Abstract
Lymphocyte activation is initiated by a global increase in mRNA synthesis. However, the mechanisms driving transcriptome amplification during the immune response are unknown. By monitoring ssDNA genome-wide, we show that the genome of naïve cells is poised for rapid activation. In G0, ~90% of promoters from genes to be expressed in cycling lymphocytes are polymerase-loaded but unmelted and support only basal transcription. Furthermore, the transition from abortive to productive elongation is kinetically limiting causing polymerases to accumulate nearer transcription start sites (TSSs). Resting lymphocytes also limit expression of the TFIIH complex, including XPB and XPD helicases involved in promoter melting and open complex extension. To date, two rate-limiting steps have been shown to control global gene expression in eukaryotes: preinitiation complex assembly and polymerase pausing. Our studies identify promoter melting as a third key regulatory step and propose that this mechanism ensures a prompt lymphocyte response to invading pathogens.
INTRODUCTION
Following active proliferation during bone marrow and thymic development, B and T cells rearrange their antigen receptors and migrate to the periphery as quiescent, G0 lymphocytes. Under these conditions RNA and protein synthesis is maintained at basal levels and energy output is limited (Sprent, 1993). Upon contact with antigens in the periphery however, resting lymphocytes drastically increase mRNA production, undergo cell division, and differentiate into shortlived effector or long-lived memory cells (Rajewsky, 1996). A successful adaptive immune response thus depends at least in part on a rapid shift from basal to fully activated gene expression. However, the molecular mechanisms controlling transcriptome amplification have not been defined.
Eukaryotic gene expression is driven by a complex series of ordered events, including RNA polymerase II (PolII) recruitment, preinitiation complex assembly, open complex formation, promoter escape, pausing, elongation, and transcriptional termination (Fuda et al., 2009; Lee and Young, 2000; Orphanides and Reinberg, 2000). Until recently, it was generally believed that expression of most protein coding genes was regulated at the level of holoenzyme recruitment to promoter regions (Margaritis and Holstege, 2008; Ptashne and Gann, 1997; Roeder, 2005). Potential exceptions to this rule were the Drosophila heat shock (Gilmour and Lis, 1986) and a small number of human genes including MYC (Bentley and Groudine, 1986), which display PolII pausing downstream of the TSS. In such cases, transcriptional regulation was proposed to occur by controlling the rate of PolII pause release. Subsequent genome-wide surveys however revealed PolII accumulation at the vast majority of transcriptionally active promoters (Core et al., 2008; Kim et al., 2005; Muse et al., 2007; Rahl et al., 2010; Seila et al., 2009; Zeitlinger et al., 2007), indicating that the rate of pause release might in fact be limiting across the genome. In addition to active sites, a fraction of unexpressed genes were also associated with paused polymerases and displayed the hallmarks of transcription initiation without elongation (Bernstein et al., 2006; Guenther et al., 2007; Lee et al., 2006; Radonjic et al., 2005; Yamane et al., 2011).
The aforementioned studies have thus provided the basis for our current view that both holoenzyme recruitment and polymerase pausing are key rate-limiting steps in eukaryote gene expression. It is important to point out however that to date genome-wide transcription studies have been by and large limited to cycling cells. By means of a novel protocol that maps ssDNA across the genome, we here show that non-cycling, G0 lymphocytes use promoter melting as an additional step to globally regulate transcription in eukaryotes.
RESULTS
A proportional increase in mRNA synthesis and histone acetylation upon lymphocyte activation
To explore transcriptional regulation during lymphocyte activation we stimulated naïve, CD43− mouse splenic B cells in the presence of lipopolysaccharide and interleukin-4 (LPS+IL4) ex-vivo. These conditions promote a drastic increase in cell size, proliferation, and plasma cell terminal differentiation (Rajewsky, 1996; Shapiro-Shelef and Calame, 2005) (Figure 1A). These morphological changes are preceded in the nucleus by global chromatin decondensation, enhanced histone acetylation, and gene activation (Fisher, 2005; Melchers and Andersson, 1984; Pogo et al., 1966, 1967; Rawlings et al., 2011). In agreement with this view, we observed a nearly 10-fold increase in total RNA and mRNA production as resting B cells differentiated for 48hs in the presence of LPS+IL4 (P < 1.5e-3, Figure 1B).
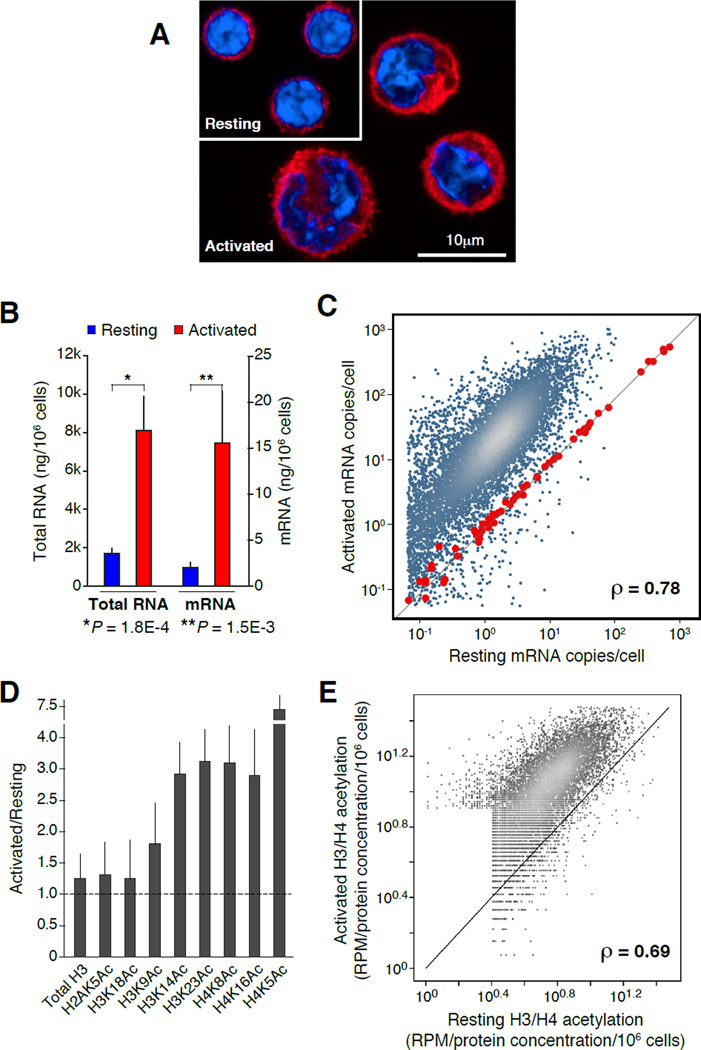
Figure 1. Proportional upregulation of mRNA synthesis and histone acetylation during B cell activation.
(A) Confocal micrograph showing representative examples of resting (G0) and activated (cycling plasmacytes) B cells. Samples were stained with anti-β-tubulin (red) and DAPI (blue). (B) Bar graph portraying total RNA (isolated with TRIzol reagent) and mRNA (isolated with oligo(dT) microbeads) from resting (blue bars) and activated (red bars) B lymphocytes (measured as ng/106 cells). Data are represented as mean +/− SEM (n = 4). (C) Comparison of resting and activated B cell transcriptomes, as determined by mRNA-Seq analysis. Values represent mRNA copies per cell, calculated based on mRNA standards (red dots) spiked-in to total RNA isolated from 106 resting or activated B cells. (D) Reverse phase protein microarray analysis of histone H3 and H4 lysine acetylation marks during B cell activation. Bars represent the ratio of total acetylation measured in activated versus naïve cells. Total histone H3 was used as “loading” control. Data are represented as mean +/− SEM (n = 4). (E) Comparison of histone H3 (K14, K23) and H4 (K5, K8, K16) acetylation in resting and activated B cells as measured by ChIP-Seq and normalized using RPMA values (panel D). Correlation in panels C and E was calculated via Spearman’s ρ.
Enhanced mRNA synthesis during activation might result from de novo transcription of a subset of genes. Alternatively, naïve lymphocytes may upregulate expression of most genes as they enter the cell cycle, a process known as transcriptome amplification (Lin et al., 2012; Loven et al., 2012; Nie et al., 2012). To directly address these possibilities we measured the absolute number of mRNA transcripts in G0 and cycling B cells by spiking-in 96 mRNA standards to total RNA isolated from 106 cells prior to transcriptome (mRNA-Seq) analysis. This normalization revealed that mRNA copy numbers of the vast majority of ORFs increased on average 11.7 fold during activation (Spearman’s ρ = 0.78, Figure 1C, and Figure S1A). Thus, rather than upregulating a specific subset of transcripts, B cell activation amplifies the entire gene expression program of G0 cells.
To determine whether transcriptome amplification correlated with epigenetic changes, we next quantified histone acetylation in resting and activated B cell populations using reverse-phase protein microarray (RPMA, (Calvo et al., 2008)). In this assay, the absolute concentration of histone modifications per million cells is calculated directly from lysates printed in microarrays. As such, protein measurements are not influenced by epitope masking or potential differences in chromatin condensation between cell types (Calvo et al., 2008). We found that LPS+IL4 activation induced a 3–7 fold increase in the total levels of acetylation (Ac) for histones H3K14, H3K23, H4K5, H4K8, and H4K16 (P < 0.003, Figure 1D). Notably, there were little or no changes in overall H2AK5Ac, H3K9Ac, or H3K18Ac under the same conditions (Figure 1D). Thus, while as a group acetylation marks are activating in nature (Wang et al., 2008; Yamane et al., 2011), lymphocyte differentiation increments global acetylation of some but not all histone lysines. Importantly, and in line with transcriptome amplification, H3K14Ac, H3K23Ac, H4K5Ac, H4K8Ac, and H4K16Ac increased proportionally during stimulation, as determined by ChIP-Seq analysis (Spearman’s ρ = 0.69, Figure 1E). Taken together, the findings show that cycling lymphocytes undergo a proportional amplification of mRNA synthesis and acetylation of specific histone lysines.
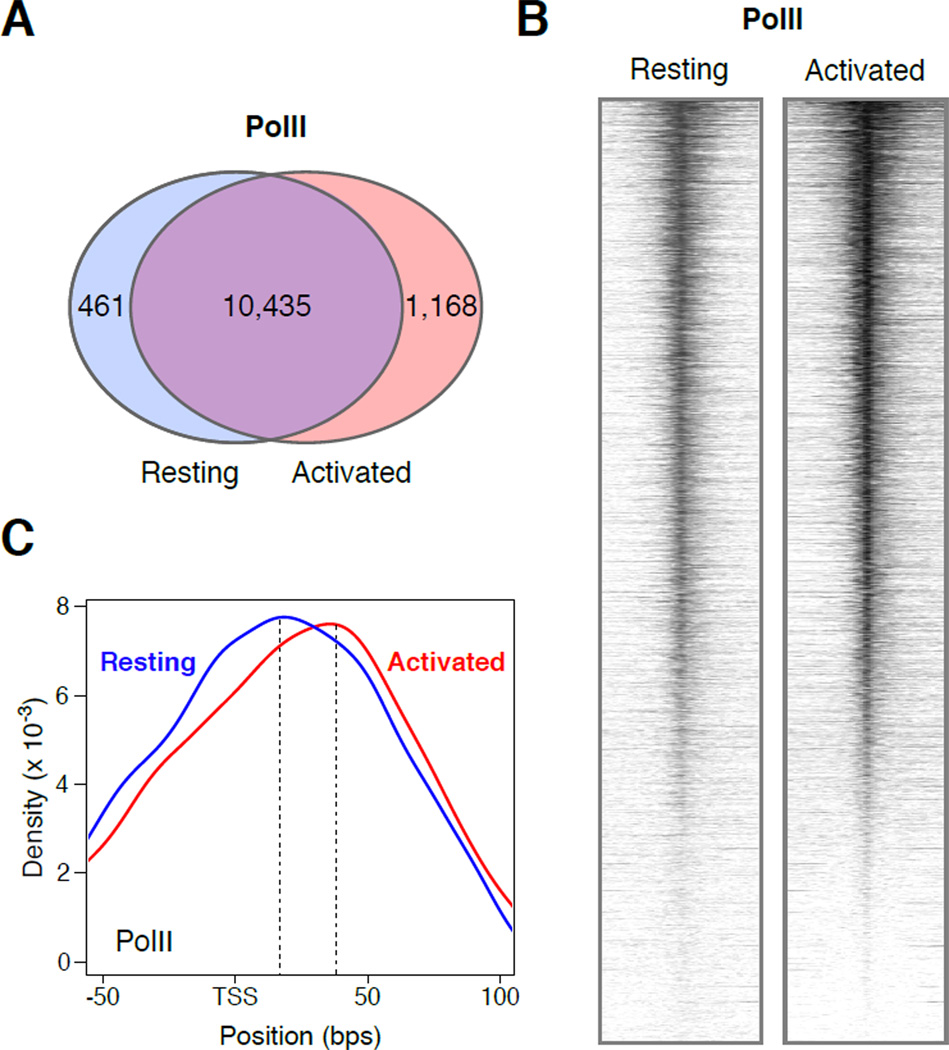
Also consistent with transcriptome amplification, the number of genes recruiting PolII was essentially unaffected during activation (Figure 2A). This result implies that few genes are transcribed de novo upon B cell differentiation. Exceptions were 461 and 1,168 genes that were preferentially expressed in resting or activated B cells (Figure 2A). Notably, although mRNA copy numbers increased on average more than 10 fold in cycling lymphocytes (Figure 1C), ChIP-Seq analysis revealed only a modest increase (1.7-fold) in PolII promoter occupancy relative to G0 (Figure 2B). Thus, the extent of transcriptome amplification during B cell activation cannot be explained on the basis of PolII promoter recruitment.
Figure 2. PolII recruitment and promoter profiles during B cell activation.
(A) Venn diagram showing the number of genes in resting (blue circle) and stimulated (red circle) B cells are associated with PolII ChIP-seq signals. (B) Heat map plots of PolII recruitment at Ensembl mouse genes (+/− 2Kb) from resting and activated B cells. (C) Density graph showing PolII occupancy near TSSs in G0 (blue line) and cycling B cells (red line). The upper value of the composite dataset is shown with a dotted line. Resting maximal PolII average density = 16bps from TSS. Activated maximal = 34bps.
To explore whether transcriptome amplification correlates with changes in promoter-proximal pausing, we next analyzed PolII composite profiles. In activated B cells, maximal polymerase density occurred ~35bps downstream of TSSs (Figure 2C), a result that is in agreement with the location of PolII pausing in mouse ES cells (Rahl et al., 2010). Notably, PolII in resting B cells displayed an average upstream shift of nearly 20 nucleotides compared to cycling counterparts (Figure 2C). This result indicates that the transition from abortive to productive elongation is kinetically limiting in G0 causing RNA polymerases to be localized nearer TSSs. Thus, basal transcription in naïve lymphocytes correlates with a delay in PolII pre-pause progression.
A genome-wide map of ssDNA
PolII promoter escape from TSSs to the pausing site is preceded by open complex formation (Margaritis and Holstege, 2008). We thus entertained the possibility that limited transcription in G0 lymphocytes might reflect inefficient promoter melting. To test this idea globally we combined chemical and enzymatic techniques to map single-stranded DNA (ssDNA) across the mammalian genome by high-throughput sequencing (ssDNA-Seq). The new approach makes use of KMnO4 treatment, which modifies ssDNA in living cells by oxidizing pyrimidine bases with a marked preference for exposed thymidine residues (Mirkovitch and Darnell, 1992). Because oxidized bases are unable to base-pair with the complementary strand, they become susceptible to mung bean nuclease cleavage (Figure 3A). The resulting double-stranded DNA ends are then 3’-tailed with terminal deoxynucleotidyl transferase (TdT) in the presence of biotinylated nucleotides. Finally, samples are sonicated to ~200bps and biotinylated DNA is streptavidin-selected prior to Illumina library preparation (Figure 3A). To avoid biotinylation of preexisting DNA breaks, we blocked samples prior to nuclease treatment by incubation with chain terminating cordycepin-5'-triphosphate and TdT (Figure 3A, and Methods). The specificity of the ssDNA-Seq protocol for melted DNA was evident from the observation that KMnO4 untreated samples yielded consistently ~20-fold less DNA than treated ones (Table S1A).
Figure 3. ssDNA-Seq approach and validation.
(A) During ssDNA-Seq ssDNA is stabilized in live cells by treatment with KMnO4, which selectively oxidizes exposed thymidines. Cordycepin and TdT are used to block pre-existing 3’ DNA free ends and ssDNA is digested with mung bean nuclease. DNA ends exposed by nuclease treatment are then biotinylated and, following sonication, streptavidin selected and deep sequenced. (B) Distribution of ssDNA signals in Raji cells. +5Kb and −5Kb represent ssDNA signals aligning within 5Kb upstream of TSS or downstream of gene stop codons respectively. Numbers in parenthesis are expected percentages if reads were randomly distributed. (C) Composite diagram showing ssDNA and PolII signals around TSSs of human genes in Raji cells. (D) Composite diagrams showing the distribution of 5’ (blue) and 3’ (red) ssDNA-Seq tags around SIDDs predicted for the mouse genome. (E) Deep-sequencing profiles at mouse SIDDs obtained with anti-PolII antibodies. (F) ssDNA-Seq analyses of relaxed (upper) or supercoiled (lower) pFLIP plasmids containing human Myc FUSE element. ssDNA-Seq 5’ and 3’ tags are represented in blue and red respectively.
We first applied ssDNA-Seq to the human Burkitt lymphoma line Raji and LPS+IL4 activated mouse B cells. Deep-sequencing generated a total of 125 million sequence reads from three independent Raji experiments (Table S1B). At this range, specific signal reached near saturation (Figure S1B). Biological replicates confirmed that ssDNA-Seq was highly reproducible (Spearman ρ > 0.9, Figure S2A). We found ssDNA island distribution biased for genic domains in Raji. Nearly 60% of ssDNA signals were directly associated with genes, with an additional 10% aligning within 5kb upstream of TSSs or downstream of stop codons (Figure 3B). The remaining 32% were aligned to intergenic domains (Figure 3B). This enrichment was higher than expected when tags were uniformly distributed (Figure 3B, values in parenthesis). Within genes, a composite analysis revealed a ssDNA peak that was maximal at TSSs and extended approximately 1.5 kb on either side (Figure 3C). This peak, which represented 12% of ssDNA signals within genes, overlapped precisely with the profile of PolII occupancy (Figure 3C).
In addition to sites of gene expression, ssDNA is expected to be associated with simple DNA repeats or AT-rich domains, which adopt thermodynamically unstable DNA conformations or non-B DNA in the presence of negative supercoiling (Kouzine et al., 2008). In spite of extensive in vitro characterization, little is known about the topology, stability and distribution of non-B DNA in living cells. To further validate the ssDNA-Seq assay, we scanned activated B cell libraries for sequence tags associated with sites of Stress-Induced DNA Duplex Destabilization (SIDD) as predicted in silico (Benham and Bi, 2004). Using a destabilization energy G(x) ≤ 3 kcal/mol, the algorithm identified a total of 1,057,816 SIDDs in the mouse genome. Similar results were obtained in Raji cells (not shown). A composite analysis of these sites revealed a remarkably well-defined ssDNA-Seq profile, with two pairs of peaks surrounding the predicted SIDDs (Figure 3D). Each peak pair, composed of 5’ (+ strand) and 3’ (− strand) sequence tags, mirrored the 200bp average size of ssDNA-Seq libraries (Figure 3A, Figure 3D, and Methods).
The extent of DNA melting at SIDDs, defined as the average distance separating the innermost boundaries of the peak pairs, was ~170bps (Figure 3D). SIDD melting was not the direct result of PolII activity because we could not detect PolII occupancy at these sites (Figure 3E). Interestingly, both the ssDNA and PolII IPs displayed a distinct depression in signal within SIDD sites. Comparable results were obtained upon immunoprecipitation of CTCF, which like PolII is not directly recruited to SIDDs (Figure S2B). This profile can be explained by non-B DNA susceptibility to sonication and T4 exonuclease activity during deep-sequencing library preparation (Figure S2C). We conclude that in addition to promoter melting, the ssDNA-Seq protocol reveals both the location and topological features of non-B DNA in mammalian cells.
As an independent confirmation of the new protocol, relaxed and supercoiled plasmids were treated with KMnO4 and DNA melting was measured by ssDNA-Seq. The plasmids carried Myc far upstream element (FUSE, (Kouzine et al., 2008)), a SIDD site that readily melts as supercoilicity at the Myc promoter builds up during transcription (Kouzine et al., 2004). The superhelical density of the topoisomers (n = 4) was monitored using agarose gels (see Methods). Deep-sequencing showed extensive melting of FUSE only in the presence of torsional stress (Figure 3F). Importantly, the melting profile of FUSE closely recapitulated that obtained from SIDD sites in vivo, with two pair of strand-specific peaks surrounding the melted domain (Figure 3F and 3D). Thus, the ssDNA-Seq protocol can detect bona-fide ssDNA with high specificity.
Melting of promoters and active enhancers in primary B cells
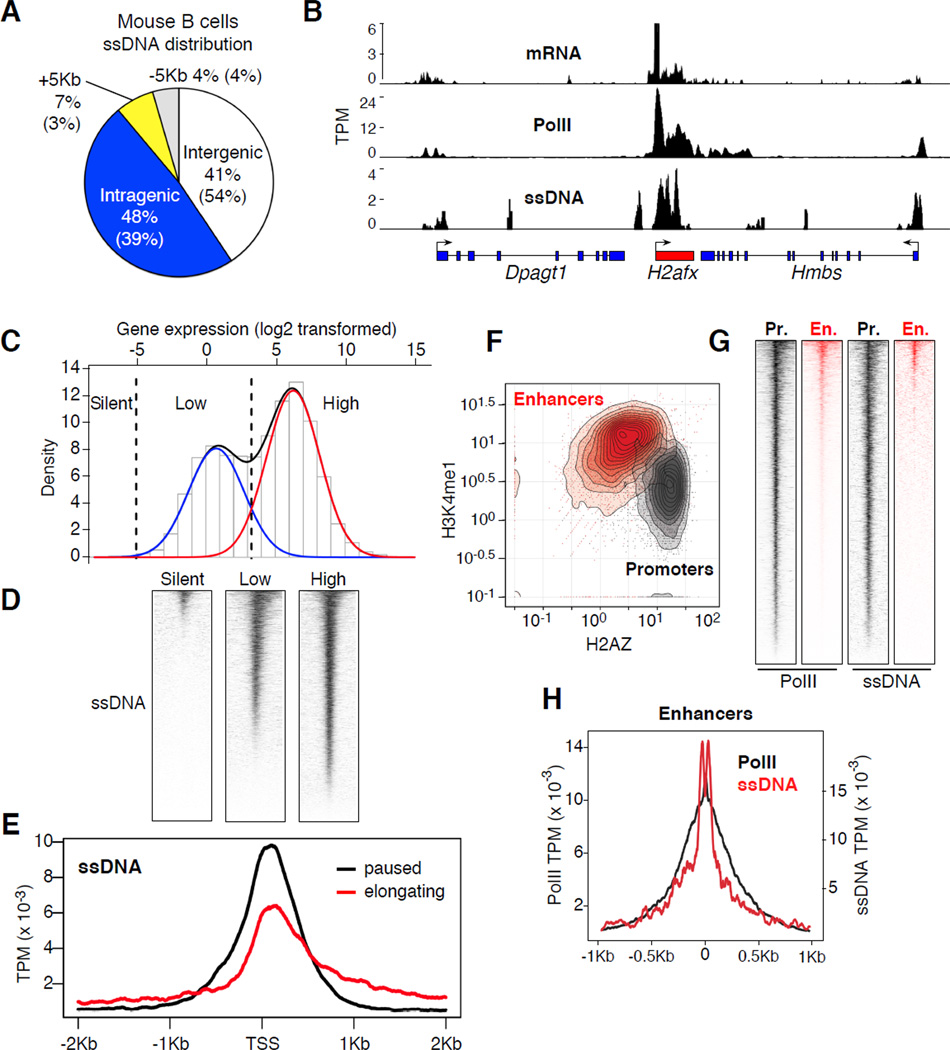
Similar to Raji, ssDNA in mouse B cells was biased for genic (+/− 5Kb) rather than intergenic domains (59% vs. 41% respectively, Figure 4A). Likewise, we found an overall correlation between ssDNA, PolII recruitment, and gene expression in primary B cells. For instance, Figure 4B shows extensive ssDNA across the highly transcribed H2afx gene, whereas the signal was less pronounced at flanking Hmbs and Dpagt1 genes, which exhibit comparatively less PolII occupancy and mRNA synthesis. To validate these observations across all genes, ssDNA was analyzed as a function of global gene expression. Based on the bimodal distribution of the activated B cell transcriptome (Figure 4C), we classified mouse genes into 3 main transcription groups: high (12,993), low (8,829), and silent (9,186). The analysis showed that the extent of ssDNA correlated with the levels of transcription in each of the groups (Figure 4D). Considering that ssDNA-Seq specifically oxidizes thymidine residues, it is important to point out that the results were not influenced by promoter CpG (or AT) contents, which in B lymphocytes are highest in low expressed genes (Figure S3A). Thus, ssDNA-Seq accurately recapitulates the location and extent of DNA melting associated with gene expression.
Figure 4. ssDNA-Seq profiles in primary B cells.
(A) Distribution of ssDNA signals in primary LPS+IL4 activated B cells. (B) Correlation between mRNA transcripts, PolII recruitment, and ssDNA at Dpagt1, H2afx, and Hmbs genes. Data represented as sequence tags per million (TPM). (C) Bimodal distribution of gene expression in activated B cells. Red and blue lines delineate first and second components respectively. Black dashed lines demarcate the threshold for highly expressed, low expressed, and silent genes. (D) Heat map of ssDNA-Seq profiles around TSSs (+/− 2Kb) for high, low, and silent gene groups defined in panel C. (E) ssDNA composite alignment at elongating (1 ≤ Pi ≤ 3, red line) and paused (Pi ≥ 10, black line) genes. (F) Density graph, p300+ islands associated with enhancers (H3K4me1highH2AZlow) or promoters (H3K4me1lowH2AZhigh). Data was normalized as fold enrichment. (G) Heat maps showing PolII and ssDNA at p300+ promoter (grey) or enhancers (red). (H) Alignment of PolII (black line, left y-axis) and ssDNA (red line, right y-axis) at p300+H3K4me1highH2AZlow enhancers.
Based on PolII binding profiles (Rahl et al., 2010), we further classified transcribed genes into paused (pausing index (Pi) > 10) and elongating (1 ≤ Pi ≤ 10). Pi is a measure of the ratio of PolII density at promoter versus gene body and directly reflects the dynamics of PolII assembly and pause release (Rahl et al., 2010). In paused genes, the rate of promoter clearance is relatively lower than that of assembly, resulting in PolII accumulation at promoter-proximal sequences (Muse et al., 2007; Zeitlinger et al., 2007). Conversely, elongating genes exhibit a higher rate of pause release and thus PolII can be readily immunoprecipitated along gene bodies (Figure S2D, (Muse et al., 2007; Zeitlinger et al., 2007)). Consistent with these dynamics, there was substantial ssDNA at promoter-proximal sequences (~1kb around TSSs) of paused genes (Figure 4E), whereas elongating genes displayed less DNA melting at promoter domains concomitant with higher melting at gene bodies (Figure 4E). These findings are thus consistent with the notion that DNA melting around TSSs is inversely proportional to the rate of PolII pause release.
In addition to promoters, PolII is recruited to a subset of active intergenic p300+ enhancers, where it has been shown to synthesize small enhancer RNAs (eRNAs, (Kim et al., 2010)). To determine whether enhancer transcription is associated with DNA melting, we compared ssDNA islands with p300 genomic occupancy in activated B cells (Kuchen et al., 2010). p300+ enhancers were distinguished from promoters by virtue of their being located distally from annotated TSSs, while being associated with high H3K4me1 (Heintzman et al., 2007; Kim et al., 2010) and low H2AZ deposition (Figure 4F and Figure S3B). Conversely, promoters were mostly p300+H3K4me1lowH2AZhigh (Figure 4F). We found PolII present at 81% of p300+ intergenic enhancer islands (3,234 of 3,994 sites) and 98% of p300+ promoter islands (4,339 of 4,392 sites, Figure 4G). By contrast, we found evidence of DNA melting in just 13% of p300+ enhancers, whereas more than 86% of p300+ promoters were associated with ssDNA (Figure 4G). This difference in DNA melting is consistent with transcriptional activity being more pronounced at promoters relative to enhancers (Kim et al., 2010). Similar to TSSs, the composite profile of ssDNA at enhancers overlapped with the peak of PolII recruitment (Figure 4H), and eRNA synthesis (Figure S4). Taken together our observations demonstrate that ssDNA-Seq reliably detects DNA melting at enhancers in a manner proportional to the extent of PolII recruitment and activity.
Limited promoter melting and TFIIH expression in G0 lymphocytes
Having validated the ssDNA-Seq approach we next analyzed promoter melting in resting splenic B cells. A total of 41 million sequence tags were obtained from two independent ssDNA-Seq experiments (Table S1B). The inter- and intra-genic distribution of ssDNA signals was similar between resting and activated B cells (Figure S5A, compare to Figure 4A). Visual inspection of the aligned data however revealed a substantial reduction in PolII-associated DNA melting across the genome of naïve cells. For instance, Figure 5A and Figure S5B show extensive ssDNA at promoters of Sfi1 and Smg9 genes in cycling cells respectively, whereas we detected little or no ssDNA in the same genes in naïve counterparts. Conversely, PolII recruitment at promoters from both genes was comparable between the two cell types (Figure 5A and Figure S5B). To extrapolate these observations to the entire genome we generated heat maps of ssDNA gene profiles from resting and activated B cells. In strike contrast to PolII recruitment which was comparable between the two cell types (Figure 2B), most promoters showed little or no melting in G0 (Figure 5B). The results demonstrate that open complex formation is limited across the genome of naïve cells. In contrast, ssDNA associated with PolI rDNA transcription, which does not directly depend on TFIIH activity for promoter melting (Assfalg et al., 2012), was readily detected in the same cells (Figure S5C). Furthermore, low levels of ssDNA in G0 cells was not due to cell cycle arrest per se because serum-starved human fibroblasts displayed similar levels of promoter melting compared to cycling counterparts (Figure S5D). In conclusion, the data argue that PolII complexes in naïve lymphocytes are found in a relatively closed configuration, whereas cell activation promotes global open complex formation and extensive gene expression.
Figure 5. Reduced promoter melting and TFIIH levels in G0.
(A) PolII and ssDNA density determined at Sfi1 gene in resting and activated B cells. Data was normalized as sequence tags per million (TPM). (B) Heat map plots of ssDNA at Ensembl mouse genes (+/− 2Kb) from activated and resting B cells. (C) Left: TFIIH subunit protein levels measured in resting [R] and 48hs LPS+IL4 activated [A] B cells by Western blot. Right: PolII subunits and the general transcription machinery. (D) XPB helicase protein levels measured in XPBYFP/YFP B cells or XPB+/+ controls. Lymphocytes were activated with LPS+IL4 for 24h and YFP expression was monitored by flow cytometry. (E) Left: Western blot measurement of PolIIS5P during activation. Gel loading was normalized per total protein content. Right: box plots represent the ratio of PolII-S5P vs. total PolII in resting (blue) and activated (red) B cells as determined by ChIP-Seq analysis. Values were normalized as TPM per total PolII-S5P and PolII per cell. Data are represented as mean +/− SEM (n = 4). (F) In vitro transcription of a linear DNA template (pG5HM(C2AT) plasmid) carrying a minimal promoter. DNA was incubated with equal amounts (80ng) of nuclear cell extracts. Reactions were complemented (+) with affinity purified TFIIH. Bar graph compares transcription levels between TFIIH-complemented reactions (+) and those without (−). Transcription in non-complemented reactions was set up to 100%. Data are represented as mean +/− SEM (n = 6).
The formation of a stable transcription bubble requires TFIIH, a 11-subunit complex that includes XPB and XPD helicases directly involved in DNA melting (Guzman and Lis, 1999; Holstege et al., 1996). Notably, expression of TFIIH subunits was substantially decreased in G0 B cells as determined by Western blot (Figure 5C). This included all members of the TFIIH core (p52, p62, XPB, and XPD) and the CAK complex (Cyclin H and CDK7). A comparison of naïve and CD3/CD28/IL-2-activated mouse T cells showed similar results (Figure S6A), indicating that TFIIH is also limiting in G0 T lymphocytes. In contrast, we found little or no differences in the expression of the basal transcription factor A (TFIIA), TATA binding protein (TBP), or total PolII during B cell activation (Figure 5C). Within the context of differentiation, TFIIH subunits were reproducibly low or undetected in resting cells (Figure S6B), even though their transcripts were moderately to highly expressed (Figure S6C). However, as early as 30’ post-activation we observed a marked increase in p62, Cyclin H, p52, and XPD transcripts and protein levels (Figure S6D and S6B respectively). At ~10h post-activation, TFIIH levels were comparable to those observed in bone marrow pro-B and pre-B cells (Figure S6B). Similar dynamics were observed by activating B lymphocytes carrying TFIIH XPB helicase fused to the yellow fluorescent protein (XPBYFP/YFP mice, (Giglia-Mari et al., 2009), Figure 5D). Based on these results we conclude that the steady-state levels of the promoter-melting complex TFIIH are substantially reduced as B cells exit the bone marrow, and rapidly restored upon activation.
In addition to promoter melting, TFIIH plays additional roles in transcription initiation, including phosphorylation of PolII C-terminal heptad repeats at serine 5 (PolII-S5P) (Feaver et al., 1994; Liao et al., 1995; Sterner et al., 1995), a feature that in turn facilitates extension of the transcription bubble from the TSS to the promoter-proximal pausing site (Dvir, 2002). Consistent with limited TFIIH expression, PolII-S5P levels in naïve B cells were ~5 times lower than in dividing counterparts, as determined both by Western blot and deep-sequencing (Wilcoxon test P < 1.0e-100, Figure 5E). This difference was readily apparent at genes programmed for abundant transcription following cellular activation (Figure S5E).
The reduction in promoter melting (Figure 5B), the low levels of PolII-S5P (Figure 5E), and the observed delay in open complex extension (Figure 2C) supports the view that TFIIH activity is limiting in G0. We then explored whether gene expression in resting cells can be complemented by addition of TFIIH. To this end we prepared nuclear cell extracts from resting and activated B cells and performed in vitro transcription of a linear DNA template containing a minimal core promoter (Ohkuma et al., 1995). Western blot analysis of the extracts confirmed little or no differences in total PolII or TBP but a substantial decrease in TFIIH p62 in G0 (not shown). Consistent with basal transcriptional activity in G0, we observed a ~20 fold difference in in vitro transcription levels between naïve and 72h–activated B cell extracts (Figure 5E). Addition of affinity purified TFIIH did not alter the transcription output of activated B cell extracts (Figure 5F). When added to G0 extracts, TFIIH consistently increased basal transcription by ~2 fold (P = 0.01, Figure 5E), however this increase was modest and never reached the levels obtained with activated B cell extracts (Figure 5E). Thus, additional activities beyond TFIIH expression are required to fully complement the G0 transcription machinery in this assay.
TTD cells show defects in promoter melting
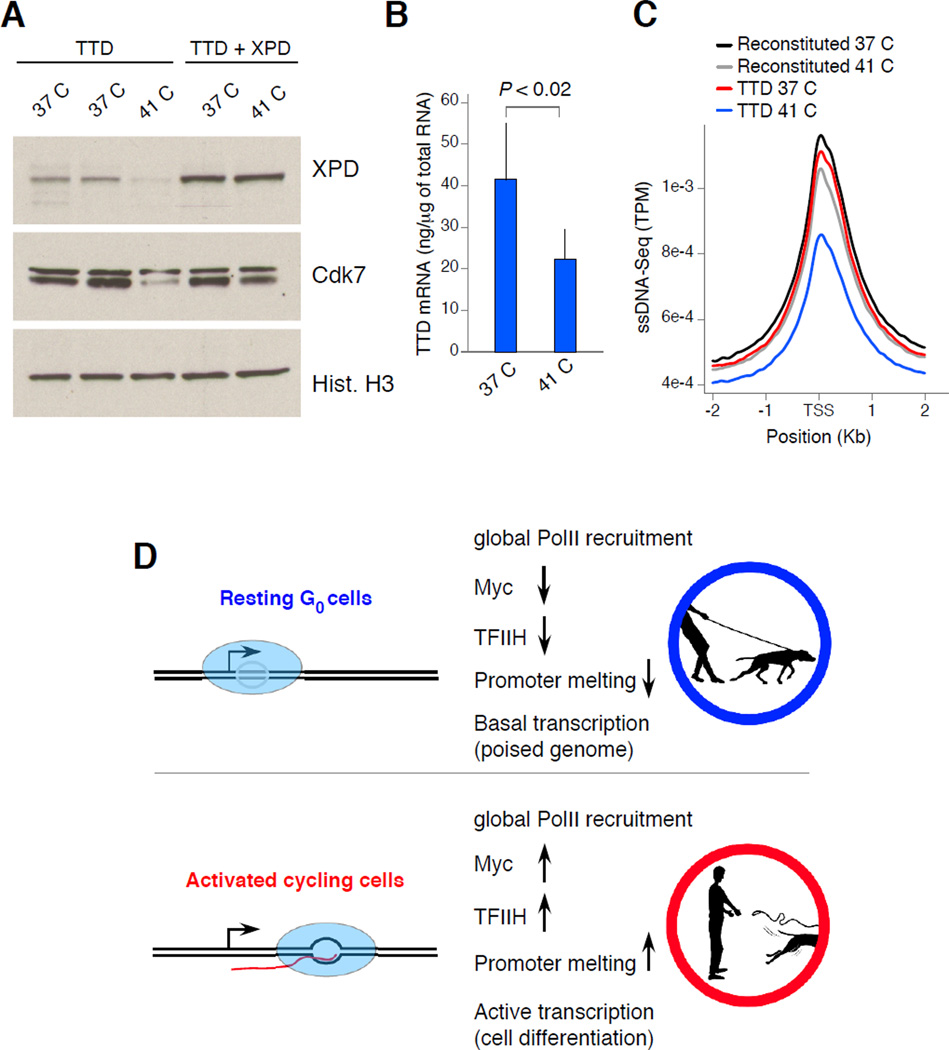
Similar to naïve lymphocytes, TFIIH expression levels are reduced in trichothiodystrophy (TTD) patient fibroblasts. The TTD syndrome is caused by mutations in TFIIH subunits, including XPB and XPD helicases, and results in ultraviolet-sensitivity, mild mental retardation, ichthyosis, and recurrent infections (Vermeulen et al., 1994). While the TTD phenotype is in part ascribed to XPD’s role in nucleotide excision repair, crippled transcription is also believed to contribute to the severity of the disease (de Boer et al., 1998; Vermeulen et al., 1994). In this context, some TTD patients carry XPD temperature sensitive mutants and display fever-dependent transcriptional defects (Vermeulen et al., 2001). In particular, fibroblasts isolated from such patients grow normally at 37°C but at higher temperatures they express low levels of TFIIH and an overall reduction in transcription (Vermeulen et al., 2001). To strengthen the link between TFIIH expression and the extent of promoter melting in vivo we analyzed TTD fibroblasts by ssDNA-Seq. Relative to 37°C, Western blot confirmed reduced expression of XPD as well as Cdk7 in TTD fibroblasts grown at 41°C but not in cells reconstituted with wild type XPD (Figure 6A). Also consistent with previous findings, TTD fibroblasts displayed reduced mRNA synthesis and defects in cell proliferation at 41°C (Figure 6B and not shown). As determined by ssDNA-Seq, these features were correlated with an overall reduction in promoter melting compared to control or TTD cells grown at 37°C (Figure 6C). The data thus demonstrate that promoter melting across the genome is tightly linked to TFIIH expression levels. Furthermore, the findings provide a mechanistic rationale for reduced mRNA synthesis and potentially the immunodeficiency observed in trichothiodystrophy syndrome patients.
Figure 6. TFIIH expression correlates with extent of transcription and promoter melting in XPD patient fibroblasts.
(A) XPD and Cdk7 protein levels in fibroblasts isolated from TTD patients carrying an XPD temperature sensitive mutant. TTD + XPD denotes fibroblasts reconstituted with wild type XPD. Cells were grown at 37°C or 41°C for at least 72 h. (B) mRNA levels in TTD fibroblasts grown at 37°C or 41°C. Data are represented as mean +/− SEM (n = 4). (C) Composite profiles of ssDNA in TTD or reconstituted fibroblasts grown at the two temperatures. (D) The findings indicate that resting lymphocytes hold tightly their transcriptome “on a leash” by allowing mostly normal recruitment and assembly of PolII, while limiting promoter melting. This scenario might be the result of low levels of TFIIH, Myc, and/or potentially additional mechanisms (upper schematics). Upon activation, extensive melting and a rapid shift from basal to full transcriptome expression correlates with stabilization of TFIIH and Myc protein levels (lower schematics). Pictures designed by Alan Hoofring from NIH Medical Arts.
DISCUSSION
Seminal work in the 1960s and 70s revealed that lymphocytes undergo a rapid transformation when exposed to mitogens, including a dramatic increase in cell size, histone acetylation, RNA synthesis, and protein production (Jaehning et al., 1975; Keller, 1975; Pogo et al., 1966, 1967). In vivo studies have since corroborated these initial observations by showing that both primary and memory responses activate G0 B and T cells in a matter of hours (Shapiro-Shelef and Calame, 2005). However, while the extracellular and intracellular signals involved have been characterized extensively, the nuclear mechanisms driving rapid activation of lymphocytes are unknown. Our findings shed new light on this issue by showing that the transcriptomes of naïve and stimulated lymphocytes are remarkably alike, in that while transcription increases ~10-fold upon activation, the number of genes and their relative expression in the two stages are largely proportional. The strong inference is that the transcriptome of naïve cells is poised, or in latent form, for rapid cellular induction.
Our studies argue that transcriptome poising in G0 cells is achieved at least in part by allowing mostly normal assembly of PolII at promoters while globally restricting open complex formation. In E. Coli, the transition from closed (RPc) to open (RPo) complex formation is facilitated by the combined action of transcriptional activators and cis-regulatory elements (Liu et al., 2004; Lutz et al., 2001). It has been proposed for some time that eukaryotes might also regulate the dynamics of transcription by controlling promoter melting. This however has been difficult to ascertain because of the challenge of quantitatively monitoring melted DNA in a large number of promoters simultaneously. Several laboratories including ours have recently used RPA- (Hakim et al., 2012; Yamane et al., 2011; Yamane et al., 2013), Rad51- (Di Virgilio et al., 2013; Khil et al., 2012; Yamane et al., 2013), and Rad52-Seq (Zhou et al., 2013) to map ssDNA in mammalian cells. These approaches however cannot detect promoter melting, but only identify sites undergoing DNA end resection during repair by homologous recombination. The ssDNA-Seq protocol now provides a means to visualize and quantify non-duplex DNA in living cells. With this new technique we were able to establish that open complex formation in eukaryotic cells can be globally regulated in response to cell cycle and environmental cues. This is in addition to holoenzyme recruitment and promoter-proximal pausing, which to date are the only other known rate-limiting steps of gene expression in higher organisms (Margaritis and Holstege, 2008).
Precisely how G0 lymphocytes inhibit open complex formation is unclear. One possibility is by controlling TFIIH protein levels, which we found to be tightly linked to the extent of promoter melting during B cell ontogeny. The role of TFIIH in promoter melting has been well established by KMnO4 footprinting analysis of several promoters (Holstege et al., 1997; Holstege et al., 1996; Kim et al., 2000), the characterization of early steps of gene expression (Guzman and Lis, 1999), and TFIIH ability to melt relaxed plasmids ex vivo (Goodrich and Tjian, 1994; Parvin and Sharp, 1993). Furthermore, we have shown that promoter melting is proportional to TFIIH expression and activity in TTD patient fibroblasts. At the same time, our in vitro transcription studies imply that the role of TFIIH in the process of transcriptome amplification is likely complemented by additional activities. Particularly, we have recently shown that the transcription factor Myc acts as a universal amplifier of transcription in B cells by associating with pre-active genes and increases overall expression (Nie et al., 2012). Similar to TFIIH, Myc protein levels peak as lymphocytes enter the cell cycle (Nie et al., 2012). We have here shown that activating histone acetylation represents yet another process that is proportionally enhanced in lymphocytes in response to mitogenic signals. Transcriptome amplification might thus result from the coordinated activity of TFIIH, general transcription factors including Myc, and epigenetic marks. We propose that within the cadre of the immune response, these processes allow quiescent lymphocytes to hold the transcriptome “on a leash” or a poised state that can be promptly reverted in the presence of pathogens (Figure 6D). Considering that the TFIIH CDK7 subunit also controls cell cycle entry by virtue of its ability to phosphorylate and activate cyclin-dependent kinases (Dvir, 2002; Klemsz et al., 1989), the model also helps explain the link between transcriptome amplification and cell cycle initiation.
In addition to the dynamics of promoter melting, the ssDNA-Seq protocol provides an unprecedented view of the location and conformation of non-B DNA. Recent computer-based thermodynamic searches have uncovered a large number of sequences across the mammalian genome that can potentially form non-B DNA in vivo (Cer et al., 2011). Intriguingly, these sequences localize preferentially to defined genomic sites and mounting evidence indicates that they may play an important physiological role. Analyses of human cells for instance have uncovered a correlation between predicted G4-quadruplexes at promoters and the extent of gene expression (Du et al., 2008). Furthermore, at skeletal muscle promoters formation of G4-quadruplexes impacts the recruitment of MyoD during differentiation (Shklover et al., 2010; Yafe et al., 2008). Similarly, transcription-induced DNA supercoiling controls the firing rate of the Myc promoter (Kouzine and Levens, 2007). From a translational point of view, DNA topology is also noteworthy because human fragile sites implicated in genomic instability and tumorigenesis often map to predicted non-B DNA structures (Barlow et al., 2013; Raghavan and Lieber, 2006). We anticipate that ssDNA-Seq will be instrumental to comprehensively study the location and dynamics of non-B DNA under physiological conditions.
EXPERIMENTAL PROCEDURES
Resting, naïve mouse B cells were isolated from splenocytes using anti-CD43 Miltenyi MicroBeads by negative selection and activated for 72hs in the presence of LPS (50µg ml−1 final concentration, E.Coli 0111:B4; Sigma) + IL4. (2.5ng ml−1 final concentration, Sigma). Apoptotic cells were removed using Miltenyi’s dead cell removal kit (#130-090–101) following by Ficoll gradient with >90% live cell purity. To minimize the influence of DNA replication on the pattern of single-stranded DNA across genome, Burkitt lymphoma Raji were arrested in G1 by treatment with 1.5% (v/v) DMSO for 96 hours as previously described (Sawai et al., 1990). To remove DMSO, cells were pelleted at 600xg for 5 min, and resuspended in fresh medium at 106 cells ml−1. Samples were processed 6 hours later when cells were synchronized at the G1 phase of the cell cycle.
Supplementary Material
Highlights.
Lymphocyte activation induces a proportional amplification of the transcriptome
ssDNA-Seq detects promoter melting and non-B DNA in living cells
Promoters in G0 lymphocytes are PolII-loaded but unmelted
TFIIH expression and activity are limited in G0 lymphocytes
ACKNOWLEDGEMENTS
We thank G. Gutierrez for technical assistance with the genome analyzer; Jim Simone and Jeff Lay for bone marrow sorts; Jason Qian and Kristina Zaal for the B cell confocal micrographs; Craig Benham for SIDD mapping. We thank the Casellas and Levens lab members for critically reading the manuscript. This work was supported in part by the Intramural Research Program of NIAMS, NCI (CCR), and NLM of the National Institutes of Health. This study utilized the high-performance computational capabilities of the Helix Systems at the National Institutes of Health, Bethesda, MD (http://helix.nih.gov).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
PolII-Seq data is under GEO under accession GSE24178, RNA-Seq is under GSE21630, and ssDNA-Seq under GSExxx.
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures, five figures, and two tables.
The authors declare no competing financial interests.
REFERENCES
- Assfalg R, Lebedev A, Gonzalez OG, Schelling A, Koch S, Iben S. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2012;40:650–659. doi: 10.1093/nar/gkr746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright GW, et al. Identification of Early Replicating Fragile Sites that Contribute to Genome Instability. Cell. 2013 doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benham CJ, Bi C. The analysis of stress-induced duplex destabilization in long genomic DNA sequences. J Comput Biol. 2004;11:519–543. doi: 10.1089/cmb.2004.11.519. [DOI] [PubMed] [Google Scholar]
- Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Calvo KR, Dabir B, Kovach A, Devor C, Bandle R, Bond A, Shih JH, Jaffe ES. IL-4 protein expression and basal activation of Erk in vivo in follicular lymphoma. Blood. 2008;112:3818–3826. doi: 10.1182/blood-2008-02-138933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cer RZ, Bruce KH, Mudunuri US, Yi M, Volfovsky N, Luke BT, Bacolla A, Collins JR, Stephens RM. Non-B DB: a database of predicted non-B DNA-forming motifs in mammalian genomes. Nucleic Acids Res. 2011;39:D383–D391. doi: 10.1093/nar/gkq1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, de Wit J, van Steeg H, Berg RJ, Morreau H, Visser P, Lehmann AR, Duran M, Hoeijmakers JH, Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang B, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. Rif1 Prevents Resection of DNA Breaks and Promotes Immunoglobulin Class Switching. Science. 2013 doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A. Promoter escape by RNA polymerase II. Biochim Biophys Acta. 2002;1577:208–223. doi: 10.1016/s0167-4781(02)00453-0. [DOI] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Fisher RP. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J Cell Sci. 2005;118:5171–5180. doi: 10.1242/jcs.02718. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Theil AF, Mari PO, Mourgues S, Nonnekens J, Andrieux LO, de Wit J, Miquel C, Wijgers N, Maas A, et al. Differentiation driven changes in the dynamic organization of Basal transcription initiation. PLoS Biol. 2009;7:e1000220. doi: 10.1371/journal.pbio.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman E, Lis JT. Transcription factor TFIIH is required for promoter melting in vivo. Mol Cell Biol. 1999;19:5652–5658. doi: 10.1128/mcb.19.8.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon K-R, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, van der Vliet PC, Timmers HT. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA, Stewart CC, Roeder RG. DNA-dependent RNA polymerase levels during the response of human peripheral lymphocytes to phytohemagglutinin. Cell. 1975;4:51–57. doi: 10.1016/0092-8674(75)90133-6. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012;22:957–965. doi: 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemsz MJ, Justement LB, Palmer E, Cambier JC. Induction of c-fos and c-myc expression during B cell activation by IL-4 and immunoglobulin binding ligands. Journal of Immunology. 1989;143:1032–1039. [PubMed] [Google Scholar]
- Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Liu J, Sanford S, Chung HJ, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJ, Young RA. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Melchers F, Andersson J. B cell activation: three steps and their variations. Cell. 1984;37:713–720. doi: 10.1016/0092-8674(84)90407-0. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Darnell JE., Jr. Mapping of RNA polymerase on mammalian genes in cells and nuclei. Mol Biol Cell. 1992;3:1085–1094. doi: 10.1091/mbc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma Y, Hashimoto S, Wang CK, Horikoshi M, Roeder RG. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol Cell Biol. 1995;15:4856–4866. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Pogo BG, Allfrey VG, Mirsky AE. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci U S A. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogo BG, Allfrey VG, Mirsky AE. The effect of phytohemagglutinin on ribonucleic acid synthesis and histone acetylation in equine leukocytes. J Cell Biol. 1967;35:477–482. doi: 10.1083/jcb.35.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Raghavan SC, Lieber MR. DNA structures at chromosomal translocation sites. Bioessays. 2006;28:480–494. doi: 10.1002/bies.20353. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Rawlings JS, Gatzka M, Thomas PG, Ihle JN. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 2011;30:263–276. doi: 10.1038/emboj.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Sawai M, Takase K, Teraoka H, Tsukada K. Reversible G1 arrest in the cell cycle of human lymphoid cell lines by dimethyl sulfoxide. Exp Cell Res. 1990;187:4–10. doi: 10.1016/0014-4827(90)90108-m. [DOI] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- Shklover J, Weisman-Shomer P, Yafe A, Fry M. Quadruplex structures of muscle gene promoter sequences enhance in vivo MyoD-dependent gene expression. Nucleic Acids Res. 2010;38:2369–2377. doi: 10.1093/nar/gkp1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J. Lifespans of naive, memory and effector lymphocytes. Curr Opin Immunol. 1993;5:433–438. doi: 10.1016/0952-7915(93)90065-z. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen W, Rademakers S, Jaspers NG, Appeldoorn E, Raams A, Klein B, Kleijer WJ, Hansen LK, Hoeijmakers JH. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat Genet. 2001;27:299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- Vermeulen W, van Vuuren AJ, Chipoulet M, Schaeffer L, Appeldoorn E, Weeda G, Jaspers NG, Priestley A, Arlett CF, Lehmann AR, et al. Three unusual repair deficiencies associated with transcription factor BTF2(TFIIH): evidence for the existence of a transcription syndrome. Cold Spring Harb Symp Quant Biol. 1994;59:317–329. doi: 10.1101/sqb.1994.059.01.036. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yafe A, Shklover J, Weisman-Shomer P, Bengal E, Fry M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008;36:3916–3925. doi: 10.1093/nar/gkn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Robbiani DF, Resch W, Bothmer A, Hirotaka N, Oliveira T, Rommel PC, Brown EJ, Nussenzweig A, Nussenzweig MC, et al. RPA accumulation during class switch recombination represents 5’–3’ DNA end resection during the S-G2/M phase of the cell cycle. Cell Reports. 2013;1 doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z-X, Zhang M-J, Peng X, Takayama Y, Xu X-Y, Huang L-Z, Du L-L. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Research. 2013 doi: 10.1101/gr.146357.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.