Abstract
In adaptating to perpetual darkness, cave species gradually lose eyes and body pigmentation and evolve alternatives for exploring their environments. Although troglodyte features evolved independently many times in cavefish, we do not yet know whether independent evolution of these characters involves common genetic mechanisms. Surface-dwelling and many cave-dwelling species make the freshwater teleost genus Sinocyclocheilus an excellent model for studying the evolution of adaptations to life in constant darkness. We compared the mature retinal histology of surface and cave species in Sinocyclocheilus and found that adult cavefish showed a reduction in the number and length of photoreceptor cells. To identify genes and genetic pathways that evolved in constant darkness, we used RNA-seq to compare eyes of surface and cave species. De novo transcriptome assemblies were developed for both species, and contigs were annotated with gene ontology. Results from cave-dwelling Sinocyclocheilus revealed reduced transcription of phototransduction and other genes important for retinal function. In contrast to the blind Mexican tetra cavefish Astyanax mexicanus, our results on morphologies and gene expression suggest that evolved retinal reduction in cave-dwelling Sinocyclocheilus occurs in a lens-independent fashion by the reduced proliferation and downregulation of transcriptional factors shown to have direct roles in retinal development and maintenance, including cone-rod homeobox (crx) and Wnt pathway members. These results show that the independent evolution of retinal degeneration in cavefish can occur by different developmental genetic mechanisms.
Keywords: RNA-seq, transcriptome, cavefish, Sinocyclocheilus, retinal degeneration, photoreceptors
Introduction
Cave animals evolve distinct troglomorphic characters in perpetual darkness, most commonly, the reduction of eyesight, pigmentation, pineal organ, and scales, countered by augmented chemoreceptors, mechanoreceptors, and lipid storage (Poulson and White 1969; Jeffery 2001, 2009). Although these traits evolve in common across many cave taxa, an open question remains: Is the evolution of troglomorphic traits due to the common and repeated evolution of orthologous genetic and developmental mechanisms? The most extensive investigation of the evolution of troglodyte traits in cavefish comes from investigations of the Mexican tetra Astyanax mexicanus. In cave Astyanax, lens apoptosis associated with overexpression of sonic hedgehog (shh) induces eye degeneration (Yamamoto et al. 2004; Alunni et al. 2007; Borowsky 2008). In Mexican tetra cavefish, mutations in oca2 and mc1r appear to be responsible for the loss of eye and body pigmentation and the “brown” body color morph, respectively, (Protas et al. 2006; Gross et al. 2009). Although Astyanax is an excellent model for understanding the molecular mechanisms of regressive evolution, research on a variety of cave-animal models is necessary to understand whether independent evolutionary lineages utilize related molecular genetic mechanisms, or if evolution can achieve similar results by different mechanisms.
The investigation of retinal degeneration in cavefish is not only important to improve our understanding of the evolutionary mechanisms of development but also because it represents a significant evolutionary mutant model for human disease (Albertson et al. 2009). Age-related macular degeneration (AMD) is the primary cause of blindness in old age and is predicted to affect approximately 3 million people in the United States by 2020 (Klein et al. 1997; Gehrs et al. 2006). AMD involves the loss of the retinal pigmented epithelium and photoreceptors, a phenotype remarkably similar to that found in some cavefishes (McCauley et al. 2004; Gross et al. 2009). Although advances have been made in treating the vascular aspects of AMD, little progress has been made for other forms of the disease (Ambati et al. 2003). The investigation of the genetic mechanisms underlying evolutionary models of retinal degeneration may provide clues that may lead to potential new therapies for AMD.
China has a rich cavefish fauna with currently 101 valid species that belong to a single order (Cypriniformes) comprising only three families; Cyprinidae (including zebrafish, goldfish, carp, and 56% of all Chinese cavefishes), Cobitidae (true loaches, 2% of cavefishes), and Balitoridae (river loaches, 42% of cavefishes) (Zhao et al. 2011). Southwestern China is one of the largest cave-rich karst geomorphologic regions in the world (Huang et al. 2008). Different karst types provide suitable conditions to support a high diversity of cavefish populations. The freshwater teleost genus Sinocyclocheilus (Cypriniformes: Cyprinidae) is endemic to the karst region of the east Yungui Plateau and northwest Guangxi in southwestern China. The genus contains over 55 known species, including many surface-dwelling species and at least 10 cave-dwelling species with different degrees of eye degeneration and melanin loss (Xiao et al. 2005; Chen, Zhang, et al. 2009). Situations such as this with gradients of visual function possess advantages for analyzing the mechanisms of retinal degeneration (Tobler et al. 2010). Mitochondrial DNA sequences show that Sinocyclocheilus species lie within the Cyprinion–Onychostoma lineage, related to rock carp (Procypris rabaudi) and common carp (Cyprinus carpio) (Wu et al. 2010) (supplementary fig. S1, Supplementary Material online). Common carp, goldfish, and related species have an extra round of whole genome duplication compared with zebrafish (Danio rerio) (Wang et al. 2012). The genus Sinocyclocheilus likely shares that tetraploid origin and fish of this genus have 96 chromosomes, about twice that of most teleosts (Xiao et al. 2002). High species diversity and phenotypic variation make Sinocyclocheilus an ideal model for the investigation of evolutionary questions. We report here integrated transcriptome-wide investigations of changes in gene expression patterns related to the evolution of eye degeneration in Sinocyclocheilus cavefish.
To evaluate morphological evolution of cavefish eyes, we used antibody markers to detect specific retinal cell types in tissue taken from adult blind cavefish (Sinocyclocheilus anophthalmus, the “blind goldenline barbel”) and compared these with a closely related surface species (S. angustiporus, the “small gill opening goldenline barbel”). To understand evolved changes in gene expression, we utilized next-generation sequencing technologies (Wang et al. 2009). The Sinocyclocheilus transcriptome, which we assembled de novo from Illumina sequencing reads, provides a valuable resource for future studies, including the facilitation of studies on other species in this genus and the construction of a gene reference for annotating an eventual Sinocyclocheilus genome assembly. In this study, we identified eye genes that were differentially expressed between surface and cave species, and found enriched signaling pathways and disease-associated genes among differentially expressed genes. Results showed that evolved retinal degeneration in cave-dwelling Sinocyclocheilus is not associated with lens disappearance, in contrast to A. mexicanus, but is more likely due to the downregulation of transcription factors shown to have direct roles in retinal development and maintenance, including cone-rod homeobox (crx) and Wnt pathway members. These results suggest that convergent troglodyte characters can evolve by different genetic mechanisms in different fish taxa. Candidate genes identified by transcriptome-wide analysis may give clues to the mechanisms controlling the evolution of eye degeneration in cavefish and provide an alternative evolutionary medical model (Albertson et al. 2009) for retinal degeneration that often accompanies old age.
Results
Sinocyclocheilus anophthalmus Cavefish Evolved Decreased Eye Size
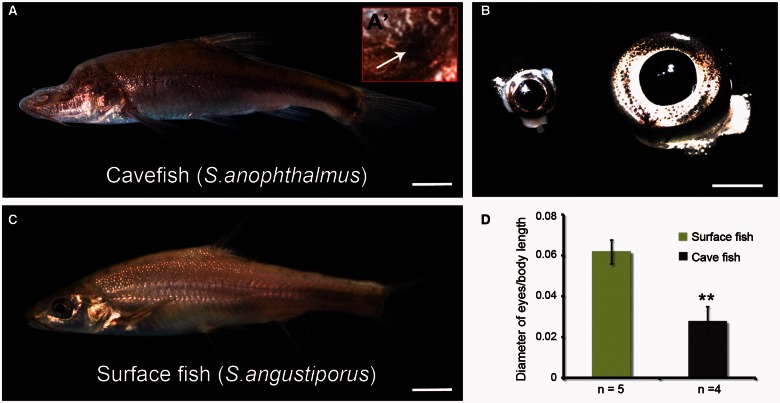
Sinocyclocheilus anophthalmus lives in constant darkness and, as its name suggests, lacks external eyes; it has a duckbill-like snout, sinking frontal bone, and long barbels and pectoral fins compared with surface fish. Sinocyclocheilus anophthalmus do not show a detectable response to natural light, even after stimulation with a laser pointer (Zhao and Zhang 2009) (fig. 1A). Consistent with the small internal eyes of adult S. anophthalmus, we discovered that its optic lobe is reduced in size compared with surface species (Meng FW, Braasch I, Zhang CG, Postlethwait JH, unpublished data). Sinocyclocheilus angustiporus is a close relative of S. anophthalmus, which has normal eyes, and although occasionally entering caves, usually occupies surface waters (fig. 1B) (Zhao and Zhang 2009). Sinocyclocheilus anophthalmus retains small eyes that are buried deeply within adipose tissue and are covered with skin (fig. 1A and A′). The examination of different developmental stages was not possible in this study due to the unavailability of cavefish embryos. But, to assess the extent of eye reduction in adult fish, we dissected the fish and measured eye diameters. Results showed that the internal eye of S. anophthalmus cavefish was about a third the size of the external eyes of adult surface fish (fig. 1C and D), although the size of retinal cell bodies, as assayed by both nuclear and cell-specific labeling, is comparable between the two species. Hematoxylin and eosin (H&E)-stained sections showed that the eyes of cavefish retained the basic vertebrate eye structure, with fully formed lens, cornea, iris and neural retina (fig. 2A and B). Although the cavefish neural retina retained its layered cellular organization, it was notably thinner than that of surface fish, with fewer cells in all three nuclear layers (fig. 2C and D). The morphological changes of photoreceptors in Sinocyclocheilus cavefish appeared to be more substantial than that of inner retinal neurons in H&E-stained sections.
Fig. 1.
Cavefish phenotypes. Cavefish Sinocyclocheilus anophthalmus (A) and surface fish S. angustiporus (B). Cavefish have no external eyes (A), but they have internal eyes (A′ and C, left) that are much smaller than the external eyes of their surface fish relatives (C, right). (D) Comparison of eye diameter normalized with total fish body length including the caudal fin. Values expressed as mean ± SD, **, P < 0.01. Scale bar in (A and B): 1 cm; (C): 2 mm.
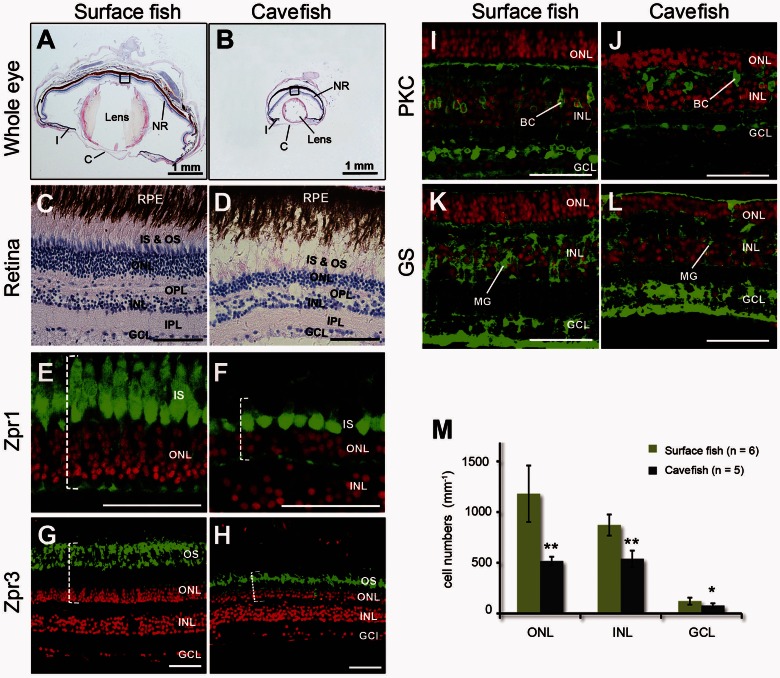
Fig. 2.
Eye degeneration in cavefish. H&E stained sections of adult surface fish eye (A, C) and cavefish eye (B, D). Representative double-labeling in the retina of surface (E, G, I) and cavefish (F, H, J) eyes; cell nuclei are dye-labeled red with ToPro3; retinal cell types are labeled green (anti-mouse Alexa Fluor-488). Cones (Zpr1; F) and rods (Zpr3; H) of cavefish were shorter than those of surface fish (E, G). Dotted brackets show the length of cones and rods. ON bipolar cells were labeled with antibodies against protein kinase C (PKC, green; I, J). Müller glial cells were labeled with antibodies against glutamine synthetase (GS, green; K, L). Cavefish had significantly fewer nuclei in the ONL, INL, and GCL (M) normalized for eye size. Values expressed as mean ± standard deviation (SD); *P < 0.05; **P < 0.01. C, cornea; I, iris; NR, neural retina; RPE, retinal pigmented epithelium; IS, inner segment; OS, outer segment; BC, biplolar cell; MG, Müller glial cells. Scale bar in (A, B): 1 mm; (C–L): 50 µm.
The vertebrate retina contains six major types of neurons organized as laminar structures, including rod and cone photoreceptors, as well as horizontal, bipolar, amacrine, and ganglion cells; these cell types form parallel microcircuits that integrate and process visual signals. To assay the distribution and numbers of different specific retinal cell types in eyes of cavefish and surface fish, we used the monoclonal antibodies Zpr1 which labels red/green double cones and Zpr3, a marker for rod outer segments and a subset of cone outer segments (Yin et al. 2012). To visualize cell types in the inner retina, we used an antibody against protein kinase C, which labels ON bipolar cells in the inner nuclear layer (INL), and an antibody against glutamine synthetase, which labels Müller glia. We found that cones, rods and bipolar cells were all present in cavefish eyes, with overall cell body sizes indistinguishable from those of surface fish. However, their numbers were significantly reduced (fig. 2E–J). In cavefish eyes, cones (fig. 2E and F) and rods (fig. 2G and H) were 49% and 59% shorter than in surface fish eyes (P < 0.001, cave n = 6, surface n = 7, t test), respectively. The presence of photoreceptor outer segments was confirmed by H&E staining (fig. 2C and D) and by zpr-3 labeling (fig. 2G and H), although the lateral organization of the outer segments was disrupted in the cavefish retinas. In the inner retina, Müller glial cells were reduced in cavefish compared with surface fish (fig. 2K–L). Counts of nuclei indicated that the cell density of the outer nuclear layer (ONL), INL, and ganglion cell layer (GCL) were all significantly decreased in cavefish relative to surface fish (fig. 2M). Cell number reduction of the ONL, which contains the photoreceptor nuclei, was more substantial than that of the INL and GCL (fig. 2M). We conclude that the evolution of Sinocyclocheilus cavefish involved a reduction in the number of photoreceptors, bipolar cells, and Müller glia, as well as substantial morphological abnormalities in photoreceptor structure.
Cell Proliferation and Apoptosis in the Cavefish Retina
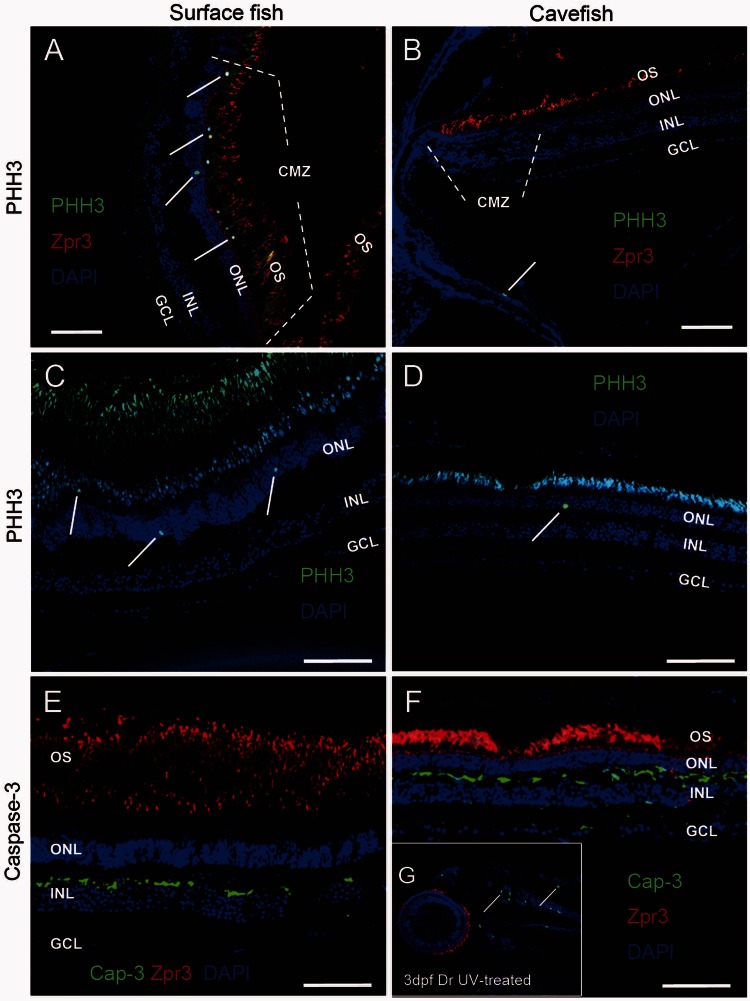
We next tested whether cell proliferation in cavefish eyes was affected during adaptive evolution, using an antibody directed against phosphorylated histone-3 (PHH3), an M-phase marker. In the eyes of surface fish (fig. 3A and C), mitotic cells were detected throughout the retina, with the highest levels of proliferation localized to the ciliary marginal zone (CMZ), in which retinal progenitors are located (Raymond et al. 2006) (1.91 ± 0.33 positive cells per linear mm of retina, n = 5). In cavefish retina, however, the overall number of proliferating cells was reduced (0.79 ± 0.31 positive cells per linear mm of retina, n = 5; P < 0.001) with significantly fewer mitotiocally active cells noted in the CMZ as compared with the surface species (fig. 3B and D). We conclude that the cavefish eye is not replenished as the fish grows as happens in surface fish.
Fig. 3.
Cavefish have reduced retinal cell proliferation. (A–D) Retinas from adult surface fish (A, C) and cavefish (B, D) were stained with anti-PHH3, a marker of the M-phase of the cell cycle. The CMZ lies at the anterior edge of the peripheral retina. Proliferating cells were located mainly in the CMZ in surface fish retinas (A). PHH3 positive cells were not detected in the CMZ of cavefish (B). PHH3 positive cell numbers per linear millimeter of retina were markedly reduced in non-CMZ of cavefish retinas (C: 0.79 ± 0.31, n = 5) relative to that of surface fish (D: 1.91 ± 0.33, n = 5, P < 0.001). (E–G) Caspase-3 staining was performed to analyze apoptosis in adult surface (E) and cave (F) eyes. Apoptosis was not apparent in the retina of either surface or cave species, although in positive controls, Caspase-3 antibody did detect apoptosis induced by UV in 3dpf zebrafish eyes (G). OS, outer segment. Scale bar in (A–F): 50 µm.
To analyze apoptosis in adult surface and cave species, we performed antibody labeling with anti-active Caspase-3, an early marker of apoptosis (Dai and Krantz 1999; Ryu et al. 2005). No apoptosis was detected in the retina of either species (fig. 3E and F). As a positive control, we investigated induced cell death in zebrafish retina; results showed that Caspase-3 antibody detected cell apoptosis induced by ultraviolet (UV) light in 3dpf zebrafish (fig. 3G). Although apoptosis was not detected in adult cavefish, we cannot exclude the possibility that excess apoptosis occurred in the embryonic or larval stages of cavefish eyes relative to surface fish eyes.
Sequencing, De Novo Assembly, and Annotation of the Sinocyclocheilus Reference Transcriptome
To help understand differences between surface and cavefish eyes on a genetic level, we profiled gene expression. We isolated total RNAs from eyes and brains of S. anophthalmus and S. anophthalmus (two individuals for each species), and constructed four cDNA libraries: cavefish eyes, cavefish brain, surface fish eyes, and surface fish brain. Two brain cDNA libraries of both species were sequenced using the Illumina Genome Analyzer IIx platform to produce 80 nucleotide (nt) paired-end reads (SRA run accession numbers: surface fish, SRR788094; cavefish, SRR788095), and two eye cDNA libraries of both species using Illumina HiSeq 2000 with 100-nt paired end reads (surface fish, SRR788096; cavefish, SRR788097).
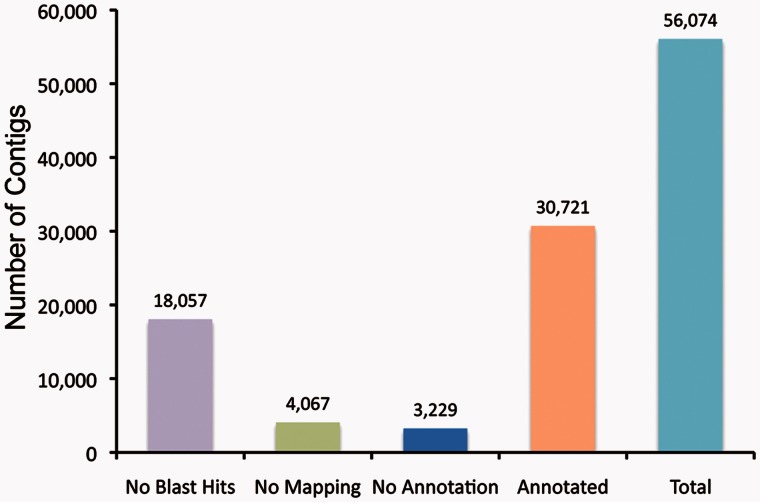
After trimming adapters and removing low quality reads, a total of 132,682,597 reads, containing 9,179,015,231 nt, were retained (supplementary table S1, Supplementary Material online). De novo assembly on surface and cave species using Trinity (Grabherr et al. 2011) assembled 66,459,580 reads into contigs with 300 nt minimum length from two species leaving 66,223,017 unassembled reads (singletons). The contig file for cavefish contained 152,464 contigs and that for surface fish contained 156,118 contigs (supplementary table S1, Supplementary Material online). To generate a common and nonredundant Sinocyclocheilus transcriptome database for further RNA-seq analyses, we merged the two contig files using Cap3 (Huang and Madan 1999). The combined Sinocyclocheilus transcriptome contained 56,074 contigs, which were on average longer than from the single-species assemblies (supplementary table S1 and fig. S2, Supplementary Material online).
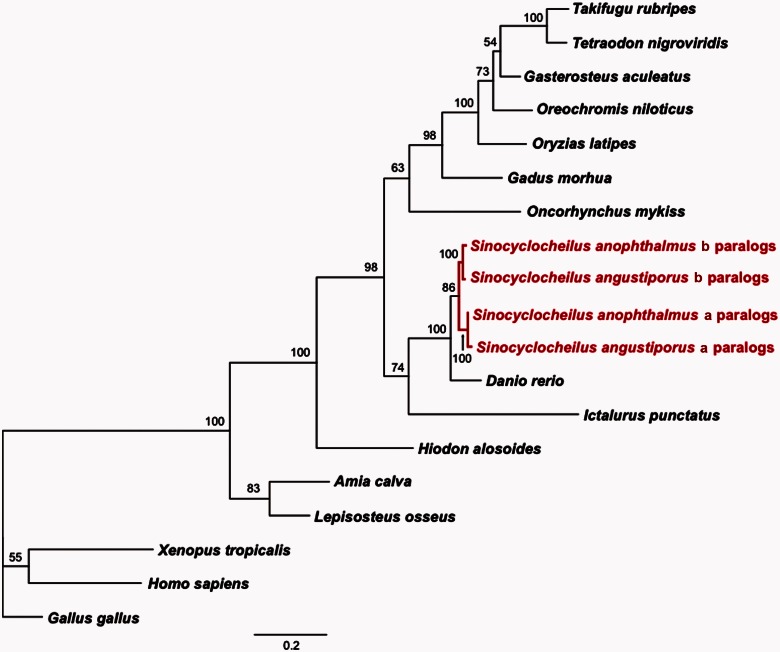
Next, we compared the combined Sinocyclocheilus transcriptome with the UniGene records for zebrafish (D. rerio), medaka (Oryzia latipes), and three-spined stickleback (Gasterosteus aculeatus). Blastn similarity searches (cut-off E value of 10−3) showed that 30,590 Sinocyclocheilus contigs matched 19,724 zebrafish UniGenes, covering 37.5% of the zebrafish UniGene records. Medaka and stickleback databases yielded fewer matches than did the zebrafish transcriptome: 1,794 Sinocyclocheilus contigs matched 1,531 medaka UniGenes (6.5%), and 1,853 Sinocyclocheilus contigs matched 1,554 stickleback UniGenes (8.3%). Two factors likely contribute to finding more UniGene hits in zebrafish than in stickleback and medaka: first, a phylogenetic tree based on de novo and downloaded sequences of zic family member-1 (zic1) and G protein-coupled receptor-85 (gpr85) shows that Sinocyclocheilus is more closely related to D. rerio than any other species in the tree (fig. 4), and second, the zebrafish UniGene database (92,836 entries) is substantially larger than that of medaka (22,128 entries) and stickleback (16,737 entries).
Fig. 4.
Phylogenetic position of Sinocyclocheilus. A phylogenetic tree generated by Maximum Likelihood analysis of the combined nucleotide sequences of the CGD paralogs of zic1 and gpr85, which have a single copy in human and zebrafish. Analysis confirms that the Sinocyclocheilus lineage experienced a genome duplication after its divergence from zebrafish and shows that zebrafish is the closest species to Sinocyclocheilus with a sequenced genome. Numbers at the nodes denote the bootstrap percentages of 100 replicates. Sequences for all taxa can be found in supplementary table S5, Supplementary Material online.
For annotation, contigs were searched against the Nr database using blastx. We found that 38,017 contigs (67.8%) had at least one hit (E-value cut-off of 10−6), while 18,057 contigs (32.2%) did not align to any known protein in the Nr database; many of these likely represent 3′ and 5′ untranslated regions (UTRs). The 38,017 contigs that hit known proteins grouped into 20,062 UniGenes. Most contigs (31,407, 82.6%) aligned best to D. rerio UniGenes. To globally characterize the transcriptome, we annotated Sinocyclocheilus contigs based on sequence homology to functionally annotated sequences in other species. We identified 30,721 contigs (54.8% of Sinocyclocheilus contigs) that had affinity to at least one Gene Ontology (GO) term. In addition, of the 38,017 contigs with good hits to the Nr database, 7,296 were classified as unknown, predicted, hypothetical, or uncharacterized proteins (fig. 5).
Fig. 5.
Annotation of the Sinocyclocheilus transcriptome. Blast2GO was used to annotate our de novo assembled transcriptome using parameters described in Materials and Methods. A total of 30,721 contigs (54.8% of total contigs) were annotated; 13% of contigs had no GO term but at least one blast hit in the Nr database. Many contigs that failed to annotate may represent 5′- or 3′-UTRs.
We assigned contigs to broad functional categories and found that 30,721 contigs were associated with 124 generic GO-Slim annotations; specifically, these included 35 categories at the Cellular Component level, 39 categories at the Molecular Function level and 50 categories at the Biological Process level.
Differential Gene Expression in Surface Fish and Cavefish Eyes
To investigate the evolution of the cavefish eye transcriptome, we compared surface fish and cavefish transcriptomes. Eye samples of both species were sequenced with Illumina HiSeq 2000, and resulting reads were trimmed for adapter and quality, aligned to the Sinocyclocheilus transcriptome, counted, and normalized to the total mapped reads. For the cavefish sample, 22,793,525 reads (49.31%) mapped to Sinocyclocheilus transcriptome contigs and an additional 412,525 unmapped reads aligned to zebrafish UniGenes. (Unmapped reads were those that had not assembled into Sinocyclocheilus contigs.) A total of 23,206,050 reads from eye samples, corresponding to 49.9% of all high quality reads in cavefish, mapped to 52,915 contigs (94.4%) in the Sinocyclocheilus transcriptome. For the surface fish sample, 29,584,957 reads (48.98%) mapped to the Sinocyclocheilus transcriptome, whereas an additional 562,988 reads mapped to zebrafish UniGenes; a total of 30,147,945 surface fish reads (50.3%) aligned to 52,656 Sinocyclocheilus reference contigs (93.9%). To analyze gene expression profiles, we converted read numbers mapped onto UniGenes to RPKM (reads per kilobase of exon per million mapped sequence reads) (Mortazavi et al. 2008).
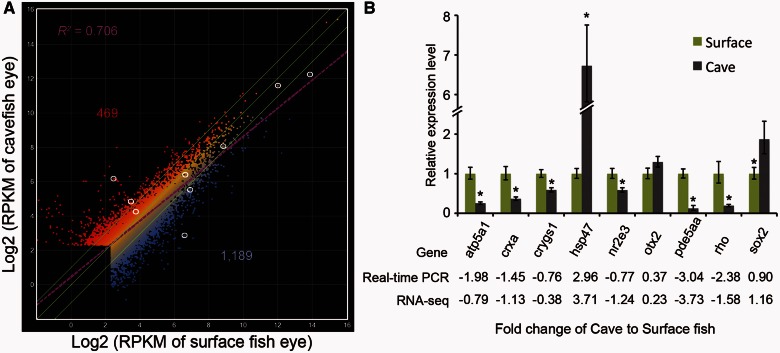
Among 9,649 unique genes whose RPKM values were over 5 in the Sinocyclocheilus eye transcriptome, we identified 1,658 genes as differentially regulated, defined as greater than a 2-fold change between cavefish and its related surface species with a P-value less than 0.05 (log2-fold changes [log2FC] > 1, P value < 0.05). Of differentially expressed genes, 1,189 (71.7%) had decreased expression and 469 (28.3%) had increased expression in cavefish eyes (fig. 6A). Supplementary table S2, Supplementary Material online, lists genes according to their change in expression level (log2FC). To validate the differential expression of genes identified by RNA-seq, we used real-time polymerase chain reaction (PCR) with primers derived from the de novo assembled Sinocyclocheilus transcriptome to quantify the mRNA levels of several genes that were selected from different change levels and different expression levels. Real-time PCR data validated the RNA-seq results (fig. 6B; supplementary fig. S3, Supplementary Material online), showing that differential gene expression in cavefish is reliable and that the de novo assembled Sinocyclocheilus transcriptome can be used for RNA-seq based gene expression analysis.
Fig. 6.
(A) Comparison of two eye RNA-seq determinations measured in reads per kilobase of exon per million mapped sequence reads (RPKM). Lateral green lines bracket genes with 2-fold differences in expression; these genes were considered to be differentially expressed in this study. A total of 469 genes were upregulated and 1,189 were downregulated in cavefish relative to surface fish. The purple line is the regression line (R2 = 0.706). White circles indicate genes verified by real-time PCR (B) using RNA isolated from eyes of each species. The expression levels of atp5a1, crxa, crygs1, hsp47, nr2e3, otx2, pde5aa, rhodopsin, and sox2 were quantified and normalized to beta-actin1. Relative expression values are mean ± SD of at least three independent experiments.
Analysis showed that the RNA-seq results confirmed conclusions from histological investigations. We found that photoreceptor genes were downregulated (rhodopsin, guanine nucleotide binding protein [G protein], alpha transducing activity polypeptide 1 [gnat1], gnat2, and opsin 1 [cone pigments], long-wave-sensitive, 1 and short-wave-sensitive 2 [opn1lw1 and opn1sw2] with log2FC = −1.5803, −1.7425, −1.7428, −2.1126, and −1.4817, respectively), as was the Müller glia gene glutamine synthetase (log2FC = −1.3598).
Identification of Enriched Pathways and Diseases among Differential Expressed Genes
Several GO categories were over- or underrepresented in the differentially expressed genes in each of the three main GO categories (supplementary fig. S4, Supplementary Material online). For example, in the biological processes category, signal transduction (GO: 0007165) and protein modification process (GO: 0006464) were overrepresented in the downregulated gene group, likely reflecting the low number of photoreceptors present in cavefish eyes (fig. 2). The proteinaceous extracellular matrix (GO: 0005578) and extracellular space categories (GO: 0005615) were overrepresented in the upregulated gene group in the cellular component category, likely reflecting the larger spaces between cells observed in our histological studies. Seven of 13 genes encoded by the mitochondrial genome were downregulated in cavefish eyes (mt-co1, 8.1×; mt-nd6, 6.8×; mt-nd5, 5.5×; mt-cyb, 5.2×; mt-nd4l, 2.7×; mt-co2, 2.5×; and mt-nd4, 2.2×), suggesting decreases in rates of energy metabolism in cavefish eyes.
To further explore altered regulation of signal pathways during cavefish evolution, we used KOBAS 2.0 (Xie et al. 2011) to identify enriched pathways and disease-associated genes among the differentially expressed genes (supplementary table S3, Supplementary Material online). Twenty-two pathways and 13 diseases had significant P values less than 0.05 after FDR correction in the downregulated gene group and 18 pathways and 23 diseases were significantly enriched in the upregulated group. Although identified pathways had some overlap among different databases within KOBAS 2.0 (Xie et al. 2011), we found that the most significantly enriched pathways were relevant to visual phototransduction in the downregulated group, including phototransduction (KEGG), visual signal transduction: cones and rods (PID Curated), synaptic transmission (Reactome), and heterotrimeric G-protein signaling pathway-rod outer segment phototransduction (PANTHER). Most of the enriched diseases were eye pathologies related to the retina. For example, retinitis pigmentosa (FunDO), retinal disease (FunDO), cone-rod dystrophy (GAD), achromatopsia (GAD), and bradyopsia (OMIM). In the upregulated group, enriched pathways and diseases were diverse, involving cytokine–cytokine receptor interaction (KEGG), muscle contraction (Reactome), cell adhesion (KEGG), pertussis (KEGG), cardiac diseases (KEGG), Wnt signaling pathway (PANTHER), and immune system diseases (KEGG). These transcriptomic results are as would be predicted from the histological analyses showing a strong reduction in photoreceptors.
Expression of Retinal Development and Maintenance Genes in Cavefish
To identify potential regulatory differences in retinal cell development and maintenance genes in adult Sinocyclocheilus cavefish, we queried genes already shown to be involved in fish retinal development. During photoreceptor development, six key transcription factors affect rod and cone differentiation and maintenance, including RAR-related orphan receptor beta (rorb), orthodenticle homolog-2 (otx2), crx (for which zebrafish has two co-orthologs of the tetrapod gene, currently called crx and otx5 but which should be called crxa and crxb[otx5], respectively [Suda et al. 2009]), neural retina leucine zipper (nrl), nuclear receptor subfamily 2 group E member 3 (nr2e3, also called retina-specific nuclear receptor) and thyroid hormone receptor beta (thrb) (Swaroop et al. 2010).
Vertebrates possess five groups of light-sensitive opsin proteins that mediate the conversion of photons into an electrochemical signal (Fain et al. 2010). Opsin expression is regulated by photoreceptor-enriched factors, including crx, retinal homeobox (rx), nr2e3, nrl, and thrb, or ubiquitously expressed factors, such as ataxin 7-like 3 (atxn7l3), K(lysine)-acetyltransferase 2B (kat2b), E1A binding protein p300 a (ep300a), and ep300b (Peng and Chen 2007; Hennig et al. 2008). Our RNA-seq experiments showed that cone opsins and rhodopsin were significantly downregulated in cavefish eyes and real-time PCR confirmed these expression differences (fig. 6; supplementary fig. S3, Supplementary Material online). Immunostaining studies of rhodopsin in tissue sections provided results expected from real-time PCR investigations: rhodopsin was distributed throughout the photoreceptor layer in surface fish eyes, but localized in a thinner layer in cavefish eyes (fig. 2E and F). In some cavefish, changes in opsin gene expression represent an early evolutionary step toward retinal degeneration (Tobler et al. 2010). Several transcription factors involved in opsin expression were downregulated more than 2-fold: rx2 (−1.09 log2FC), nr2e3 (−1.24), crxa (−1.45), and crxb (otx5, −1.17). In contrast, rx1 (−0.03 log2FC), nrl (0.32), thrb (−0.32), and ubiquitously expressed transcriptional factors, atxn7l3 (0.03), kat2b (−0.08), ep300a (−0.15), and ep300b (0.35), were expressed at similar levels in both species (fig. 7; supplementary table S2, Supplementary Material online).
Fig. 7.
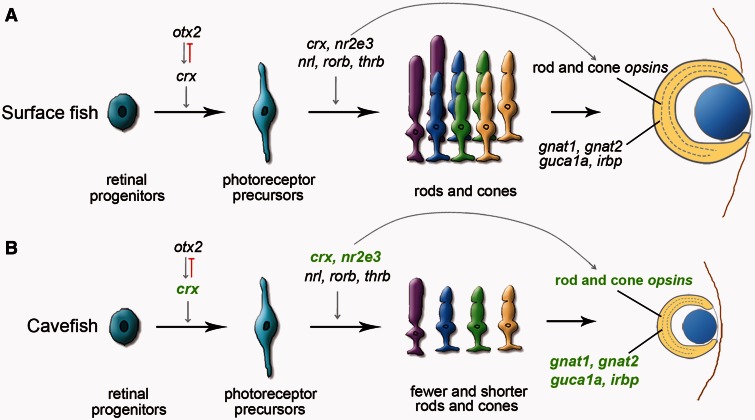
Proposed model for the evolution of eye degeneration in the cavefish Sinocyclocheilus anophthalmus. (A) Surface fish. (B) Cavefish. In normal vertebrate eyes (A), otx2 upregulates crx, which controls the differentiation of photoreceptor precursors; a number of other factors, including nr2e3, nrl, thrb, and crx itself, help to regulate the differentiation of precursor cells into mature rods and cones that express differentiation products, including gnat1, gnat2, guca1a, and irbp (Hennig et al. 2008; Swaroop et al. 2010). In cavefish eyes (B), transcriptomic data showed downregulation of crx, which would lead to decreased photoreceptor cell differentiation, fewer and malformed photoreceptors, and diminished expression of a series of photoreceptor-specific genes (rod opsins and cone opsins, otx5, gnat1, gnat2, guca1a, and irbp). Because otx2 expression was unchanged in cavefish eyes, another regulatory factor may lie upstream of crx, and/or evolved changes in the promotors of crx paralogs in cavefish may dampen the response of crx to normal otx2 signal. Because fish can regenerate photoreceptors from the CMZ and from Müller glia cells, this pathway likely works throughout life and is thus involved in retinal maintenance. Downregulated genes in cavefish are indicated by green letters. Upregulated genes included several immune-related genes, which may represent response to inflammation associated with eye degeneration.
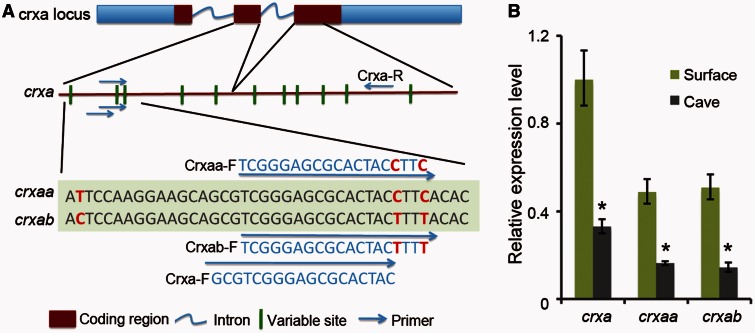
Experiments knocking down crxa gene expression in zebrafish had shown that many genes in the visual transduction cascade were downregulated, including rhodopsin, cone opsins, interphotoreceptor retinoid-binding protein (irbp), otx5(crxb), gnat1, gnat2, and guanylate cyclase activator 1A(guca1a) (Shen and Raymond 2004). Similarly, in S. anophthalmus cavefish, the expression levels of a series of phototransduction genes, including genes reduced in crxa morpholino treated zebrafish, were also decreased (fig. 7; supplementary table S2, Supplementary Material online). These data support the hypothesis that the reduction of crxa expression that we observed in cavefish eyes gives rise to the downregulation of phototransduction genes in cavefish eyes. We also designed primers to detect the expression level of the two different crxa paralogs in Sinocyclocheilus that derived from the carp genome duplication (CGD), crxaa and crxab, with real-time PCR (fig. 8A). Results showed that both crxaa and crxab were downregulated in cavefish relative to surface fish, but the ratio between crxaa and crxab was similar in each species (fig. 8B).
Fig. 8.
(A) Our de novo assembled Sinocyclocheilus transcriptome identified two co-orthologs, crxaa and crxab, of the single zebrafish crxa gene; these arose in the tetraploid origin of Sinocyclocheilus. The crxaa and crxab genes have several identifying nucleotide variations. To distinguish the expression level of different crxa paralogs, we designed primers to detect crxaa and crxab with real-time PCR using RNA isolated from eyes of each species. (B) The expression levels of crxa paralogs were quantified and normalized to beta-actin-1. Relative expression values are mean ± SD of at least three independent experiments. Results showed that both crxaa and crxab were downregulated in cavefish, but that there was no obvious difference between crxaa and crxab within each species.
Comparison between Sinocyclocheilus and Astyanax Cavefish
Eye degeneration in A. mexicanus cavefish involves lens cell apoptosis induced by shh overexpression (Yamamoto et al. 2004). In contrast, the lens did not show obvious morphological changes in adult S. anophthalmus cavefish, (fig. 2A and B). Lens measurements in histological sections of Sinocyclocheilus eyes revealed no substantial difference between the species regarding the relative areas of the lens normalized by the area of eye sections (35.8 ± 8.4% in surface fish; 35.2 ± 9.0% in cavefish). Although the expression of shh also did not change significantly in the eyes of adult S. anophthalmus relative to the eyes of surface species (log2FC = −0.01), it is the expression of shh from surrounding tissues during embryonic development, not shh expression in the eye itself, that is important for eye development in Astyanax (Yamamoto et al. 2004; Pottin et al. 2011).
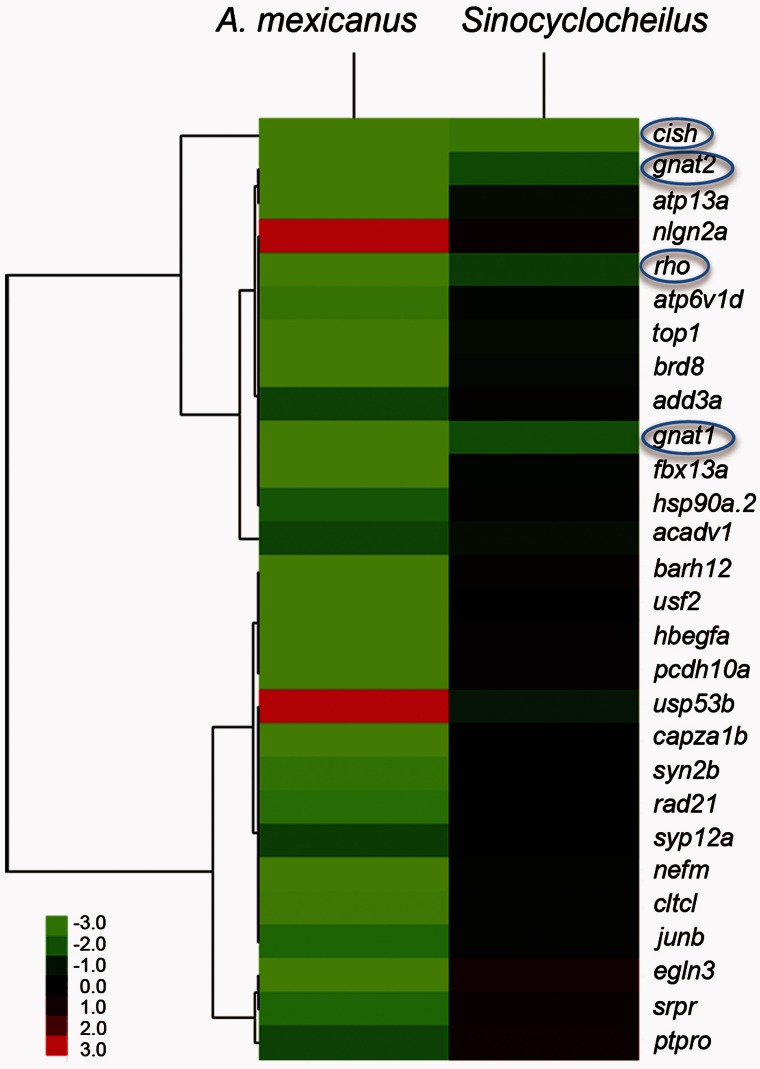
Next, we compared our RNA-seq results from adult Sinocyclocheilus cavefish and surface fish with expression studies that used a zebrafish microarray to query gene expression from whole bodies of 3-day-old larvae of A. mexicanus cavefish and surface fish (Strickler and Jeffery 2009). Comparisons revealed that 28 of the 67 differentially expressed genes in A. mexicanus larval bodies were represented in our RNA-seq adult eye transcriptome gene set (fig. 9). Some of the 39 missing genes may have been expressed in the many cell types other than the eye found in the whole body of Astyanax larvae. Of the 28 genes present in both data sets, only rhodopsin, cytokine inducible SH2-containing protein (cish), gnat1, and gnat2, had greater than 2-fold change between cave and surface species in our experiments (boxed in fig. 9). Several crystallin genes were expressed in the epithelial cells of the lens, and downregulated in Astyanax cavefish larvae (Strickler and Jeffery 2009). The anti-apoptotic activity of αA-crystallin was implicated in Astyanax eye degeneration (Strickler et al. 2007; Strickler and Jeffery 2009), but the αA-crystallin gene was not present in the Sinocyclocheilus transcriptome, presumably because it may be expressed only in early development (Strickler et al. 2007). The expression levels of several crystallins, including γM2-crystallins and βB-crystallins, were actually higher in Sinocyclocheilus cavefish than in surface fish (supplementary table S2, Supplementary Material online), providing further evidence that lens degeneration is unlikely to be driving retinal degeneration in Sinocyclocheilus cavefish.
Fig. 9.
Cluster analysis of genes expressed differentially between cavefish and surface fish comparing Astyanax and Sinocyclocheilus. The Astyanax transcriptome data came from the cross-species hybridization of Astyanax larval whole body cDNA to a microarray of zebrafish sequences (Strickler and Jeffery 2009), which identified 67 genes with differential expression. Comparisons between Astyanax and Sinocyclocheilus revealed that, of 28 genes in the Astyanax data set that were also detected in the Sinocyclocheilus RNA-seq data, only four were differentially expressed in both species of cavefish. These four genes (rhodopsin, cytokine inducible SH2-containing protein [cish], gnat1, and gnat2), are indicated by blue circles. Color intensity represents upregulation and downregulation, respectively.
Discussion
Retinal Defects in Cave Sinocyclocheilus
We analyzed morphological and transcriptomic differences in the eyes of closely related cavefish and surface fish in the genus Sinocyclocheilus to suggest genes, genetic pathways, and developmental mechanisms that evolved during the adaptation of S. anophthalmus cavefish to constant darkness. Results showed that cavefish eyes were imbedded in fat under the skin, and were reduced in overall size. Retinal layers were normally patterned in cavefish, but were reduced in thickness and cell density, especially in the photoreceptor layer. Although retinal cell bodies were about the same size in cavefish and surface fish, the apical-basal length of photoreceptor cells was significantly shorter in cavefish compared with surface fish (fig. 2E–H), and photoreceptor outer segments were disorganized. In addition, retinal cell proliferation was greatly reduced in cavefish eyes relative to surface fish eyes, and gene expression patterns were substantially altered in cave and surface species, especially the expression of genes previously shown to regulate retinal development.
Phylogenetic analyses that combined morphology and mitochondrial gene sequences divided genus Sinocyclocheilus into four clades (Zhao and Zhang 2009). The cave and surface species studied here, S. anophthalmus and S. angustiporus, both belong to Clade tingi, which diverged from other clades before the Second Act of the Tibetan Orogeny about 2.6 million years ago. Some species within this clade inhabit caves and others occupy surface streams belonging to the same water system, located only a few kilometers from each other. S. anophthalmus is one of the few troglophile species in Clade tingi, and likely separated from other species in this clade about 1 million years ago (Zhao and Zhang 2009).
Histological investigations showed that, although cavefish eyes retained the gross retinal architecture of vertebrate eyes with lenses of approximately normal size relative to the size of the eyeball, they had thin retinas, short photoreceptor cells, and more sparsely populated retinal cell layers compared with surface fish. Although we were not able to obtain cavefish at early developmental stages for this study, we detected several classes of differentiated retinal cells organized into distinct retinal cell layers in adult cavefish, indicating that embryonic eye induction, differentiation, and patterning signals are intact during cavefish development.
In contrast to mammals, the eyes of adult individuals of cyprinids and other teleost fish grow continuously and can regenerate in response to retinal damage (Otteson and Hitchcock 2003). In mature zebrafish, new photoreceptors arise from two sources, the CMZ at the periphery of the retina and the Müller glia cells in the INL (Raymond et al. 2006). Although no obvious apoptosis was found in cavefish (fig. 3E and F), cell proliferation rates were reduced in cavefish photoreceptors (fig. 3B and D) compared with surface fish, most notably in the CMZ region (fig. 3A and C). This observation suggests the hypothesis that the reduction in cavefish eye size results from changes in the genetic regulation of proliferative signals in the retina during in postembryonic stages. The morphology of the adult cavefish eye, in which individual retinal cell body size is comparable with that of surface fish, but cell height, cell number, and overall eye size are dramatically reduced, provides further evidence for evolutionary changes specifically affecting growth and proliferation factors.
Data from our transcriptomics experiments further support conclusions inferred from morphological analyses. Among 9,649 unique genes, we identified 1,189 that were downregulated and 469 that were upregulated in cavefish eyes relative to surface fish eyes. The major pathways and disease-associated genes that were downregulated in eyes of cavefish versus surface fish involved visual transduction of the retina (including protein modification, visual signal transduction, G-protein signaling, respiratory electron transport, and synaptic transmission). These results were consistent with our observations on eye morphology showing abnormal retinal cell morphology. Genetic factors associated with retinal disease in humans, including retinitis pigmentosa, cone–rod dystrophy, achromatopsia, and macular degeneration, were also notably downregulated.
We conclude that developing cavefish do not lack embryonic eye induction signals, because all normal histological cell types are present in adult cavefish eyes and these layers enjoy nearly normal spatial relationships to each other. Although cavefish eyes possess the same cell types as surface fish, cell morphologies are altered and cell numbers are diminished. We conclude that the proliferation and/or maintenance of retinal cells, especially photoreceptor cells, is defective in cavefish. This conclusion suggests that the degeneration of cavefish eyes results from changes in genetic regulation during late stages of retinal development and/or in the maintenance and postembryonic growth of eyes rather than in events specifying early stages of eye development.
Gene Pathways Altered in the Cavefish Eye Transcriptome
Several gene pathways were altered in cavefish eyes relative to those of surface fish. Results showed that wnt2 (4.03×), lef1 (3.16×) and, importantly, several Wnt antagonists, including axin2 (2.06×), naked cuticle homolog 1 (nkd1, 2.13×), sfrp1a (1.55×), and sfrp2 (2.64×), were upregulated in cavefish eyes. The Wnt signaling pathway directs growth and differentiation in retina and other tissues (Van Raay and Vetter 2004; Clevers 2006). Analysis of model systems suggests that the Wnt pathway plays an anti-apoptotic role in the retina (Sundin et al. 2005; Yi et al. 2007; Chen, Hu, et al. 2009). The Wnt antagonists Sfrp1, 2, 3, and 5 are upregulated in human retinas affected by retinitis pigmentosa, an inherited form of photoreceptor degeneration that generally begins in the peripheral retina with rod cell dysfunction. Upregulation of Sfrp2 reduces Wnt signaling, leading to increased cell death in the retina (Jones et al. 2000a, 2000b; Yi et al. 2007). Although we did not detect apoptosis in the retinas of adult cavefish, we cannot rule out the possibility that cell death at an earlier time point might play a role in reducing retinal size in adult Sinocyclocheilus cavefish. The observed upregulation of sfrp2 presents the possibility that reduced Wnt signaling in cavefish retinas may lead to photoreceptor degeneration at an earlier stage of life. The significance of the upregulation of wnt2 in cavefish eyes in the face of the upregulation of Wnt inhibitors warrants further study.
The photoreceptor-specific nuclear receptor nr2e3 was downregulated in Sinocyclocheilus cavefish eyes. Mutations of the human NR2E3 gene are responsible for several retinopathies, including clumped pigmentary retinal degeneration, which involves night blindness, rudimentary rod function, and hyperfunction of the “blue” S-cones (Schorderet and Escher 2009). NR2E3 activates transcription of several rod-specific genes, including rhodopsin (Cheng et al. 2004), and suppresses cone genes (Chen et al. 2005; Hennig et al. 2008; Swaroop et al. 2010). The downregulation of nr2e3 in Sinocyclocheilus cavefish could contribute to the reduction in opsin expression we observed in RNA-seq analysis.
Results showed increased expression of zic1, zic2a, and zic2b in cavefish eyes by 5.7x, 2.2x, and 7.5x, respectively, whereas zic3 expression remained unchanged. The embryonic mouse retina expresses Zic1, Zic2, and Zic3 but expression of Zic1 and Zic2 gradually disappears during retinal development, while Zic3 expression continues into adulthood (Watabe et al. 2011). Evidence that Zic factors are important for retinal development comes from overexpression of Zic genes in retinal progenitors, which decreases the number of rod photoreceptors (Watabe et al. 2011); thus, the increase in zic1 and zic2 expression in cavefish eyes may also contribute to the observed decrease in photoreceptors.
Another category of genes that were downregulated in S. anophthalmus cavefish eyes involve catabolism, energy metabolism and ATP synthesis. This suggests that the degenerated eye of Sinocyclocheilus cavefish has greatly reduced metabolic rate compared with surface fish eyes. Given that a functioning eye uses energy at a rate that even exceeds that of the brain (Wangsa-Wirawan and Linsenmeier 2003; Protas et al. 2007), selection may act to reduce this energy sink in species inhabiting environments in which retinal function is unnecessary for survival (Protas et al. 2007; Jeffery 2009).
Downregulation of crx and Retinal Reduction in Cavefish
Several studies have helped to improve our understanding of gene regulatory networks that influence photoreceptor development and homeostasis (Hennig et al. 2008; Hu et al. 2010; Swaroop et al. 2010). RNA-seq data can test whether the regulation of genes in these networks was modified during cavefish evolution. The transcription factor crx is expressed in photoreceptors and other retinal neurons, and appears to be near the top of the hierarchy of retinal gene regulation. Expression of crx enhances the expression of photoreceptor-specific genes and events downstream of otx2 (Furukawa et al. 1997; Nishida et al. 2003; Shen and Raymond 2004; Swaroop et al. 2010) (fig. 7) and both crxa and its duplicate from the teleost genome duplication (TGD) crxb(otx5) are downregulated in cavefish eyes (fig. 6; supplementary fig. S3, Supplementary Material online). Knockdown of crxa in zebrafish results in eyes that are 10% smaller than control eyes and reduces expression of several phototransduction genes, including rhodopsin, cone opsins, irbp, otx5, gnat1, gnat2, and guca1a (Shen and Raymond 2004). In Crx knockout mice, photoreceptors developed but failed to express many phototransduction genes, resulting in the deficiency of photoreceptor outer segments and eventually, retinal degeneration (Furukawa et al. 1999). Bipolar cells also express Crx, and their differentiation and maintenance is impaired in Crx knockout mice (Furukawa et al. 1999; Hennig et al. 2008). This evidence indicates that crx is near the top of the pathway for photoreceptor development and differentiation (Hennig et al. 2008; Swaroop et al. 2010). Like crx knockdown zebrafish and mice, Sinocyclocheilus cavefish also exhibit small eyes, short photoreceptor cells, and reduced numbers of retinal cells (figs. 1C and 2E–J). These features, combined with low crx expression and downregulation of genes downstream of crx (fig. 7; supplementary table S2, Supplementary Material online), are consistent with the mechanistic hypothesis that much of the histological eye phenotype of Sinocyclocheilus cavefish lies downstream of evolved decrease of crx activity that leads to shorter and fewer photoreceptor cells and reduced retinal cell gene expression. Experiments to critically test the hypothesis that crx plays a direct role in retinal degeneration of cavefish requires further investigation.
What causes the downregulation of crx expression in Sinocyclocheilus cavefish eyes? Although Otx2 is the only transcription factor yet known to directly interact with the promoter of crx and to regulate crx expression (Furukawa et al. 1997; Nishida et al. 2003), otx2 expression was unchanged in the eyes of cavefish relative to surface fish. One hypothesis to explain these data is that cis evolution in the promotors of crx genes decreased responsiveness to Otx2 or other regulatory proteins. Arguing against this hypothesis is that both crxa and crxb(otx5) were downregulated in cavefish eyes, and so mutations with similar effects would have had to occur in promotors of multiple paralogs, which is not parsimonious. An alternative hypothesis is that cavefish have an evolutionarily mutated version of an as yet unidentified regulator of crx genes, perhaps an inhibitor, that is responsible for the downregulation of crx paralogs in the cavefish S. anophthalmus. In ongoing research, we are looking for factors that might regulate crxa expression during retinal development and maintenance.
Comparing Sinocyclocheilus and Astyanax Cavefish
Like Sinocyclocheilus, adult Astyanax cavefish have degenerated eyes that are covered with skin and have sunken into their orbits (Jeffery et al. 2000; Yamamoto and Jeffery 2000). Also like Sinocyclocheilus, eye degeneration in A. mexicanus cavefish evolved several times independently in different cave populations (Tobler et al. 2010; Strecker et al. 2012). In Astyanax, retinal degeneration is initiated by apoptosis of the lens, which forms during embryogenesis and then degenerates (Yamamoto et al. 2004). Our adult S. anophthalmus cavefish also have internal eyes (fig. 1C), which preserve basic but abnormal eye structures, including neural retina, iris, cornea, and lens (fig. 2B). In contrast to Astyanax, however, we did not find obvious abnormalities in either the morphology or transcriptomics of the lens in S. anophthalmus. Instead, the major morphological changes in Sinocyclocheilus cavefish eyes occurred in the neural retina. Although lens dysfunction is the causative factor in Astyanax cavefish eye degeneration (Yamamoto and Jeffery 2000), we find no detectable lens degeneration in Sinocyclocheilus cavefish.
For transcriptomics, the Sinocyclocheilus transcriptome was annotated primarily based on the zebrafish genome while Astyanax transcriptomics used cross-hybridization to a zebrafish microarray (Strickler and Jeffery 2009). Phylogenically, Sinocyclocheilus (Cyprinidae) and zebrafish (Cyprinidae) are more closely related to each other than either species is to Astyanax (Characidae) (supplementary fig. S1, Supplementary Material online). Although some genes may have been missed in the cross-species microarray for Astyanax, for genes that did hybridize, comparisons between surface and cave dwelling Astyanax will be valid. Comparisons of gene expression profiles of cavefish/surface fish pairs for Astyanax (Strickler and Jeffery 2009) and Sinocyclocheilus revealed only four genes that were differentially expressed in both genera of cavefish, including cish, rhodopsin, gnat1, and gnat2. Cish (cytokine inducible SH2-containing protein), is an anti-inflammatory molecule that is upregulated in the cornea after interleukin stimulation (Ueta et al. 2011); in cavefish eyes, it is likely responding to inflammation associated with retinal dysfunction. The visual pigment rhodopsin and the transducin alpha subunits of rods and cones, Gnat1 and Gnat2, respectively, are members of the visual phototransduction cascade (Shen and Raymond 2004). These comparisons suggest that retinal degeneration in A. mexicanus cavefish and S. anophthalmus share downstream effectors in retinal degeneration, although their reduced expression in S. anophthalmus might be caused by a lens-independent process, such as regulation by crx or other factors.
Comparative analysis of morphology and gene expression patterns in two evolutionarily independent cavefish lineages suggest that eye reduction in these two taxa appears to be controlled by different mechanisms, a lens-dependent process in Astyanax but a lens-independent process in Sinocyclocheilus. We do not know, however, in either case, the upstream evolutionary genetic changes or the selective regime, if any, that cause the retinal degeneration.
Conclusions
In this study, we contrasted histology and gene expression in the whole eye transcriptome of two closely related species: the surface species S. angustiporus and the cave species S. anophthalmus. Results showed that cavefish developed distinct troglomorphic characters in the evolutionary adaptation to perpetual darkness, including eye reduction, duckbill-like snout and long barbels and pectoral fins. Cavefish eyes showed reduction in eye size, retinal cell density, photoreceptor cell height, and cell proliferation, and downregulation of genes encoding photoreceptor protein products and several retinal cell regulators, including crx, although expression of otx2, the only known direct crx upstream regulator, was unchanged in cavefish eyes. These findings suggest the hypothesis that the evolution of crx dysregulation by a novel trans-acting factor is a major contributor to the downregulation of retina-specific genes and reduced eye size in Sinocyclocheilus cavefish.
Our analyses of Sinocyclocheilus cavefish retinas were limited to samples from adult time points. As such, although we did not detect pyknotic nuclei or the presence of apoptotic markers in retinal tissue, we cannot exclude the possibility that cell death at an earlier time point contributes to the overall reduction in retinal cells observed. Our results, however, do show substantial reductin in cell proliferation in adult retinas compared with surface fish, presenting the possibility that there are multiple mechanisms affecting the retinal changes we observe in Synocyclocheilus cavefish. Future investigations will focus on determining which of the observed changes in gene regulation might influence reduced proliferation or retinal degeneration, and at which developmental stage these phenomena initiate. Finally, the retinal reduction observed in Sinocyclocheilus cavefish appears to be independent of lens degeneration, in contrast to Astyanax cavefish. These results show that evolution can tinker with different developmental mechanisms to achieve similar adaptations to cave environments.
Materials and Methods
Animals
Sinocyclocheilus fish were collected in Yunnan province, China. Cavefish (S. anophthalmus) were collected in Jiuxiang cave (in perpetual darkness), Yiliang County, and surface fish (S. angustiporus) were collected from Huangnihe River in Agang Town, Luoping County (supplementary fig. S5, Supplementary Material online). Both collecting sites are in the Nanpanjiang River drainage, the largest tributary of Xijiang River of the Pearl River basin. Although the two collecting sites are only approximately 30 km apart, they are isolated by a mountain range (supplementary fig. S5, Supplementary Material online). Cavefish were maintained in the laboratory in a dark environment and surface fish were exposed to a natural daylight cycle. All experimental procedures involving animals were conducted and approved by the Animal Care and Use Committee of Institute of Zoology, Chinese Academy of Sciences.
Histological Analysis and Immunohistochemistry
For H&E staining and immunohistochemistry, adult S. angustiporus and S. anophthalmus fish were euthanized, eyes were removed, cleaned of associated skin and adipose tissue, fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), and equilibrated in 30% sucrose prior to cryosectioning. To improve fixative infiltration to the retina, the cornea was perforated with a small pin. Slides were stained with H&E using standard procedures. For immunohistochemistry, slide edges were lined with a Gnome PAP Pen liquid blocker and washed three times for 5 min with PBS before blocking for 1 h in blocking solution (10% normal goat serum and 2% bovine serum albumen in 1× PBS). We used the monoclonal antibodies Zpr1 and Zpr3 (Zebrafish International Resource Center, Eugene, Oregon, USA) (Vihtelic et al. 1999) to label unknown epitopes found in red/green double cones inner segments and the outer segments of rods and red/green double cones, respectively; antibodies against protein kinase C (Millipore, 05-983) to label ON bipolar cells in the INL; and antibodies against glutamine synthetase (Millipore, MAB302) to label Müller glial cells in the INL (Shkumatava et al. 2004). Antibodies were diluted 1:200 with blocking solution and incubated overnight at 4°C. Slides were then washed three times for 15 min with phosphate buffered saline with Tween 20 at 25°C. We then applied anti-mouse Alexa Fluor-488 (Invitrogen) secondary antibody solution (diluted 1:200 in blocking solution) to slides and incubated at room temperature for 2–3 h in a dark humid chamber. Slides were counterstained with ToPro3 (Invitrogen, Cat no.: 642/661) to label DNA in cell nuclei, mounted in Vectashield (Vector, H-1000, stored at 4°C), and coverslips were sealed with clear nail polish. Fluorescent images were analyzed using a Bio-Rad Radiance 2100MP confocal microscope system.
For analysis of proliferation and apoptosis, primary antisera were rabbit antibodies (Cell Signaling Technology, Inc.), as follows: for proliferation, anti-Phospho-Histone H3 (no. 3377); for apoptosis, anti-Cleaved Caspase-3 (no. 9661), which used at the dilution of 1:1,000 and 1:400, respectively. Zpr3 labeled the photoreceptors. The secondary antibodies, Alexa Fluor-488-coupled anti-rabbit IgG and Alexa Fluor-594-coupled anti-mouse IgG (Invitrogen), were used at 1:200. Slides were mounted in Vectashield mounting medium with DAPI (Vector, H-1200).
Quantification of nuclei and photoreceptor length measurements were performed with ImageJ 1.43u software. Nuclear densities and cell length were compared between surface and cave species using a t-test. All data met the assumption of normality.
Constructing the Reference Transcriptome
Total RNA was isolated using TRIzol reagent (Invitrogen) from whole eye and brain of adult surface and cave species, respectively. Two individuals were used for each species. Oligo(dT) selection was performed using MicroPoly(A) PuristTM (Ambion) according to the manufacturer’s protocol. Superscript III First-Strand Synthesis System (Invitrogen) was used to create first-strand cDNA. Second-strand cDNA was synthesized using random primers and Klenow exo-polymerase (Epicentre). cDNA was sheared to short fragments with Biorupter Sonicator, to which sequencing adapters were ligated. After size selection, suitable fragments were used as templates for PCR amplification, which, although it might occasionally introduce some artifacts, is necessary to obtain sufficient quantities for analysis. Four libraries were prepared, brain and eye libraries for each of two species. Finally, two brain cDNA libraries of both species (200–400 bp insert) were sequenced using 80-nt paired-end reads from the Illumina sequencer GA-II, and two libraries of eye samples (200–400 bp insert) using 100-nt paired end reads on the HiSeq 2000 (Illumina Inc., San Diego, CA, USA).
De novo assembly of the Sinocyclocheilus transcriptome was carried out using the short read assembly program Trinity (http://trinityrnaseq.sourceforge.net/) (Grabherr et al. 2011). After removing adapter sequences and low quality base calls, raw reads of surface fish and cavefish were assembled using default parameters with 300 nt minimum contig length. We made separate contig files of surface and cave species.
To generate a common and nonredundant Sinocyclocheilus transcriptome database for further RNA-seq analyses, we merged contig files from the two species using Cap3 (Huang and Madan 1999). If contigs were more than 95% identical and overlapped by more than 80 bp, they were merged. The longest contig of the group was retained in the transcriptome database, and redundant contigs were removed. Libraries of merged contigs were defined as nonredundant transcriptomes of Sinocyclocheilus for this article.
Contigs were aligned by BLASTX to the NCBI Nr protein database using an E-value cut-off of 10−6 (E value < 0.000001). Alignment results were used to retrieve gene names from gene2accession provided by NCBI. Annotation of the Sinocyclocheilus contigs was accomplished using the Blast2GO program by mapping the BLAST results to the GO database and finally selecting GO annotation using Blast2GO rules with default values (http://www.blast2go.com/) (Conesa et al. 2005). We also mapped the GO annotations to the Generic GO-Slim terms using Blast2GO. This Transcriptome Shotgun Assembly project for S. angustiporus has been deposited at DDBJ/EMBL/GenBank under the accession GAHO00000000. The version described in this article is the first version, GAHO01000000. This Transcriptome Shotgun Assembly project for Sinocyclocheilus anophthalmus has been deposited at DDBJ/EMBL/GenBank under the accession GAHL00000000. The version described in this paper is the first version, GAHL01000000.
Differential Gene Expression Using RNA-seq Analysis
After removing barcodes and low quality base calls, we mapped trimmed Illumina reads from surface fish and cavefish eyes to our nonredundant Sinocyclocheilus transcriptome using Bowtie (Langmead 2010). Because of sequence variation between paralogs and orthologs, the maximum difference in a read was set to three nucleotides, which is the maximum number of differences supported by Bowtie; other parameters were default.
Relative to some other cypriniforms, including zebrafish, the lineage leading to Sinocyclocheilus, to goldfish (Carassius auratus auratus), and to carps (including C. carpio) experienced a whole-genome duplication (Ohno et al. 1968; Leggatt and Iwama 2003; Mable et al. 2011; Wang et al. 2012; Braasch and Postlethwait 2013) that we call the CGD. The lineage leading to teleosts, including zebrafish, which is the closest relative to Sinocyclocheilus with a sequenced genome, experienced an earlier genome duplication event, the TGD (Amores et al. 1998; Postlethwait et al. 2000; Taylor et al. 2003; Jaillon et al. 2004). In annotating the Sinocyclocheilus eye transcriptome, we collapsed the two copies from the CGD into one annotation unit corresponding to the zebrafish ortholog.
To assess variation between Sinocylocheilus paralogs and their zebrafish orthologs, we randomly picked 16 genes that were abundantly expressed in Sinocyclocheilus eyes and analyzed the average identities of paralogs or orthologs between surface and cave species. Results showed that the average identity of paralogs duplicated in the CGD within Sinocyclocheillus species was 97.6 ± 0.96%, and the average identity of orthologs between the two species was 98.1 ± 0.90% (supplementary fig. S6, Supplementary Material online). Current high-throughput methods have difficulties distinguishing paralogs with such high identities from short-read RNA-seq experiments; thus, when counting gene expression levels, we ignored the CGD paralogs in these two species so the relative expression value was the sum of the two CGD paralogs. In addition, identities between paralog-aa (or paralog-ab) and either of the two paralog-b genes (paralog-ba or paralog-bb) was about 81% in Sinocyclocheilus, similar to that of zebrafish paralog-a and paralog-b (supplementary fig. S6, Supplementary Material online), which were produced by the TGD (Amores et al. 1998). With these similarities, Sinocyclocheilus reads could be aligned to the correct paralogs at the level of the zebrafish genome.
The average identity of all Sinocyclocheilus contigs with their best hits to zebrafish (D. rerio) was 90.5 ± 4.2% (supplementary fig. S6, Supplementary Material online). When we aligned transcriptome reads to the D. rerio transcriptome, the number of mapped reads was about half the number that mapped to the Sinocyclocheilus transcriptome. Because of evolution in the Sinocyclocheilus lineage after the CGD and to sequencing errors, we inferred that some valid Sinocyclocheilus sequence reads might not have been consolidated into the de novo assembly. By BLASTing transcriptome contigs against the zebrafish UniGene database, we identified gaps in the Sinocyclocheilus transcriptome contigs. To account for this problem, we first mapped trimmed RNA-seq reads to the de novo Sinocyclocheilus transcriptome assembly using Bowtie; then, we rescued reads that failed to map to the Sinocyclocheilus transcriptome by aligning them to the zebrafish UniGene database.
To assess differential expression, we merged the number of reads of surface fish or of cavefish that mapped to the same UniGene of either our Sinocyclocheilus transcriptome or the zebrafish UniGene database, and converted mapped read numbers (including reads mapped on both Sinocyclocheilus and D. rerio) to RPKM values (Mortazavi et al. 2008). To enhance statistical robustness, we excluded from analysis genes with fewer than five RPKM in either species, although these genes do appear in supplementary table S2, Supplementary Material online. Differential expression analysis for RNA-seq data used a rigorous algorithm that computes a conditional probability for observing N1 reads for a gene in a treatment given that N2 reads were observed in controls and experimentals with no biological replicates (Audic and Claverie 1997). P values correspond to the significance of differential expression. Log2FC and P values were computed for each gene. If log2FC more than 1 and P value less than 0.05, genes were considered to be differentially expressed (Schartl et al. 2012).
Real-Time PCR
cDNA samples were created using the Superscript III First-Strand Synthesis System (Invitrogen) from the mRNAs of the eyes of different surface and cavefish adults than those used for the RNA-seq experiments. Real-time PCR was conducted using SYBR Green (TaKaRa) chemistry. Real-time PCR primers were designed based on locations with identical sequences of the relevant gene in both species derived from our de novo assembled Sinocyclocheilus transcriptome using PerlPrimer design software (http://perlprimer.sourceforge.net/) (supplementary table S4, Supplementary Material online). The beta-actin1 gene was used as an internal control. All real-time PCR assays were run in a 96-well block with 20 μl reactions in each well. In each assay, reference samples were set up, and the expression levels of target genes were then quantified relative to this sample. A total of 16 genes were tested with Real-time PCR. Each reaction (samples and primers) was repeated at least three times.
Phylogenetic Analysis
In our transcriptomes, we identified nucleotide sequences of zic1 (zic family member-1) and gpr85 (G protein-coupled recepto-85 also known as sreb2), both of which are present in single copy in zebrafish and were previously used as nuclear phylogenetic markers in ray-finned fish (Li et al. 2007, 2008). For both Sinocyclocheilus species, two CGD paralogs of both genes were found and categorized as “a” and “b” paralogs based on shared nucleotide substitutions. For other ray-finned fish and for tetrapods, zic1 and gpr85 sequences were obtained from Genbank or Ensembl. See supplementary table S5, Supplementary Material online, for accession numbers.
Sequence alignments were generated with MUSCLE (Edgar 2004) for both genes individually and then concatenated. For Sinocyclocheilus, “a” paralogs (zic1a + gpr85a) and “b” paralogs (zic1b + gpr85b) were concatenated. A maximum likelihood phylogenetic tree based on 1,857 bp (zic1: 903 bp; gpr85: 954 bp) was generated with PhyML (Guindon et al. 2010) using the GTR model and 100 bootstrap replications.
Identification of Enriched Pathways and Diseases
We used KOBAS 2.0 (Xie et al. 2011) to identify enriched pathways and diseases from among genes that were differentially expressed in cave and surface fishes. The sequences of differentially expressed genes were compared with the Homo sapiens database using the “annotate” feature in KOBAS 2.0 to allow inferences from the abundant data on human pathways and diseases. We then used “identify” to find significantly enriched pathways and disease-associated genes; “inputs” were the two “annotate” results of upregulated and downregulated gene sets and the “background” was the entire set of 9,649 unique genes expressed in Sinocyclocheilus eyes identified by RNA-seq. Data were statistically analyzed using the hypergeometric test and Benjamini-Hochberg FDR (false discovery rate) correction, and only pathways or diseases with corrected P value less than 0.05 were considered to be enriched.
Supplementary Material
Supplementary tables S1–S5 and figures S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
F.W.M., C.G.Z., and J.H.P. conceived this study. F.W.M. and J.H.P. designed the experiments. F.W.M. and C.G.Z. collected the fish samples. F.W.M. and J.B.P. carried out the H&E staining and immunohistochemistry. F.W.M. and T.T. prepared the cDNA libraries for RNA-seq. F.W.M., I.B., L.X.W., and J.H.P. performed computer analysis of RNA-seq data. I.B. generated the maximum likelihood phylogenetic tree. F.W.M. performed real-time PCR. F.W.M. generated all images and F.W.M. and J.H.P. wrote the manuscript. All authors read, revised, and approved the final manuscript. The authors thank Dr Dante B. Fenolio for photographs shown in figure 1A and B and Dr Yahui Zhao for helpful discussion. This work was supported by National Program on Key Basic Research Project (2011CB943800) and National Institute of Health grants R01 RR020833-06 (alias R01 OD011116) to J.H.P. and R24 RR032670 to J.H.P. and W. Cresko.
References
- Albertson RC, Cresko W, Detrich HW, 3rd, Postlethwait JH. Evolutionary mutant models for human disease. Trends Genet. 2009;25:74–81. doi: 10.1016/j.tig.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunni A, Menuet A, Candal E, Penigault JB, Jeffery WR, Retaux S. Developmental mechanisms for retinal degeneration in the blind cavefish Astyanax mexicanus. J Comp Neurol. 2007;505:221–233. doi: 10.1002/cne.21488. [DOI] [PubMed] [Google Scholar]
- Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol. 2003;48:257–293. doi: 10.1016/s0039-6257(03)00030-4. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, et al. (13 co-authors) Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Borowsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–R24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Braasch I, Postlethwait JH. Polyploidy in fish and the teleost genome duplication. In: Soltis PS, Soltis D, editors. Polyploidy and genome evolution. Berlin (Germany): Springer-Verlag; 2013. [Google Scholar]
- Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Zhang RD, Feng JG, Xiao H, Li WX, Zan RG, Zhang YP. Exploring factors shaping population genetic structure of the freshwater fish Sinocyclocheilus grahami (Teleostei, Cyprinidae) J Fish Biol. 2009;74:1774–1786. doi: 10.1111/j.1095-8649.2009.02204.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hu Y, Moiseyev G, Zhou KK, Chen D, Ma JX. Photoreceptor degeneration and retinal inflammation induced by very low-density lipoprotein receptor deficiency. Microvasc Res. 2009;78:119–127. doi: 10.1016/j.mvr.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Dai C, Krantz SB. Interferon gamma induces upregulation and activation of caspases 1, 3, and 8 to produce apoptosis in human erythroid progenitor cells. Blood. 1999;93:3309–3316. [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain GL, Hardie R, Laughlin SB. Phototransduction and the evolution of photoreceptors. Curr Biol. 2010;20:R114–R124. doi: 10.1016/j.cub.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration—emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. (21 co-authors) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wan J, Hackler L, Jr, Zack DJ, Qian J. Computational analysis of tissue-specific gene networks: application to murine retinal functional studies. Bioinformatics. 2010;26:2289–2297. doi: 10.1093/bioinformatics/btq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Cai Y, Xing X. Rocky desertification, antidesertification, and sustainable development in the karst mountain region of Southwest China. Ambio. 2008;37:390–392. doi: 10.1579/08-s-493.1. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, et al. (61 co-authors) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231:1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Regressive evolution in Astyanax cavefish. Annu Rev Genet. 2009;43:25–47. doi: 10.1146/annurev-genet-102108-134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, Strickler A, Guiney S, Heyser D, Tomarev S. Prox 1 in eye degeneration and sensory organ compensation during development and evolution of the cavefish Astyanax. Dev Genes Evol. 2000;210:223–230. doi: 10.1007/s004270050308. [DOI] [PubMed] [Google Scholar]
- Jones SE, Jomary C, Grist J, Stewart HJ, Neal MJ. Altered expression of secreted frizzled-related protein-2 in retinitis pigmentosa retinas. Invest Ophthalmol Vis Sci. 2000a;41:1297–1301. [PubMed] [Google Scholar]
- Jones SE, Jomary C, Grist J, Stewart HJ, Neal MJ. Modulated expression of secreted frizzled-related proteins in human retinal degeneration. Neuroreport. 2000b;11:3963–3967. doi: 10.1097/00001756-200012180-00012. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Jensen SC, Meuer SM. The five-year incidence and progression of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology. 1997;104:7–21. doi: 10.1016/s0161-6420(97)30368-6. [DOI] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11:Unit 11.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev Fish Biol Fisheries. 2003;13:237–246. [Google Scholar]
- Li C, Lu G, Orti G. Optimal data partitioning and a test case for ray-finned fishes (Actinopterygii) based on ten nuclear loci. Syst Biol. 2008;57:519–539. doi: 10.1080/10635150802206883. [DOI] [PubMed] [Google Scholar]
- Li C, Orti G, Zhang G, Lu G. A practical approach to phylogenomics: the phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol Biol. 2007;7:44. doi: 10.1186/1471-2148-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK, Alexandrou MA, Taylor MI. Genome duplication in amphibians and fish: an extended synthesis. J Zool. 2011;284:151–182. [Google Scholar]
- McCauley DW, Hixon E, Jeffery WR. Evolution of pigment cell regression in the cavefish Astyanax: a late step in melanogenesis. Evol Dev. 2004;6:209–218. doi: 10.1111/j.1525-142X.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Ohno S, Wolf U, Atkin NB. Evolution from fish to mammals by gene duplication. Hereditas. 1968;59:169–187. doi: 10.1111/j.1601-5223.1968.tb02169.x. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Hitchcock PF. Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Res. 2003;43:927–936. doi: 10.1016/s0042-6989(02)00400-5. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007;16:2433–2452. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Pottin K, Hinaux H, Retaux S. Restoring eye size in Astyanax mexicanus blind cavefish embryos through modulation of the Shh and Fgf8 forebrain organising centres. Development. 2011;138:2467–2476. doi: 10.1242/dev.054106. [DOI] [PubMed] [Google Scholar]
- Poulson TL, White WB. The cave environment. Science. 1969;165:971–981. doi: 10.1126/science.165.3897.971. [DOI] [PubMed] [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17:452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, Holzschuh J, Erhardt S, Ettl AK, Driever W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc Natl Acad Sci U S A. 2005;102:18467–18472. doi: 10.1073/pnas.0506187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Kneitz S, Wilde B, Wagner T, Henkel C, Spaink H, Meierjohann S. Conserved expression signatures between medaka and human pigment cell tumors. PLoS One. 2012;7:e37880. doi: 10.1371/journal.pone.0037880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet DF, Escher P. NR2E3 mutations in enhanced S-cone sensitivity syndrome (ESCS), Goldmann-Favre syndrome (GFS), clumped pigmentary retinal degeneration (CPRD), and retinitis pigmentosa (RP) Hum Mutat. 2009;30:1475–1485. doi: 10.1002/humu.21096. [DOI] [PubMed] [Google Scholar]
- Shen YC, Raymond PA. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Dev Biol. 2004;269:237–251. doi: 10.1016/j.ydbio.2004.01.037. [DOI] [PubMed] [Google Scholar]
- Shkumatava A, Fischer S, Muller F, Strahle U, Neumann CJ. Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development. 2004;131:3849–3858. doi: 10.1242/dev.01247. [DOI] [PubMed] [Google Scholar]
- Strecker U, Hausdorf B, Wilkens H. Parallel speciation in Astyanax cave fish (Teleostei) in Northern Mexico. Mol Phylogenet Evol. 2012;62:62–70. doi: 10.1016/j.ympev.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Strickler AG, Byerly MS, Jeffery WR. Lens gene expression analysis reveals downregulation of the anti-apoptotic chaperone alphaA-crystallin during cavefish eye degeneration. Dev Genes Evol. 2007;217:771–782. doi: 10.1007/s00427-007-0190-z. [DOI] [PubMed] [Google Scholar]
- Strickler AG, Jeffery WR. Differentially expressed genes identified by cross-species microarray in the blind cavefish Astyanax. Integr Zool. 2009;4:99–109. doi: 10.1111/j.1749-4877.2008.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y, Kurokawa D, Takeuchi M, Kajikawa E, Kuratani S, Amemiya C, Aizawa S. Evolution of Otx paralogue usages in early patterning of the vertebrate head. Dev Biol. 2009;325:282–295. doi: 10.1016/j.ydbio.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Sundin OH, Leppert GS, Silva ED, et al. (14 co-authors) Extreme hyperopia is the result of null mutations in MFRP, which encodes a Frizzled-related protein. Proc Natl Acad Sci U S A. 2005;102:9553–9558. doi: 10.1073/pnas.0501451102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler M, Coleman SW, Perkins BD, Rosenthal GG. Reduced opsin gene expression in a cave-dwelling fish. Biol Lett. 2010;6:98–101. doi: 10.1098/rsbl.2009.0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta M, Sotozono C, Kinoshita S. Expression of interleukin-4 receptor alpha in human corneal epithelial cells. Jpn J Ophthalmol. 2011;55:405–410. doi: 10.1007/s10384-011-0030-6. [DOI] [PubMed] [Google Scholar]
- Van Raay TJ, Vetter ML. Wnt/frizzled signaling during vertebrate retinal development. Dev Neurosci. 2004;26:352–358. doi: 10.1159/000082277. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Doro CJ, Hyde DR. Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis Neurosci. 1999;16:571–585. doi: 10.1017/s0952523899163168. [DOI] [PubMed] [Google Scholar]
- Wang JT, Li JT, Zhang XF, Sun XW. Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio) BMC Genomics. 2012;13:96. doi: 10.1186/1471-2164-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangsa-Wirawan ND, Linsenmeier RA. Retinal oxygen: fundamental and clinical aspects. Arch Ophthalmol. 2003;121:547–557. doi: 10.1001/archopht.121.4.547. [DOI] [PubMed] [Google Scholar]
- Watabe Y, Baba Y, Nakauchi H, Mizota A, Watanabe S. The role of Zic family zinc finger transcription factors in the proliferation and differentiation of retinal progenitor cells. Biochem Biophys Res Commun. 2011;415:42–47. doi: 10.1016/j.bbrc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Wu X, Wang L, Chen S, Zan R, Xiao H, Zhang YP. The complete mitochondrial genomes of two species from Sinocyclocheilus (Cypriniformes: Cyprinidae) and a phylogenetic analysis within Cyprininae. Mol Biol Rep. 2010;37:2163–2171. doi: 10.1007/s11033-009-9689-x. [DOI] [PubMed] [Google Scholar]
- Xiao H, Chen SY, Liu ZM, Zhang RD, Li WX, Zan RG, Zhang YP. Molecular phylogeny of Sinocyclocheilus (Cypriniformes: Cyprinidae) inferred from mitochondrial DNA sequences. Mol Phylogenet Evol. 2005;36:67–77. doi: 10.1016/j.ympev.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Xiao H, Zhang RD, Feng JG, Ou YM, Li WX, Chen SY, Zan RG. Nuclear DNA content and ploidy of seventeen species of fishes in Sinocyclocheilus. Zool Res. 2002;23:195–199. [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Jeffery WR. Central role for the lens in cave fish eye degeneration. Science. 2000;289:631–633. doi: 10.1126/science.289.5479.631. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signalling controls eye degeneration in blind cavefish. Nature. 2004;431:844–847. doi: 10.1038/nature02864. [DOI] [PubMed] [Google Scholar]
- Yi H, Nakamura RE, Mohamed O, Dufort D, Hackam AS. Characterization of Wnt signaling during photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2007;48:5733–5741. doi: 10.1167/iovs.07-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Brocher J, Linder B, Hirmer A, Sundaramurthi H, Fischer U, Winkler C. The 1D4 antibody labels outer segments of long double cone but not rod photoreceptors in zebrafish. Invest Ophthalmol Vis Sci. 2012;53:4943–4951. doi: 10.1167/iovs.12-9511. [DOI] [PubMed] [Google Scholar]
- Zhao YH, Gozlan RE, Zhang CG. Out of sight out of mind: current knowledge of Chinese cave fishes. J Fish Biol. 2011;79:1545–1562. doi: 10.1111/j.1095-8649.2011.03066.x. [DOI] [PubMed] [Google Scholar]
- Zhao YH, Zhang CG. Endemic fishes of Sinocyclocheilus (Cypriniformes: Cyprinidae) in China—species diversity, cave adaptation, systematics and zoogeography. Beijing (China): Science Press; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.