Summary
Arp2/3 complex is a seven-subunit assembly that nucleates branched actin filaments. Small molecule inhibitors CK-666 and CK-869 bind to Arp2/3 complex and inhibit nucleation, but their modes of action are unknown. Here we use biochemical and structural methods to determine the mechanism of each inhibitor. Our data indicate that CK-666 stabilizes the inactive state of the complex, blocking movement of the Arp2 and Arp3 subunits into the activated filament-like (short pitch) conformation, while CK-869 binds to a serendipitous pocket on Arp3 and allosterically destabilizes the short pitch Arp3-Arp2 interface. These results provide key insights into the relationship between conformation and activity in Arp2/3 complex, and will be critical for interpreting the influence of the inhibitors on actin filament networks in vivo.
Introduction
Small molecule inhibitors play important roles as drugs and as research tools, and increasing the types of macromolecules that can be targeted by small molecules is an important focus of pharmacological and biochemical studies. While active sites of enzymes are frequently targeted, proteins lacking a catalytic site can also be inhibited using molecules that disrupt protein-protein interfaces critical for function. Small molecules can also be used to inhibit large molecular assemblies that serve as multi-functional molecular machines (Pommier and Marchand, 2011). In such assemblies, small molecules can selectively block one of multiple active sites, protein-protein interfaces or conformational changes critical for discrete aspects of macromolecular function. Therefore, small molecule inhibitors can become powerful tools to dissect the mechanism of macromolecular assemblies in vitro and in vivo. However, realization of this potential requires a precise determination of how the small molecules inhibit function.
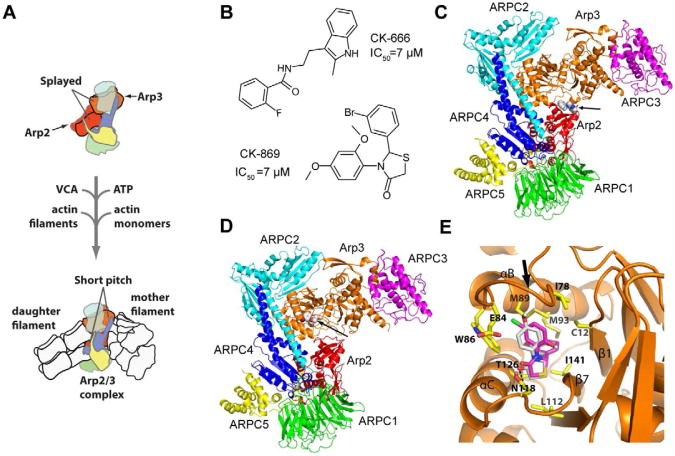
Arp2/3 (actin-related protein 2/3) complex is a seven subunit macromolecular machine that regulates the actin cytoskeleton. It catalyzes the kinetically slow process of actin filament nucleation (Cooper, 1983; Sept and McCammon, 2001), a reaction regulated in cells to allow precise spatial and temporal control of the assembly of actin filament networks (Firat-Karalar and Welch, 2011). Arp2/3 complex binds to the sides of pre-existing (mother) actin filaments and nucleates a new (daughter) filament in a complex reaction that requires interaction of Arp2/3 complex with filaments, actin monomers, ATP, and activating proteins called nucleation promoting factors (NPFs) (Goley and Welch, 2006; Pollard, 2007) (Fig. 1A). WASp/Scar family proteins, the best characterized NPFs, have a conserved region called VCA. The CA motif of VCA binds Arp2/3 complex and the V region binds actin monomers, tethering them to the complex (Boczkowska et al., 2008; Marchand et al., 2001; Miki and Takenawa, 1998). High-resolution x-ray crystal structures show that in the absence of NPFs, Arp2 and Arp3, the two actin-related subunits in the complex, are splayed apart (Nolen and Pollard, 2007; Robinson et al., 2001). Electron microscopy reconstructions show that upon activation, Arp2 moves ∼25 Å to form a dimer with Arp3 that mimics a short pitch actin dimer within a filament, thereby providing a template for nucleation of a new filament (Rouiller et al., 2008). How the activating factors initiate short pitch conformation is not known, and whether small molecules could be exploited to block this large conformational change is not clear.
Fig. 1.
CK-666 and CK-869 bind to different sites on Arp2/3 complex. (A) Overview of branching nucleation reaction showing proposed conformational change of Arp2 and Arp3 into the activated short pitch conformation. (B) Structures of CK-666 and CK-869. Previously reported IC50 values are indicated (Nolen et al., 2009). (C) Overall binding mode of CK-666 from a previously reported 2.5 Å x-ray crystal structure of CK-666 bound to Bos taurus Arp2/3 complex (3UKR) (Baggett et al., 2012). CK-666 (marked with arrow) binds at the interface of Arp3 (orange) and Arp2 (red). The other subunits of the complex are ARPC1-5, and are colored green, cyan, magenta, blue and yellow, respectively. CK-666 binding does not change the position of the subunits compared to inhibitor-free structures, but appears to stabilize the splayed (inactive) conformation of the Arp2 and Arp3 subunits. (D) Overall binding mode of CK-869 from the 2.75 Å x-ray crystal structure reported here. CK-869 (marked with arrow) binds to a hydrophobic pocket in Arp3 (orange). Color scheme is identical to panel (C). (E) Close up of the binding pocket of CK-869. The binding site for CK-869 (grey) is identical to the site for CK-548 (magenta) and is exposed when the sensor loop (arrow) flips into an open conformation. See also Figure S1 and Table S1.
Previously, two distinct classes of small molecule Arp2/3 complex inhibitors were discovered, CK-636 and CK-548, which block nucleation of actin filaments by Arp2/3 complex in vitro (Nolen et al., 2009). Treatment of cultured cells with these inhibitors blocks formation of actin structures known to require Arp2/3 complex, including Listeria actin comet tails, podosomes, and yeast endocytic actin patches (Nolen et al., 2009; Rizvi et al., 2009). Because they provide a simple, fast-acting and reversible method of inhibition, these compounds can be powerful tools to probe the role of Arp2/3 complex in other actin remodeling processes. Crystal structures of CK-636 and CK-548 bound to Arp2/3 complex provided preliminary clues as to how they might function, but the molecular mechanism of inhibition has not been determined.
Here we use a combination of biochemical and biophysical methods to determine the mechanisms of CK-666 and CK-869, more potent versions of parent compounds CK-636 and CK-548. Despite their distinct binding sites, our data suggest that both CK-666 and CK-869 inhibit nucleation by blocking the movement of Arp2 into the short pitch conformation. Remarkably, conformational trapping by each inhibitor is accomplished by a different mechanism. CK-666 functions as a classical allosteric effector, stabilizing the inactive state of the complex, while CK-869 appears to directly disrupt key protein-protein interfaces in the short pitch Arp2-Arp3 dimer to destabilize the active state. By measuring the influence of the inhibitors on interactions of the complex with NPFs, ATP, actin monomers and filaments, we provide insight into the relationship between conformation and activation and a basis for understanding the effects of the inhibitors on branched actin networks in vivo. These results support the feasibility of using small molecules to both allosterically disrupt protein-protein interfaces and to lock multi-protein complexes in inactive conformations by targeting and stabilizing subunit interfaces.
Results
Crystal structure of CK-869 bound to Arp2/3 complex
CK-666 and CK-869 are commercially available compounds derived from CK-636 and CK-548, respectively (Nolen et al., 2009) (Fig. 1B). The recently reported crystal structure of CK-666 bound to Arp2/3 complex showed that like CK-636, it binds to a pocket at the interface of Arp2 and Arp3, suggesting both inhibitors may block formation of the Arp2-Arp3 short pitch dimer (Baggett et al., 2012) (Fig. 1C). While chemically similar to its parent compound, it is not known if CK-869 occupies the same binding pocket as CK-548. Therefore, we solved the crystal structure of CK-869 bound to Bos taurus (Bt) Arp2/3 complex. A 2.75 Å resolution crystal structure showed that CK-869, like CK-548, binds to a hydrophobic cleft in subdomain 1 of Arp3, making a single hydrogen bond with the amide group of Asn118 (Fig. 1D,E, Fig. S1, Table S1). As with CK-548, binding of CK-869 locks the sensor loop into an open position. Similarity between this structure and the CK-548-bound structure indicates that CK-548 and CK-869 use a common mechanism of inhibition.
CK-869 causes structural changes in ATP-bound Arp3 that may contribute to complex inactivation
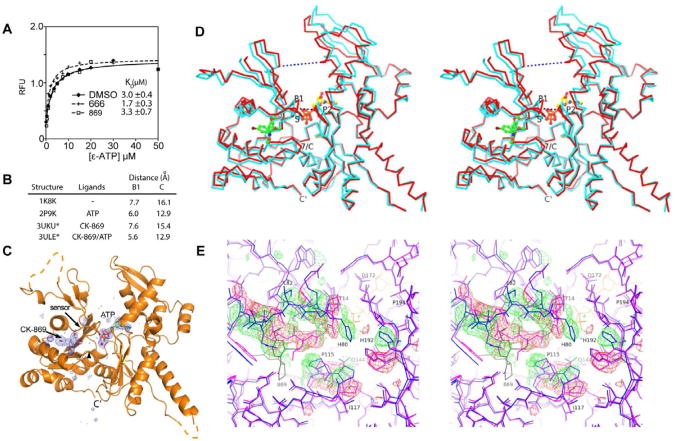
Arp2/3 complex requires ATP to nucleate actin filaments (Dayel et al., 2001), and mutations in the nucleotide binding pockets (NBP) of Arp2 or Arp3 cause defects in nucleation (Goley et al., 2004; Martin et al., 2005) and branched network turnover (Ingerman et al., 2013). Because neither inhibitor binds to the NBP of Arp3 or Arp2 we ruled out direct competition with ATP as an inhibition mechanism. However, the sensor loop in actin and actin-related proteins is allosterically linked to the nucleotide binding pocket (Nolen and Pollard, 2007; Otterbein et al., 2001), so we reasoned that the sensor loop flip caused by CK-869 might influence ATP binding to Arp3. Therefore, we measured the affinity of 1-N6-etheno-ATP (ε-ATP) to BtArp2/3 complex in the presence and absence of inhibitors (Fig. 2A). Previous experiments showed that the signal of this assay is predominantly due to binding of ε-ATP to Arp3 (Martin et al., 2005). ε-ATP bound with a KD = 3.0 ± 0.4 μM in the absence of inhibitor, and the KD did not change significantly in the presence of saturating CK-666 or CK-869, indicating that neither compound inactivates Arp2/3 complex by blocking ATP binding to Arp3. To determine if CK-869 influences ATP-induced conformational changes, we solved the 2.5 Å co-crystal structure of Arp2/3 with bound ATP and CK-869. Importantly, CK-869 does not prevent the cleft of Arp3 from closing, a conformational change observed in other ATP-bound Arp2/3 complex structures and thought to be required for activation of the complex (Goley et al., 2004; Nolen and Pollard, 2007; Rouiller et al., 2008) (Fig. 2B-D). However, binding of CK-869 to ATP-loaded Arp3 caused several significant structural changes. First, Pro115 and Pro116 in the loop between β7 and αC move ∼1.5 Å toward the space previously occupied by the sensor loop (Fig. 2E, Fig. S2). This movement causes Gln144, a conserved residue in the nucleotide cleft, to adopt a position in which it directly contacts the calcium in the nucleotide cleft, as opposed to making a water-mediated contact, as observed in other ATP-bound actin and Arp2/3 complex structures (Fig. S2) (Nolen et al., 2004; Nolen and Pollard, 2007). These changes do not occur in the CK-869-bound structure without ATP, and may be critical for inhibition by CK-869, as discussed below.
Fig. 2.
CK-869 influences ATP-induced conformational changes in Arp3. (A) ε-ATP binding assays with BtArp2/3 complex (0.4 μM) and 200 μM of either CK-666 or CK-869 was titrated with ε-ATP and the fluorescence measured. (B) Binding of CK-869 does not block ATP-induced closure of the nucleotide cleft. Distances across nucleotide cleft as described in D. Asterisked structures are reported here. (C) Difference electron density maps calculated without phase contributions from ATP, Ca2+ (red sphere), or CK-869 contoured at 3.0 σ show strong density for both ATP and CK-869 bound to Arp3 subunit. (D) Stereoview of Cα trace of Arp3 from the CK-869/ATP structure (red) with overlaid Arp3 from the apo-enzyme CK-869 structure. CK-869 is shown in stick representation with green carbons and ATP with yellow carbons. Black dotted line indicates the B1 distance (Thr14 CA to Gly173 CA) and the blue dotted line indicates the C distance (Gly67 CA - Glu202 CA). P1: P1 phosphate binding loop, P2: P2 phosphate binding loop, 7/C: β7/αC loop, S: sensor loop. (E) Conformational changes caused by binding of CK-869 to the ATP-bound complex. Close up stereo view of Arp3 from ATP bound inhibitor-free complex (2P9K.pdb, purple sticks) overlaid onto Arp3 from the CK-869/ATP structure (blue sticks, ATP in yellow lines). Positive 3.0 σ (green) and negative 3.5 σ (red) electron density from a difference map were calculated by modeling the sensor loop and β7/αC loop from the CK-869/ATP structure in the conformation observed in the 2P9K.pdb structure. CK-869 is labeled and rendered as grey sticks. See also Figure S2 and Table S1.
Neither inhibitor blocks the low or high affinity NPF binding sites on the complex
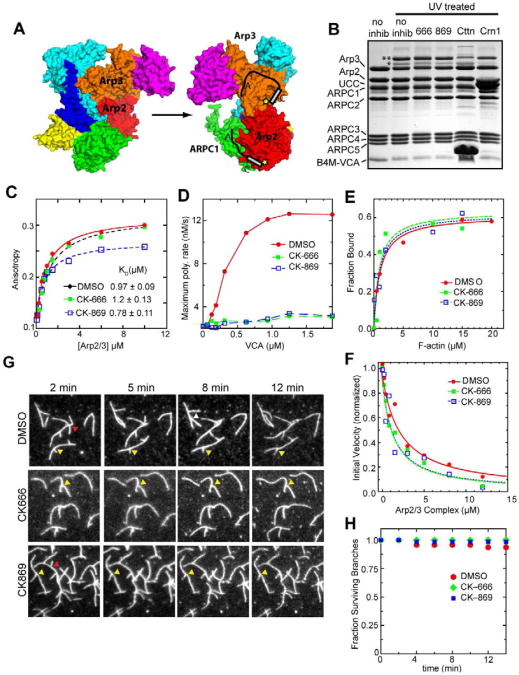
We next investigated the effect of the inhibitors on activator binding. Chemical crosslinking and analytical ultracentrifugation experiments demonstrated that Arp2/3 complex binds to two VCA molecules (Liu et al., 2011; Padrick et al., 2011). Isothermal titration calorimetry measurements showed that VCA binds more tightly to one of the two sites, consistent with a recent observation that only a 1:1 and not a 2:1 VCA:Arp2/3 assembly can be isolated by gel filtration (Gaucher et al., 2012; Ti et al., 2011). Initial characterization of the inhibitors showed that neither compound significantly affects binding of monomeric N-WASp-VCA to the high affinity NPF site, hypothesized to be on Arp2 and ARPC1 (Fig. 3A) (Nolen et al., 2009; Padrick et al., 2011; Ti et al., 2011). The second NPF site binds VCA 100-fold more weakly and is hypothesized to be on Arp3 (Ti et al., 2011). Crosslinking, x-ray crystallography and molecular modeling suggest the second NPF site is on the “back side” of Arp3 relative to the mother filament binding face (Padrick et al.; Ti et al.) (Fig. 3A), although density from recent single particle averaged electron microscopy structures suggest VCA binds to the pointed ends of Arp3 and Arp2 (Xu et al., 2011). Both NPF binding sites are critical for activation of the complex (Ti et al.), but it is not known if the inhibitors affect interactions of VCA with the low affinity site. Therefore, we used a chemical cross-linking assay to test the effect of the inhibitors on VCA binding at each site. N-WASp-VCA labeled with benzophenone-4-maleimide (B4M) at position 464 (N-terminal to its C region) forms a UV-induced covalent crosslink with Arp2 or Arp3 subunits, resulting in the appearance of two higher molecular weight bands (Fig. 3B). Addition of 200 μM CK-869 or CK-666 did not significantly influence crosslinking to Arp2 or Arp3, suggesting neither inhibitor functions by blocking VCA from binding to either site. In contrast, addition of cortactin or Crn1, proteins that compete with VCA at only the Arp3 or Arp2 site, respectively (Liu et al.; Weaver et al., 2002), significantly reduced crosslinking at their expected sites, demonstrating the sensitivity of the assay. To quantitatively probe the binding to Arp3, we also tested the effect of the inhibitors on the affinity of the N-terminal acidic region (NTA) of cortactin in a fluorescence anisotropy binding assay. Alexa-546 labeled NTA bound to the complex with a KD of 0.98 ± 0.09 μM (Fig. 3C). Neither CK-666 nor CK-869 had a significant effect on the binding of Alexa-546-NTA (KD = 1.2 ± 0.1 and 0.8 ± 0.1, respectively), however the anisotropy saturates at a lower value in the presence of CK-869, indicating the complex could adopt a different conformation. Together, these data show that neither inhibitor has a significant influence at either NPF site, and that neither inhibitor functions by blocking activator binding. Actin polymerization assays support this conclusion since high concentrations of VCA could not overcome inhibition by CK-666 and CK-869 (Fig. 3D).
Fig. 3.
Neither inhibitor significantly influences NPF binding or interactions with actin. (A) Hypothetical model of two CAs binding to Arp2/3 complex in the inactive conformation (1K8K.pdb). Yellow stars indicate relative position of T464C-B4M label in crosslinking assays. (B) Photoactivatable crosslinking assay in which BtArp2/3 complex (2.5 μM) and 8 μM N-WASP-VCA-T464C-B4M (B4M-VCA) were cross linked in the presence of 200 μM CK-666, 200 μM CK-869, 80 μM cortactin-NTA (NTA), or 20 μM Crn1-UCC (UCC). Double and single asterisks indicate Arp3-VCA and Arp2-VCA crosslinked adducts, respectively. (C) Binding isotherm showing fluorescence anisotropy of 100 nM Alexa-546-cortactin-NTA titrated with BtArp2/3 complex. Addition of 200 μM CK-666 or 150 μM CK-869 did not affect the binding affinity. (D) Plot of maximum polymerization rate of 3 μM 15 % pyrene-labeled actin with 20 nM BtArp2/3 complex versus N-WASP-VCA concentration in the presence and absence of 200 μM inhibitors. (E) Copelleting assay in which BtArp2/3 complex was copelleted with 0-20 μM actin filaments and DMSO or 200 μM CK-666 or CK-869. (F) Plot of initial polymerization rate versus concentration of BtArp2/3 complex for a reaction containing 20 nM gelsolin-capped actin filament seeds, 2 μM 15% pyrene labeled actin, and DMSO or 200 μM inhibitor. (G) TIRF debranching reactions in which Oregon green-labeled actin was polymerized in the presence of 20 nM BtArp2/3 complex and 100 nM GST-VCA for 6 min before flushing the chamber with buffer (t=0) containing either DMSO or 200 μM of inhibitor. Red arrowheads indicate debranching events. Yellow arrowheads indicate stable branches. (H) Quantification of debranching for DMSO and inhibitor-containing reactions. Fraction of surviving branches was calculated from at least two separate movies, with n= 47, 33, and 20 for the DMSO, CK-666, and CK-869 conditions, respectively.
Inhibitors do not influence interactions with sides or ends of filaments or disassemble preformed branches
Branching nucleation requires binding of the complex to the sides of filaments (Achard et al.; Goley et al., 2010; Machesky et al., 1999), so we measured the influence of the inhibitors on copelleting of the complex with preformed actin filaments. BtArp2/3 complex bound to actin filaments with a KD = 0.9 ± 0.3 μM (Fig. 3E). CK-666 and CK-869 each increased the KD to 2 ± 1 μM, which is within the range of affinities measured for uninhibited Arp2/3 complex from a range of species (Beltzner and Pollard, 2007; Goley et al., 2010). Therefore, defects in actin filament side binding cannot account for inhibition.
Arp2/3 complex also binds to the pointed ends of filaments, and binding has been hypothesized to require adoption of the short pitch dimer (Dayel and Mullins, 2004; LeClaire et al., 2008; Rouiller et al., 2008). We measured pointed end capping by titrating gelsolin-capped actin filaments with increasing concentrations of the complex and measuring polymerization from free pointed ends. The apparent KD of Arp2/3 complex was 1.3 ± 0.2 μM without inhibitors, and 0.6 ± 0.1 μM and 0.5 ± 0.05 μM in the presence of 200 μM CK-666 and CK-869, respectively (Fig. 3F). Therefore, the inhibitors do not block pointed end binding. To determine if the inhibitors influence interactions of the complex with actin filaments in the context of a branch junction, we used TIRF microscopy to monitor branch dissociation in the presence and absence of the inhibitors. Branches were formed in the absence of inhibitor then washed with a buffer containing inhibitor or DMSO as a control. Time-lapse movies show that neither inhibitor increased the rate of branch dissociation over the timescale measured (Fig. 3G,H). This suggests that neither inhibitor actively disassembles branches.
CK-869 and CK-666 block a conformational change caused by VCA-recruited actin monomers binding to Arp2/3 complex
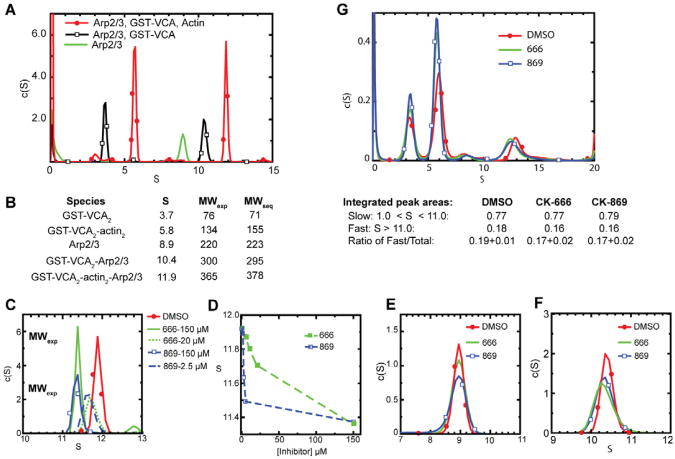
Because neither inhibitor affected the interaction of Arp2/3 complex with actin filaments, NPFs, or ATP, we next asked if the inhibitors influence actin monomer recruitment. Padrick, et. al., recently used sedimentation velocity analytical ultracentrifugation (AUC) to demonstrate two VCA peptides, each with an actin monomer bound, can simultaneously bind to Arp2/3 complex to form a stable 2:2:1 VCA:actin:Arp2/3 complex (Padrick et al.). We repeated this experiment using GST-dimerized VCA, and showed that the sedimentation coefficient distribution profiles of a mixture of BtArp2/3 complex with excess GST-VCA and latrunculin B-bound actin yielded peaks at 5.8 S and 11.9 S. The experimentally determined molecular weights of these peaks are consistent with a 2:2 GST-VCA2:actin2 complex and a 2:2:1 actin2:GST-VCA2:Arp2/3 complex (Fig. 4A,B). This demonstrates that GST-VCA can simultaneously recruit two actin monomers to the complex. We found that saturating concentrations of inhibitors caused significant differences in sedimentation of the 2:2:1 assembly, with the peak shifting from 11.9 S to 11.4 S in the presence of CK-666 or CK-869 (Fig. 4C-F). Sub-saturating concentrations of each inhibitor resulted in broader peaks with intermediate S- values, indicating rapid conversion between fast and slower sedimenting species (Fig. 4C). Neither inhibitor influenced the sedimentation profiles of Arp2/3 complex alone, and both caused only small changes in the S values of Arp2/3 complex with GST-VCA bound (Fig. 4E,F).
Fig. 4.
CK-666 and CK-869 significantly decrease the sedimentation rate of the GST-VCA2:actin2:Arp2/3 complex in sedimentation velocity analytical ultracentrifugation experiments. (A) Sedimentation coefficient distribution for three sedimentation velocity runs containing 1 μM BtArp2/3 complex with or without 10 μM GST-VCA or 10 μM GST-VCA plus 10 μM actin with 20 μM latrunculin B. (B) Tabulation of sedimentation coefficients and molecular weights (exp = experimentally calculated; seq = calculated from sequence) for peaks observed in experiments in (A). (C, E-F). Sedimentation coefficient distribution of 1 μM BtArp2/3 complex alone (E), with 10μM GST-VCA (F), or with 10 μM GST-VCA, 10 μM actin and 20 μM latrunculin (C) in the presence or absence of CK-666 or CK-869. (D) Plot of sedimentation coefficient versus inhibitor concentration for a range of concentrations of CK-666 or CK-869. (G) Sedimentation coefficient distributions for three sedimentation velocity runs monitored by absorbance at 491 nm containing 1 μM BtArp2/3 complex, 10 μM GST-VCA, 10 μM 70% labeled OG-actin, 20 μM latrunculin B with or without 150 μM CK-666 or CK-869. Integrated peak areas averaged from three separate runs are reported, along with the ratio of the fast sedimenting species compared to the total.
The decreased sedimentation coefficient of the fast pelleting species could be due to dissociation of components of the 2:2:1 assembly or a large conformational change caused by the inhibitors. The reduction in S is not large enough to be consistent with dissociation of either the entire GST-VCA2:actin2 hetero-tetramer or two actin monomers from the assembly, so we reasoned that the inhibitors might cause dissociation of a single actin monomer. To test this directly, we repeated the sedimentation velocity experiments using Oregon green-labeled actin to track its sedimentation. If the inhibitors cause one actin monomer to dissociate, the integrated area of the rapidly sedimenting peak will decrease by one half. Instead, we observed that area of the peak does not change in the presence of the inhibitors (Fig. 4G). In addition, the frictional coefficient ratios of the 2:2:1 assemblies increased from 1.6 to 2.1 or 2.0 in the presence of CK-666 or CK-869, respectively, indicating the inhibitor-bound assembly adopts a less spherical conformation. Using the frictional coefficients and the S values, we calculated the experimental mass of the assemblies in the presence of the inhibitors, and found that they were also consistent with a 2:2:1 assembly (Fig 4C). These observations indicate that inhibitors do not cause actin monomers to dissociate from the 2:2:1 assembly, but instead cause a large conformational change that slows its sedimentation.
Developing a biochemical assay to directly probe for the short pitch conformation
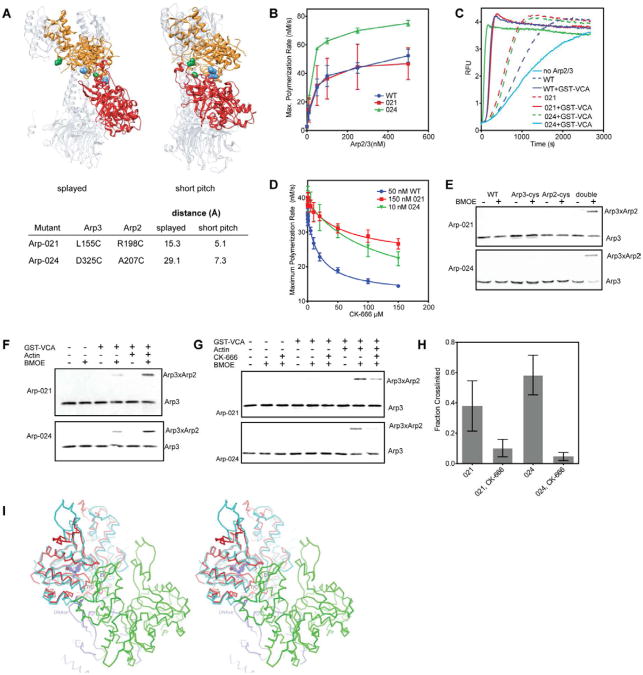
Our AUC data suggests that GST-VCA and actin monomers favor a conformation of the complex that is blocked by the inhibitors. While no high-resolution structures of the 2:2:1 assembly are available, low resolution EM reconstructions of Arp2/3 complex with bound NPFs suggest that binding of N-WASp alone stimulates formation of the short pitch Arp2-Arp3 dimer (Xu et al., 2011). Therefore, we hypothesized that the 2:2:1 assembly may adopt the short pitch conformation in the absence of inhibitors, and that this conformation is blocked when inhibitors are bound. To test this hypothesis, we developed a biochemical assay to directly detect the splayed (inactive) to short pitch (filament-like) structural change. We engineered cysteines in budding yeast Arp2/3 complex to create a crosslinking probe sensitive to the relative position of Arp2 and Arp3. We created two mutant complexes, Arp-021 and Arp-024, in which cysteine pairs on Arp2 and Arp3 are predicted to be within crosslinking distance in the short pitch Arp2-Arp3 dimer, but not in the splayed conformation (Fig. 5A) (Robinson et al., 2001; Rouiller et al., 2008). While we observed some variability in preparations, Arp-021 showed similar activity to the wild type complex in pyrene actin polymerization assays (Fig. 5B,C). In contrast, Arp-024 consistently showed increased activity compared to wild type. Both mutant complexes were inhibited by CK-666, though inhibition was weaker than in wild type complex (Fig. 5D). We are currently dissecting the molecular basis for the differences in activity and in susceptibility to CK-666. We note that because both complexes are active, stimulated by NPF (Fig. 5C), and inhibited by CK-666, they can be used to probe for inhibition of NPF-induced activating conformational change by the inhibitor.
Fig. 5.
CK-666 blocks formation of the short pitch Arp2-Arp3 dimer. (A) Structures of inactive (“splayed”, 2P9K) and active (“short-pitch”)(Rouiller et al., 2008) Arp2/3 complex showing relative orientation of Arp3 (orange) and Arp2 (red). Distance between engineered cysteine residues in mutants Arp-021 (green residues) and Arp-024 (blue atoms) in each conformation is indicated in table. (B) Time courses of actin polymerization containing 3 μM 15% pyrene actin and 150 nM GST-VCA with a range of concentrations of wild type or mutant Arp2/3 complex. Error bars are standard deviations from three separate experiments using either two (Arp-024, wt) or three (Arp-021) different preparations of Arp2/3 complex. (C) Time courses of actin polymerization showing activation of mutant and wild type complexes by 150 nM GST-VCA. Concentrations of complex were adjusted to give similar maximum polymerization rates. (wt: 50 nM, Arp-021: 150 nM, Arp-024: 10nM). The “no Arp” reaction contained 150 nM GST-VCA but no Arp2/3 complex. (D) Plot of maximum polymerization rate versus concentration of CK-666. Reactions contained 150 nM GST-VCA plus inhibitor and were otherwise identical to conditions in panel C. IC50 values were as follows: wild type, 20 ± 2 μM; Arp-021, 63 ± 23 μM; Arp-024, 110 ± 35 μM. (E) Both cysteines are required (double) for crosslinked product to form. Anti-Arp3 western blot of crosslinking reactions containing either double cysteine mutant Arp-021 or Arp-024, or the single mutant version of each, plus 10 μM GST-VCA and 10 μM Latrunculin-B actin. (F) Anti-Arp3 western blots of crosslinking reactions containing 1 μM Arp2/3 complex, 100 μM BMOE, 10 μM GST-VCA and 10 μM Latrunculin-B actin, as indicated. (G) Anti-Arp3 western blot of crosslinking reactions containing 10 μM GST-VCA, 10 μM Latrunculin-B actin, and CK-666 as indicated. (H) Quantification of CK-666 inhibition of the short pitch crosslink. The fraction crosslinked was calculated by measuring the ratio of crosslinked to uncrosslinked Arp3. Error bars represent standard error from three replicates. (I) Model showing structural basis for inhibition of Arp2/3 complex by CK-869. Stereo view diagram showing Arp3 (red) from the CK-869-bound Arp2/3 complex structure superposed with an actin subunit (cyan) from a cryo EM actin filament structure (3G37.pdb). CK-869 is shown as ball-and-sticks with purple carbon atoms. “S” marks the sensor loop and “7/C” marks the β7/αC loop in Arp3. “E/F” marks the αE/αF loop in the green actin subunit in the short pitch position relative to Arp3. See also Figure S3.
In the presence of GST-VCA and latrunculin B-bound actin monomers, both double cysteine mutants produced a high molecular weight band reactive to an anti-Arp3 antibody when treated with bis(maleimido)ethane (BMOE), an 8 Å sulfhydryl cross linking reagent (Fig. 5E). The cross linked product also reacted with an Arp2 antibody (Fig. S3A) and did not form in the single cysteine mutant controls indicating that the cross link is between the engineered cysteines. Small molecule CK-869 does not inhibit the budding yeast Arp2/3 complex and did not decrease formation of the Arp3-Arp2 crosslink, showing there is a correlation between inhibition and the reduction in crosslinking (Fig. S3B). Therefore, both Arp-021 and Arp-024 Arp2/3 complexes can be used to probe for the short pitch conformation.
Actin monomers directly contribute to formation of the short pitch conformation
Binding of VCA alone causes conformational changes in Arp2/3 complex that have been investigated by FRET and two-and three-dimensional electron microscopy reconstructions, but less is known about the influence of VCA bound to actin monomers (Boczkowska et al., 2008; Goley et al., 2004; Rodal et al., 2005; Xu et al., 2011). Therefore, to better understand how the inhibitors influence the conformation of the 2:2:1 assembly, we first used the cysteine double mutants to test the relative contribution of actin monomers and GST-VCA to formation of the short pitch conformation. Unexpectedly, we found that a reaction containing GST-VCA but not actin monomers had decreased crosslinking compared to a reaction containing both actin monomers and GST-VCA (Fig. 5F). These data demonstrate that actin monomers recruited by VCA play an active role in favoring the short pitch conformational change. In some reactions, we observed crosslinking even in the absence of activators, though it was always significantly less than with VCA or actin monomers and VCA (Fig. S3C). This observation is consistent with the reported levels of constitutive activity in budding yeast Arp2/3 complex (Wen and Rubenstein, 2005), and with single particle EM studies that show a significant fraction of budding yeast Arp2/3 complex adopts a closed (and presumably active) conformation in the absence of NPFs (Rodal et al., 2005). Together, our results demonstrate the short pitch conformation is sampled even without activators, but that GST-VCA and actin monomers cooperate to skew the conformational equilibrium toward the short pitch conformation.
CK-869 and CK-666 block formation of the short pitch dimer
We next tested the effect of CK-666 on crosslinking. CK-666 at 200 μM eliminated the Arp2-Arp3 crosslink formed in the absence of activating factors, and reduced the amount of the cross linked band formed in the presence GST-VCA or GST-VCA and actin (Fig. 5G,H). These data demonstrate that CK-666 blocks formation of the short pitch conformation, explaining how it inhibits nucleation by Arp2/3 complex. Because crosslinking is blocked both in the presence or absence of activators (Fig. S3C), we conclude that the effect of CK-666 is not limited to the 2:2:1 assembly, but that it also influences the conformational equilibria of Arp2/3 alone or Arp2/3 complex bound to GST-VCA.
The same crosslinking assay could not be used to directly test if CK-869 affects formation of the short pitch dimer because CK-869 does not inhibit ScArp2/3 complex (Nolen et al., 2009). However, the crosslinking data for CK-666 indicate that conformational differences in the 2:2:1 assembly detected by AUC are caused by a failure to adopt the short pitch conformation. CK-869 had a similar influence on the complex as CK-666 in the AUC experiments, suggesting CK-869 may also prevent formation of the short pitch dimer. We used the structure of the complex with CK-869 and ATP bound to investigate this hypothesis. Overlaying the structure onto Arp3 from the electron tomography branch junction model showed that CK-869-induced changes in Arp3 did not affect the short pitch interface. However, in this model the centers of mass of Arp2 and Arp3 are 46.8 Å apart and share only ∼25 Å2 of buried surface area (Rouiller et al., 2008). In contrast, in the recently published ∼8 Å cryo-EM structure of an actin filament, the centers of mass of two short pitch actin monomers are separated by only 40.6 Å (Murakami et al., 2010) and share 817 Å2 of buried surface area. In this structure, the β7/αC loop and the sensor loop contact the αE/αF loop from the actin monomer in the short pitch position (Fig. 5I). These contacts require the β7/αC loop to bend downward toward the barbed end relative to its position in actin monomer structures (Murakami et al., 2010). When we superimposed Arp2 and CK-869 bound Arp3 onto this structure, we found that the β7/αC loop in Arp3 sterically clashes with the αE/αF loop in Arp2 (Fig. 5I). Together with the AUC data, these models suggest that the conformational changes in the sensor and β7/αC loop caused by CK-869 may disrupt the short pitch interface of Arp2 and Arp3. Therefore, our data are consistent with a model in which CK-869 inhibits the Arp2/3 complex by destabilizing the short pitch conformation.
Discussion
CK-666 and CK-869 use distinct structural mechanisms to inhibit Arp2/3 complex: implications for inhibiting protein-protein interactions
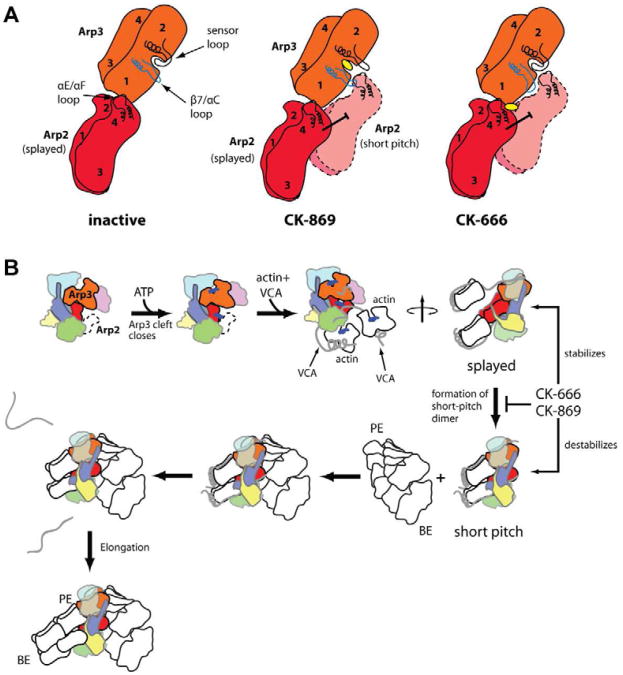
Here we demonstrate that CK-666 inhibits Arp2/3 complex by blocking formation of the short pitch Arp2-Arp3 dimer. The structural basis for this conformational control by CK-666 is evident in x-ray crystal structures, which showed that CK-666 stabilizes the splayed conformation by contacting residues from Arp3 and Arp2 that are aligned only in the inactive state (Baggett et al., 2012). Our data show that this stabilization is sufficient to block formation of the short pitch dimer, thereby inhibiting the complex. Binding of CK-666 requires only minor side chain movements, so CK-666 inhibits by trapping a native inactive conformation of the complex. Analogous trapping mechanisms are commonly used by naturally occurring allosteric inhibitors, but there are relatively few examples of synthetic compounds that exploit serendipitous allosteric sites for conformational trapping (Hardy and Wells, 2004; Lee and Craik, 2009). These results raise the intriguing possibility that other serendipitous sites on Arp2/3 complex might be exploited to lock it into the short pitch conformation, providing a new tool for investigating both the mechanistic details of activation and in vivo function of the complex.
Our data suggest that CK-869 also blocks formation of the short pitch dimer. While CK-666 accomplishes this by stabilizing the splayed conformation, CK-869 appears to destabilize the short pitch interface of Arp2 and Arp3. Interestingly, it does not bind to and directly block one side of this protein-protein interface (PPI), the typical mode of action for PPI inhibitors (Wilson, 2009). Instead, it indirectly disrupts two key loops at the interface by binding to a pocket near the PPI that normally anchors one of the loops (Fig. 6A). These results highlight the importance of using computational methods that incorporate loop flexibility into computational screening (B-Rao et al., 2009), even in cases where surface loops are well-ordered and appear to be locked into place in crystal structures, as observed for the sensor loop in Arp3 structures without CK-869 (Nolen and Pollard, 2007). Uncovering such serendipitous pockets near other important PPIs could provide a general way to circumvent the problems inherent in directly targeting the extensive and relatively flat surfaces typical of PPIs (Wells and McClendon, 2007).
Fig. 6.
Cartoon showing proposed structural bases for inhibition of Arp2/3 complex by CK-869 and CK-666. In the inhibitor-free inactive state the sensor loop is closed over the CK-869 binding pocket and the β7/αC loop of Arp3 is in the same conformation observed in crystal structures of monomeric actin (left panel). CK-869 binding locks the sensor loop in an open conformation and the β7/αC loop moves toward subdomain 2 (middle panel). This structural change destabilizes the Arp3-Arp2 short pitch interface because the β7/αC loop of Arp3 clashes with the αE/αF loop of Arp2 in the short pitch position. In contrast, CK-666 stabilizes the splayed conformation of Arp2/3 complex by binding to the interface of Arp2 and Arp3 in the inactive conformation (right panel). B. Simplified potential reaction pathway for Arp2/3 complex activation. Steps affected by CK-666 and CK-869 and highlighted and the hypothesized conformational state of the complex at each step is depicted. ATP binding causes cleft closure in Arp3 (Nolen et al., 2004) and VCA binding causes a compaction or closure of the entire complex (Goley et al., 2004; Rodal et al., 2005), which we depict here as a reordering and closure of subdomains 1 and 2 of Arp2 from the disordered (dotted line) state observed in inactive Arp2/3 complex crystal structures. The switch from the splayed to short pitch conformation involves a ∼25 Å movement of Arp2 which exposes the barbed end of Arp3 for interaction with a VCA-tethered actin monomer. This conformational change is stimulated by VCA-recruited actin monomers and blocked by both CK-666 and CK-869.
Implications for understanding the mechanism of branching nucleation by Arp2/3 complex
While actin filaments, actin monomers, ATP, and NPFs are all required for activation, how biochemical states of the complex are related to its conformation is still poorly understood. Our data provides several important new insights into the relationship between activation and conformation.
First, binding of VCA alone is not sufficient to lock the complex into the short pitch dimer conformation. Crosslinking assays showed that NPF alone does not completely shift the equilibrium to the short-pitch state, since actin monomers were required to maximize formation of cross linked Arp2-Arp3 dimer. The extent to which an NPF alone can promote the conformational change may depend on multiple factors, including whether it engages one or both sites and on the species of Arp2/3 complex. For instance, our crosslinking data demonstrated that GST-VCA increased the population of the short pitch state over no NPF in the budding yeast complex. In contrast, the fluorescence anisotropy binding data show binding of monomeric VCA to the bovine complex is not influenced by the inhibitors, so VCA cannot cause the short pitch dimer conformation to be significantly populated, since this would result in thermodynamic micro irreversibility. A precise understanding of the relationship between NPF binding and the conformation of the complex will be critical for understanding the fundamental mechanisms of regulation of the complex.
Second, our crosslinking data demonstrate that actin monomers stimulate formation of the short pitch Arp2-Arp3 dimer. While previous data showed the V region of VCA is required for activation of Arp2/3 complex by WASp/Scar family proteins (Marchand et al., 2001), whether actin monomers are recruited to the complex solely to bypass slow formation of an Arp2-Arp3-actin hetero-oligomers or if they play a direct role in activating the complex has been uncertain. Small angle x-ray scattering data showed that VCA recruits the first actin monomer to the barbed end of Arp2 (Boczkowska et al., 2008). Steric clash prevents the second actin from being delivered to the barbed end of Arp3 when the complex is in the splayed conformation (Fig. 6B) (Padrick et al., 2011). In contrast, the barbed end of Arp3 is exposed in the short pitch conformation, leading to the hypothesis that delivery of actin to Arp3 and binding of CA cooperatively induce the short pitch conformation (Boczkowska et al., 2008; Padrick et al., 2011). Our data strongly support this model, and the crosslinking assay reported here will allow us to directly test requirements for forming the short pitch dimer, including recruitment of single versus two VCA-actins to the complex.
Third, our data demonstrate that binding of the complex to actin filaments is not required to stimulate the short pitch conformation. Actin filaments are required for Arp2/3 complex to nucleate a filament (Achard et al., 2010; Higgs et al., 1999), so our data suggest that formation of the short pitch dimer is not sufficient for nucleation. One possibility is that the V region of VCA blocks the barbed end of actin subunits in the 2:2:1 assembly (Chereau et al., 2005; Padrick et al., 2011), and that binding of the assembly to actin filaments is required for release of VCA (Fig. 6B). While this model is consistent with experiments that suggest that VCA is released from branch junctions (Egile et al., 2005; Martin et al., 2006), additional experiments will be required to test this model.
One unexpected result of this work is that neither inhibitor influenced binding of the Arp2/3 complex to the pointed end of preformed actin filaments. Pointed end binding has been hypothesized to shift the conformational equilibrium of the complex to favor the short pitch dimer (Dayel and Mullins, 2004; LeClaire et al., 2008). Our data suggest that the complex remains splayed when bound to the pointed end. Because the barbed end of Arp3 is blocked in the splayed conformation, we speculate that the barbed end of Arp2 mediates interactions with the pointed end, and that this interaction is sufficient to shift the nucleotide cleft of Arp2 into a hydrolysis-competent conformation (Dayel and Mullins, 2004). We note that the failure of Arp2/3 complex to adopt the short pitch conformation at the pointed end is consistent with a recent high resolution cryo electron microscopy structure, which shows that the terminal pointed end actin subunit is titled and does not form the canonical short pitch dimer with the penultimate actin subunit (Narita et al., 2011).
Implications for understanding influence of inhibitors on in vivo branched actin networks
The molecular mechanism of each inhibitor has important implications for interpreting its influence on branched actin networks in vivo. For instance, we have shown that the inhibitors do not stimulate dissociation of preformed branches in vitro. Therefore, the rate of disassembly of in vivo actin networks upon treatment with the inhibitors reflects the rate of turnover of Arp2/3-branched networks in the absence of inhibitors. Second, we showed that the inhibitors block the nucleation activity of the complex without altering its other biochemical activities. This functional specificity has advantages over genetic ablation methods, considering that knock out or knock downs of Arp2/3 complex subunits result in destabilization and often complete loss of the complex (Gournier et al., 2001). For example, neither inhibitor significantly influenced NPF binding, indicating that the inhibitors will not affect NPF-dependent localization of the complex in vivo. Therefore, both inhibitors will be useful in investigating the recruitment of Arp2/3 complex to sites of actin network initiation in the absence of branching nucleation.
Significance
Here we use biochemical and structural methods to dissect the mechanism of Arp2/3 complex by small molecules CK-666 and CK-869. This work provides several important insights into the relationship between conformation and activity of the Arp2/3 complex. We show that actin monomers recruited by WASp-VCA stimulate the short-pitch conformation of the complex without requiring binding of the complex to the sides of actin filaments, as previously posited. Future mechanistic studies will be aimed at how binding to actin filaments is coupled to activation so that the complex creates only branched actin filament networks. By demonstrating that the inhibitors do not influence the interaction of the complex with NPFs, ATP or actin filaments, nor cause active disassembly of preformed branches, we provide a mechanistic framework for understanding the influence of the inhibitors on actin networks in vivo. Finally, by dissecting the structural/biochemical mechanisms of two distinct inhibitors of the Arp2/3 complex, this work provides new conceptual bases for understanding how to inhibit protein-protein interaction in macromolecular assemblies.
Experimental Procedures
X-ray Crystallography
Crystals of Bos taurus Arp2/3 complex were grown by hanging drop vapor diffusion as previously described (Nolen et al., 2004). Crystals were transferred to soaking solution containing 18% PEG 8000, 50 mM Hepes pH 7.5, 100 mM potassium thiocyanate, 20 % glycerol and either 0.5 mM CK-666, 0.5 mM CK-869, or 0.5 mM CK-869 plus 2 mM ATP and 2 mM CaCl2 and soaked at 4 °C for 16 hrs. Data was collected at beamline 5.0.1 or 4.2.2 at the Advanced Light Source in Berkeley, CA. Phases were solved by molecular replacement using the apo-Arp2/3 complex as a starting model (1K8K.pdb) and the structures were refined using crystallography and NMR system (Brunger et al., 1998) with inhibitor parameter files generated using the prodrg server (Schuettelkopf and van Aalten, 2004). Structures were deposited in the protein data bank with accession codes 3UKU and 3ULE.
Analytical Ultracentrifugation
Sedimentation velocity experiments were carried out in a Beckman XL-I analytical ultracentrifuge. Samples were prepared with a final concentration of 1 μM BtArp2/3 complex, 10 μM GST-VCA, and 10 μM actin (or 10 μM 70 % Oregon green actin) with 20 μM Latrunculin B in AUC buffer (5 mM HEPES pH 7.0, 50 mM KCl, 1 mM EGTA, 1 mM MgCl2 with or without 150 μM inhibitor). Latrunculin B was added to actin prior to mixing with the other protein components. Actin and the other protein components were mixed 1:1 with a final volume of 410 μL. Samples were loaded into a two-channel ultracentrifuge cell and a blank buffer consisting of the protein storage buffers, AUC buffer and latrunculin B mixed identically to the protein solutions was loaded into the blank channel. For interference experiments, sapphire windows were used and for absorbance experiments, quartz windows were used. Cells were loaded in an An-60Ti rotor and centrifuged at 50,000 RPM at 20 °C. Data were analyzed using SEDFIT using a continuous c(s) with bimodal f/f0 (Schuck, 2000). Fits were considered satisfactory if the root mean square deviation was less than 0.009 and the residuals were randomly distributed.
Dual cysteine crosslinking assays
1 μM Arp2/3 complex, 10 μM activator, 20 μM Latrunculin B and 10 μM actin (as indicated) were incubated in buffer (10 mM imidazole pH 7.0, 50 mM KCl, 10mM EGTA, 10mM MgCl2, 10 mM ATP, 10 mM CaCl2). Latrunculin B was added to actin first in order to prevent spontaneous polymerization of the actin. 100 μM BMOE was added at room temperature to initiate the crosslinking reaction. After 10 sec, 10 mM DTT was added to quench the reaction. Western blots were probed for Arp2 (antibody sc-11969) or Arp3 (antibody sc-11973). Additional experimental details can be found in the supplemental information.
Supplementary Material
Highlights.
CK-666 blocks formation of short pitch dimer by stabilizing the splayed conformation
CK-869 disrupts the Arp2-Arp3 short pitch interface required for activation
Actin monomers stimulate the short pitch conformation
Neither inhibitor actively disassembles branched actin filaments
Acknowledgments
We thank Rong Li for budding yeast strains, Matt Lord for help with budding yeast strain construction, Steve Weitzel and Pete von Hippel for assistance with AUC, and Karen Needham and Su-Ling Liu for help with reagent preparation. We are grateful to Matt Welch for providing CFP- and YFP-tagged Arp2/3 complex for FRET experiments and for comments on the manuscript. We also thank Ken Prehoda for critically reading the manuscript. We thank Dave Kovar for assistance with TIRF assays and Bruce Bowerman and Chris Doe for use of their microscope. This work was supported by a National Institutes of Health Grant RO1-GM092917 and an American Heart Association Grant 10SDG2610189 (to B.J.N). B.H. is funded by NIH F32-GM097913.
Footnotes
Author Contributions: B.H., B.J.N., and M.S.H designed the experiments. B.H., B.J.N., M.S.H, and L.A.H performed the experiments. B.H. and B.J.N wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achard V, Martiel JL, Michelot A, Guerin C, Reymann AC, Blanchoin L, Boujemaa-Paterski R. A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 2010;20:423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- B-Rao C, Subramanian J, Sharma SD. Managing protein flexibility in docking and its applications. Drug Discov Today. 2009;14:394–400. doi: 10.1016/j.drudis.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Baggett AW, Cournia Z, Han MS, Patargias G, Glass AC, Liu SY, Nolen BJ. Structural characterization and computer-aided optimization of a small-molecule inhibitor of the arp2/3 complex, a key regulator of the actin cytoskeleton. ChemMedChem. 2012;7:1286–1294. doi: 10.1002/cmdc.201200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2007;283:7135–7144. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- Boczkowska M, Rebowski G, Petoukhov MV, Hayes DB, Svergun DI, Dominguez R. X-Ray Scattering Study of Activated Arp2/3 Complex with Bound Actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Buhle EL, Jr, Walker SB, Tsong TY, Pollard TD. Kinetic evidence for a monomer activation step in actin polymerization. Biochemistry. 1983;22:2193–2202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- Dayel MJ, Holleran EA, Mullins RD. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc Natl Acad Sci U S A. 2001;98:14871–14876. doi: 10.1073/pnas.261419298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Mullins RD. Activation of Arp2/3 Complex: Addition of the First Subunit of the New Filament by a WASP Protein Triggers Rapid ATP Hydrolysis on Arp2. PLoS Biol. 2004;2:E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Rouiller I, Xu XP, Volkmann N, Li R, Hanein D. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:e383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher JF, Mauge C, Didry D, Guichard B, Renault L, Carlier MF. Interactions of Isolated C-terminal Fragments of Neural Wiskott-Aldrich Syndrome Protein (N-WASP) with Actin and Arp2/3 Complex. J Biol Chem. 2012;287:34646–34659. doi: 10.1074/jbc.M112.394361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Rammohan A, Znameroski EA, Firat-Karalar EN, Sept D, Welch MD. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci U S A. 2010;107:8159–8164. doi: 10.1073/pnas.0911668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell. 2001;8:1041–1052. doi: 10.1016/s1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr Opin Struct Biol. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Ingerman E, Hsiao JY, Mullins RD. Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J Cell Biol. 2013;200:619–633. doi: 10.1083/jcb.201211069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Didry D, Carlier MF, Pantaloni D. Activation of Arp2/3 complex by Wiskott-Aldrich Syndrome protein is linked to enhanced binding of ATP to Arp2. J Biol Chem. 2001;276:46689–46692. doi: 10.1074/jbc.C100476200. [DOI] [PubMed] [Google Scholar]
- LeClaire LL, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J Cell Biol. 2008;182:647–654. doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GM, Craik CS. Trapping moving targets with small molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Needham KM, May JR, Nolen BJ. Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J Biol Chem. 2011;286:17039–17046. doi: 10.1074/jbc.M111.219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch M, Drubin DG. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem Biophys Res Commun. 1998;243:73–78. doi: 10.1006/bbrc.1997.8064. [DOI] [PubMed] [Google Scholar]
- Murakami K, Yasunaga T, Noguchi TQ, Gomibuchi Y, Ngo KX, Uyeda TQ, Wakabayashi T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- Narita A, Oda T, Maeda Y. Structural basis for the slow dynamics of the actin filament pointed end. Embo J. 2011;30:1230–1237. doi: 10.1038/emboj.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci U S A. 2004;101:15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhbitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci U S A. 2011;108:E472–479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Marchand C. Interfacial inhibitors: targeting macromolecular complexes. Nat Rev Drug Discov. 2011;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettelkopf AW, van Aalten DMF. PRODRG- a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci U S A. 2011;108:E463–471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Wen KK, Rubenstein PA. Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require an Arp2/3-activating protein. J Biol Chem. 2005;280:24168–24174. doi: 10.1074/jbc.M502024200. [DOI] [PubMed] [Google Scholar]
- Wilson A. Inhibition of protein-protein interactions using designed molecules. Chemical Society reviews. 2009;38:3289–3300. doi: 10.1039/b807197g. [DOI] [PubMed] [Google Scholar]
- Xu XP, Rouiller I, Slaughter BD, Egile C, Kim E, Unruh JR, Fan X, Pollard TD, Li R, Hanein D, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. Embo J. 2011 doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.