Sample Sites and Virus Sampling
Viruses were collected from bulk hot spring water by tangential flow filtration as described previously (Schoenfeld et al. 2008). The following three springs (supplementary fig. S1, Supplementary Material online) were sampled: 1) OCT, White Creek Group, Lower Geyser Basin, YNP, sampled in 2003 (OCT03) and 2007 (OCT07); 2) LHC, near Mammoth Lakes, CA, sampled in 2001; and 3) GBS, near Gerlach, NV, sampled in 2007. OCT samples were collected about 10 m down the outflow channel from the source pool, where abundant filamentous growth was easily observable. The feature at LHC, designated LHC1 (Vick et al. 2010), was sampled directly at the small source pool, which contained no obvious filamentous growth. GBS was sampled in the large source pool. Detailed information on these springs have been previously published, including GPS coordinates, bulk water geochemistry, solid phase carbon and nitrogen geochemistry, clay mineralogy, and microbial community composition and function (Breitbart et al. 2004; Costa et al. 2007; Schoenfeld et al. 2008; Vick et al. 2010; Dodsworth et al. 2011, 2012; Hedlund et al. 2011).
Sequence-Based Screen for Polymerases
Viral libraries from LHC and OCT03 samples were constructed and sequenced using Sanger chemistry as described (Schoenfeld et al. 2008). Sequences were assembled at 95% and 75% NAID over 20 nucleotides using SeqMan software (DNAStar, Madison, WI). Clones were chosen for further study based on BLASTx expect values of less than 0.001 to PolA sequences in the nr database in GenBank. Clones with paired-end reads suggesting they were likely to contain complete genes were tested for thermostable DNA polymerase activity (Moser et al. 2012). Lysates positive for thermostable Pol activity were confirmed using a radioactive incorporation assay (Hogrefe et al. 2001).
PCR Amplification of Entire polA orf from OCT Virus Preparation
OCT 2007 and GBS DNA samples were amplified with the GenomiPhi kit (GE Healthcare, Waukesha, WI). The amplified OCT07 DNA was used as a template for PCR amplification with primers derived from the 75% OCT03 assembly (primers 2137 and 6768r, fig. 1). The amplicon was inserted into the pET28 vector and was propagated in 10G cells for sequence analysis and in BL21 HiControl vector for protein expression (Lucigen, Middleton, WI).
Functional Screen for Polymerases
For functional screening and 454 sequencing, amplified GBS DNA was debranched with S1 nuclease (Zhang et al. 2006). The GBS DNA was sheared to between 2 and 5 kb with a Hydroshear device (GeneMachine), inserted into the cloning site of a pETite vector (Lucigen Corporation) and used to transform BL21 HiControl cells. The amplified OCT07 DNA was inserted directly into a pJAZZ vector (Godiska et al. 2010) and used to transform 10G cells. Both libraries were screened using a 96-well format variation of the radioactive assay.
Bioinformatics and Phylogenetic Inference
DNA sequences of inserts of all clones showing polymerase activity were determined in their entirety and assembled using either Sequencher (Gene Codes, Ann Arbor, MI) or SeqMan (DNAStar) software. HMMER (http://hmmer.org; last accessed May 11, 2013) search was done using “hmmscan” using curated, high-quality PFAM-A entries. Multiple sequence alignment was done using MUSCLE Ver. 3.8.31 (Edgar 2004) with the default parameters, manually corrected using Seaview Ver. 4.0 (Gouy et al. 2010) and visualized using TeXshade (Beitz 2000). Masks were created using Gblocks (ver 0.91b) (Castresana 2000) using default command line parameters except the minimum percentage of sequences for a flank position (“-b2=" option) was changed from the default 85% to 50%. Maximum likelihood analyses were carried out using RAxML Ver. 7.2.6 (Stamatakis 2006) with 1,000 replicates and iTOL (Letunic and Bork 2011).
Genomic regions surrounding the presumed prophage polA genes were examined for evidence of lateral gene transfer using a 10 kb window, 5 kb up- and down-stream of the polA genes to identify GC% anomalies, and to search for prophage-like genes in Artemis (supplementary table S1, Supplementary Material online) (Carver et al. 2008). Potential domains were searched using NCBI’s Web CD-Search Tool (Marchler-Bauer et al. 2011). GC anomalies were defined as a departure of more than 2.5 standard deviations from the genomic mean.
Supplementary Material
Supplementary table S1 and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank David and Sandy Jamieson for generous access to GBS and Penny Custer and Christie Hendrix for assisting with access to LHC1 and OCT, respectively. They acknowledge the assistance of Forest Rohwer and Mya Breitbart in collecting the LHC sample and that of Stephen Techtman and Kui Wang in collecting the GBS sample. Octopus Spring (OCT), YNP, was sampled with permission from National Park Service under permit YELL-5240. Little Hot Creek (LHC) was sampled with permission form the National Forest Service, Mammoth Lakes office. Great Boiling Spring (GBS) was sampled with permission granted by a private landowner, David Jamieson. This work was supported by National Science Foundation (grant numbers IIP-0109756, IIP-0215988 IIP-0839404 to T.W.S. and MCB-0546865 and OISE-0968421 to B.P.H.) and National Institutes of Health (grant numbers R43HG002714 and R44HG002714 to T.W.S. and R43HG006078 to D.A.M.) and Department of Energy Joint Genome Institute project (CSP-182 to B.P.H. and J.A.D.). Additional support was provided by a UNLV Faculty Opportunity Award and a donation from Greg Fullmer through the UNLV Foundation.
References
- Anderson K, Nelson S, Mayo A, Tingey D. Interbasin flow revisited: the contribution of local recharge to high-discharge springs, Death Valley, CA. J Hydrol. 2006;323:276–302. [Google Scholar]
- Baker T, Kornberg A. DNA replication. Sausalito (CA): University Science Books; 1992. [Google Scholar]
- Bao K, Cohen SN. Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex. Proc Natl Acad Sci U S A. 2004;101(40):14361–14366. doi: 10.1073/pnas.0404386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese LS, Steitz TA. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitz E. TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics. 2000;16(2):135–139. doi: 10.1093/bioinformatics/16.2.135. [DOI] [PubMed] [Google Scholar]
- Bolduc B, Shaughnessy DP, Wolf YI, Koonin EV, Roberto FF, Young M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol. 2012;86(10):5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Gueguen L, Gouy M. Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of bacteria. BMC Evol Biol. 2008;8:272. doi: 10.1186/1471-2148-8-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Wegley L, Leeds S, Schoenfeld T, Rohwer F. Phage community dynamics in hot springs. Appl Environ Microbiol. 2004;70(3):1633–1640. doi: 10.1128/AEM.70.3.1633-1640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6(4):417–424. doi: 10.1016/s1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24(23):2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chang JR, Choi JJ, Kim HK, Kwon ST. Purification and properties of Aquifex aeolicus DNA polymerase expressed in Escherichia coli. FEMS Microbiol Lett. 2001;201(1):73–77. doi: 10.1111/j.1574-6968.2001.tb10735.x. [DOI] [PubMed] [Google Scholar]
- Chan YW, Mohr R, Millard AD, Holmes AB, Larkum AW, Whitworth AL, Mann NH, Scanlan DJ, Hess WR, Clokie MR. Discovery of cyanophage genomes which contain mitochondrial DNA polymerase. Mol Biol Evol. 2011;28(8):2269–2274. doi: 10.1093/molbev/msr041. [DOI] [PubMed] [Google Scholar]
- Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa KC, Navarro JB, Shock EL, Zhang CL, Soukup D, Hedlund BP. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles. 2009;13(3):447–459. doi: 10.1007/s00792-009-0230-x. [DOI] [PubMed] [Google Scholar]
- Costa MJ, Pedro L, de Matos AP, Aires-Barros MR, Belo JA, Goncalves J, Ferreira GN. Molecular construction of bionanoparticles: chimaeric SIV p17-HIV I p6 nanoparticles with minimal viral protein content. Biotechnol Appl Biochem. 2007;48(Pt 1):35–43. doi: 10.1042/BA20060208. [DOI] [PubMed] [Google Scholar]
- Daubin V, Ochman H. Start-up entities in the origin of new genes. Curr Opin Genet Dev. 2004;14(6):616–619. doi: 10.1016/j.gde.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Deschavanne P, DuBow MS, Regeard C. The use of genomic signature distance between bacteriophages and their hosts displays evolutionary relationships and phage growth cycle determination. Virol J. 2010;7:163. doi: 10.1186/1743-422X-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio M. The universal ancestor was a thermophile or a hyperthermophile: tests and further evidence. J Theor Biol. 2003;221(3):425–436. doi: 10.1006/jtbi.2003.3197. [DOI] [PubMed] [Google Scholar]
- Dodsworth JA, Hungate B, de la Torre JR, Jiang H, Hedlund BP. Measuring nitrification, denitrification, and related biomarkers in terrestrial geothermal ecosystems. Methods Enzymol. 2011;486:171–203. doi: 10.1016/B978-0-12-381294-0.00008-0. [DOI] [PubMed] [Google Scholar]
- Dodsworth JA, McDonald AI, Hedlund BP. Calculation of total free energy yield as an alternative approach for predicting the importance of potential chemolithotrophic reactions in geothermal springs. FEMS Microbiol Ecol. 2012;81(2):446–454. doi: 10.1111/j.1574-6941.2012.01369.x. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2(3):e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134(1–2):221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filee J, Forterre P. Viral proteins functioning in organelles: a cryptic origin? Trends Microbiol. 2005;13(11):510–513. doi: 10.1016/j.tim.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Filee J, Forterre P, Laurent J. The role played by viruses in the evolution of their hosts: a view based on informational protein phylogenies. Res Microbiol. 2003;154(4):237–243. doi: 10.1016/S0923-2508(03)00066-4. [DOI] [PubMed] [Google Scholar]
- Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117(1):5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Garrett RA, Prangishvili D, Shah SA, Reuter M, Stetter KO, Peng X. Metagenomic analyses of novel viruses and plasmids from a cultured environmental sample of hyperthermophilic neutrophiles. Environ Microbiol. 2010;12(11):2918–2930. doi: 10.1111/j.1462-2920.2010.02266.x. [DOI] [PubMed] [Google Scholar]
- Glansdorff N, Xu Y, Labedan B. The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct. 2008;3:29. doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiska R, Mead D, Dhodda V, Wu C, Hochstein R, Karsi A, Usdin K, Entezam A, Ravin N. Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli. Nucleic Acids Res. 2010;38(6):e88. doi: 10.1093/nar/gkp1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold T. The deep, hot biosphere. Proc Natl Acad Sci U S A. 1992;89(13):6045–6049. doi: 10.1073/pnas.89.13.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. Molecular signatures in protein sequences that are characteristics of the phylum Aquificae. Int J Syst Evol Microbiol. 2006;56(Pt 1):99–107. doi: 10.1099/ijs.0.63927-0. [DOI] [PubMed] [Google Scholar]
- Happonen LJ, Redder P, Peng X, Reigstad LJ, Prangishvili D, Butcher SJ. Familial relationships in hyperthermo- and acidophilic archaeal viruses. J Virol. 2010;84(9):4747–4754. doi: 10.1128/JVI.02156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedlund BP, McDonald AI, Lam J, Dodsworth JA, Brown JR, Hungate BA. Potential role of Thermus thermophilus and T. oshimai in high rates of nitrous oxide (N2O) production in approximately 80 degrees C hot springs in the US Great Basin. Geobiology. 2011;9(6):471–480. doi: 10.1111/j.1472-4669.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11(2):457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- Hogrefe HH, Cline J, Lovejoy AE, Nielson KB. DNA polymerases from hyperthermophiles. Methods Enzymol. 2001;334:91–116. doi: 10.1016/s0076-6879(01)34461-0. [DOI] [PubMed] [Google Scholar]
- Homer MJ, Bruinsma ES, Lodes MJ, et al. (12 co-authors) A polymorphic multigene family encoding an immunodominant protein from Babesia microti. J Clin Microbiol. 2000;38(1):362–368. doi: 10.1128/jcm.38.1.362-368.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Eder W, Heldwein S, Wanner G, Huber H, Rachel R, Stetter KO. Thermocrinis ruber gen. nov., sp. nov., A pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl Environ Microbiol. 1998;64(10):3576–3583. doi: 10.1128/aem.64.10.3576-3583.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugler M, Huber H, Molyneaux SJ, Vetriani C, Sievert SM. Autotrophic CO2 fixation via the reductive tricarboxylic acid cycle in different lineages within the phylum Aquificae: evidence for two ways of citrate cleavage. Environ Microbiol. 2007;9(1):81–92. doi: 10.1111/j.1462-2920.2006.01118.x. [DOI] [PubMed] [Google Scholar]
- Kazlauskas D, Venclovas C. Computational analysis of DNA replicases in double-stranded DNA viruses: relationship with the genome size. Nucleic Acids Res. 2011;39(19):8291–8305. doi: 10.1093/nar/gkr564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SR, Chen CY, Schmitt MW, Bower CN, Loeb LA. The biochemistry and fidelity of synthesis by the apicoplast genome replication DNA polymerase Pfprex from the malaria parasite Plasmodium falciparum. J Mol Biol. 2011;410(1):27–38. doi: 10.1016/j.jmb.2011.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H, Overgaard-Hansen K. Proteolytic cleavage of DNA polymerase from Escherichia coli B into an exonuclease unit and a polymerase unit. FEBS Lett. 1970;6(1):25–27. doi: 10.1016/0014-5793(70)80032-1. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Kanehisa M. Systematic survey for novel types of prokaryotic retroelements based on gene neighborhood and protein architecture. Mol Biol Evol. 2008;25(7):1395–1404. doi: 10.1093/molbev/msn081. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol Direct. 2006;1:39. doi: 10.1186/1745-6150-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Haridon S, Reysenbach AL, Tindall BJ, Schonheit P, Banta A, Johnsen U, Schumann P, Gambacorta A, Stackebrandt E, Jeanthon C. Desulfurobacterium atlanticum sp. nov., Desulfurobacterium pacificum sp. nov. and Thermovibrio guaymasensis sp. nov., three thermophilic members of the Desulfurobacteriaceae fam. nov., a deep branching lineage within the Bacteria. Int J Syst Evol Microbiol. 2006;56(Pt 12):2843–2852. doi: 10.1099/ijs.0.63994-0. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39(Web Server issue):W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Llinas M, Keck JL, Kappe SH. The primase domain of PfPrex is a proteolytically matured, essential enzyme of the apicoplast. Mol Biochem Parasitol. 2011;180(2):69–75. doi: 10.1016/j.molbiopara.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Gingery M, Doulatov SR, et al. (17 co-authors) Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J Bacteriol. 2004;186(5):1503–1517. doi: 10.1128/JB.186.5.1503-1517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, et al. (27 co-authors) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Russell MJ. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci. 2003;358(1429):59–83. doi: 10.1098/rstb.2002.1183. discussion 83–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. The apicoplast. Protoplasma. 2011;248(4):641–650. doi: 10.1007/s00709-010-0250-5. [DOI] [PubMed] [Google Scholar]
- Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10(4):388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Coleman RL, Dodsworth JA, Ross CA, Shock EL, Williams AJ, Hartnett HE, McDonald AI, Havig JR, Hedlund BP. Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning. PLoS One. 2012;7(5):e35964. doi: 10.1371/journal.pone.0035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MJ, Difrancesco RA, Gowda K, Klingele AJ, Sugar DR, Stocki S, Mead DA, Schoenfeld TW. Thermostable DNA polymerase from a viral metagenome is a potent rt-PCR enzyme. PLoS One. 2012;7(6):e38371. doi: 10.1371/journal.pone.0038371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25(24):5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Chen CY, Doerig C, Henriquez FL, Roberts CW, Barrett MP. The Toxoplasma gondii plastid replication and repair enzyme complex, PREX. Parasitology. 2009;136(7):747–755. doi: 10.1017/S0031182009006027. [DOI] [PubMed] [Google Scholar]
- Nordstorm DK, Ball JW, McCleskey RB. Ground water to surface water: chemistry of thermal outflows in Yellowstone National Park. In: Inskeep WP, McDermott TR, editors. Geothermal biology and geochemistry in Yellowstone National Park: proceeding of the Thermal Biology Institute Workshop; October 2003; Yellowstone National Park (WY) Bozeman (MT): Montana State University Publications; 2005. pp. 73–94. [Google Scholar]
- Oshima K, Chiba Y, Igarashi Y, Arai H, Ishii M. Phylogenetic position of aquificales based on the whole genome sequences of six aquificales species. Int J Evol Biol. 2012;2012:859264. doi: 10.1155/2012/859264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Basta T, Haring M, Garrett RA, Prangishvili D. Genome of the Acidianus bottle-shaped virus and insights into the replication and packaging mechanisms. Virology. 2007;364(1):237–243. doi: 10.1016/j.virol.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Perez LE, Merrill GA, Delorenzo RA, Schoenfeld TW, Vats A, Moser MJ. Evaluation of the specificity and sensitivity of a potential rapid influenza screening system. Diagn Microbiol Infect Dis. 2013;75:77–80. doi: 10.1016/j.diagmicrobio.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011;35(6):1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006;4(11):837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975;72(3):784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Schoenfeld T. Genome signature analysis of thermal virus metagenomes reveals Archaea and thermophilic signatures. BMC Genomics. 2008;9:420. doi: 10.1186/1471-2164-9-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Wassenaar TM, Ghose C, Blaser MJ. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics. 2006;7:8. doi: 10.1186/1471-2164-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recker M, Buckee CO, Serazin A, Kyes S, Pinches R, Christodoulou Z, Springer AL, Gupta S, Newbold CI. Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathog. 2011;7(3):e1001306. doi: 10.1371/journal.ppat.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Wickham GS, Pace NR. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60(6):2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter D, Jr, Mobley ML. Environment. Monitoring Earth's critical zone. Science. 2009;326(5956):1067–1068. doi: 10.1126/science.1179117. [DOI] [PubMed] [Google Scholar]
- Rovira-Graells N, Gupta AP, Planet E, Crowley VM, Mok S, Ribas de Pouplana L, Preiser PR, Bozdech Z, Cortes A. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 2012;22(5):925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S. The apicomplexan plastid and its evolution. Cell Mol Life Sci. 2011;68(8):1285–1296. doi: 10.1007/s00018-011-0646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- Schoenfeld T, Liles M, Wommack KE, Polson SW, Godiska R, Mead D. Functional viral metagenomics and the next generation of molecular tools. Trends Microbiol. 2010;18(1):20–29. doi: 10.1016/j.tim.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld T, Patterson M, Richardson PM, Wommack KE, Young M, Mead D. Assembly of viral metagenomes from yellowstone hot springs. Appl Environ Microbiol. 2008;74(13):4164–4174. doi: 10.1128/AEM.02598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman DW, Lineweaver CH. The hyperthermophilic origin of life revisited. Biochem Soc Trans. 2004;32(Pt 2):168–171. doi: 10.1042/bst0320168. [DOI] [PubMed] [Google Scholar]
- Seow F, Sato S, Janssen CS, Riehle MO, Mukhopadhyay A, Phillips RS, Wilson RJ, Barrett MP. The plastidic DNA replication enzyme complex of Plasmodium falciparum. Mol Biochem Parasitol. 2005;141(2):145–153. doi: 10.1016/j.molbiopara.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Setlow P, Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. II. A proteolytic fragment containing the 5' leads to 3' exonuclease function. Restoration of intact enzyme functions from the two proteolytic fragments. J Biol Chem. 1972;247(1):232–240. [PubMed] [Google Scholar]
- Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Snyder JC, Young MJ. Advances in understanding archaea-virus interactions in controlled and natural environments. Curr Opin Microbiol. 2011;14(4):497–503. doi: 10.1016/j.mib.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Spear JR, Walker JJ, McCollom TM, Pace NR. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc Natl Acad Sci U S A. 2005;102(7):2555–2560. doi: 10.1073/pnas.0409574102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159(5):349–357. doi: 10.1016/j.resmic.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5(10):801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Vick TJ, Dodsworth JA, Costa KC, Shock EL, Hedlund BP. Microbiology and geochemistry of Little Hot Creek, a hot spring environment in the Long Valley Caldera. Geobiology. 2010;8(2):140–154. doi: 10.1111/j.1472-4669.2009.00228.x. [DOI] [PubMed] [Google Scholar]
- Villarreal LP, DeFilippis VR. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol. 2000;74(15):7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, McFadden GI. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7(1):57–79. [PubMed] [Google Scholar]
- Wang C, Villion M, Semper C, Coros C, Moineau S, Zimmerly S. A reverse transcriptase-related protein mediates phage resistance and polymerizes untemplated DNA in vitro. Nucleic Acids Res. 2011;39(17):7620–7629. doi: 10.1093/nar/gkr397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol Rev. 2006;30(3):321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301(5635):976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witmer K, Schmid CD, Brancucci NM, Luah YH, Preiser PR, Bozdech Z, Voss TS. Analysis of subtelomeric virulence gene families in Plasmodium falciparum by comparative transcriptional profiling. Mol Microbiol. 2012;84(2):243–259. doi: 10.1111/j.1365-2958.2012.08019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu MX, Slater MR, Ackermann HW. Isolation and characterization of Thermus bacteriophages. Arch Virol. 2006;151(4):663–679. doi: 10.1007/s00705-005-0667-x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kasciukovic T, White MF. The CRISPR associated protein Cas4 is a 5' to 3' DNA exonuclease with an iron-sulfur cluster. PLoS One. 2012;7(10):e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24(6):680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.