Abstract
In the title compound, [Ni(C5H4N3O2)2(H2O)2]·2H2O, the NiII ion lies on an inversion center and is coordinated in an slightly distorted octahedral environment by two N,O-chelating 3-aminopyrazine-2-carboxylate (APZC) ligands in the equatorial plane and two trans-axial aqua ligands. In the crystal, O—H⋯O, N—H⋯O and O—H⋯N hydrogen bonds involving the solvent water molecules, aqua and APZC ligands form layers parallel to (010). These layers are linked further via O—H⋯O, N—H⋯O and C—H⋯O hydrogen bonds involving the axial aqua ligands, amino groups and the carboxylate groups of the APZC ligands, forming a three-dimensional network.
Related literature
For background to hybrid compounds, see: Bouchene et al. (2013 ▶); Bouacida et al. (2007 ▶, 2009 ▶). For the structure of the non-hydrated analogue, see: Ptasiewicz-Bak & Leciejewicz (1999 ▶). For 3-aminopyrazine-2-carboxylate–metal (M) complexes, see: Bouchene et al. (2013 ▶) [M = Co(II)]; Leciejewicz et al. (1997 ▶) [M = Ca(II)]; Leciejewicz et al. (1998 ▶) [M = Sr(II)]; Ptasiewicz-Bak & Leciejewicz (1997 ▶) [M = Mg(II)]; Tayebee et al. (2008 ▶) [M = Na(I)]; Ptasiewicz-Bak & Leciejewicz (1999 ▶). For proprieties and applications of pyrazine-2-carboxylic acid, see: Zhang & Mitchison (2003 ▶); Manju & Chaudhary, (2010 ▶); Chanda & Sangeetika (2004 ▶).
Experimental
Crystal data
[Ni(C5H4N3O2)2(H2O)2]·2H2O
M r = 406.98
Monoclinic,

a = 9.7939 (15) Å
b = 5.1123 (9) Å
c = 16.776 (3) Å
β = 115.838 (11)°
V = 756.0 (2) Å3
Z = 2
Mo Kα radiation
μ = 1.34 mm−1
T = 150 K
0.18 × 0.16 × 0.15 mm
Data collection
Bruker APEXII CCD diffractometer
6200 measured reflections
1326 independent reflections
1121 reflections with I > 2σ(I)
R int = 0.051
Refinement
R[F 2 > 2σ(F 2)] = 0.028
wR(F 2) = 0.073
S = 1.04
1326 reflections
127 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.36 e Å−3
Δρmin = −0.59 e Å−3
Data collection: APEX2 (Bruker, 2011 ▶); cell refinement: SAINT (Bruker, 2011 ▶); data reduction: SAINT; program(s) used to solve structure: SIR2002 (Burla et al., 2005 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and DIAMOND (Brandenburg & Berndt, 2001 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813012208/lh5610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813012208/lh5610Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1W—H1A⋯O2W i | 0.83 (3) | 1.97 (3) | 2.789 (3) | 169 (3) |
| O1W—H1B⋯O1ii | 0.75 (3) | 1.94 (3) | 2.690 (3) | 176 (4) |
| N3—H1N⋯O2W iii | 0.86 | 2.27 | 3.117 (3) | 168 |
| O2W—H2A⋯O2W iv | 0.77 (3) | 2.12 (3) | 2.867 (3) | 164 (4) |
| O2W—H2B⋯N2v | 0.78 (3) | 2.03 (3) | 2.792 (3) | 168 (3) |
| N3—H2N⋯O2 | 0.86 | 2.10 | 2.733 (3) | 130 |
| N3—H2N⋯O2vi | 0.86 | 2.20 | 2.871 (3) | 135 |
| C5—H5⋯O1W vii | 0.93 | 2.54 | 3.377 (4) | 150 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  ; (vii)
; (vii)  .
.
Acknowledgments
We are grateful to all personal of the LCATM laboratory, Université Oum El Bouaghi, Algeria, for their assistance. Thanks are due to MESRS and ATRST (Ministére de l’Enseignement Supérieur et de la Recherche Scientifique et l’Agence Thématique de Recherche en Sciences et Technologie - Algeria) via the PNR programm for financial support.
supplementary crystallographic information
Comment
The pyrazine-2-carboxylic acid bridging ligand, owing to its ability to act in acidic environments (Zhang & Mitchison, 2003), has been extensively studied for biological applications, such as anti-tubercular (Manju et al., 2010), antipyretic, antitumor, and anticancer (Chanda et al., 2004). An additional amino substitution on 3-amino-2-pyrazine carboxylic acid could be expected to enhance crystal packing through extensive hydrogen bonding. APZC has a large variety of coordination geometries in metal complexes (Leciejewicz et al., 1997, 1998; Ptasiewicz-Bak & Leciejewicz, 1997; Tayebee et al., 2008).
In continuation of our search to enrich the variety of such kinds of hybrid compounds and to investigate the influence of hydrogen bonds on the structural features (Bouacida et al., 2007, 2009), we report here the synthesis and crystal structure of the title compound, (I), as a extention of our earlier work on N,O chelated ligands (Bouchene et al.2013) which can be involved in covalent interactions in metal coordination chemistry.
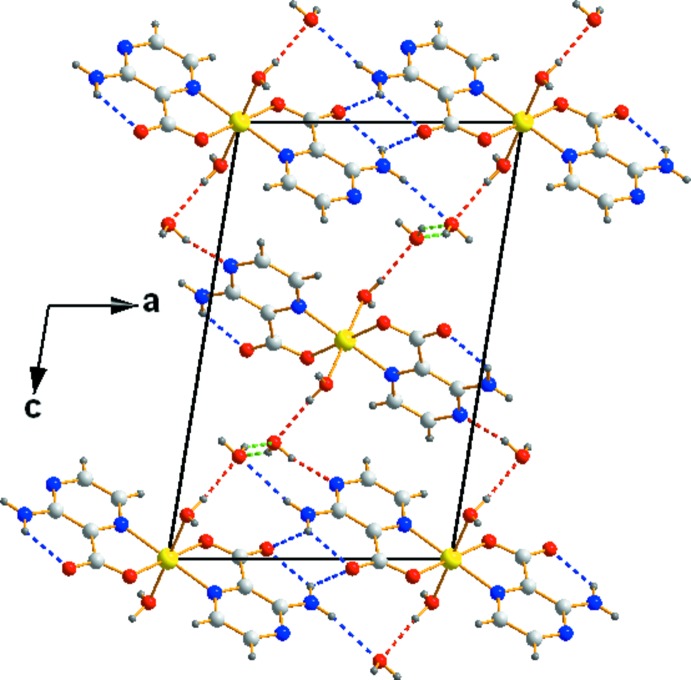
The asymmetric unit of (I) consists of one-half of the molecule, with the other half generated by a crystallographic inversion center. The molecular structure is shown in Fig. 1. The NiII ion is coordinated by two 3-amino-2-pyrazine carboxylate ligands via N,O-chelating groups in the equatorial plane and two aqua O atoms in the axial sites forming a slightly distorted octahedral coordination environment. The Ni—N, Ni—O and Ni—Oaqua distances are consistent with the reported data for the anhydrous Ni(II)(APZC)2(H2O)2 complex (Ptasiewicz-Bak & Leciejewicz, 1999). In the crystal, the solvent water molecules and complex molecules are involved in intermolecular O—H···O, O—H···N and N—H···O hydrogen bonds forming two-dimensional layers parallel to (010) (Fig.2). Further O—H···O hydrogen bonds (Fig.3) involving the aqua ligands, N—H···.O hydrogen bonds the carboxylate groups of the APZC ligands form a three-dimensional network.
Experimental
Nickel dichloride hexahydrate (0.2 mmol) and 3-aminopyrazine-2-carboxylic acid (0.02 mmol) were dissolved in acidified water with concentrated hydrogen chloride acid (37%). Light green crystals, suitable for X-ray diffraction study, were obtained from evaporation of obtained solution for three days.
Refinement
The H atoms bonded to C and N were located in differnce Fourier maps but subsequently introduced in calculated positions and treated as riding on their parent atoms (C or N) with C—H = 0.93 Å and N—H = 0.86 Å with Uiso(H) = 1.2Ueq(C or N). Atoms H1W and H2W were located in a difference Fourier map and refined isotropically wirh Uiso(H) = 1.5Ueq(O).
Figures
Fig. 1.
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level. H atoms are represented as small spheres of arbitrary radii. Symmetry code: (i)-x+1, -y+1, -z+2.
Fig. 2.
Partial packing plot viewed along the b axis showing the hydrogen bonds (dashed lines) forming layers.
Fig. 3.

Partial packing of (I) showing only the O—H···O hydrogen bonds which connect the layers in Fig. 2 into a three-dimensional network.
Crystal data
| [Ni(C5H4N3O2)2(H2O)2]·2H2O | F(000) = 420 |
| Mr = 406.98 | Dx = 1.788 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.7939 (15) Å | Cell parameters from 2285 reflections |
| b = 5.1123 (9) Å | θ = 2.7–25° |
| c = 16.776 (3) Å | µ = 1.34 mm−1 |
| β = 115.838 (11)° | T = 150 K |
| V = 756.0 (2) Å3 | Cube, white |
| Z = 2 | 0.18 × 0.16 × 0.15 mm |
Data collection
| Bruker APEXII CCD diffractometer | 1121 reflections with I > 2σ(I) |
| Radiation source: sealed tube | Rint = 0.051 |
| Graphite monochromator | θmax = 25.1°, θmin = 2.7° |
| φ and ω scans | h = −11→11 |
| 6200 measured reflections | k = −6→6 |
| 1326 independent reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.028 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.073 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0363P)2 + 0.4886P] where P = (Fo2 + 2Fc2)/3 |
| 1326 reflections | (Δ/σ)max < 0.001 |
| 127 parameters | Δρmax = 0.36 e Å−3 |
| 0 restraints | Δρmin = −0.59 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.2428 (3) | 0.2367 (5) | 0.99519 (16) | 0.0089 (5) | |
| C2 | 0.2750 (3) | 0.4494 (4) | 1.06347 (15) | 0.0079 (5) | |
| C3 | 0.1864 (3) | 0.4915 (5) | 1.11094 (15) | 0.0090 (5) | |
| C4 | 0.3430 (3) | 0.8353 (5) | 1.18295 (16) | 0.0098 (5) | |
| H4 | 0.3692 | 0.9711 | 1.2239 | 0.012* | |
| C5 | 0.4304 (3) | 0.7942 (5) | 1.13805 (15) | 0.0097 (5) | |
| H5 | 0.5141 | 0.8999 | 1.1492 | 0.012* | |
| N1 | 0.3937 (2) | 0.6010 (4) | 1.07831 (13) | 0.0074 (4) | |
| N2 | 0.2227 (2) | 0.6890 (4) | 1.17019 (13) | 0.0102 (4) | |
| N3 | 0.0696 (2) | 0.3396 (4) | 1.10092 (14) | 0.0125 (5) | |
| H1N | 0.0202 | 0.3689 | 1.1316 | 0.015* | |
| H2N | 0.0436 | 0.2122 | 1.0637 | 0.015* | |
| O1 | 0.33555 (17) | 0.2240 (3) | 0.96010 (11) | 0.0090 (4) | |
| O2 | 0.13487 (18) | 0.0885 (3) | 0.97784 (12) | 0.0137 (4) | |
| O1W | 0.6426 (2) | 0.2542 (4) | 1.10017 (12) | 0.0102 (4) | |
| H1A | 0.726 (3) | 0.312 (6) | 1.1356 (19) | 0.015* | |
| H1B | 0.652 (3) | 0.124 (6) | 1.083 (2) | 0.015* | |
| O2W | 0.0735 (2) | 0.6137 (4) | 0.76616 (12) | 0.0133 (4) | |
| H2A | 0.046 (4) | 0.472 (6) | 0.754 (2) | 0.02* | |
| H2B | 0.104 (3) | 0.663 (6) | 0.733 (2) | 0.02* | |
| Ni1 | 0.5 | 0.5 | 1 | 0.00664 (16) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0073 (11) | 0.0071 (12) | 0.0116 (12) | 0.0010 (9) | 0.0036 (10) | 0.0014 (10) |
| C2 | 0.0067 (12) | 0.0078 (12) | 0.0087 (11) | 0.0002 (9) | 0.0029 (10) | 0.0009 (9) |
| C3 | 0.0078 (11) | 0.0088 (12) | 0.0098 (11) | 0.0029 (10) | 0.0031 (9) | 0.0029 (11) |
| C4 | 0.0114 (12) | 0.0077 (12) | 0.0105 (12) | −0.0014 (9) | 0.0050 (10) | −0.0024 (10) |
| C5 | 0.0068 (12) | 0.0089 (12) | 0.0137 (13) | −0.0011 (9) | 0.0048 (10) | −0.0017 (10) |
| N1 | 0.0040 (10) | 0.0067 (10) | 0.0102 (10) | 0.0004 (8) | 0.0019 (8) | 0.0009 (8) |
| N2 | 0.0097 (10) | 0.0101 (11) | 0.0123 (10) | −0.0008 (8) | 0.0061 (9) | 0.0001 (8) |
| N3 | 0.0094 (10) | 0.0139 (11) | 0.0195 (11) | −0.0052 (9) | 0.0113 (9) | −0.0056 (9) |
| O1 | 0.0069 (8) | 0.0093 (8) | 0.0137 (9) | −0.0018 (7) | 0.0073 (7) | −0.0032 (7) |
| O2 | 0.0092 (9) | 0.0135 (9) | 0.0213 (10) | −0.0055 (7) | 0.0092 (8) | −0.0059 (8) |
| O1W | 0.0077 (9) | 0.0081 (9) | 0.0147 (9) | −0.0015 (7) | 0.0046 (8) | −0.0030 (8) |
| O2W | 0.0148 (10) | 0.0129 (9) | 0.0169 (10) | −0.0035 (8) | 0.0115 (8) | −0.0018 (8) |
| Ni1 | 0.0049 (2) | 0.0062 (2) | 0.0106 (2) | −0.00089 (17) | 0.00506 (18) | −0.00108 (18) |
Geometric parameters (Å, º)
| C1—O2 | 1.228 (3) | N1—Ni1 | 2.0657 (19) |
| C1—O1 | 1.281 (3) | N3—H1N | 0.86 |
| C1—C2 | 1.510 (3) | N3—H2N | 0.86 |
| C2—N1 | 1.327 (3) | O1—Ni1 | 2.0233 (16) |
| C2—C3 | 1.427 (3) | O1W—Ni1 | 2.0755 (18) |
| C3—N3 | 1.331 (3) | O1W—H1A | 0.83 (3) |
| C3—N2 | 1.351 (3) | O1W—H1B | 0.74 (3) |
| C4—N2 | 1.332 (3) | O2W—H2A | 0.77 (3) |
| C4—C5 | 1.381 (3) | O2W—H2B | 0.78 (3) |
| C4—H4 | 0.93 | Ni1—O1i | 2.0233 (16) |
| C5—N1 | 1.340 (3) | Ni1—N1i | 2.066 (2) |
| C5—H5 | 0.93 | Ni1—O1Wi | 2.0755 (18) |
| O2—C1—O1 | 124.7 (2) | H1N—N3—H2N | 120 |
| O2—C1—C2 | 119.8 (2) | C1—O1—Ni1 | 115.72 (15) |
| O1—C1—C2 | 115.5 (2) | Ni1—O1W—H1A | 118 (2) |
| N1—C2—C3 | 120.2 (2) | Ni1—O1W—H1B | 113 (2) |
| N1—C2—C1 | 116.0 (2) | H1A—O1W—H1B | 110 (3) |
| C3—C2—C1 | 123.7 (2) | H2A—O2W—H2B | 108 (3) |
| N3—C3—N2 | 117.7 (2) | O1—Ni1—O1i | 180 |
| N3—C3—C2 | 122.7 (2) | O1—Ni1—N1 | 80.54 (7) |
| N2—C3—C2 | 119.6 (2) | O1i—Ni1—N1 | 99.46 (7) |
| N2—C4—C5 | 122.8 (2) | O1—Ni1—N1i | 99.46 (7) |
| N2—C4—H4 | 118.6 | O1i—Ni1—N1i | 80.54 (7) |
| C5—C4—H4 | 118.6 | N1—Ni1—N1i | 180.0000 (10) |
| N1—C5—C4 | 119.5 (2) | O1—Ni1—O1Wi | 89.81 (7) |
| N1—C5—H5 | 120.3 | O1i—Ni1—O1Wi | 90.19 (7) |
| C4—C5—H5 | 120.3 | N1—Ni1—O1Wi | 90.92 (7) |
| C2—N1—C5 | 119.9 (2) | N1i—Ni1—O1Wi | 89.08 (7) |
| C2—N1—Ni1 | 112.20 (15) | O1—Ni1—O1W | 90.19 (7) |
| C5—N1—Ni1 | 127.92 (16) | O1i—Ni1—O1W | 89.81 (7) |
| C4—N2—C3 | 117.9 (2) | N1—Ni1—O1W | 89.08 (7) |
| C3—N3—H1N | 120 | N1i—Ni1—O1W | 90.92 (7) |
| C3—N3—H2N | 120 | O1Wi—Ni1—O1W | 180.00 (9) |
| O2—C1—C2—N1 | 180.0 (2) | N3—C3—N2—C4 | 177.5 (2) |
| O1—C1—C2—N1 | 1.0 (3) | C2—C3—N2—C4 | −1.1 (3) |
| O2—C1—C2—C3 | 0.2 (4) | O2—C1—O1—Ni1 | 178.74 (18) |
| O1—C1—C2—C3 | −178.8 (2) | C2—C1—O1—Ni1 | −2.3 (3) |
| N1—C2—C3—N3 | −177.5 (2) | C1—O1—Ni1—N1 | 2.15 (16) |
| C1—C2—C3—N3 | 2.3 (4) | C1—O1—Ni1—N1i | −177.85 (16) |
| N1—C2—C3—N2 | 1.0 (3) | C1—O1—Ni1—O1Wi | −88.81 (16) |
| C1—C2—C3—N2 | −179.2 (2) | C1—O1—Ni1—O1W | 91.19 (16) |
| N2—C4—C5—N1 | 0.5 (4) | C2—N1—Ni1—O1 | −1.50 (15) |
| C3—C2—N1—C5 | 0.0 (3) | C5—N1—Ni1—O1 | 179.2 (2) |
| C1—C2—N1—C5 | −179.9 (2) | C2—N1—Ni1—O1i | 178.50 (15) |
| C3—C2—N1—Ni1 | −179.41 (17) | C5—N1—Ni1—O1i | −0.8 (2) |
| C1—C2—N1—Ni1 | 0.8 (2) | C2—N1—Ni1—O1Wi | 88.16 (16) |
| C4—C5—N1—C2 | −0.7 (3) | C5—N1—Ni1—O1Wi | −91.1 (2) |
| C4—C5—N1—Ni1 | 178.57 (17) | C2—N1—Ni1—O1W | −91.84 (16) |
| C5—C4—N2—C3 | 0.4 (3) | C5—N1—Ni1—O1W | 88.9 (2) |
Symmetry code: (i) −x+1, −y+1, −z+2.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H1A···O2Wi | 0.83 (3) | 1.97 (3) | 2.789 (3) | 169 (3) |
| O1W—H1B···O1ii | 0.75 (3) | 1.94 (3) | 2.690 (3) | 176 (4) |
| N3—H1N···O2Wiii | 0.86 | 2.27 | 3.117 (3) | 168 |
| O2W—H2A···O2Wiv | 0.77 (3) | 2.12 (3) | 2.867 (3) | 164 (4) |
| O2W—H2B···N2v | 0.78 (3) | 2.03 (3) | 2.792 (3) | 168 (3) |
| N3—H2N···O2 | 0.86 | 2.10 | 2.733 (3) | 130 |
| N3—H2N···O2vi | 0.86 | 2.20 | 2.871 (3) | 135 |
| C5—H5···O1Wvii | 0.93 | 2.54 | 3.377 (4) | 150 |
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) −x+1, −y, −z+2; (iii) −x, −y+1, −z+2; (iv) −x, y−1/2, −z+3/2; (v) x, −y+3/2, z−1/2; (vi) −x, −y, −z+2; (vii) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH5610).
References
- Bouacida, S., Belhouas, R., Kechout, H., Merazig, H. & Bénard-Rocherullé, P. (2009). Acta Cryst. E65, o628–o629. [DOI] [PMC free article] [PubMed]
- Bouacida, S., Merazig, H., Benard-Rocherulle, P. & Rizzoli, C. (2007). Acta Cryst. E63, m379–m381.
- Bouchene, R., Bouacida, S., Berrah, F., Belhouas, R. & Merazig, H. (2013). Acta Cryst. E69, m129–m130. [DOI] [PMC free article] [PubMed]
- Brandenburg, K. & Berndt, M. (2001). DIAMOND Crystal Impact, Bonn, Germany.
- Bruker (2011). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Chanda, S. & Sangeetika, J. (2004). J. Indian Chem. Soc. 81, 203–206.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Leciejewicz, J., Ptasiewicz-Bak, H. & Paluchowska, B. (1997). Pol. J. Chem. 71, 1339–1364.
- Leciejewicz, J., Ptasiewicz-Bak, H. & Zachara, J. (1998). Pol. J. Chem. 72, 1994–1998.
- Manju, A. & Chaudhary, D. (2010). Asian J. Chem. Environ. Res. 3, 13–17.
- Ptasiewicz-Bak, H. & Leciejewicz, J. (1997). Pol. J. Chem. 71, 1350–1358.
- Ptasiewicz-Bak, H. & Leciejewicz, J. (1999). Pol. J. Chem. 73, 717–725.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tayebee, R., Amani, V. & Khavasi, H. P. (2008). Chin. J. Chem. 26, 500–504.
- Zhang, Y. & Mitchison, D. A. (2003). Int. J. Tuberc. Lung Dis. 7,6–21. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813012208/lh5610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813012208/lh5610Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report