Abstract
Root system architecture (RSA) – the spatial configuration of a root system – is an important developmental and agronomic trait, with implications for overall plant architecture, growth rate and yield, abiotic stress resistance, nutrient uptake, and developmental plasticity in response to environmental changes. Root architecture is modulated by intrinsic, hormone-mediated pathways, intersecting with pathways that perceive and respond to external, environmental signals. The recent development of several non-invasive 2D and 3D root imaging systems has enhanced our ability to accurately observe and quantify architectural traits on complex whole-root systems. Coupled with the powerful marker-based genotyping and sequencing platforms currently available, these root phenotyping technologies lend themselves to large-scale genome-wide association studies, and can speed the identification and characterization of the genes and pathways involved in root system development. This capability provides the foundation for examining the contribution of root architectural traits to the performance of crop varieties in diverse environments. This review focuses on our current understanding of the genes and pathways involved in determining RSA in response to both intrinsic and extrinsic (environmental) response pathways, and provides a brief overview of the latest root system phenotyping technologies and their potential impact on elucidating the genetic control of root development in plants.
Keywords: root growth, root development, hormone interactions, root system architecture, genetics, rice, O. sativa
INTRODUCTION
The exploration of root biology lags far behind above-ground vegetative and reproductive growth and development in plants. There is a vast array of studies on root biology, but the literature is dispersed, highly fragmented, and difficult to search because there are no comprehensive phenotypic databases for plants. Many studies of root genes have been classified based on discovery technique [i.e., mutant, quantitative trait loci (QTL), transgenic analyses] or response variable (hormones, microbial populations, insects, nutrients, water levels), but they have not been joined into a systemic understanding of root genetics. Furthermore, comprehensive ontology terms pertaining to root biology have yet to be established, let alone adopted, and gene functional annotation linking phenotypic characteristics into mechanistic pathways and networks is incomplete. Recently, genome-wide association study (GWAS) approaches both advance and demand better integration of genetic studies, annotations, and pathways into a more complete and searchable data network.
Effective GWAS requires the efficient integration of genotyping, phenotyping, and informatics capabilities. The continued development of increasingly rapid, low-cost, high-throughput genotyping and sequencing technologies, such as second and third generation sequencing and high density single-nucleotide polymorphism (SNP) arrays, have made it straightforward for researchers to generate massive amounts of genotypic data on individuals and populations of interest. The speed, efficiency, and cost of high-throughput precision phenotyping of those same populations lags far behind, requiring significant investments of money, time, and labor to generate the data needed for large-scale mapping studies. The selection of traits measured may be limited due to a lack of quantitative measurement resolution and/or accuracy, leading to the frequent description of traits in qualitative classes that combine multiple biological processes, as opposed to specific quantifiable traits that each measure a distinct biological step or the result of a particular process. Furthermore, existing database resources that seek to compile and integrate phenotypic and physiological data with genotypic data, such as the Database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) and PhenomicDB (Groth et al., 2010), are limited by low data submission and limited curation capacity. While these databases are emerging as useful resources for human and bacterial data, plant-related datasets, particularly in relation to root system biology, are still woefully underrepresented.
Lack of comprehensive phenotypic and informatics resources is currently one of the most limiting factors for leveraging the power of GWAS. Although much about gene function, expression, and pathway or network interaction remains to be discovered, the plant genetics community has accumulated phenotypic data from both field and controlled environments during the last half-century. If properly structured and organized, these data could be interrogated to assist with candidate gene identification and interpretation of GWAS output. The problem is that there is no efficient way to access, parse, and cross-reference these data and therefore, they remain fragmented, dispersed and incompletely indexed. Because the collection, curation, and biological application of phenomic data is much more complicated and multi-dimensional than genotypic data, it has yet to be standardized and streamlined into automated processing modules. As a result, finding, integrating and interrogating the components of complex phenotypes, particularly those associated with plant root system architecture (RSA), requires the intervention of expert biologists who manually search through the literature to discover relevant QTLs, pathways and candidate genes. The annotation process is a complex, multi-step, iterative adventure for the scientist interested in defining relevant genes and networks for association or linkage mapping analyses.
This review was motivated by the need to identify a priori candidate genes involved in rice RSA, morphology, growth, and development related to the interpretation of an association mapping study based on a rice diversity panel that had been genotyped with 700,000 SNPs and screened for 19 components of seedling 3D RSA [unpublished data, McCouch and Kochian labs, Cornell University and United States Department of Agriculture-Agricultural Research Service (USDA-ARS)]. We identified from the literature known genes involved in RSA, which encompasses a range of heterogenous traits involved in many different aspects of plant growth architecture, morphology and phenology. After narrowing the search space using GWAS, we integrated positional information about candidate genes found through mutant analysis, orthologous gene identification, comparative mapping, trait similarity, pathway, and network extension, with the our candidate gene regions identified by GWAS. This was aided by the use of ontology and synteny-related informatics to find genes underlying GWAS peaks and QTLs (Lawrence and Harper, 2008; Vilella et al., 2009; Chen et al., 2012; Lamesch et al., 2012). This article provides a comprehensive review of the genetics underlying root growth, development, and response to environmental stimuli. We provide tables of genes that have been associated experimentally and in silico by sequence homology with root development in rice, along with positional information and gene ontology (GO) evidence codes to facilitate database population and curation (Tables S2 and S3 in Supplementary Material).
DEFINING ROOT SYSTEM ARCHITECTURE
Root system architecture is a complex trait and refers to the spatial configuration of the root system in terms of the precise geometric arrangement of all root axes as laid down in the rooting medium. Root architecture is comprised of a whole system set of descriptors, and as such is senior to and distinct from, though naturally dependent on, the secondary fields of root anatomy, morphology, topology, and distribution; however, individual root architecture components may draw on or overlap with these fields. To clarify, root anatomy refers to the internal cellular structure and arrangement of a root; root morphology, the surface features, including diameter, root hair and cap characteristics, and contorsion; root topology, the hierarchical description of the connection of root axes to one another; and root distribution, the presence and distribution of roots in a positional gradient or grid along a horizontal and/or vertical axis.
As proposed by Fitter (1991), there are five main components of root architecture, each of which may be comprised of several specific traits or parameters. These components are: (1) branch magnitude – the number of interior links (internode segments between two branching points or nodes) or exterior links (internode segments between a branching point and an endpoint, i.e., root apical meristem (RAM); (2) topology, the pattern of branch distribution, which is usually herringbone (alternate lateral branching off a parent root), dichotomous (opposite, bifurcating branches), or radial (whorls of branches around a parent root (Hochholdinger, 2009; Lynch and Brown, 2012); (3) link/internode lengths, the distance between branch points among different root orders of an individual root, which may be averaged across a system; (4) root angles, specifically the azimuth (radial angle) of a lateral root’s (LR) emergence around the circumference of a parent root, the branching angle or departure rate of a LR from a parent root, and the spreading angle of the entire system; and (5) link radius, the diameter of any given root (Fitter, 1991).
PATHWAYS AND NETWORKS INFLUENCING ROOT ARCHITECTURE TRAITS
As with any phenotypic manifestation, all of these simple root architecture components: branch number, branching pattern, length, orientation, angle, and diameter are developmentally controlled by complex interacting genetic pathways, which also modulate growth and developmental responses in response to the perception of environmental cues. Malamy and Ryan (2001) refer to these familiar factors – genetics, environment, and the interaction between the two – as belonging to either “intrinsic pathways” or extrinsic “environmental response pathways.”
Hormones, their receptors, signaling components, and transcription factors (TFs) make up the main chemical and molecular components of the intrinsic pathways. Extrinsic response pathways involve similar networks of receptors for environmental stimuli and their downstream signal transduction and TFs. Many components of the environmental perception and response networks are shared with or interregulated by intrinsic response pathways, and are also mediated by hormonal regulation in order to effect a growth response to external signals (see Table 1 for a review of the major hormones and their role in modulating root architectural traits; Table 2 for a review of the major extrinsic factors, their effects on root growth and development, and the major genes and hormones involved, and Table S1 in Supplementary Material for the key genes involved in root growth and development covered in this review). Recent studies have also identified micro-interfering RNAs (miRNAs) and small-interfering RNAs (siRNAs) which affect RSA by the post-transcriptional regulation of components involved in root growth and environmental perception and response and are themselves transcriptionally interregulated by feedback loops within the same intrinsic and extrinsic pathways (see reviews in Meng et al., 2010; Khan et al., 2011).
Table 1.
Hormones and their involvement in root growth and development.
| Hormone | Chemical compounds | Function | Hormone source | Species | Reference |
|---|---|---|---|---|---|
| Auxin | IAA | Promotes lateral root initiation by specifying lateral root founder cells | Endogenous, root tip | At | Casimiro et al. (2001), De Smet et al. (2007), Dubrovsky et al. (2008) |
| IAA | Promotes lateral root emergence | Endogenous, shoot | At | Bhalerao et al. (2002) | |
| NAA | Increases lateral root primordia initiation and outgrowth | Exogenous | Os, Nt | Campanoni and Nick (2005), Sreevidya et al. (2010) | |
| 2,4-D | Increases lateral root primordia initiation through cell division (but does not promote cell elongation and root outgrowth) | Exogenous | Os, Nt | Campanoni and Nick (2005), Sreevidya et al. (2010) | |
| IAA | Promotes primary root elongation by facilitating the response of root cells to GA3 | Exogenous | At | Fu and Harberd (2003) | |
| Cytokinins | Kinetin, BAP | Inhibits lateral root primordia formation by perturbing PIN gene expression and disrupting formation of a RAM auxin gradient controlling cell division to maintain the QC and neighboring initials | Increased endogenous | At | Laplaze et al. (2007), Dello Ioio et al. (2008); reviewed in Péret et al. (2009b) |
| Kinetin, trans-zeatin | Stimulates lateral root elongation | Exogenous | Os | Rani Debi et al. (2005), Laplaze et al. (2007), Dello Ioio et al. (2008), reviewed in Bishopp et al. (2009) | |
| Kinetin, trans-zeatin | Stimulates crown root primordia formation | Exogenous | Os | Rani Debi et al. (2005), Hirose et al. (2007), Zhao et al. (2009) | |
| Zeatins, other endogenous cytokinins | Inhibits primary root elongation by reducing cell division in RAM, thus regulating RAM size | Increased endogenous | At | Kuderová et al. (2008), Ruzicka et al. (2009) | |
| Gibberellins | GA3 | Interacts with ethylene to promote crown root primordia outgrowth and elongation | Exogenous | Os | Steffens et al. (2006) |
| GA3 | Promotes primary root elongation in the presence of auxin by repressing growth-repressing DELLA proteins | Decreased endogenous and increased exogenous | At | Fu and Harberd (2003) | |
| GA3 | Inhibits lateral root primordia initiation | Exogenous | Pt | Gou et al. (2010) | |
| Ethylene | Ethylene | Promotes crown root formation at submerged nodes | Internode | Os | Lorbiecke and Sauter (1999) |
| Ethylene | Promotes crown root emergence at submerged nodes through induction of epidermal cell death over sites of lateral root primordia formation | Internode | Os | Mergemann and Sauter (2000) | |
| Ethylene | |||||
| Jasmonates | MeJA | Promotes lateral root formation through interaction with auxin pathway | Increased endogenous | At, Gm | Xue and Zhang (2007), Sun et al. (2009) |
| MeJA | Inhibits primary root growth | Increased endogenous | Gm | Xue and Zhang (2007) | |
| Abscisic acid | ABA | Induces lateral root primordia formation under non-stress conditions by modulating the auxin response | Endogenous | At | Brady et al. (2003) |
| ABA | Maintains primary root elongation under drought stress | Endogenous | Zm | Saab et al. (1990) | |
| ABA | Inhibits lateral root outgrowth prior to lateral root meristem formation under non-stress conditions | Exogenous | At | De Smet et al. (2006) | |
| Brassinosteroids | BL | May induce lateral root initiation in the presence of auxin, through modulating auxin signaling | Exogenous | At | Bao et al. (2004) |
| BL | Induces primary root elongation in the presence of exogenous auxin (IAA) by affecting ethylene biosynthesis and the gravitropic response | Exogenous | Zm, At | Chang et al. (2004), Kim et al. (2007), Yun et al. (2009) | |
| HBR | Induces primary and crown root elongation possibly through modulating auxin signaling | Exogenous | Hv | Kartal et al. (2009) | |
| Strigolactone | GR24 (synthetic strigolactone analog) | May either inhibit primary root elongation in low concentrations, or stimulate primary root growth in high concentrations, in the presence of auxin, by putative regulation of auxin efflux carriers | Exogenous | At | Koltai et al. (2010), Kapulnik et al. (2011), Ruyter-Spira et al. (2011) |
| GR24 (synthetic strigolactone analog) | Induces primary root curving in high concentrations, in the presence of no-low auxin by inducing asymmetric cell elongation | Exogenous | At | Koltai et al. (2010) | |
| SLs | Promote crown root elongation by inducing meristematic cell division, possibly through the modulation of local auxin concentrations that regulate meristem cell number | Endogenous | Os | Arite et al. (2012) | |
| SLs | Putatively modulates auxin sensitivity by downregulating auxin efflux carrier expression to inhibit lateral root formation under low auxin levels by reducing auxin accumulation in roots, or inducing lateral root formation under high auxin concentrations by allowing optimal auxin levels to be met | Endogenous | At | Kapulnik et al. (2011), Ruyter-Spira et al. (2011) |
2,4-D, 2,4-dichlorophenoxyacetic acid; ABA, abscisic acid; At, Arabidopsis thaliana; BAP, 6-benzylaminopurine; BL, brassinolide; GA3, gibberellic acid; Gm, Glycine max; HBL, homobrassinolide; Hv, Hordeum vulgare; IAA, indole-3-acetic acid; MeJA, methyl jasmonate; NAA, 1-naphthaleneacetic acid; Nt, Nicotiana tabacum; Os, Oryza sativa; Pt, Populus tremulus; QC, quiescent center; RAM, root apical meristem; SLs, endogenous strigolactones; Zm, Zea mays.
Table 2.
Effects of extrinsic factors in modulating root system architecture.
At, Arabidopsis thaliana; BRs, brassinosteroids; CR/CRP, crown root/crown root primordia; Lj, Lotus japonicus; LR/LRP, lateral root/lateral root primordia; Nt, Nicotiana tabacum; Os, Oryza sativa; PR, primary root; Ps, Pisum sativum; QC, quiescent center; RAM, root apical meristem; SA, salicylic acid; SLs, strigolactones; Zm, Zea mays.
To date, the vast majority of research elucidating the genes and pathways involved in root architecture development has been done with the simple, dicot taproot system of Arabidopsis thaliana (Scheres et al., 1996; Ueda et al., 2005; Péret et al., 2009a). This has allowed for the gradual application of this knowledge in discerning conserved developmental pathways shared with monocot crown root (CR) systems, primarily studied in cereal crops such as rice (Oryza sativa L.) and maize (Zea mays L.).
THE IMPORTANCE OF ROOT ARCHITECTURE
The 3D configuration of a root system is important mechanically, providing physical anchorage of the plant in soil, and physiologically, in nutrient and water sensing and uptake, and in response to soil biota. The rate of root system growth and its vertical and horizontal spread can affect seedling vigor, neighbor competition, and exploitation of different limiting resources, such as phosphorus, nitrogen, and water, through root growth or support of symbioses, and can be highly specific to environmental conditions – a root architecture which may favor the growth of a plant under low water conditions, may impede its growth in flooded soil. The specific growth and development characteristics of a plant’s root system also confers some degree of developmental plasticity to the organism in dealing with nutrient and water availability, seasonal and climate changes, beneficial or disease causing organisms, or toxic compounds in soil. Together, these qualities of anchorage, soil nutrient exploitation, and developmental plasticity as determined by root architecture can have far-reaching effects on maximal yield, especially under stress, and yield stability, and a greater understanding of the genes and pathways involved in root architectural development may be translated into the breeding of improved crop varieties.
INTRINSIC PATHWAYS – GENETIC AND HORMONAL REGULATION OF ROOT ARCHITECTURE
PRIMARY ROOT INITIATION, DEVELOPMENT, AND ELONGATION
The primary root (PR), derived from the radicle and laid down during embryogenesis, grows to form the foundation of the dicotyledonous taproot system, and is the first root of the fibrous, CR-based root system of monocots. Establishment of the RAM of the PR involves cell identity differentiation and the formation and maintenance of a quiescent center (QC) and stem cell population. In Arabidopsis, auxin signaling and its antagonistic feedback by cytokinins (CKs) have been implicated in the development of a root stem cell niche ( Muller and Sheen, 2008; Kartal et al., 2009; Moubayidin et al., 2009; Pernisova et al., 2009; Ruzicka et al., 2009). The secondary regulation of auxin signaling by gibberellins, and brassinosteroids has also been implied (Sabatini et al., 1999; Frigerio et al., 2006). Polar auxin transport by the AUXIN1/LIKE AUXIN (AUX1/LAX) family of auxin influx transporters and the PIN-FORMED 3 (PIN3) and PIN7 auxin efflux transporters lead to the creation and maintenance of an auxin concentration gradient with a root tip maximum (Bennett et al., 1996; Parry et al., 2001; Kramer, 2004; Blilou et al., 2005; Carraro et al., 2006; Swarup et al., 2008; Liu et al., 2009; Wang et al., 2009; see reviews in Petrásek and Friml, 2009; Overvoorde et al., 2010; Figure 1). Several multidrug resistant/P-glycoprotein (MDR-PGP) subfamily members of the ATP-binding cassette subfamily B (ABCB) are also key auxin influx and efflux membrane transporters (Noh et al., 2001, 2003; Luschnig, 2002).
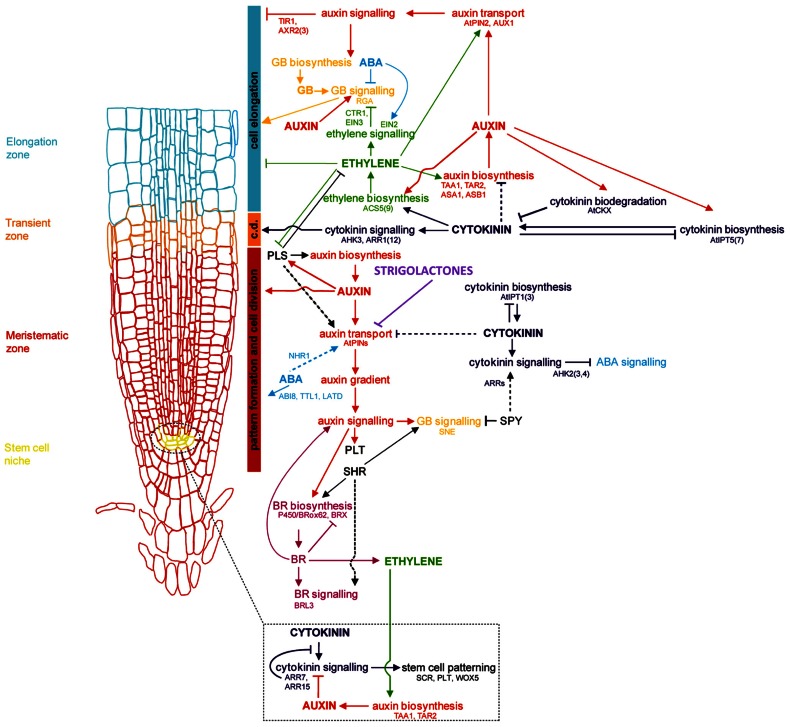
FIGURE 1.
Genetic and hormonal control of primary root development in Arabidopsis Model of the current understanding of hormone interaction and genetic regulation of primary root and general root apical meristem growth and development in Arabidopsis. Important genes involved in integrating signals from different hormone pathways are shown in black; hormone networks are color-coded; dashed lines represent unresolved or indirect relations. The fundamental role of auxin-mediated signaling in controlling all major aspects of root growth, from cell division, differentiation, and elongation, can be visualized, as well as the antagonistic regulation of auxin by cytokinins, and secondary regulation by other hormones, including ABA, ethylene, GA, brassinosteroids (BRs), and strigolactones. c.d. is “cell differentiation,” in reference to the transition zone where cell differentiation is initiated (modified from Benkova and Hejatko, 2009).
Strigolactones (SLs), a new class of plant hormones and rhizosphere signaling molecules have also been implicated in PR development based on crosstalk with auxin signaling. In the presence of auxin, exogenous application of the synthetic SL analog GR24 has been found to either inhibit Arabidopsis PR elongation in low concentrations, or stimulate PR growth in high concentrations by putative regulation of the auxin efflux carriers PIN1, PIN3, and PIN7 (Aida et al., 2002; Kapulnik et al., 2011; Ruyter-Spira et al., 2011). GR24 has also been found to induce PR curving in high concentrations, in the presence of no or low auxin levels by inducing asymmetric cell elongation (Koltai et al., 2010). It should be noted, however, that due to the increased stability of GR24 in aqueous solution, as compared with natural SLs, the effects of this synthetic strigolactone on root growth may be misrepresented (Akiyama et al., 2010).
The presence or absence of auxin transcriptionally regulates many genes involved in general root growth and development through the action of auxin/indole-acetic acid (Aux/IAA) and Auxin Response Factor (ARF) modules (De Smet et al., 2010; Goh et al., 2012b). When not bound to Aux/IAA proteins, ARFs are free to recognize and bind to auxin-responsive elements (AREs) in the promoters of target genes, activating or repressing their transcription. In the absence of auxin or under low auxin concentrations, AUX/IAA proteins, negative regulators of auxin response genes (Abel, 1994) bind with their ARFs, inactivating ARF activity. Under high auxin concentrations, AUX/IAA proteins are targeted for degradation by the SCFTIR E3 ubiquitin ligase complex (Farrás et al., 2001; Gray et al., 2001; Reed, 2001; Gagne et al., 2002; Jebanathirajah et al., 2002; Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Badescu and Napier, 2006; Maraschin et al., 2009; Figure 1).
Other layers of ARF regulation involve miRNAs. The miR160 family has been found to play a role in Arabidopsis PR and LR development through its regulation of the ARF TFs, ARF10 and ARF16, which are functionally redundant but both required for root cap cell formation and development (Wang et al., 2005). Transgenic overexpression of miR160 in rice also induced severe root cap defects, suggesting the presence of a similar regulatory pathway in monocots, although the target(s) of miR160 in rice have not yet been determined (unpublished data as cited in Meng et al., 2010). Normal root cap formation in all roots is necessary for normal root system development and impinges on multiple downstream RSA components, specifically, root elongation, LR production, and root growth angle as dictated by the gravitropic response through root tip sensing (Wang et al., 2005; Band et al., 2012).
In Arabidopsis, a second set of TFs: SHORTROOT (SHR) and its target, SCARECROW (SCR), both GAI, RGA, SCR (GRAS) TFs, are involved in the specification and localization of stem cells and the QC, as well as root radial patterning. They affect not only PR initiation, but also root diameter, and the regulation of cell division and differentiation necessary for downstream LR development (Di Laurenzio et al., 1996; Helariutta et al., 2000; Sabatini et al., 2003; Paquette and Benfey, 2005; Lucas et al., 2011). SCR is also suggested to have a possible role in mediating a cross-response between gibberellic acid (GA), brassinosteroid, and auxin signaling involved in stem cell maintenance (Muller and Sheen, 2008; Ruzicka et al., 2009; reviewed in Benkova and Hejatko, 2009). The maize SCR homolog, ZmSCR, was shown to be essential for the development of the maize radicle during the formation of the coleorhizae, the unique grass structure that sheathes and protects the PR meristem (PRM) during embryogenesis and germination (Tillich, 1977; reviewed in Hochholdinger and Zimmermann, 2008).
A third set of TFs, related to the second set, are the DELLA proteins, including the Arabidopsis GIBBERELLIN INSENSITIVE (GAI), REPRESSOR OF GA1 (RGA) and RGA-LIKE 1, RGA-LIKE 2, and RGA-LIKE 3 (RGL1, RGL2, and RGL3), rice SLENDER RICE (SLR), and its barley homolog, SLENDER1 (SLN1), are negative regulators of GA-mediated root growth, and appear to be negatively regulated by auxin. The ubiquitination and destruction of these DELLA TFs in the presence of auxin and GA thus allow for root cell division and elongation (Dill and Sun, 2001; Ikeda et al., 2001, 2002; Chandler et al., 2002; Fleet and Sun, 2005; Perez-Perez, 2007; Figure 1).
LATERAL ROOT GROWTH – FROM PRIMORDIA INITIATION TO ELONGATION
First order (or primary) LRs are roots that branch off of the taproot or adventitious roots in dicots, and the primary seminal root or CRs in monocots. These first order laterals may be short and determinate, or they may develop higher orders of ramification (second, third, fourth-order, etc. laterals). LRs account for the majority of the root mass in most plant root systems, and perform key functions in soil exploration, nutrient and water uptake, and symbiosis development. While LR production is generally developmental, it may also be adaptive, in response to environmental influences within the rhizosphere. LRs are similar in anatomy, but usually smaller in diameter than their parent root, due to a reduced number of cortical cell layers and xylem and phloem poles (Coudert et al., 2010).
Lateral root growth may be organized into four stages with different implications for RSA: (1) LR initiation, (2) LR primordia (LRP) formation, (3) LR meristem (LRM) outgrowth and emergence from the parent root, and (4) LR elongation (Malamy and Benfey, 1997, Figure 2). The first three stages all affect the potential number and radial orientation of LRs. Development may be halted at any stage during this process which, prior to emergence would reduce the number, position, and pattern of mature LRs; LR elongation affects LR branching angle, branch length, development rate, and whole system topology.
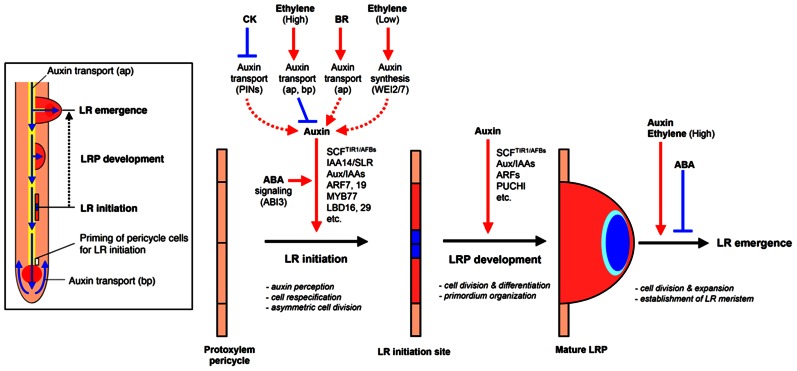
FIGURE 2.
Hormonal and genetic control of lateral root formation in Arabidopsis. LR formation is a three-stage process consisting of LR initiation, LRP development, and LR emergence. LR initiation is positively regulated by auxin but negatively regulated by CK and high concentrations of ethylene [high concentrations of exogenous 1-aminocyclopropane-1-carboxylic acid (ACC)]. The polar auxin transport with a balance of influx and efflux in both acropetal and basipetal directions is necessary for LR initiation and setting up auxin gradient to organize LR primordium (LRP; blue color in LR initiation site and primordium). CK inhibits auxin maxima by altering the expression of PINs, thereby inhibiting auxin gradient for LR initiation. High concentrations of ethylene or exogenous ACC, an ethylene precursor, inhibited LR initiation by enhancing acropetal (ap) and basipetal (bp) auxin transport. BR promotes LR initiation by increasing acropetal (ap) auxin transport. Low concentrations of ethylene (low concentrations of exogenous ACC) promote LR initiation by increasing Trp-dependent auxin synthesis mediated by WEI2 and WEI7. Normal ABA signaling mediated by ABI3 is necessary for proper auxin responsiveness for LR initiation. Auxin also promotes LR primordium development but CK inhibits LR primordium development and affects auxin maxima by altering the expression of PINs. ABA inhibits LR emergence whereas auxin and ethylene (via high concentrations of exogenous ACC) promotes LR emergence (modified from Fukaki and Tasaka, 2009).
LATERAL ROOT INITIATION
The first stage in LR development takes place in the parent root pericycle in Arabidopsis, and the pericycle and endodermis layers in crop cereals like maize and rice (Fahn, 1990; Casimiro et al., 2001). This process is characterized by founder cell identity priming and fate fixation by auxin, cell cycle activation of the founder cells, and asymmetric cell division (Malamy and Benfey, 1997; De Smet et al., 2007; reviewed in Fukaki and Tasaka, 2009). The IAA28-ARFs module, the first of three known AUX/IAA-ARF modules regulating LR development is active in this LR initiation stage for LR founder cell specification (De Rybel et al., 2010). Cell cycle reactivation and control is fundamental to LR initiation and is partially induced by the accumulation of high auxin levels in quiescent xylem pole pericycle or endodermal cells (Beeckman et al., 2001; Casimiro et al., 2001; Malamy, 2005), and the priming of specific xylem pole or endodermal cells to become LR founder cells by 15-h oscillations in the auxin level (De Smet et al., 2007). In Arabidopsis, this root tip-synthesized auxin gradient was found to promote asymmetric cell division of xylem pole pericycle founder cells (Casimiro et al., 2001; De Smet et al., 2007) by the auxin-induced upregulation of cell cycle genes, including cyclins and cyclin-dependent kinases (CDKs; Soni, 1995; Meijer and Murray, 2000; Boniotti and Gutierrez, 2001), and the synchronous downregulation of CDK repressors, such as KRP1 and KRP2, which inhibit the G1 to S transition phase in LRP (Himanen et al., 2002; reviewed in Fukaki et al., 2007; Figure 2).
Further research has suggested that cyclic changes in auxin concentration are insufficient as the sole trigger of LR initiation, and that molecular clock-coordinated oscillating gene expression within the so-called “oscillation zone,” a region encompassing the PR basal meristem and elongation zone, is also necessary for the spatial and temporal definition of LR pre-branching sites. These pre-branching sites develop LRP, but may not always grow out into fully emerged LRs (Moreno-Risueno et al., 2010). In Arabidopsis, two sets of 2084 and 1409 genes were found to oscillate either in phase or in antiphase, respectively, with specific waves of each phase being associated with increased expression of particular genes, mostly notably members of the ARF, NAC, myeloblastosis (MYB), and SOMBRERO TF families. T-DNA insertions in several of these genes also showed defects in LR pre-branching site initiation and reduced LR number (Moreno-Risueno et al., 2010).
LATERAL ROOT PRIMORDIA FORMATION
The formation of LRP is characterized by several rounds of anticlinal and periclinal cell division (Malamy and Benfey, 1997). As modeled in Arabidopsis, this process generates a patterned LRP similar to the PR tip (DiDonato et al., 2004). Mutant and transgenic studies in Arabidopsis suggest that the formation of both the LRM and the PRM are driven by equivalent, if not the same, hormonal and genetic factors (Malamy and Benfey, 1997). Auxin is the primary signaling hormone regulating LRP development through the formation of an auxin gradient (Péret et al., 2009a). This gradient is modulated upstream by low levels of antagonistic CKs which would otherwise repress LRP formation via the disruption of auxin efflux PIN protein localization, which itself is partly responsible for creating the auxin gradient (Laplaze et al., 2007). CK specifically affects the rate of cell differentiation between the cell division and elongation/differentiation zones but does not affect the rate of cell division in the meristem (Dello Ioio et al., 2008). SLs may also regulate LRP formation, possibly by altering auxin sensitivity through downregulating the expression of auxin efflux carriers such as PIN1, thus inhibiting LR formation under low auxin levels by reducing auxin accumulation in roots, or inducing LR formation under high auxin concentrations by allowing optimal auxin levels to be met (Ruyter-Spira et al., 2011).
The Arabidopsis GTP:GDP ANTIPORTER/PROTEIN HOMODIMERIZATION (GNOM) protein also appears to play an essential role in regulating PIN protein trafficking for auxin gradient formation (Steinmann et al., 1999; Geldner et al., 2003; Laplaze et al., 2007). The accumulation of auxin in the central cells and later in the tip of the LRP signals the targeted degradation of AUX/IAA proteins, repressors of auxin-induced transcription. Furthermore, the auxin gradient enables ARF7/NPH4 and ARF19 module-upregulated transcription of target genes for cell ID and pattern formation, including other downstream TFs, such as LATERAL ORGAN BOUNDARIES DOMAIN 16/ASYMMETRIC LEAVES2-LIKE 18 (LBD16/ASL18) and LBD29/ASL16 (Okushima et al., 2005; Lee et al., 2009; Goh et al., 2012a; Figure 2).
LATERAL ROOT OUTGROWTH
Lateral root primordia emergence through the overlying tissues of the parent root involves both further growth, in terms of cell elongation and division, and further differentiation, particularly the development and activation of the LRM, the definitive feature of a newly formed LR (Malamy and Benfey, 1997). Primordia emergence requires the coordinated separation of the overlying cells in the parent root in order to avoid excessive damage and infection risk (Laskowski et al., 2006; Swarup et al., 2008). In Arabidopsis, only three single-cell tissue layers have to be penetrated; in rice as many as 15 cell layers must be penetrated for LRP emergence (Osmont et al., 2007; Péret et al., 2009b).
This process of root cell separation for root primordial emergence is regulated by basipetal, shoot-derived auxin (Bhalerao et al., 2002) and LRP-derived auxin (Swarup et al., 2008), promoting cell separation and upregulating the expression of cell wall-remodeling genes in the endodermal, cortical, and epidermal cells layers overlaying the LRP (Swarup et al., 2008). LAX3, a high-affinity auxin influx transporter, upregulated in response to LRP-derived auxin, and specifically expressed in the epidermal and cortical cells overlaying LRP, facilitates auxin influx in these cells, spatially regulating the subsequent expression of auxin-induced genes involved in cell wall remodeling (Swarup et al., 2008). These cell wall-modification genes encode a suite of enzymes, including pectate lyases such as phospholipase A2 (PLA2), pectin methylesterases (PMEs), polygalacturonase (PG), an expansin (EXP17), and at least one known glycosyl hydrolase, GLH17, all of which are implicated in facilitating cell wall loosening and separation for LRP outgrowth to occur (Henrissat and Davies, 1997; Cosgrove, 2000; Marin-Rodriguez et al., 2002; Laskowski et al., 2006; Swarup et al., 2008; Ogawa et al., 2009; Figure 2).
The activation of the LRM is also thought to occur during LRP emergence from the parent root (Laskowski et al., 1995). While the genes and pathways involved in this process have yet to be elucidated, a shift in auxin signaling or source of synthesis from the parent root to the new LRM is implicated, as the arrested post-emergence growth of the Arabidopsis aberrant lateral root formation3 (alf3) mutant can be rescued with the application of exogenous auxin, suggesting that the ability of the new LR to synthesize its own auxin may coincide or cause lateral meristem (LM) activation (Celenza et al., 1995; Péret et al., 2009b; Figure 2). Multiple Aux/IAA–ARF modules, including the SHY2/IAA3–ARF module (Goh et al., 2012b), may play a role in the complex networks regulating LRP development and LR emergence. These networks may also be mediated post-transcriptionally by the downregulation of LR emergence through the auxin-induced expression of miRNA164a and miR164b which target for degradation the mRNAs of NAM/ATAF/CUC1 (NAC1; Guo et al., 2005), a TF involved in transmitting auxin signals for LR emergence (Xie, 2000). Preliminary research shows this miR164-NAC1 regulatory module may also be conserved in tomato (Zeng et al., 2010) and rice (Meng et al., 2010).
LATERAL ROOT ELONGATION
The genetic control of post-emergence LR elongation affects the rate and angle of LR growth, LRM determinancy and branching potential, all of which are important considerations in RSA. Not much is known about the genetic control of these traits; however, these are areas under active research. The Arabidopsis PLETHORA1 and 2 (PLT1 and 2) and CLAVATA3 (CLV3) genes are implicated in both primary and LRM maintenance of the root stem cell niche and QC, as mutants of these genes fail to maintain the QC and root stem cells, and thus stop root elongation (Aida et al., 2004; Fiers et al., 2004). In vitro application of the artificially synthesized, mature CLV3 peptide, a 12-amino acid ligand, processed from the conserved 14-amino acid CLE (CLV3/ESR) domain of a larger peptide (Fiers et al., 2006), and peptide synthesis or overexpression of other members of its greater CLE family of related proteins sharing the conserved and essential CLE motif, all caused the termination of root development (Strabala and O’Donnell, 2006; Kinoshita et al., 2007), suggesting other CLE genes could be involved in regulating RAM identity (reviewed in Miwa et al., 2009). Cell division and elongation, particularly elongation or expansion is one of the primary drivers of root growth rate, and while the genes involved have not yet been cloned, the maize mutants short lateral root1 and 2 (slr1 and slr2) display short, slow-growing LRs on their primary and embryonic CRs, which microscopy studies haves attributed to a decrease in cell elongation (Hochholdinger et al., 2001). Hormonal interactions also play a role in LR growth: auxins, ethylene, and abscisic acid (ABA) have been shown to inhibit LR elongation, while CKs promote elongation (Rani Debi et al., 2005; Iwama et al., 2007; Figure 2, Table 1). Amongst the many auxin transporters potentially involved in LR elongation, ABCB19/MDR1, an important shoot basipetal auxin transporter, has also been shown to be important for root acropetal auxin transport and necessary for maintenance of a high enough auxin concentration to support post-emergence LR elongation at a normal rate (Wu et al., 2007).
The angle of LR growth is thought to be at least partially under genetic control due to tropic responses, as different Arabidopsis and rice accessions display variations in LR angle (Mullen and Hangarter, 2003; Iyer-Pascuzzi et al., 2010), which may be attributable to differences in intrinsically programed LR gravitropic setpoint angle (GSA), the angle of growth relative to the gravity vector (Digby and Firn, 2002). Mutant analyses of Arabidopsis lines with a normal PR gravitropic response, but variations in LR GSA suggest that the genetic control of GSA may be independent between LR and PR, and that GSA may be mediated by auxin signaling and a root phototropic response (Mullen and Hangarter, 2003).
CROWN ROOTS – FROM INITIATION TO ELONGATION
Crown roots, also called nodal or shoot-borne roots, are adventitious roots unique to monocots and part of normal monocot root system development. Along with their associated LRs, CRs make up the bulk of the fibrous monocot root system. CRs may be developmentally separated into two different types: the embryonic CRs – seminal roots which form around the coleoptilar node along with the PR (radicle) during embryogenesis, and the post-embryonic CRs that arise during germination and throughout the life of the plant (Hochholdinger and Tuberosa, 2009). Along with dicot root and the monocot seminal PR, all CRs, both embryonic and post-embryonic, can be considered primary order roots, as like the radicle they arise from the main stem of the plant and not from another root as do LRs.
CROWN ROOT PRIMORDIA INITIATION AND DEVELOPMENT
Most root development research has focused on PR and LR, thus much if the current knowledge about the genetic control of CR development is deduced from studies of maize and rice mutants or based on comparative analysis with Arabidopsis PR, LR, and adventitious root studies. The overarching hormonal regulation and the gene families regulating PR, LR, CR (in monocots), and adventitious (in dicots) root growth appear to be largely conserved (Hochholdinger et al., 2004; Coudert et al., 2010). The functions of individual genes in the genetic pathways regulating the development may, however, be slightly different.
Crown root primordia (CRP) initials are produced from periclinal divisions of parenchyma cells which give rise to the pattern arrangement of differentiated epidermis/endodermis initials, central cylinder cells, and root cap initial cells (Itoh et al., 2005). This is followed by the establishment of epidermis and endodermis by periclinal divisions of the endodermis–endodermis initials, and then the formation of the cortical cells and central metaxylem (Itoh et al., 2005).
Similar to early processes in PR and LR development, the initiation and development of CRs is also controlled by auxin-mediated signaling (reviewed in Rebouillat et al., 2009). OsGNOM1, an ortholog of Arabidopsis GNOM1, was found to be involved in regulating proper PIN1 auxin efflux protein trafficking, and thus the polar auxin transport necessary for auxin gradient formation to signal the proper asymmetrical division of parenchyma cells for CRP development (Geldner et al., 2003; Liu et al., 2009; Péret et al., 2009b; Richter et al., 2010). Maize and rice homologs of the Arabidopsis SHR and SCR genes, GRAS TFs, also have been shown to be essential for the radial patterning necessary for CRP development. With a similar endogenous expression pattern to the Arabidopsis genes and in vitro evidence of the capacity for interaction between each species pair, it is likely that in monocots the two TFs share a similar role in CR, as opposed to LRP development and interact with each other to restrict the formation of the endodermis to a single-cell layer (Cui et al., 2007).
There is also evidence to suggest that the monocot radicle/primary seminal root, the embryonic CRs, and the post-embryonic CRs may be under different genetic control. The monogenic maize mutant rootless concerning crown and seminal roots (rtcs) does not form any CRs, just the PR and its associated laterals (Hetz et al., 1996). Other monogenic maize mutants display less severe root developmental phenotypes: lateral rootless 1 (lrt1) does not develop CRs at the coleoptilar node or any LRs on the PR or remaining embryonic CRs (Hochholdinger and Feix, 1998), whereas the rum1 mutant has no embryonic CRs, and few, late-developing LRs and post-embryonic CRs (Woll et al., 2005). Rice mutants crown rootless1 (crl1) and adventitious rootless1 (arl1), found to be allelic, have no CRs or CRP, fewer LRs off the PR, and an abnormal gravitropic response (Inukai et al., 2001). Rice ARL1/CRL1 and RTCS have been shown to encode LBD (Lateral organ Boundary Domain) proteins similar to those encoded by the Arabidopsis LBD16 and 29 genes (Inukai et al., 2005; Liu et al., 2005; Taramino et al., 2007). All genes are members of the same family and are probably auxin responsive, having AREs; however, they each have different functions. LBD16 and 29 are involved in LR formation in Arabidopsis, the maize RTCS gene is involved only in CR development, and the rice ARL1/CRL1 gene in both LR and CR development (Figure 3; Inukai et al., 2005; Liu et al., 2005; Taramino et al., 2007).
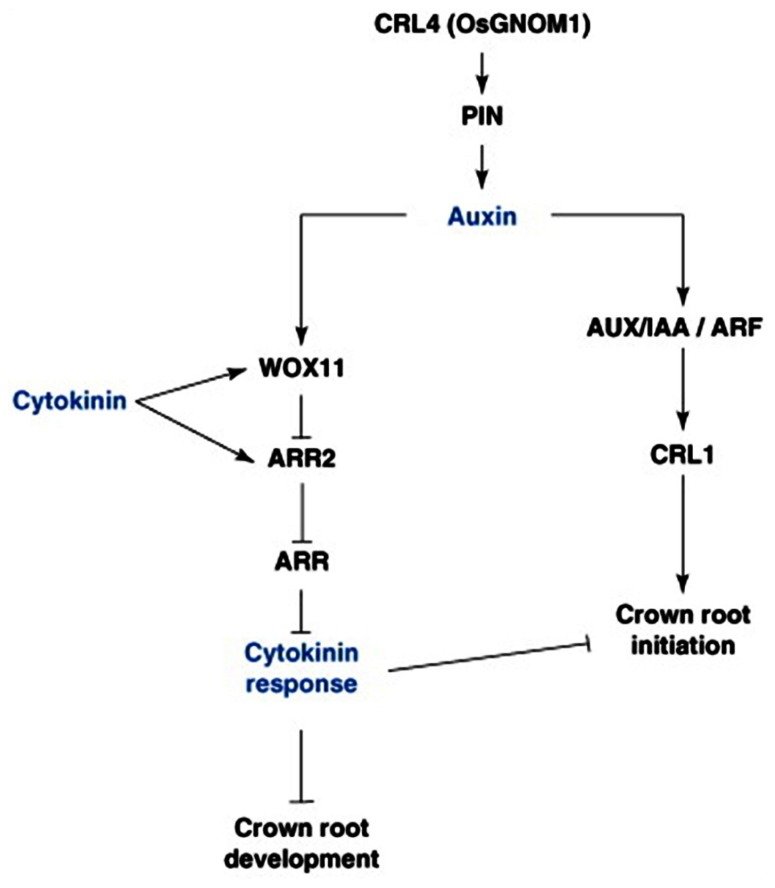
FIGURE 3.
Hormonal and genetic control of crown root formation in rice. Crown root initiation in rice is promoted by auxin, and regulated by the inhibitory influence of cytokinin. Arrows represent the positive regulatory action of one element of the network on another one. A line ending with a bar represents the negative regulatory action of one element of the network on another one. ARF, Auxin Response Factor; ARL, ARR, type-A RESPONSE REGULATOR; AUX/IAA, AUXIN/INDOLE-3-ACETIC ACID; CRL4, CROWN ROOTLESS4; GNOM1, GTP:GDP ANTIPORTER/PROTEIN HOMODIMERIZATION1; PIN1, PIN-FORMED1; WOX11, WUSHEL-Related Homeobox 1 (Coudert et al., 2010).
Similar to LR formation in Arabidopsis, CKs also plays a secondary role in mediating CR development in monocots through antagonism of auxin-related signaling pathways. The rice WUSCHEL-RELATED HOMEOBOX11 (WOX11) gene encodes an auxin and CK-induced TF expressed in early CRP and the actively dividing regions of the shoot apical meristem (Zhao et al., 2009) and found to repress the CK-upregulated type-A response regulator gene, RR2 (Jain et al., 2006), which may function as a negative regulator of CK signaling and may repress cell proliferation in the CR meristem, thus repressing CR emergence (Zhao et al., 2009). Knockout mutants of WOX11 exhibited inhibited CR growth, while overexpression of the gene increased rates of CR cell division, leading to precocious CR growth. Additionally both mutant and overexpressor lines also showed altered transcription of auxin and CK-responsive genes, suggesting that WOX11 may play a pivotal role in integrating auxin and CK signaling to control cell division rates in the CRP (Figure 3; Zhao et al., 2009).
CROWN ROOT OUTGROWTH AND ELONGATION
While the formation of CRP is under genetic and physiological control, the emergence of developing CRs from stem nodes is at least partially influenced by the environment. Mergemann and Sauter (2000) found that in accessions of deep-water rice, the buildup of ethylene caused by submergence induces the death of epidermal cells above CRP, thus promoting emergence of CRs through the epidermis of the submerged nodal branches.
Recent studies on this phenomenon have shown that GA is also involved as a non-essential but synergistic upregulator of CRP emergence and elongation rate in the presence of ethylene, and ABA as a likely inhibitor of both ethylene and GA signaling pathways (Steffens and Sauter, 2005; Steffens et al., 2006). While the specific hormone biosynthesis, signaling, and target genes implicated in this H2O2 programed cell death pathway have not yet been identified, it has been shown that the epidermal cells overlying CRP may be predestined to die, exhibiting a lower transcription level of METALLOTHIONEIiN2b (MT2b), which encodes a reactive oxygen scavenger that, in higher levels, would prevent cell damage by H2O2 (Steffens and Sauter, 2009). It is possible that CRP emergence may also be auxin-regulated, as rice RNAi-knockdown lines of the OsPIN1 gene, which encodes an auxin efflux carrier, show arrested CRP emergence (Xu et al., 2005); however, the physiological mechanism by which auxin signaling influences CRP emergence is yet unknown.
Strigolactones may play a role in positively regulating CR elongation through promoting root meristematic cell division (Arite et al., 2012), potentially through modulating auxin flux. Rice dwarf mutants for genes involved in SL biosynthesis (SL-deficient rice mutants max3/rms5/d17, max4/rms1/d10, and d27) or SL signaling (SL-insensitive rice mutants max2/rms4/d3 and d14) were found to have a short CR phenotype due to an apparent decrease in cell division, leading to a narrower meristematic zone (Arite et al., 2012). This decreased cell division may be due to SL-modulation of local auxin levels, affecting meristem cell number as seen in PRs of homologous Arabidopsis SL-deficient and SL-insensitive mutants (Kapulnik et al., 2011; Ruyter-Spira et al., 2011); however, the specific mechanism of SL effect on root growth has yet to be fully elucidated.
EXTRINSIC PATHWAYS – ROOT SYSTEM ARCHITECTURE CHANGES IN RESPONSES TO ENVIRONMENTAL STIMULI
The intrinsic genetic pathways detailed previously control the normal development of plant root systems by directing the primordia initiation, outgrowth, and elongation of various root types. Modulation of these pathways in response to the environment allow plants the phenotypic plasticity to modify specific components of their RSA to exploit limiting nutrient resources and respond to a constantly fluctuating complex of biotic and abiotic stresses. Even different ecotypes or varieties from the same species that are adapted for growth in dissimilar rhizosphere environments can vary widely in intrinsic root system development schemes and plasticity responses, resulting in heritably different RSAs (Malamy, 2005; Suralta et al., 2008; Clark et al., 2011; Gowda et al., 2011; Pacheco-Villalobos and Hardtke, 2012; Figure 4). While the genes and pathways involved in environmental perception and signaling may be unique to a particular stimulus, root growth response pathways often feed into the underlying genetic pathways by co-opting hormonal regulation. Current understanding of the genetic and hormonal regulation of RSA changes induced by tropisms, nutrient availability, toxic compounds, symbioses, and abiotic stresses are reviewed here and in Table 2.
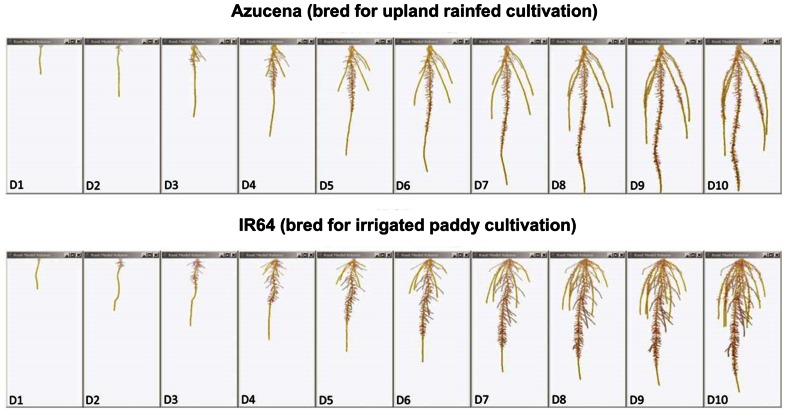
FIGURE 4.
Root system models of two rice varieties bred for contrasting agricultural systems. These root system models, generated from image series of seedling rice root systems of cv. Azucena (top), and cv. IR64 (bottom) over 10 days of growth (D1–D10) in a clear, gellan-gum nutrient media show how the breeding of crop varieties adapted to particular cultivation systems and agroecological environments has resulted in inadvertent selection for different crop root architectures. Azucena, a rice variety bred for an upland rainfed growing environment develops a deeply rooted primary and crown root architecture consistent with rapid growth in search of water, whereas IR64, bred for a water-sufficient, irrigated paddy system is more shallowly rooted, but develops longer, highly branched lateral roots in the top part of the root system to scavenge nutrient resources, such as nitrogen and phosphorus, from near the soil surface. Primary and crown roots are shown in yellow; the root system skeleton is shown in red (modified from Clark et al., 2011; models were generated using RootReader3D software).
GRAVITY
The downward growth of roots influences RSA traits such as root angle, length, and depth, is primarily controlled by a positive gravitropic response, hypothesized to be perceived through the sedimentation of statoliths (amyloplasts – starch-containing plastids, or other plastids, such as chloroplasts) within statocytes, specialized gravity-sensing cells in the root tip (reviewed in Morita, 2010). The mechanism of gravity sensing is yet unknown, but is postulated to be through statolith pressure or movement receptor perception, or pressure-based opening of mechanosensitive ion channels (reviewed in Perrin et al., 2005). In Arabidopsis, ALTERED RESPONSE TO GRAVITY1 (ARG1) and ALTERED RESPONSE TO GRAVITY-LIKE2 (ARL2), J-domain proteins localized to endomembrane organelles, are thought to intact with one another to form a gravity signal transduction complex, promoting rapid, transient cytoplasmic alkalinization through Ca2+ influx, and the redistribution of auxin efflux carrier PIN3 to the lower membrane of the statocytes (Boonsirichai et al., 2003; Harrison and Masson, 2008a,b). The relocalization of PIN3 results in the asymmetric redistribution of auxin along the new, lowest side of the root tip (Young and Evans, 1996; Lucas et al., 2008; Nishimura et al., 2009), followed by basipetal auxin transport to the root distal elongation zone, mediated by the auxin influx carrier AUX1 and efflux carrier ETHYLENE INSENSITIVE ROOT1 (EIR1; Blancaflor and Masson, 2003; Swarup et al., 2005; Band et al., 2012; Brunoud et al., 2012). This new auxin gradient is thought to signal the upregulation of genes leading to cell elongation along the top end of the distal elongation zone, thus producing root tip curvature downward (Blancaflor and Masson, 2003; reviewed in Petrásek and Friml, 2009). In addition to auxin, other phytohormones or signaling molecules, including CKs (Aloni et al., 2006), reactive oxygen species (Cervantes, 2001; Joo et al., 2001), flavonoids and ethylene (Buer et al., 2006; Edelmann and Roth, 2006) may be involved in gravitropic root tip curvature growth response by controlling differential cell elongation in parallel with auxin or as regulators of the auxin-mediated signaling pathway.
The aforementioned concept of a genetically controlled measure of gravitropism, the GSA – the equilibrium angle (or range of angles) from vertical at which an organ shows no gravity-induced differential growth (Digby and Firn, 1995), has bearing on RSA traits such as CR and LR angle of growth. Mutant analyses of Arabidopsis lines with a normal PR gravitropic response but variations in LR GSA suggest that the genetic control of GSA may be independent between LR and PR, and that GSA may be mediated by auxin signaling and a root phototropic response (Mullen and Hangarter, 2003).
LIGHT
Although the root systems of most plants are largely underground and not exposed to light, plant roots may be exposed to light through ambient diffusion or soil upheaval and have been found to possess phytochromes, phototropins, and cryptochromes, including both red and blue light photoreceptors (Ruppel et al., 2001; Mullen et al., 2002; Galen et al., 2007; Molas and Kiss, 2008). Root responses to light have been studied mostly in Arabidopsis, which is found to display a negative phototropic response to blue light, mediated by the root phototropin (PHOT1; Huala et al., 1997; Christie et al., 1998; Galen et al., 2007), and a positive phototropic response to red light, mediated by the root-expressed phytochromes A and B (PhyA and PhyB; Kiss et al., 2003). PhyA also promotes root elongation under exposure to far red (Kurata and Yamamoto, 1997; Correll et al., 2003; Costigan et al., 2011) and blue light (Costigan et al., 2011). Auxin concentration differentials may be partially responsible for root growth responses to shoot light exposure, as the proper plasma membrane localization of the auxin efflux transporter PIN2 was found to be greatly increased in light-grown, but the protein was targeted for vesicular degradation in dark-grown seedlings (Laxmi et al., 2008). Jasmonic acid (JA) is also implicated in a root-localized light response, as one study has demonstrated that phytochromes, or more specifically, phytochrome chromophores are necessary for the JA-mediated root growth inhibition (Costigan et al., 2011).
WATER AVAILABILITY
Given that one of the main functions of the root system is water uptake, soil water availability and soil hydraulic conductivity, especially in the extreme conditions of drought leading to water deficiency or flooding leading to soil saturation and hypoxia, is arguably the most important environmental factor influencing root growth and development. Studies in Arabidopsis have shown that decreasing osmotic potential as a representation of drought stress reduces the LR outgrowth and emergence from LRP of plants grown on agar plates (Deak and Malamy, 2005). Similar research in maize has shown that small increases in negative water potential stimulate PR elongation, but further water stress decreases the rate of PR growth (Sharp et al., 1988; Wiegers et al., 2009).
Hormonal signaling controlling root growth responses to water availability is not yet fully elucidated, however, ABA has been shown to stimulate PR elongation and LR emergence in response to drought (De Smet et al., 2006). In contrast, in flooded deep-water rice plants, a decreased internode ABA level and the concurrent accumulation of GA and the ethylene produced as a response to hypoxia and flooding stress, initiates the programed cell death of adventitious root primordia epidermal cells, allowing the adventitious root development and outgrowth (Mergemann and Sauter, 2000; Steffens et al., 2006). Similarly, the Arabidopsis LONG CHAIN FATTY ACID SYNTHETASE2 (LACS2) gene essential for cutin biosynthesis was shown to be required in order for plants to be able to synthesize a cutin layer that suppresses LR emergence under low water availability (Macgregor et al., 2008). The rice ethylene response factor (ERF)-like TF SUBMERGENCE1 (SUB1; Xu et al., 2006), a TF involved in mediating responses to both plant submergence and drought, may also be one of many genes involved in regulating root growth under water stress, as osmotic stress-induced inhibition of root growth was found to be slightly suppressed in rice varieties with a functional copy of the SUB1 gene (Fukao et al., 2011).
GROWTH IN RESPONSE TO SOIL NUTRIENTS
Plant root adaptive growth in response to soil macro and micronutrients depends on a wide array of variables: nutrient forms, availability, concentration, localization, and nutrient behavior in soil, as well as the nutrient status of the plant. Similar to the tropic responses above, plant root growth in response to a nutrient stimulus requires four main steps: stimulus perception, signal transduction, target gene regulation, gene product mediation of growth response.
NITROGEN
Nitrogen, the most limiting nutrient to plant growth is an interesting example of these highly plastic plant responses to nutrient availability, as it can inhibit LR outgrowth, development and elongation under high N conditions, or in soil with low inorganic nitrogen, soil patches with high inorganic nitrogen can have a local, stimulatory effect on LR elongation and branching within the high N area. Arabidopsis senses nitrate through the PR tip, with downstream components of the nitrate LR growth response pathway include high and low-affinity Arabidopsis NITRATE TRANSPORTERS1.1 and 2.1 (AtNRT1.1 and 2.1; Zhang et al., 1999; Malamy and Ryan, 2001), and nitrate-responsive TFs, including the MADS box TF ARABIDOPSIS NITRATE REGULATED1 (ANR1; Zhang and Forde, 1998).
The nitrate transporters may be either nitrate sensors or, transporters that facilitate N movement for detection via another protein. AtNRT2.1 is necessary for LR growth repression in plants with a high external carbon to nitrogen value (Malamy and Ryan, 2001; Little et al., 2005; Remans et al., 2006b), and AtNRT1.1 is a dual-affinity transporter induced by both auxin and nitrate and important for nitrate uptake under high N conditions (Liu et al., 1999; Guo et al., 2002; Munos et al., 2004). AtNRT1.1 is also an auxin influx facilitator, decreasing its auxin transport activity in response to nitrate sensing, and is proposed to repress LR development by promoting basipetal auxin transport out of LRP under low external nitrate conditions (Krouk et al., 2010). ANR1 mediates the localized N response, regulating the increased proliferation of LRs in N-dense patches, and may be a direct or indirect target of the signal perception/transduction pathway involving AtNRT1.1 (Zhang and Forde, 1998; Remans et al., 2006a). ABA may also act in the same pathway as nitrate by inhibiting LR growth under high N conditions (Signora et al., 2001; De Smet et al., 2003). SLs appear to be upregulated in plants under low N conditions (Yoneyama et al., 2007b); however, whether increased these SL levels have a definite impact on root growth has yet to be determined.
Changes in RSA may also be induced depending on the prevailing available organic form of nitrogen, such as L-glutamate or carnitine. In Arabidopsis seedlings, the sensing of L-glutamate by the PR tip inhibits cell division in the PRM and induces LR formation and outgrowth. L-glutamate may act more as a signaling molecule as opposed to a nitrogen source, as several Arabidopsis auxin-signaling mutants display varying levels of sensitivity to L-glutamate (Walch-Liu et al., 2006), and a rice glutamate receptor mutant displays a host of RSA changes, with short PR and LR, reduced cell division and RAM cell death (Li et al., 2006). Carnitine, transported in Arabidopsis by AtOCT1, has been shown to stimulate LR formation, perhaps by locally affecting the C:N ratio important in modulating LR development (Lelandais-Briere et al., 2007).
PHOSPHORUS
Phosphorus is the second most limiting nutrient because of its high affinity to bind metals in acidic and alkaline topsoil layers, forming insoluble substrates. Phosphorus is taken up by plants as phosphate (Pi), either directly by the root system or, in arbuscular mycorrhizae hostplants, may also be transferred through the fungal symbiont – the genetic control of which will be explored in detail later in this paper.
Under high Pi conditions in Arabidopsis, PR growth is promoted, while LR growth is inhibited (Linkohr et al., 2002). Under natural conditions where Pi is limiting, plants adopt a root foraging strategy to explore topsoil layers for phosphorus. This Pi foraging strategy may be accomplished through one of several different RSA and physiological changes. In Arabidopsis and rice, growth shifts to favor an increased root:shoot ratio, with a higher initiation and outgrowth of LRs, forming a shallow, highly branched root system (Williamson et al., 2001; López-Bucio et al., 2002; Shimizu et al., 2004). Under low Pi conditions, Arabidopsis PR growth is inhibited (Williamson et al., 2001; Linkohr et al., 2002; López-Bucio et al., 2002), while root hairs increase in density and length (Bates and Lynch, 1996, 2000). In legumes, including soybean, pea, and common bean (Phaseolus vulgaris), basal root growth angle is shifted from a downward to a more horizontal direction (Bonser et al., 1996), though a recent study shows the opposite effect in Arabidopsis, with LR GSAs shifting to a steeper, downward orientation under low Pi conditions (Bai et al., 2013). Several different families of plants develop proteoid or cluster roots – highly branched bunches of LRs just below the soil surface that secrete phosphatases and organic acids which solubilize bound phosphate for uptake (Shane and Lambers, 2005; Schulze et al., 2006).
In Arabidopsis, the PR tip is the key organ involved in phosphate sensing, and the initial effect of low external Pi perception is the inhibition of PR growth by the loss of meristem activity and cell elongation (Williamson et al., 2001; Sanchez-Calderon et al., 2005). While a plant Pi-receptor has yet to be identified, studies suggest that the P5 type ATPase PHOSPHATE DEFICIENCY RESPONSE2 (PDR2), and multicopper oxidase LOW PHOSPHATE ROOT1 (LPR1) function in an endoplasmic reticulum-localized Pi-signaling pathway (Ticconi et al., 2004, 2009; Reymond et al., 2006). PHOSPHATE STARVATION RESPONSE1 (PHR1; Bari et al., 2006), an Arabidopsis MYB-like TF that binds the promoter sequences of low Pi-induced genes, and its regulator SMALL UBIQUITIN-LIKE MODIFIER1 (AtSIZ1; Miura et al., 2005), a small ubiquitin modified E3 ligase, and the downstream PHOSPHATE2 (PHO2), an E2 conjugase, and the microRNA miR-399, which regulates PHO2 expression, are all involved in Pi-deficiency-related transcriptional changes (Bates and Lynch, 2000; Bari et al., 2006). The Arabidopsis WRKY75 TF is also induced during Pi-deprivation and may modulate both phosphate and non-phosphate induced LR development and control the transcription of genes such as high-affinity Pi transporters important for Pi uptake (Devaiah and Raghothama, 2007; Devaiah et al., 2007). The Pi-induced tobacco bZIP TF PHOSPHATE INDUCED2 (PHI2; Sano and Nagata, 2002) and rice bHLH TF PI STARVATION-INDUCED TRANSCRIPTION FACTOR1 (OsPTF1; Yi et al., 2005) may also have a role in modulating low Pi-induced changes in RSA.
Increased auxin sensitivity, decreased CK sensitivity, and changes in auxin transport and localization appear to be at least partially responsible for Pi stress-induced LR development. A shift in auxin overaccumulation from the PR apex to the LRP, or an increased sensitivity of LRP to auxin have been suggested as proposed mechanisms for increases in LRP emergence and LR density (Franco-Zorrilla et al., 2002; López-Bucio et al., 2005; Nacry et al., 2005). TIR1 auxin receptor-dependent degradation of TF-repressing AUX/IAA proteins is essential for LR development in Pi-stressed seedlings (Perez-Torres et al., 2008). The effect of auxin under low Pi conditions is also regulated by CK signaling, which represses auxin-induced gene transcription Pi-starved Arabidopsis plants display a decreased response to CK, partly due to the reduced expression of the CR receptor CRE1 (Franco-Zorrilla et al., 2002). Ethylene perception is likely also necessary for increased root hair development and LR elongation and decreased PR elongation under low Pi conditions (Schmidt and Schikora, 2001; López-Bucio et al., 2002; Ma et al., 2003) and has additionally been shown to affect Pi stress-induced changes in basal root growth angle in bean (Lynch and Brown, 2001). Similar to CK, GA acts as a negative repressor of Pi-induced root architecture changes under low Pi conditions; Pi-deficient plants accumulate DELLA proteins, which repress GA-induced root growth suppression and thus allow for auxin-mediated LR initiation and elongation (Jiang et al., 2007). SL production is induced by low Pi in many species including tomato, Arabidopsis, pea, and rice (López-Ráez and Bouwmeester, 2008; Umehara et al., 2010; Kohlen et al., 2011; Foo et al., 2012; Mayzlish-Gati et al., 2012). Some studies suggest that increased production and exudation of SLs under soil Pi or N deficiency is dependent on whether the plant (1) is an arbuscular mycorrhizal fungi (AMF)-compatible host, and (2) whether it is dependent on the arbuscular mycorrhizal symbiosis (AMS) for Pi and N uptake (Yoneyama et al., 2007a,b, 2008; Umehara et al., 2010); however, what effect, if any, this increased SL exudation has on root growth is unclear. Exogenously applied GR24 appears to increase LR formation under low Pi or decrease LR formation under sufficient Pi though the F-box protein MORE AXILLARY GROWTH2 (MAX2), a putative component of the SL-signaling pathway (Kapulnik et al., 2011; Ruyter-Spira et al., 2011).
SULFUR
Sulfur, taken up by plant roots as sulfate, is another limiting plant macronutrient, and is essential for the synthesis of methionine and cysteine. Sulfur deficiency can have significant effects on RSA; sulfate limited Arabidopsis and maize plants increase their LR production, developing an extensive, highly branched root system, often at the expense of shoot growth (Kutz et al., 2002; Bouranis et al., 2008). Another conflicting Arabidopsis study found a decrease in LRP and emerged LR under low-sulfate growth conditions (Dan et al., 2007). To rectify these two opposing developmental outcomes, a two-state model was proposed wherein short-term sulfur limitation let to increased LR growth for sulfate foraging, but longer-term sulfate deficiency led to overall decreased growth and photosynthesis, ending in premature senescence (Hoefgen and Nikiforova, 2008; Lewandowska and Sirko, 2008).
While the genes involved in internal and external sulfate sensing and transcriptional regulation have not yet been cloned and characterized, several components of root sulfate import and signal transduction have been identified. Of the five major classes of sulfate transporters identified in Arabidopsis and rice (Takahashi et al., 1999; Buchner et al., 2004; reviewed in Takahashi, 2010), the group 1 high-affinity transporters are essential for root sulfate uptake. Arabidopsis SULFATE TRANSPORTER1;2 (SULTR1;2) is expressed under both sulfate-sufficient and low-sulfate conditions and transcriptionally regulated by the ETHYLENE-INSENSITIVE3-LIKE3 TF SLIM1, whereas the SULTR1;1 gene induced only under sulfate stress (Takahashi et al., 2000; Yoshimoto et al., 2002; Maruyama-Nakashita et al., 2004) and upregulated by O-acetylserine (thiol) lyase (OAS), a rate-limiting enzyme involved in sulfate assimilation into cysteine (Leustek et al., 2000; Saito, 2000).
Auxin may play a central role in LR production under sulfate stress. In Arabidopsis, sulfate deficiency activates the transcription of NITRILASE3 (NIT3), which converts indole-3-acetonitrile to the auxin IAA (Kutz et al., 2002). However, while NIT3 activity is especially upregulated in LRP under sulfate limitation, increased concentrations of auxin have not been proven (Kutz et al., 2002; Lewandowska and Sirko, 2008). Studies of sulfur-limitation regulated auxin signaling genes such as BIG, named for the huge 560 kD calossin-like protein it encodes, required for the polar transport of auxin (Gil et al., 2001), as well as the auxin TF genes IAA13, IAA28, and ARF-2, indicate that auxin is likely involved in the indirect regulation of sulfur homeostasis and short to long-term sulfur deficiency responses (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003; Hoefgen and Nikiforova, 2008; Lewandowska and Sirko, 2008). JA may also play a role in sulfur regulation, as demonstrated by research in Arabidopsis finding low sulfur JA biosynthesis genes upregulated under low sulfur in (Maruyama-Nakashita et al., 2003), exogenous application of JA promoted sulfur assimilation and there is also evidence to suggest that CKs and sucrose may affect sulfur responsive gene transcription (Ohkama et al., 2002).
TOXIC COMPOUNDS
High soil concentrations of naturally occurring soluble salts, aluminum, and heavy metals, such as cadmium, lead, and chromium, can be highly phytotoxic and seriously impair plant root growth. Plants exhibit two main strategies to manage toxic soil compounds: (1) producing root exudates that bind and neutralize the toxin in the rhizosphere, and (2) actively transporting the compound into the root, but neutralizing and sequestering it in vacuoles for safe accumulation, or eliminating it through exudation.
ALUMINUM TOXICITY
Aluminum is the third most abundant element and the most abundant metal in the Earth’s crust. Aluminum toxicity is one of the major constraints to yield productivity worldwide, especially in the acid soils of the tropics and subtropics that comprise almost 50% of all non-irrigated arable land in those regions (Uexküll and Mutert, 1995). At a soil pH of 5.5 or less, Al is solubilized into Al3+, its phytotoxic form, which has a high plant uptake affinity through diffusion (Kochian, 1995). Al3+ is highly toxic to plant growth, causing a rapid inhibition of root apical cell expansion and elongation, and the eventual cessation of cell division, resulting in a stunted, brittle root system with swollen malformed tips, inhibited LR initiation and outgrowth, deformed root hairs, and a poor nutrient and water uptake capacity (Foy, 1984; Delhaize et al., 1993; Kochian, 1995; Matsumoto, 2000).
In addition to Arabidopsis, several cereal crops, such as, maize, rice, sorghum, and wheat have been used to examine the physiological and molecular mechanisms of aluminum tolerance, as members of the grass family appear to be among the most resistant to aluminum toxicity (Delhaize et al., 1993; Magalhaes et al., 2004; Mao et al., 2004; Caniato et al., 2007). The two most well-studied mechanisms of aluminum tolerance include external avoidance, through root secretion of organic acids such as malate, citrate, and oxalate, which chelate Al3+ ions in the rhizosphere, preventing their diffusion into roots (Miyasaka et al., 1991; Delhaize et al., 1993; Ma and Furukawa, 2003), and true, internal tolerance, by the uptake, organic acid chelation, and sequestration of bound aluminum substrates (Matsumoto et al., 1996; Ma et al., 2001; Huang et al., 2009b; Klug and Horst, 2010); however, only the molecular pathways involved in Al3+-stress-induced RSA changes will be discussed below.
The site of Al3+ sensitivity in maize is the root apex (Ryan et al., 1993); however, exposure of only the distal transition zone of maize roots to Al3+ was found to reduce cell elongation in the elongation zone (Sivaguru and Horst, 1998), suggesting the presence of a diffusible signal between the zones, later found to be the ethylene-mediated basipetal transport of auxin (Kollmeier et al., 2000; Sun et al., 2010). In Arabidopsis, the ethylene receptor gene ETHYLENE RECEPTOR1 (ETR1I; O’Malley et al., 2005) and the ethylene signal transducer ETHYLENE INSENSITIVE2 (EIN2; Alonso et al., 1999) were found to be necessary to the Al3+ induced inhibition of root elongation (Sun et al., 2010). These genes, likely along with other members of the ethylene signaling pathway, are essential for Al3+ induced upregulation of the Arabidopsis ethylene synthesis genes 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE2, 6, and 8 (AtACS2, 6, 8) and 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID OXIDASE1 and 2 (AtACO1, and 2; Tsuchisaka and Theologis, 2004), followed by the upregulation of auxin transporters AtPIN2 and AUX1, leading to auxin distribution changes that are likely responsible for the inhibition of root elongation (Sun et al., 2010).
The binding of Al3+ to negative binding sites on root cell walls and plasma membranes, has also been proposed to inhibit root elongation by increasing wall and membrane rigidity leading to transverse ruptures between the dermal and outer cortical cell layers from the inner cortex, and causing root tip damage (Kopittke et al., 2008), as well as impaired membrane function (Miyasaka et al., 1989; Ahn et al., 2001; Jones et al., 2006; Sun et al., 2010). Al3+ binds primarily to negatively charged pectin in cell walls; the degree of binding has been found to be determined not by the amount of pectin, but by its negative charge as modulated by methylation (Eticha et al., 2005) by PME (Schmohl et al., 2000).
Interestingly enough, the blocking of Al3+ cell wall binding sites (Huang et al., 2009a) may be a major mechanism of aluminum resistance in rice, which does not appear to secrete enough chelating organic acids to rely on an Al3+ external avoidance strategy (Ma et al., 2002). Two genes, SENSITIVE TO ALUMINUM RHIZOTOXICITY1 and 2 (STAR1 and 2) encode the nuclear binding domain and transmembrane domain, respectively, of an ABC transporter with specificity for uridine diphosphate (UDP) glucose that is upregulated following root exposure to Al3+ (Huang et al., 2009a). Both STAR genes are upregulated by the constitutively-expressed rice root ALUMINUM RESISTANT TRANSCRIPTION FACTOR1 (ART1), which also upregulates several other genes implicated in different aluminum tolerance mechanisms (Yamaji et al., 2009). Among these are rice homologs of genes encoding proteins implicated in modulating root elongation and cell wall elasticity: namely an α-expansin EXPA10, members of which have been shown to decrease cell wall extension potential when exposed to Al3+ (Gao et al., 2008), and are additionally downregulated in response to Al3+ (Lee and Kende, 2002), and an Arabidopsis cell wall-associated putative endochitinase CHITINASE A (AtCHIA; Yokoyama and Nishitani, 2004), likely involved in modulating cell wall extension by regulating chitin levels (Kwon et al., 2005).
The upregulation of 1,3-β-D-glucan synthase (Bhuja et al., 2004), resulting in callose deposition in root apices, especially in endodermal and cortical cell walls (Budíková, 1999), is another signal of Al3+-induced injury (Jones et al., 2006; Sivaguru et al., 2006). It is proposed that this abnormal callose deposition may inhibit both symplastic and apoplastic flow (Sivaguru et al., 2000, 2006; reviewed in Horst et al., 2010), causing inhibition of root growth. It is not yet understood whether callose deposition actually represents Al3+-induced injury, is a secondary cell-strengthening response to aluminum damage, or possibly even a defense response to block further Al3+ binding.
SALINITY
Salinity is estimated to affect at much as 20% of the world’s agricultural land and 20% of the world’s irrigated cropland (Flowers and Yeo, 1995) due to a number of natural and man-made factors, including natural salinity and coastal proximity, poor water or fertilizer management, the clearing of vegetation, and prolonged cycles of drought and flooding. In most saline soils, sodium chloride (NaCl) is the most soluble and abundant salt, with calcium and magnesium chloride in lesser concentrations. The dominant causes of plant saline toxicity are complicated by the effects of saline soils on external root osmotic stress, which affects water and nutrient uptake, especially in competition with potassium (K+) and calcium (Ca2+), and internal ionic stress most frequently from the buildup of high sodium (Na+) concentrations (Munns and Tester, 2008).
Different species of plants have varying levels of salt tolerance, from the highly halophilic saltbush (Atriplex spp.) to highly sensitive species, such as rice and Arabidopsis (Munns and Tester, 2008). RSA is generally not affected as severely as shoot branching and leaf expansion under salt stress; in many plants, root growth decreases under NaCl treatment due to reduced epidermal cell division and elongation rates, likely in response to the osmotic stress (Kurth et al., 1986; Zidan et al., 1990). Salt stress also was shown to increase LR production and suppress PR elongation in Arabidopsis (He et al., 2005), induce programed cell death in rice root tips (Li et al., 2007), as well as raise the root death rate in sensitive tomato accessions (Snapp and Shennan, 1992).
Of the many mechanisms of salt tolerance – uptake inhibition, internal sequestration, leaf exclusion, root efflux, and osmotic stress tolerance (reviewed in Munns and Tester, 2008) – root uptake inhibition, efflux, and osmotic stress tolerance have probably the greatest local effect in mediating RSA changes and root growth responses. Na+ is thought to enter the root by passive diffusion through either high-affinity K+ transporters (HKTs), such as the rice OsHKT2;1 (Horie et al., 2007), or through non-selective cation channels (NSCCs); possibly glutamate activated receptors (GLRs), which complex with glutamate to form a channel (Demidchik et al., 2010), or cyclic nucleotide-gated channels (CNGC; Leng et al., 2002; Tester and Davenport, 2003). In the current Arabidopsis model of Na+ stress signaling, internal Na+ presence is perceived by a yet unknown sensor, triggering cytosolic Ca2+ flux sensed by the Ca2+ sensor Salt Overly Sensitive3 (SOS3; Liu and Zhu, 1998), which complexes with and activates SOS2, CBL-interacting protein kinase (Quintero et al., 2002). The SOS2/SOS3 complex is involved in controlling three different Na+ transporters to maintain a low cytoplasmic [Na+]. These include: SOS1, a plasma membrane Na+/H+ antiporter that increases Na+ efflux out of the cell (Zhu et al., 1998; Quintero et al., 2002), a vacuolar Na+/H+ exchanger (NHX1), which facilitates N+ sequestration in vacuoles (Apse et al., 1999; Gaxiola et al., 1999) and may negatively regulate HKTs, such as Arabidopsis HKT1, restricting Na+ buildup in the cytoplasm (Uozumi et al., 2000; Rus et al., 2001; Zhu, 2002; reviewed in Mahajan and Tuteja, 2005). Ionic balance between Na+, H+, Ca2+, and K+ is essential; under low K+ conditions in rice, moderate levels of Na+ influx into the roots through OsHKT2;1 transporters were found to be beneficial in partially maintaining root elongation otherwise inhibited under low K+; however, the biochemical advantage to this phenomenon is not yet understood (Horie et al., 2007, 2009).
Symbiotic interaction with plant rhizobacteria and arbuscular or ectomycorrhizal fungi have also been shown to mitigate saline toxicity and alleviate salt stress, perhaps by modulation of root ion and nutrient levels (Sheng et al., 2008; Dimkpa et al., 2009; Evelin et al., 2009; Luo et al., 2009; Shilev et al., 2010). Internal fluctuations in the concentrations and transport of several hormones, including the stress-induced ABA, as well as ethylene, auxin, CKs, and possibly GAs, are observed in response to salinity stress and are mostly linked to shoot-to-root Na+ stress signaling (Kuiper et al., 1990; He et al., 2005; Khadri et al., 2006; Cao et al., 2008; Bano, 2010). Ethylene and auxin signaling were, however, found to be required for increased LR production in salt-stressed Arabidopsis seedlings in connection with the TF AtNAC2, induced by upstream EIN2 transduced ethylene signaling (He et al., 2005). Interestingly enough, auxin and ABA are also implicated in the opposite RSA response of Medicago truncatula under salt stress: decreased PR elongation, LRP initiation, and LR emergence. In this study, ABA and salt stress both induced upregulation of HOMEOBOX 1 (HB1), a TF found to represses LRP emergence by repressing the downstream TF LBD1, which would otherwise activate downstream genes promoting LRP outgrowth (Ariel et al., 2010). Microarray comparative analysis of rice, Arabidopsis and ice plant (Mesembryanthemum crystallinum) revealed several dozen common genes with salinity-induced transcriptional changes, including genes involved in stress perception and osmotic regulation (Pareek et al., 2007). The precise identity of root architecture-related genes regulated by salt stress-induced TFs have yet to be determined.
SYMBIOSES
Plant root symbiotic associations with microbes, most notably the mycorrhizal and rhizobial symbioses, have long been known to promote plant nutrient uptake efficiency.
In order to support these symbioses, host plant root architecture may undergo a number of significant changes throughout the pre-contact root–microbe signaling, symbiosis development, and establishment processes detailed in the following sections on mycorrhizal and rhizobial symbioses below. Although both symbioses induce different changes in root architecture and plant nutrient status, they share some similar components in their signaling and early developmental pathways, the so-called “SYM pathway” (Parniske, 2008). Recently, a set of seven common SYM genes/proteins required for both symbioses were identified (Parniske, 2008). These include: the Leu-rich repeat receptor kinase SYMRK/DOES NOT MAKE INFECTION2 (DMI2), activated after Nod factor signal perception (Endre et al., 2002; Yoshida and Parniske, 2005); two nuclear membrane-localized cation channels, CASTOR (Imaizumi-Anraku et al., 2004) and POLLUX/DMI1 (Ané et al., 2004; Imaizumi-Anraku et al., 2004); two nucleoporins, NUP85 (Saito et al., 2007) and NUP133 (Kanamori et al., 2006), all necessary for inducing the Ca2+ spike signal (Kosuta et al., 2008); the calcium/calmodulin-dependent protein kinase (CCaMK; Lévy et al., 2004; Mitra et al., 2004; Tirichine et al., 2007), which acts downstream of Ca2+ spiking and is thought to transduce the calcium signals, partly through the physical interaction and phosphorylation of CYCLOPS, a protein with a nuclear localization signal and carboxy-terminal coiled-coil domain protein of unknown function (Yano et al., 2008; Horváth et al., 2011). Intersecting research on the arbuscular mycorrhizal (AM) and rhizobial symbioses have largely been carried out on the model legumes Lotus japonicus and Medicago truncatula, as neither Arabidopsis, nor any of the other non-leguminous model plants have the ability to host the rhizobial symbiosis.
MYCORRHIZAL SYMBIOSES
Over 90% of land plants form symbioses with mycorrhizal fungi. These symbioses improve plant nutrient capture through fungal mineral scavenging and transfer to the plant, and can be linked to significant changes in plant root architecture. Most of the research in this field, and subsequently in this review, is focused on the AMS, the most common type of mycorrhizal symbiosis, found in over 80% of plant species and involving the ~200 obligate biotroph fungal species of the Glomeromycota phylum (Schüßler et al., 2001; Strack et al., 2003). The AMS has ancient origins – estimated to be 400 million years old, it is suggested to have played a major role in the early colonization of land by plants (Pirozynski and Malloch, 1975; Simon et al., 1993). The AMS is characterized by pre-contact plant–fungal signaling, fungal contact and entry of the host plant root system, and the formation of arbuscules, highly branched fungal structures within root cortical cells that are the site of nutrient (primarily P, but also N, Zn, and Fe) transfer from the fungus to the plant and carbohydrate transfer from the plant to the fungus (reviewed in Parniske, 2008).
Pre-contact signaling, development, and maturation phases of the AMS all may induce changes in RSA, however, separating these changes from those induced indirectly as a result of improvements in plant nutrient status is challenging. Previous studies have generally reported increases in root branching as a result of colonization, yet a review of these studies reveal further complicating factors: plant root systems do not respond to AM fungal colonization in the same ways. Colonization-induced root responses appear to differ depending on host plant species, types (woody vs. non-woody; monocot vs. dicot), or varieties, soil water and nutrient status, especially of P, and possibly even the species of AM fungi (Hetrick et al., 1988; Berta et al., 1990, 1995; Olah et al., 2005; Gutjahr et al., 2009; reviewed in Hetrick, 1991; Berta et al., 2002; Parniske, 2008). Strigolactone synthesis and exudation from the roots triggers AM fungal hyphal branching, a key step in root colonization (Akiyama et al., 2005); however, the direct effect of Sls on AMS-related root growth and development is unclear and highly dependent on plant Pi and N status and concentration in the rhizosphere (see prior sections on nitrogen and phosphorus).
In maize, root thickness and overall root mass, but not LR formation, are increased by AM colonization, which also partially restores the LR growth completely absent in the lrt1 mutant, possibly indicating the involvement of auxin signaling (Paszkowski and Boller, 2002). A partial hormonal influence in AM colonization-induced RSA changes may well be possible; studies have reported altered levels of auxin (Fitze et al., 2005), ethylene (Vierheilig et al., 2002), CK (Dixon, 1990; Barker and Tagu, 2000), and ABA in colonized roots (Herrera-Medina et al., 2007), as well as specific roles for auxin, CK, and ABA in AMS development (Barker and Tagu, 2000; Fitze et al., 2005; Ludwig-Muller and Guther, 2007). In contrast with maize, in which the AMS stimulates an increase in root thickness, but not root number, AM colonization in rice was found to induce CR elongation and both fine, determinate and long, indeterminate LR number (Gutjahr et al., 2009). Interestingly enough, while AMF-exposed three monogenic essential rice SYM gene mutants, pollux-2, ccamk-2, and cyclops-1, did not develop colonized roots, they showed a decrease in CRs and an increase in LRs over non-AMF-mutant controls, indicating the presence of root growth pathways induced by AM fungi, but independent of the SYM pathway (Gutjahr et al., 2009).
The only definite example of AM fungi-induced RSA development is in the legume Medicago truncatula, where pre-fungal contact LR formation was discovered to be induced by a diffusible factor from AM fungi, the so-called “Myc” factor of AM fungi that affects plant host signaling pathways (Olah et al., 2005). Induction of LR development by this pathway requires the proper function of two SYM pathway components, DMI1 and 2 (Endre et al., 2002; Stracke et al., 2002; Hogg et al., 2006), as well as the novel MtENOD11 protein, all of which have necessary but yet undetermined roles in pre-symbiont contact AM and Rhizobium symbiosis signaling (Kosuta et al., 2003; Olah et al., 2005).
Rhizobium–LEGUME SYMBIOSIS
The Rhizobium–legume symbiosis is the most prominent and well-studied of plant associations with N-fixing bacteria, and consists of a symbiotic association between the roots of legumes (Fabaceae) and root nodule-forming, N-fixing soil bacteria of the family Rhizobiaceae. Another similar, though lesser-studied, root nodule symbiosis is the actinorhizal symbiosis between plant species in three rosid orders, the Fagales, Cucurbitales, and Rosales, and N-fixing actinobacteria of the genus Frankia (Swensen, 1996). Host plants in both symbioses benefit by gaining an internal supply of fixed-N, as well as potential increases in resistance to some disease and abiotic stresses, while the endosymbiotic bacteria gain a protected living environment and a carbon source supplied by plant photosynthate. Similar to the AMS, the Rhizobium–legume symbiosis starts with pre-contact signaling between the bacteria and host plant, followed by bacterial infection of root hairs, root hair curling, infection thread and nodule development, and bacterial colonization of nodules (reviewed in Provorov, 2000).
Colonization of legume roots may affect RSA in two ways: root nodule formation and changes in PR or LR growth. The two types of symbiotic nodules – determinate and indeterminate – differ both structurally and developmentally, and are dependent on the host plant species. Cells of the tip meristem of determinate nodules fully differentiate at maturity and are not maintained resulting in spherical nodules at uniform developmental stages, whereas the tip of the meristem of indeterminate nodules is continuously active and producing new infected tissue, creating larger and longer cylindrical or bulbous nodules with different developmental zones (reviewed in Markmann and Parniske, 2009). Studies also suggest that there is a balance between LR and nodule formation, with nodule primordia initiation dependent on the suppression of LR emergence (Nutman, 1948; Lohar et al., 2004).
Given the ancient origin and near-universality of the AMS in the plant kingdom, and the familial specificity of the rhizobial symbiosis to only the Leguminosae, it has been proposed that the rhizobial symbiosis has recruited much of the key symbiotic development pathway from the AMS, then modified and evolved genes and pathways for nodulation specific functions (Markmann and Parniske, 2009). Although the functioning alleles of the seven aforementioned known genes in the shared SYM pathway are necessary for the development of both the AM and rhizobial symbioses (Kistner et al., 2005), none of these are directly involved in symbiosis-related RSA changes. Each of these seven gene products is involved in only the early stages of the SYM signal reception and transduction pathway. The downstream, symbiosis-activated genes and networks feeding into intrinsic hormone-controlled and nutrient-modulated root growth pathways are what is actually involved in regulating Rhizobium-induced nodulation and LR development to balance plant nitrogen fixation needs with its carbon budget.
Cytokinin accumulation in root hairs and cortical cells after Rhizobium inoculation has been implicated as a key differentiation signal in stimulating root nodule organogenesis in response to Nod factor signaling (Lohar et al., 2004; Ferguson et al., 2010). CK suppresses pericycle cell division for LRP initiation, promotes cortical cell division for nodule primordia formation, and stimulates the expression of early NODULIN (Nod) genes (Bauer et al., 1996; Fang and Hirsch, 1998; Svistoonoff et al., 2010), a broad array of genes found to be transcriptionally activated or upregulated during nodulation, many of which are involved in cell wall synthesis (reviewed in Nap and Bisseling, 1990; Frugier et al., 2008). The prominent role of CK presence and/or perception in nodule formation is emphasized by studies showing pseudonodule formation in both legumes and non-legumes due to exogenously applied CK (Arora et al., 1959; Rodriguez-Barrueco and De Castro, 1973; Relic et al., 1993) and a CK-like purine derivative secreted by a Bradyrhizobium strain that does not produce Nod factors (Giraud et al., 2007), as well as a gain-of-function mutation in a lotus histidine kinase CK receptor lhk1 that results in Rhizobium and CK-independent, spontaneous root nodule formation (Tirichine et al., 2007). CK receptors implicated in nodule development in Medicago truncatula include MtCRE1 (Gonzalez-Rizzo et al., 2006), an ortholog of Arabidopsis cytokinin receptor 1/Arabidopsis histidine kinase 4 (AHK4; Yamada et al., 2001), and CK-response regulators similar to the Arabidopsis CK-response proteins ARR4–5 (Gonzalez-Rizzo et al., 2006) and ARR10–12 (Lohar et al., 2006). TFs activated downstream of CK signaling in root cortical cells include NODULATION SIGNALING PATHWAY1 and 2 (NSP1 and NSP2; Kalo et al., 2005; Smit et al., 2005) and NODULE INCEPTION (NIN; Catoira, 2000; Borisov et al., 2003; Marsh et al., 2007). All three of these TFs are essential for nodulation, and may regulate and coordinate nodule development by regulating the expression of downstream NODULINs – genes expressed specifically during nodulation (Nap and Bisseling, 1990; Verma et al., 1992), such as EARLY NODULIN11 (ENOD11), a putative cell wall repetitive hydroxyl-proline-rich protein (Journet et al., 2001; Charron et al., 2004).
In addition to CK, a hormone network including auxin, JA, ABA, GA, salicylic acid (SA), brassinosteroids, and ethylene are also tightly regulated during nodule organogenesis (reviewed in Ferguson et al., 2010). Auxin, brassinosteroids, and GA are reported to be positive regulators of nodule formation, while ABA, JA, and ethylene are reported to be negative regulators, possibly by their involvement in plant stress and defense response pathways (reviewed in Ding and Oldroyd, 2009). Several Medicago truncatula ERFs have been found to be associated with Nod factor signal transduction, including the ERF REQUIRED FOR NODULATION (ERN; Middleton et al., 2007) and ERF REQUIRED FOR NODULE DIFFERENTIATION (EFD; Vernie et al., 2008). ABA is also thought to modulate the CK response by promoting LR growth, suppressing nodule formation, and inhibiting Rhizobium and Nod factor-induced gene expression (Ding et al., 2008). Most studies done on hormones and nodulation to date have only involved one to two hormone classes, thus a system-wide view of the interactions and effects of the major plant hormones on nodule organogenesis regulation has yet to be assembled.
PHENOTYPING PLATFORMS FOR FURTHER UNDERSTANDING OF ROOT ARCHITECTURE TRAITS
High power, high resolution GWAS and sequencing methods have far outpaced phenotyping methods necessary for the discovery of regions and underlying genes involved in plant growth and development (McNally et al., 2006; Yan et al., 2009; Huang et al., 2010; Tung et al., 2010; Zhang et al., 2010). Precise, single-trait elucidation and accurate, efficient measurement are an absolute requirement for the replicated phenotyping of large panels of individuals necessary to resolve trait–genotype associations using GWAS. Traditional methods used for root growth and architecture evaluation, such as field excavation, root bagging, plate culture, core sampling, and rhizotrons (reviewed in Shashidhar et al., 2012) are poorly suited for the large number of individuals required by GWAS due to a range of issues including low volume and sampling size, poor trait complexity resolution and measurement accuracy, and high labor, time, space, and material costs. However, these traditional approaches provide invaluable information about plant growth and yield under relevant field conditions and can be productively integrated with results from newer phenotyping platforms to provide a strong rationale for prioritizing future research.
A host of new, minimally intrusive, non-destructive, whole-root system growth systems and imaging platforms have now been developed that should revolutionize our ability to explore the genetic basic of RSA. Of these, hydroponics (Famoso et al., 2010) and gel (Fang et al., 2009; Iyer-Pascuzzi et al., 2010; Clark et al., 2011) growth systems are currently amongst those best suited for RSA trait measurement and analysis for their highly controlled and standardized rooting environments, ease in whole-root system visualization and adaptability for the imposition of environmental stresses and nutrient profiles. Both of these systems involve root growth in a non-natural, liquid or semi-solid rooting environment, however, they can require tailored adjustment for use with different plant species, and are somewhat spatially and thus developmentally limited to relatively simple root systems from small or young plants. X-ray computed tomography (Lontoc-Roy et al., 2006; Perret et al., 2007; Tracy et al., 2010), nuclear magnetic resonance (NMR; Menzel et al., 2007), laser (Braga et al., 2009), ground penetrating radar (GPR) and infrared (IR) and near-infrared (NIR) imaging systems (Tirlapur and Konig, 1999; Dokken and Davis, 2007) are advantageous in their ability to visualize plant root systems grown in soil or solid rooting media, but are currently limited by their small analysis volume and often low resolution and precision, as well as their cost, accessibility, and low-throughput.
With further advancements, NMR, GPR, and IR/NIR technologies have the greatest scale-up potential for the eventual non-destructive imaging and phenotyping of field-grown plant root systems. Although these current root growth systems and imaging technologies are still unable to accurately visualize and quantify complex, mature plant root systems grown under field conditions, they have contributed greatly to increase the precision and efficiency of 2D and 3D spatial and temporal imaging crucial for obtaining information about natural development of RSA in a solid rooting media (reviewed in Danjon and Reubens, 2007; Gregory et al., 2009). Comparative data analysis and integration, especially across controlled environment and field studies is necessary to determine whether QTLs detected by different phenotyping approaches are colocalized along the chromosomes. These regions can be targeted for further investigation to elucidate the genes and molecular mechanisms underlying the trait or phenotype(s) of interest.
The concurrent design of automated or semiautomated image capture systems and software for automated image processing, analysis, and root phenotype quantification (Armengaud et al., 2009; French et al., 2009; Famoso et al., 2010; Clark et al., 2011, 2013) are absolutely essential for simple, precise, and efficient root phenotyping with whole-root system growth platforms. These automated image capture and quantification software systems are also often easily adaptable to an array of low and high-tech growth systems, providing the potential to enhance the throughput and accuracy of root trait measurement from plants grown in a variety of growth systems. Sustained innovation inaccurate, efficient, large-scale, high-throughput root growth and analysis systems, especially those tailored toward more the complex and natural soil and field environments will continue to be essential for future studies on the association and linkage mapping of RSA traits.
UNDERSTANDING THE GENETIC AND ENVIRONMENTAL CONTROL OF WHOLE SYSTEM ARCHITECTURE
Recent development of new, non-invasive, controlled, root phenotyping techniques and the ability to accurately visualize and quantify RSA paves the way for the further development of higher throughput technologies to assist with linkage and association mapping and mutant analysis. Concurrent advances in the development of informative populations and use of the latest genotyping/sequencing techniques can allow for the faster determination of genes involved in root architectural components and the molecular mechanisms underlying the intrinsic and extrinsic pathways which control root growth and development.
The next step will be to look at this new root phenotypic data in combination with the well-studied above-ground shoot and yield related traits to determine whether any correlations may be made between root architectural traits and plant performance in different environments. Progress is being made on root–shoot hormone synthesis and signaling pathways (De Kroon et al., 2009; Puig et al., 2012), but the elucidation and integration of the complexes of molecular and hormonal networks that coordinate the developmental regulation with environmental perception and response remains an intriguing opportunity for the plant biology community and a compelling goal for plant breeders who seek new strategies for enhancing crop performance in the face of water and land shortages in the decades to come.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Janelle K. H. Jung is supported by funding from NSF Plant Genome Research Program Award #10265555, and previously supported by a USDA National Needs Fellowship from the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, under Agreement No. 2007-38420-17748. The authors would like to thank Randy Clark for developing the root trait search terms for homology searches, help in populating the rice root gene list, and for helpful discussion, Sam Leiboff for developing and running the scripts to search maize and Arabidopsis genome databases for genes involved in root growth, Charles Chen for running PICARA and Compara to search for rice homologs of maize and Arabidopsis root growth and development genes, and Cheryl Utter for assistance with manuscript submission. The authors would also like to thank the two reviewers for their helpful corrections and suggestions for revision, especially regarding recent literature on strigolactones.
SUPPLEMENTARY MATERIAL
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/Plant_Genetics_and_Genomics/10.3389/fpls.2013.00186/abstract
REFERENCES
- Abel S. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. U.S.A. 91 326–330 10.1073/pnas.91.1.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. J., Sivaguru M., Osawa H., Chung G. C., Matsumoto H. (2001). Aluminum inhibits the H(+)-ATPase activity by permanently altering the plasma membrane surface potentials in squash roots. Plant Physiol. 126 1381–1390 10.1104/pp.126.4.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V. A., Blilou I., Reis Galinha C. I., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Aida M., Vernoux T., Furutani M., Traas J., Tasaka M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129 3965 [DOI] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 824–827 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Akiyama K., Ogasawara S., Ito S., Hayashi H. (2010). Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51 1104–1117 10.1093/pcp/pcq058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R., Aloni E., Langhans M., Ullrich C. I. (2006). Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 97 883–893 10.1093/aob/mcl027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M., Hirayama T., Roman G., Nourizadeh S., Ecker J. R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152 10.1126/science.284.5423.2148 [DOI] [PubMed] [Google Scholar]
- Ané J., Kiss G., Riely B. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367 10.1126/science.1092986 [DOI] [PubMed] [Google Scholar]
- Apse M. P., Aharon G. S., Snedden W. A., Blumwald E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258 10.1126/science.285.5431.1256 [DOI] [PubMed] [Google Scholar]
- Ariel F., Diet A., Verdenaud M., Gruber V., Frugier F., Chan R., et al. (2010). Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 22 2171–2183 10.1105/tpc.110.074823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T., Kameoka H., Kyozuka J. (2012). Strigolactone positively controls crown root elongation in rice. J. Plant Growth Regul. 31 165–172 10.1007/s00344-011-9228-6 [DOI] [Google Scholar]
- Armengaud P., Zambaux K., Hills A., Sulpice R., Pattison R. J., Blatt M. R., et al. (2009). EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J. 57 945–956 10.1111/j.1365-313X.2008.03739.x [DOI] [PubMed] [Google Scholar]
- Arora N., Skoog F., Allen O. N. (1959). Kinetin-induced pseudonodules on tobacco roots. Am. J. Bot. 46 610–613 10.2307/2439306 [DOI] [Google Scholar]
- Badescu G. O., Napier R. M. (2006). Receptors for auxin: will it all end in TIRs? Trends Plant Sci 11 217–223 10.1016/j.tplants.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Bai H., Murali B., Barber K., Wolverton C. (2013). Low phosphate alters lateral root setpoint angle and gravitropism. Am. J. Bot. 100 175–182 10.3732/ajb.1200285 [DOI] [PubMed] [Google Scholar]
- Band L., Wells D., Larrieu A. (2012). Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. U.S.A. 109 4668–4673 10.1073/pnas.1201498109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano A. (2010). Root-to-shoot signal transduction in rice under salt stress. Pak. J. Bot. 42 329–339 [Google Scholar]
- Bao F., Shen J., Brady S. R., Muday G. K., Asami T., Yang Z. (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134 1624–1631 10.1104/pp.103.036897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Pant B. D., Stitt M., Scheible W.-R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141 988–999 10.1104/pp.106.079707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker S. J., Tagu D. (2000). The roles of auxins and cytokinins in mycorrhizal symbioses. J. Plant Growth Regul. 19 144–154 10.1007/s003440000021 [DOI] [PubMed] [Google Scholar]
- Bates T. R., Lynch J. P. (1996). Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 19 529–538 10.1111/j.1365-3040.1996.tb00386.x [DOI] [Google Scholar]
- Bates T. R., Lynch J. P. (2000). Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 87 958–963 10.2307/2656994 [DOI] [PubMed] [Google Scholar]
- Bauer P., Ratet P., Crespi M. D., Schultze M., Kondorosi A. (1996). Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and MsEnod12A expression patterns in alfalfa roots. Plant J. 10 91–105 10.1046/j.1365-313X.1996.10010091.x [DOI] [Google Scholar]
- Beeckman T., Burssens S., Inze D. (2001). The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 52 403–411 10.1093/jexbot/52.suppl_1.403 [DOI] [PubMed] [Google Scholar]
- Benkova E., Hejatko J. (2009). Hormone interactions at the root apical meristem. Plant Mol. Biol. 69 383–396 10.1007/s11103-008-9393-6 [DOI] [PubMed] [Google Scholar]
- Bennett M. J., Marchant A., Green H. G., May S. T., Millner P. A., Walker A. R., et al. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273 948–950 10.1126/science.273.5277.948 [DOI] [PubMed] [Google Scholar]
- Berta G., Fusconi A., Hooker J. E. (2002). “Arbuscular mycorrhizal modifications to plant root systems: scale, mechanisms and consequences,” in Mycorrhizal Technology in Agriculture: from Genes to Bioproducts eds Gianinazzi S., Schüepp H., Barea J. M., Haselwandter K. (Basel: Birkhäuser Verlag) 71–85 [Google Scholar]
- Berta G., Fusconi A., Trotta A., Scannerini S. (1990). Morphogenetic modifications induced by the mycorrhizal fungus glomus strain E3 in the root system of Allium porrum L. New Phytol. 114 207–215 10.1111/j.1469-8137.1990.tb00392.x [DOI] [Google Scholar]
- Berta G., Trotta A., Fusconi A., Hooker J. E., Munro M., Atkinson D., et al. (1995). Arbuscular mycorrhizal induced changes to plant growth and root system morphology in Prunus cerasifera. Tree Physiol. 15 281–293 10.1093/treephys/15.5.281 [DOI] [PubMed] [Google Scholar]
- Bhalerao R. P., Eklof J., Ljung K., Marchant A., Bennett M., Sandberg G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29 325–332 10.1046/j.0960-7412.2001.01217.x [DOI] [PubMed] [Google Scholar]
- Bhuja P., McLachlan K., Stephens J., Taylor G. (2004). Accumulation of 1,3-beta-D-glucans, in response to aluminum and cytosolic calcium in Triticum aestivum. Plant Cell Physiol. 45 543–549 10.1093/pcp/pch068 [DOI] [PubMed] [Google Scholar]
- Bishopp A., Help H., Helariutta Y. (2009). Cytokinin signaling during root development. Int. Rev. Cell Mol. Biol. 276 1–48 10.1016/S1937-6448(09)76001-0 [DOI] [PubMed] [Google Scholar]
- Blancaflor E. B., Masson P. H. (2003). Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol. 133 1677–1690 10.1104/pp.103.032169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Boniotti M. B., Gutierrez C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28 341–350 10.1046/j.1365-313X.2001.01160.x [DOI] [PubMed] [Google Scholar]
- Bonser A. M., Lynch J., Snapp S. (1996). Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris. New Phytol. 132 281–288 10.1111/j.1469-8137.1996.tb01847.x [DOI] [PubMed] [Google Scholar]
- Boonsirichai K., Sedbrook J. C., Chen R., Gilroy S., Masson P. H. (2003). ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell 15 2612–2625 10.1105/tpc.015560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov A. Y., Madsen L. H., Tsyganov V. E., Umehara Y., Voroshilova V. A., Batagov A. O., et al. (2003). The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 131 1009–1017 10.1104/pp.102.016071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouranis D., Buchner P., Chorianopoulou S. N., Hopkins L., Protonotarios V. E., Siyiannis V. F., et al. (2008). “Responses to sulfur limitation in maize,” in Sulfur Assimilation and Abiotic Stress in Plants eds Khan N. A., Singh S., Umar S. (Berlin: Springer; ) 1–19 10.1007/978-3-540-76326-0_1 [DOI] [Google Scholar]
- Brady S. M., Sarkar S. F., Bonetta D., McCourt P. (2003). The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34 67–75 10.1046/j.1365-313X.2003.01707.x [DOI] [PubMed] [Google Scholar]
- Braga R. A., Dupuy L., Pasqual M., Cardoso R. R. (2009). Live biospeckle laser imaging of root tissues. Eur. Biophys. J. 38 679–686 10.1007/s00249-009-0426-0 [DOI] [PubMed] [Google Scholar]
- Brunoud G., Wells D. M., Oliva M., Larrieu A., Mirabet V., Burrow A. H., et al. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482 103–106 10.1038/nature10791 [DOI] [PubMed] [Google Scholar]
- Buchner P., Takahashi H., Hawkesford M. J. (2004). Plant sulfate transporters: co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55 1765–1773 10.1093/jxb/erh206 [DOI] [PubMed] [Google Scholar]
- Budíková S. (1999). Structural changes and aluminium distribution in maize root tissues. Biol. Plant. 42 259–266 10.1023/A:1002116803679 [DOI] [Google Scholar]
- Buer C. S., Sukumar P., Muday G. K. (2006). Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 140 1384–1396 10.1104/pp.105.075671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanoni P., Nick P. (2005). Auxin-dependent cell division and cell elongation. 1-naphthaleneacetic acid and 2,4-dichlorophenoxyacetic acid activate different pathways. Plant Physiol. 137 939–948 10.1104/pp.104.053843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caniato F. F., Guimaraes C. T., Schaffert R. E., Alves V. C. M., Kochian L. V., Borem A., et al. (2007). Genetic diversity for aluminum tolerance in sorghum. Theor. Appl. Genet. 114 863–876 10.1007/s00122-006-0485-x [DOI] [PubMed] [Google Scholar]
- Cao Y. R., Chen S.-Y., Zhang J.-S. (2008). Ethylene signaling regulates salt stress response: an overview. Plant. Signal. Behav. 3 761–763 10.4161/psb.3.10.5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N., Forestan C., Canova S., Traas J., Varotto S. (2006). ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol. 142 254–264 10.1104/pp.106.080119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R. P., Beeckman T., Dhooge S., Swarup R., et al. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13 843–852 10.1105/tpc.13.4.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666 10.1105/tpc.12.9.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Grisafi P. L., Fink G. R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142 10.1101/gad.9.17.2131 [DOI] [PubMed] [Google Scholar]
- Cervantes E. (2001). ROS in root gravitropism: the auxin messengers? Trends Plant Sci. 6 556–556 10.1016/S1360-1385(01)02197-5 [DOI] [Google Scholar]
- Chandler P. M., Marion-Poll A., Ellis M., Gubler F. (2002). Mutants at the slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 129 181–190 10.1104/pp.010917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Kim Y., Lee J. (2004). Brassinolide interacts with auxin and ethylene in the root gravitropic response of maize (Zea mays). Physiol. Plant. 121 666–673 10.1111/j.0031-9317.2004.00356.x [DOI] [Google Scholar]
- Charron D., Pingret J.-L., Chabaud M., Journet E.-P., Barker D. G. (2004). Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 136 3582–3593 10.1104/pp.104.051110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., DeClerck G., Tian F., Spooner W., McCouch S., Buckler E. (2012). PICARA, an analytical pipeline providing probabilistic inference about a priori candidates genes underlying genome-wide association QTL in plants. PLoS ONE 7:e46596 10.1371/journal.pone.0046596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J. M., Reymond P., Powell G. K., Bernasconi P., Raibekas A. A., Liscum E., et al. (1998). Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282 1698–1701 10.1126/science.282.5394.1698 [DOI] [PubMed] [Google Scholar]
- Clark R. T., Famoso A. N., Zhao K., Shaff J. E., Craft E. J., Bustamante C. D., et al. (2013). High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ. 36 454–466 10.1111/j.1365-3040.2012.02587.x [DOI] [PubMed] [Google Scholar]
- Clark R. T., MacCurdy R. B., Jung J. K., Shaff J. E., McCouch S. R., Aneshansley D. J., et al. (2011). Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 156 455–465 10.1104/pp.110.169102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll M. J., Coveney K. M., Raines S. V., Mullen J. L., Hangarter R. P., Kiss J. Z. (2003). Phytochromes play a role in phototropism and gravitropism in Arabidopsis roots. Adv. Space Res. 31 2203–2210 10.1016/S0273-1177(03)00245-X [DOI] [PubMed] [Google Scholar]
- Cosgrove D. J. (2000). Loosening of plant cell walls by expansins. Nature 407 321–326 10.1038/35030000 [DOI] [PubMed] [Google Scholar]
- Costigan S. E., Warnasooriya S. N., Humphries B. A., Montgomery B. L. (2011). Root-localized phytochrome chromophore synthesis is required for photoregulation of root elongation and impacts root sensitivity to jasmonic acid in Arabidopsis. Plant Physiol. 157 1138–1150 10.1104/pp.111.184689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert Y., Perin C., Courtois B., Khong N. G., Gantet P. (2010). Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15 219–226 10.1016/j.tplants.2010.01.008 [DOI] [PubMed] [Google Scholar]
- Cui H., Levesque M. P., Vernoux T., Jung J. W., Paquette A. J., Gallagher K. L., et al. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316 421–425 10.1126/science.1139531 [DOI] [PubMed] [Google Scholar]
- Dan H., Yang G., Zheng A.-L. (2007). A negative regulatory role for auxin in sulfate deficiency response in Arabidopsis thaliana. Plant Mol. Biol. 63 221–235 10.1007/s11103-006-9084-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danjon F., Reubens B. (2007). Assessing and analyzing 3D architecture of woody root systems, a review of methods and applications in tree and soil stability, resource acquisition and allocation. Plant Soil 303 1–34 10.1007/s11104-007-9470-7 [DOI] [Google Scholar]
- Deak K. I., Malamy J. (2005). Osmotic regulation of root system architecture. Plant J. 43 17–28 10.1111/j.1365-313X.2005.02425.x [DOI] [PubMed] [Google Scholar]
- De Kroon H., Visser E. J. W., Huber H., Mommer L., Hutchings M. J. (2009). A modular concept of plant foraging behaviour: the interplay between local responses and systemic control. Plant Cell Environ. 32 704–712 10.1111/j.1365-3040.2009.01936.x [DOI] [PubMed] [Google Scholar]
- Delhaize E., Ryan P. R., Randall P. J. (1993). Aluminum tolerance in wheat (Triticum aestivum L.) (II. Aluminum-stimulated excretion of malic acid from root apices). Plant Physiol. 103 695–702 10.1104/pp.103.3.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F. S., Sabatini S. (2008). Emerging role of cytokinin as a regulator of cellular differentiation. Curr. Opin. Plant Biol. 11 23–27 10.1016/j.pbi.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Cuin T. A., Svistunenko D., Smith S. J., Miller A. J., Shabala S., et al. (2010). Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci. 123 1468–1479 10.1242/jcs.064352 [DOI] [PubMed] [Google Scholar]
- De Rybel B., Vassileva V., Grunewald W., Naudts M., Levesque M. P., Ehrismann J. S., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107 2705–2710 10.1073/pnas.0915001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Lau S., Voss U., Vanneste S., Benjamins R., Rademacher E. H., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107 2705–2710 10.1073/pnas.0915001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Signora L., Beeckman T., Inze D., Foyer C., Zhang H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33 543–555 10.1046/j.1365-313X.2003.01652.x [DOI] [PubMed] [Google Scholar]
- De Smet I., Tetsumura T., De Rybel B., Frei dit Frey N., Laplaze L., Casimiro I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134 681–690 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- De Smet I., Zhang H., Inzé D., Beeckman T. (2006). A novel role for abscisic acid emerges from underground. Trends Plant Sci. 11 434–439 10.1016/j.tplants.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Devaiah B. N., Karthikeya A. S., Raghothama K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143 1789–1801 10.1104/pp.106.093971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B. N., Raghothama K. G. (2007). Transcriptional regulation of Pi starvation responses by WRKY75. Plant Signal. Behav. 2 424–425 10.4161/psb.2.5.4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- DiDonato R. J., Arbuckle E., Buker S., Sheets J., Tobar J., Totong T., et al. (2004). Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J. 37 340–353 10.1046/j.1365-313X.2003.01964.x [DOI] [PubMed] [Google Scholar]
- Digby J., Firn R. D. (1995). The gravitropic set-point angle (GSA): the identification of an important developmentally controlled variable governing plant architecture. Plant Cell Environ. 18 1434–1440 10.1111/j.1365-3040.1995.tb00205.x [DOI] [PubMed] [Google Scholar]
- Digby J., Firn R. D. (2002). Light modulation of the gravitropic set-point angle (GSA). J. Exp. Bot. 53 377–381 10.1093/jexbot/53.367.377 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J. E., Pysh L., Helariutta Y., Freshour G., et al. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433 10.1016/S0092-8674(00)80115-4 [DOI] [PubMed] [Google Scholar]
- Dill A., Sun T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimkpa C., Weinand T., Asch F. (2009). Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32 1682–1694 10.1111/j.1365-3040.2009.02028.x [DOI] [PubMed] [Google Scholar]
- Ding Y., Kalo P., Yendrek C., Sun J., Liang Y., Marsh J. F., et al. (2008). Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20 2681–2695 10.1105/tpc.108.061739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Oldroyd G. E. (2009). Positioning the nodule, the hormone dictum. Plant Signal. Behav. 4 89–93 10.4161/psb.4.2.7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. K. (1990). Cytokinin activity in citrus jambhiri seedlings colonized by mycorrhizal fungi. Agric. Ecosyst. Environ. 29 103–106 10.1016/0167-8809(90)90262-C [DOI] [Google Scholar]
- Dokken K. M., Davis L. C. (2007). Infrared imaging of sunflower and maize root anatomy. J. Agric. Food Chem. 55 10517–10530 10.1021/jf072052e [DOI] [PubMed] [Google Scholar]
- Dubrovsky J. G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M. G., Friml J., Shishkova S., et al. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. U.S.A. 105 8790–8794 10.3410/f.1115286.571292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann H. G., Roth U. (2006). Gravitropic plant growth regulation and ethylene: an unsought cardinal coordinate for a disused model. Protoplasma 229 183–191 10.1007/s00709-006-0205-z [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G. B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966 10.1038/nature00842 [DOI] [PubMed] [Google Scholar]
- Eticha D., Stass A., Horst W. J. (2005). Cell-wall pectin and its degree of methylation in the maize root-apex: significance for genotypic differences in aluminium resistance. Plant Cell Environ. 28 1410–1420 10.1111/j.1365-3040.2005.01375.x [DOI] [Google Scholar]
- Evelin H., Kapoor R., Giri B. (2009). Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Ann. Bot. 104 1263–1280 10.1093/aob/mcp251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahn A. (1990). Plant Anatomy. 4th Edn New York: Pergamon [Google Scholar]
- Famoso A. N., Clark R. T., Shaff J. E., Craft E., McCouch S. R., Kochian L. V. (2010). Development of a novel aluminum tolerance phenotyping platform used for comparisons of cereal aluminum tolerance and investigations into rice aluminum tolerance mechanisms. Plant Physiol. 153 1678–1691 10.1104/pp.110.156794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Yan X., Liao H. (2009). 3D reconstruction and dynamic modeling of root architecture in situ and its application to crop phosphorus research. Plant J. 60 1096–1108 10.1111/j.1365-313X.2009.04009.x [DOI] [PubMed] [Google Scholar]
- Fang Y., Hirsch A. M. (1998). Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 116 53–68 10.1104/pp.116.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrás R., Ferrando A., Jásik J., Kleinow T., Okrész L., Tiburcio A., et al. (2001). SKP1–SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20 2742–2756 10.1093/emboj/20.11.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. J., Indrasumunar A., Hayashi S., Lin M.-H., Lin Y.-H., Reid D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52 61–76 10.1111/j.1744-7909.2010.00899.x [DOI] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., Van der Schors R., Van der Geest L., Li K. W., Stiekema W. J., et al. (2006). The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 141 1284–1292 10.1104/pp.106.080671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Hause G., Boutilier K., Casamitjana-Martinez E., Weijers D., Offringa R., et al. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327 37–49 10.1016/j.gene.2003.11.014 [DOI] [PubMed] [Google Scholar]
- Fitter A. H. (1991). “The ecological significance of root system architecture: an economic approach,” in Plant Root Growth: An Ecological Perspective ed Atkinson D. (Oxford: Blackwell Scientific Publications) 229–243 [Google Scholar]
- Fitze D., Wiepning A., Kaldorf M., Lidwig-Muller J. (2005). Auxins in the development of an arbuscular mycorrhizal symbiosis in maize. J. Plant Physiol. 162 1210–1219 10.1016/j.jplph.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Fleet C. M., Sun T. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85 10.1016/j.pbi.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Flowers T. J., Yeo A. R. (1995). Breeding for salinity resistance in crop plants: where next? Aust. J. Plant Physiol. 22 875–884 10.1071/PP9950875 [DOI] [Google Scholar]
- Foo E., Yoneyama K., Hugill C. J., Quittenden L. J., Reid J. B. (2012). Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant 6 76–87 10.1093/mp/sss115 [DOI] [PubMed] [Google Scholar]
- Foy C. D. (1984). Adaptation of plants to mineral stress in problem soils. Ciba Found. Symp. 102 20–39 10.1002/9780470720837.ch3 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Martin A. C., Solano R., Rubio V., Leyva A., Paz-Ares J. (2002). Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 32 353–360 10.1046/j.1365-313X.2002.01431.x [DOI] [PubMed] [Google Scholar]
- French A., Ubeda-Tomas S., Holman T. J., Bennett M. J., Pridmore T. (2009). High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol. 150 1784–1795 10.1104/pp.109.140558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio M., Alabadi D., Perez-Gomez J., Garica-Carcel L., Phillips A. L., Hedden P., et al. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142 553–563 10.1104/pp.106.084871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier F., Kosuta S., Murray J. D., Crespi M., Szczyglowshi K. (2008). Cytokinin: secret agent of symbiosis. Trends Plant Sci. 13 115–120 10.1016/j.tplants.2008.01.003 [DOI] [PubMed] [Google Scholar]
- Fu X., Harberd N. P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743 10.1038/nature01387 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Okushima Y., Tasaka M. (2007). Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 256 111–137 10.1016/S0074-7696(07)56004-3 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tasaka M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69 437–449 10.1007/s11103-008-9417-2 [DOI] [PubMed] [Google Scholar]
- Fukao T., Yeung E., Bailey-Serres J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23 412–427 10.1105/tpc.110.080325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. M., Downes B. P., Shiu S.-H., Durski A. M., Vierstra R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99 11519–11524 10.1073/pnas.162339999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen C., Rabenold J. J., Liscum E. (2007). Functional ecology of a blue light photoreceptor: effects of phototropin-1 on root growth enhance drought tolerance in Arabidopsis thaliana. New Phytol. 173 91–99 10.1111/j.1469-8137.2006.01893.x [DOI] [PubMed] [Google Scholar]
- Gao Q., Zhao M., Li F., Guo Q., Xing S., Wang W. (2008). Expansins and coleoptile elongation in wheat. Protoplasma 233 73–81 10.1007/s00709-008-0303-1 [DOI] [PubMed] [Google Scholar]
- Gaxiola R. A., Rao R., Sherman A., Grisafi P., Alper S. L., Fink G. R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. U.S.A. 96 1480–1485 10.1073/pnas.96.4.1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., et al. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230 10.1016/S0092-8674(03)00003-5 [DOI] [PubMed] [Google Scholar]
- Gil P., Dewey E., Friml J., Zhao Y., Snowden K. C., Putterill J., et al. (2001). BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15 1985–1997 10.1101/gad.905201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Moulin L., Vallenet D., Barbe V., Cytryn E., Avarre J.-C., et al. (2007). Legumes symbioses: absence of nod genes in photosynthetic bradyrhizobia. Science 316 1307–1312 10.1126/science.1139548 [DOI] [PubMed] [Google Scholar]
- Goh T., Joi S., Mimura T., Fukaki H. (2012a). The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139 883–893 10.1242/dev.071928 [DOI] [PubMed] [Google Scholar]
- Goh T., Kasahara H., Mimura T., Kamiya Y., Fukaki H. (2012b). Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 1461–1468 10.1098/rstb.2011.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693 10.1105/tpc.106.043778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J., Strauss S. H., Tsai C. J., Fang K., Chen Y., Jiang X., et al. (2010). Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 22 623–639 10.1105/tpc.109.073239PMCid:150761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda V. R. P. V., Henry A., Yamauchi A., Shashidhar H. E., Serraj R. (2011). Root biology and genetic improvement for drought avoidance in rice. Field Crops Res. 122 1–13 10.1016/j.fcr.2011.03.001 [DOI] [Google Scholar]
- Gray W. M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414 271–276 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Gregory P. J., Bengough A. G., Grinev D., Schmidt S., Thomas W. T. B., Wojciechowski T., et al. (2009). Root phenomics of crops: opportunities and challenges. Funct. Plant Biol. 36 922–929 10.1071/FP09150 [DOI] [PubMed] [Google Scholar]
- Groth P., Kalev I., Kirov I., Traikov B., Leser U., Weiss B. (2010). Phenoclustering: online mining of cross-species phenotypes. Bioinformatics 26 1924–1925 10.1093/bioinformatics/btq311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F. Q., Wang R., Crawford N. M. (2002). The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J. Exp. Bot 53 835–844 10.1093/jexbot/53.370.835 [DOI] [PubMed] [Google Scholar]
- Guo H.-S., Xie Q., Fei J.-F., Chua N.-H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17 1376–1386 10.1105/tpc.105.030841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Casieri L., Paszkowski U. (2009). Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytol. 184 829–837 10.1111/j.1469-8137.2009.02839.x [DOI] [PubMed] [Google Scholar]
- Harrison B., Masson P. H. (2008a). ARG1 and ARL2 form an actin-based gravity-signaling chaperone complex in root statocytes? Plant Signal. Behav. 3 650–653 10.4161/psb.3.9.5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B. R., Masson P. H. (2008b). ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 53 380–392 10.1111/j.1365-313X.2007.03351.x [DOI] [PubMed] [Google Scholar]
- He X. J., Mu R.-L., Cao W.-H., Zhang Z.-G., Zhang J.-S., Chen S.-Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44 903–916 10.1111/j.1365-313X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 555–567 10.1016/S0092-8674(00)80865-X [DOI] [PubMed] [Google Scholar]
- Henrissat B., Davies G. (1997). Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7 637–644 10.1016/S0959-440X(97)80072-3 [DOI] [PubMed] [Google Scholar]
- Herrera-Medina M. J., Steinkellner S., Vierheilig H., Bote J. A. O, Garrido J. M. G. (2007). Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 175 554–564 10.1111/j.1469-8137.2007.02107.x [DOI] [PubMed] [Google Scholar]
- Hetrick B. A. D. (1991). Mycorrhizas and root architecture. Cell. Mol. Life Sci. 47 355–362 10.1007/BF01972077PMCid:151221 [DOI] [Google Scholar]
- Hetrick B. A. D., Leslie J. F., Thompson Wilson G., Gerschefske Kitt D. (1988). Physical and topological assessment of effects of a vesicular-arbuscular mycorrhizal fungus on root architecture of big bluestem. New Phytol. 110 85–96 10.1111/j.1469-8137.1988.tb00240.x [DOI] [Google Scholar]
- Hetz W., Hochholdinger F., Schwall M., Feix G. (1996). Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J. 10 845–857 10.1046/j.1365-313X.1996.10050845.x [DOI] [Google Scholar]
- Himanen K., Boucheron E., Vanneste S., De Almeida Engler J., Inze D., Beechmann T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351 10.1105/tpc.004960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Fujiwara T., Awazuhara M., Kimura T., Noji M., Siato K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33 651–663 10.1046/j.1365-313X.2003.01658.x [DOI] [PubMed] [Google Scholar]
- Hirose N., Makita N., Kojima M., Kamada-Nobusada T., Sakakibara H. (2007). Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48 523–539 10.1093/pcp/pcm022 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F. (2009). “The maize root system: morphology, anatomy and genetics,” in Handbook of Maize: Its Biology eds Bennetzen J. L., Hake S. C. (New York: Springer) 145–160 10.1007/978-0-387-79418-1_8 [DOI] [Google Scholar]
- Hochholdinger F., Feix G. (1998). Early post-embryonic root formation is specifically affected in the maize mutant Irt1. Plant J. 16 247–255 10.1046/j.1365-313x.1998.00280.x [DOI] [PubMed] [Google Scholar]
- Hochholdinger F., Park W. J., Feix F. H. (2001). Cooperative action of SLR1 and SLR2 is required for lateral root-specific cell elongation in maize. Plant Physiol. 125 1529–1539 10.1104/pp.125.3.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F., Park W. J., Sauer M., Woll K. (2004). From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci. 9 42–48 10.1016/j.tplants.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F., Tuberosa R. (2009). Genetic and genomic dissection of maize root development and architecture. Curr. Opin. Plant Biol. 12 172–177 10.1016/j.pbi.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F., Zimmermann R. (2008). Conserved and diverse mechanisms in root development. Curr. Opin. Plant Biol. 11 70–74 10.1016/j.pbi.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Hoefgen R., Nikiforova V. J. (2008). Metabolomics integrated with transcriptomics: assessing systems response to sulfur-deficiency stress. Physiol. Plant. 132 190–198 10.1111/j.1399-3054.2007.01012.x [DOI] [PubMed] [Google Scholar]
- Hogg B. V., Cullimore J. V., Ranjeva R., Bono J.-J. (2006). The DMI1 and DMI2 early symbiotic genes of Medicago truncatula are required for a high-affinity nodulation factor-binding site associated to a particulate fraction of roots. Plant Physiol. 140 365–373 10.1104/pp.105.068981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Costa A., Kim T. H., Han M. J., Horie R., Leung H.-Y., et al. (2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 26 3003–3014 10.1038/sj.emboj.7601732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Hauser F., Schroeder J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14 660–668 10.1016/j.tplants.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst W. J., Wang Y., Eticha D. (2010). The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: a review. Ann. Bot. 106 185–197 10.1093/aob/mcq053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B., Yeun L. H., Domonkos A., Halász G., Gobbato E., Ayaydin F., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24 1345–1358 10.1094/MPMI-01-11-0015 [DOI] [PubMed] [Google Scholar]
- Huala E., Oeller P. W., Liscum E., Han I.-S., Larsen E., Briggs W. R. (1997). Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278 2120–2123 10.1126/science.278.5346.2120 [DOI] [PubMed] [Google Scholar]
- Huang C. F., Yamaji N., Mitani N., Yano M., Nagamura Y., Ma F. (2009a). A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21 655–667 10.1105/tpc.108.064543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Xia H., Li Z., Xiong Y., Kong G. (2009b). Soil aluminum uptake and accumulation by Paspalum notatum. Waste Manag. Res. 27 668–675 10.1177/0734242X09103835 [DOI] [PubMed] [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42 961–967 10.1038/ng.695 [DOI] [PubMed] [Google Scholar]
- Ikeda A., Sonoda Y., Vernieri P., Perata P., Hirochika H., Yamaguchi J. (2002). The slender rice mutant, with constitutively activated gibberellin signal transduction, has enhanced capacity for abscisic acid level. Plant Cell Physiol. 43 974–979 10.1093/pcp/pcf115 [DOI] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., et al. (2001). Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010 10.1105/tpc.13.5.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., Takeda N., Charpentier M. (2004). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531 10.1038/nature03237 [DOI] [PubMed] [Google Scholar]
- Inukai Y., Miwa M., Nagato Y., Kitano H., Yamauchi A. (2001). Characterization of rice mutants deficient in the formation of crown roots. Breed. Sci. 51 123–129 10.1270/jsbbs.51.123 [DOI] [Google Scholar]
- Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., et al. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17 1387–1396 10.1105/tpc.105.030981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J., Nonomura K.-I., Ikeda K., Yamaki S., Inukai Y., Yamagushi H., et al. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46 23–47 10.1093/pcp/pci501 [DOI] [PubMed] [Google Scholar]
- Iwama A., Yamashino T., Tanaka Y., Sakakibara H., Kakimoto T., Sato S., et al. (2007). AHK5 histidine kinase regulates root elongation through an ETR1-dependent abscisic acid and ethylene signaling pathway in Arabidopsis thaliana. Plant Cell Physiol. 48 375–380 10.1093/pcp/pcl065 [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A. S., Symonova O., Mileyko Y., Hao Y., Belcher H., Harer J., et al. (2010). Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 152 1148–1157 10.1104/pp.109.150748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Tyagi A. K., Khurana J. P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). Plant Biol. 6 1 10.1146/annurev.arplant.52.1.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebanathirajah J. A., Peri S., Pandey A. (2002). Toll and interleukin-1 receptor (TIR) domain-containing proteins in plants: a genomic perspective. Trends Plant Sci. 7 388–391 10.1016/S1360-1385(02)02309-9 [DOI] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N. P., Fu X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145 1460–1470 10.1104/pp.107.103788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. L., Blancaflor E. B., Kochian L. V., Gilroy S. (2006). Spatial coordination of aluminium uptake, production of reactive oxygen species, callose production and wall rigidification in maize roots. Plant Cell Environ. 29 1309–1318 10.1111/j.1365-3040.2006.01509.x [DOI] [PubMed] [Google Scholar]
- Joo J. H., Bae Y. S., Lee J. S. (2001). Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol. 126 1055 10.1104/pp.126.3.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E. P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., et al. (2001). Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14 737–748 10.1094/MPMI.2001.14.6.737 [DOI] [PubMed] [Google Scholar]
- Kalo P., Gleason C., Edwards A., Marsh J. M., Mitra R. M., Hirsch S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789 10.1126/science.1110951 [DOI] [PubMed] [Google Scholar]
- Kanamori N., Madsen L. H., Radutoiu S., Frantescu M., Quistgaard E. M. H., Miwa H., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103 359–364 10.1073/pnas.0508883103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y., Delaux P. P.-M., Resnick N., Mayzlish-Gati E., Wininger S., Bhattacharya C., et al. (2011). Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233 209–216 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- Kartal G., Temel A., Arican E., Gozukirmizi N. (2009). Effects of brassinosteroids on barley root growth, antioxidant system and cell division. Plant Growth Regul. 58 261–267 10.1007/s10725-009-9374-z [DOI] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Khadri M., Tejera N. A., Lluch C. (2006). Alleviation of salt stress in common bean (Phaseolus vulgaris) by exogenous abscisic acid supply. J. Plant Growth Regul. 25 110–119 10.1007/s00344-005-0004-3 [DOI] [Google Scholar]
- Khan G., Declerck M., Sorin C. (2011). MicroRNAs as regulators of root development and architecture. Plant Mol. Biol. 77 47–58 10.1007/s11103-011-9793-x [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Lee S. M., Joo S.-H., Yun H. S., Lee Y., Kaufman P. B., et al. (2007). Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ. 30 679–689 10.1111/j.1365-3040.2007.01659.x [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., Sawa S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48 1821–1825 10.1093/pcp/pcm154 [DOI] [PubMed] [Google Scholar]
- Kiss J. Z., Mullen J. L., Correll M. J., Hangarter R. P. (2003). Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 131 1411–1417 10.1104/pp.013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., et al. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17 2217–2229 10.1105/tpc.105.032714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug B., Horst W. J. (2010). Spatial characteristics of aluminum uptake and translocation in roots of buckwheat (Fagopyrum esculentum). Physiol. Plant. 139 181–191 10.1111/j.1399-3054.2010.01355.x [DOI] [PubMed] [Google Scholar]
- Kochian L. V. (1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46 237 10.1146/annurev.pp.46.060195.001321 [DOI] [Google Scholar]
- Kohlen W., Charnikhova T., Liu Q. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host. Plant Physiol. 155 974–987 10.1104/pp.110.164640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmeier M., Felle H. H., Horst W. J. (2000). Genotypical differences in aluminum resistance of maize are expressed in the distal part of the transition zone. Is reduced basipetal auxin flow involved in inhibition of root elongation by aluminum? Plant Physiol. 122 945–956 10.1104/pp.122.3.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H., Dor E., Hershenhorn J., Joel D. M., Weininger S., Lekalla S., et al. (2010). Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J. Plant Growth Regul. 29 129–136 10.1007/s00344-009-9122-7 [DOI] [Google Scholar]
- Kopittke P., Blamey F. P. C., Menzies N. W. (2008). Toxicities of soluble Al, Cu, and La include ruptures to rhizodermal and root cortical cells of cowpea. Plant Soil 303 217–227 10.1007/s11104-007-9500-5 [DOI] [Google Scholar]
- Kosuta S., Chabaud M., Lougnon G., Cough C., Denarie J., Barker D. G., et al. (2003). A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol. 131 952–962 10.1104/pp.011882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R. J., Downie J. A., et al. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. U.S.A. 105 9823–9828 10.1073/pnas.0803499105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E. M. (2004). PIN and AUX/LAX proteins: their role in auxin accumulation. Trends Plant Sci. 9 578–582 10.1016/j.tplants.2004.10.010 [DOI] [PubMed] [Google Scholar]
- Krouk G., Lacombe B., Bielach A., Perrine-Walker F., Malinska K., Mounier E., et al. (2010). Nitrate-regulated Auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18 927–937 10.1016/j.devcel.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Kuderová A., Urbánková I., Válková M., Malbeck J., Brzobohaty B., Némethová D., et al. (2008). Effects of conditional IPT-dependent cytokinin overproduction on root architecture of Arabidopsis seedlings. Plant Cell Physiol. 49 570–582 10.1093/pcp/pcn073 [DOI] [PubMed] [Google Scholar]
- Kuiper D., Schuit J., Kuiper P. J. C. (1990). Actual cytokinin concentrations in plant tissue as an indicator for salt resistance in cereals. Plant Soil 123 243–250 10.1007/BF00011276 [DOI] [Google Scholar]
- Kurata T., Yamamoto K. T. (1997). Light-stimulated root elongation in Arabidopsis thaliana. J. Plant Physiol. 151 346–351 10.1016/S0176-1617(97)80263-5 [DOI] [Google Scholar]
- Kurth E., Cramer G. R., Lauchli A., Epstein E. (1986). Effects of NaCl and CaCl(2) on cell enlargement and cell production in cotton roots. Plant Physiol. 82 1102–1106 10.1104/pp.82.4.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz A., Muller A., Hennig P., Kaiser W. M., Piotrowski M., Weiler E. W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30 95–106 10.1046/j.1365-313X.2002.01271.x [DOI] [PubMed] [Google Scholar]
- Kwon H.-K., Yokoyama R., Nishitani K. (2005). A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 46 843–857 10.1093/pcp/pci089 [DOI] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., Sasidharan R., et al. (2012). The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40 D1202–D1210 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L., Benkova E., Casimiro I., Maes L., Vanneste S., Swarup R., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 3889–3900 10.1105/tpc.107.055863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P. B., Cancel J., Rounds M., Ochoa V. (2007). Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225 1447–1458 10.1007/s00425-006-0452-4 [DOI] [PubMed] [Google Scholar]
- Laskowski M., Biller S., Stanley K., Kajstura T., Prusty R. (2006). Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 47 788–792 10.1093/pcp/pcj043 [DOI] [PubMed] [Google Scholar]
- Laskowski M. J., Williams M. E., Nusbaum H. C., Sussex I. E. (1995). Formation of lateral root meristems is a two-stage process. Development 121 3303–3310 [DOI] [PubMed] [Google Scholar]
- Lawrence C., Harper L. (2008). MaizeGDB: the maize model organism database for basic, translational, and applied research. Int. J. Plant Genomics 2008 496957 10.1155/2008/496957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE 3:e1510 10.1371/journal.pone.0001510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. W., Kim N. Y., Lee D. J., Kim J. (2009). LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151 1377–1389 10.1104/pp.109.143685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kende H. (2002). Expression of alpha-expansin and expansin-like genes in deepwater rice. Plant Physiol. 130 1396–1405 10.1104/pp.008888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais-Briere C., Jovanovic M., Torres G. M., Perrin Y., Lemoine R., Corre-Menguy F., et al. (2007). Disruption of AtOCT1, an organic cation transporter gene, affects root development and carnitine-related responses in Arabidopsis. Plant J. 51 154–164 10.1111/j.1365-313X.2007.03131.x [DOI] [PubMed] [Google Scholar]
- Leng Q., Mercier R. W., Hua B.-G., Fromm H., Berkowitz G. A. (2002). Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 128 400–410 10.1104/pp.010832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T., Martin M. N., Bick J.-A., Davies J. P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 141–165 10.1146/annurev.arplant.51.1.141 [DOI] [PubMed] [Google Scholar]
- Lévy J., Bres C., Geurts R. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364 10.1126/science.1093038 [DOI] [PubMed] [Google Scholar]
- Lewandowska M., Sirko A. (2008). Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim. Pol. 55 457–471 [PubMed] [Google Scholar]
- Li J., Zhu S., Song X., Shen Y., Chen H., Yu J., et al. (2006). A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell 18 340–349 10.1105/tpc.105.037713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Jiang A.-L., Zhang W. (2007). Salt stress-induced programmed cell death in rice root tip cells. J. Integr. Plant Biol. 49 481–486 10.1111/j.1744-7909.2007.00445.x [DOI] [Google Scholar]
- Linkohr B. I., Williamson L. C., Fitter A. H., Leyser O. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29 751–760 10.1046/j.1365-313X.2002.01251.x [DOI] [PubMed] [Google Scholar]
- Little D. Y., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J. E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. U.S.A. 102 13693–13698 10.1073/pnas.0504219102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang S., Yu X., Yu J., He X., Zhang S., et al. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43 47–56 10.1111/j.1365-313X.2005.02434.x [DOI] [PubMed] [Google Scholar]
- Liu J., Zhu J. K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945 10.1126/science.280.5371.1943 [DOI] [PubMed] [Google Scholar]
- Liu K.-H., Huang C.-Y., Tsay Y.-F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11 865–874 10.1105/tpc.11.5.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wang J., Wang L., Wang X., Xue Y., Wu P., et al. (2009). Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 19 1110–1119 10.1038/cr.2009.70 [DOI] [PubMed] [Google Scholar]
- Lohar D. P., Schaff J. E., Laskey J. G., Kieber J. J., Bilyeu K. D., Bird D. M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 38 203–214 10.1111/j.1365-313X.2004.02038.x [DOI] [PubMed] [Google Scholar]
- Lohar D. P., Sharopova N., Endre G., Penuela S., Samac D., Town C., et al. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140 221–234 10.1104/pp.105.070326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontoc-Roy M., Dutilleul P., Prasher S. O., Han L., Brouillet T., Smith D. L. (2006). Advances in the acquisition and analysis of CT scan data to isolate a crop root system from the soil medium and quantify root system complexity in 3-D space. Geoderma 137 231–241 10.1016/j.geoderma.2006.08.025 [DOI] [Google Scholar]
- López-Bucio J., Hernandez-Abreu E., Sanchez-Calderon L., Nieto-Jacobo M. F., Simpson J., Herrera-Estrella L. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129 244–256 10.1104/pp.010934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J., Hernandez-Abreu E., Sanchez-Calderon L., Perez-Torres A., Rampey R. A., Bartel B., et al. (2005). An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 137 681–691 10.1104/pp.104.049577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez J. A., Bouwmeester H. (2008). Fine-tuning regulation of strigolactone biosynthesis under phosphate starvation. Plant Signal. Behav. 3 963–965 10.1111/j.1469-8137.2008.02406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorbiecke R., Sauter M. (1999). Adventitious root growth and cell-cycle induction in deepwater rice. Plant Physiol. 119 21–30 10.1104/pp.119.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M., Godin C., Jay-Allemand C., Laplaze L. (2008). Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J. Exp. Bot. 59 55–66 10.1093/jxb/erm171 [DOI] [PubMed] [Google Scholar]
- Lucas M., Swarup R., Paponov I. (2011). Short-root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 155 384–398 10.1104/pp.110.165126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Muller J., Guther M. (2007). Auxins as signals in arbuscular mycorrhiza formation. Plant Signal. Behav. 2 194–196 10.4161/psb.2.3.4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. B., Janz D., Jiang X., Gobel C., Wildhagen H., Tan Y., et al. (2009). Upgrading root physiology for stress tolerance by ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol. 151 1902–1917 10.1104/pp.109.143735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C. (2002). Auxin transport: ABC proteins join the club. Trends Plant Sci. 7 329–332 10.1016/S1360-1385(02)02292-6 [DOI] [PubMed] [Google Scholar]
- Lynch J. P., Brown K. M. (2001). Topsoil foraging – an architectural adaptation of plants to low phosphorus availability. Plant Soil 237 225–237 10.1023/A:1013324727040 [DOI] [Google Scholar]
- Lynch J. P., Brown K. M. (2012). New roots for agriculture: exploiting the root phenome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 1598–1604 10.1098/rstb.2011.0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. F., Furukawa J. (2003). Recent progress in the research of external Al detoxification in higher plants: a minireview. J. Inorg. Biochem. 97 46–51 10.1016/S0162-0134(03)00245-9 [DOI] [PubMed] [Google Scholar]
- Ma J. F., Ryan P. R., Delaize E. (2001). Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6 273–278 10.1016/S1360-1385(01)01961-6 [DOI] [PubMed] [Google Scholar]
- Ma J. F., Shen R., Zhao Z., Wissuwa M., Takeuchi Y., Ebitani T., et al. (2002). Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol. 43 652–659 10.1093/pcp/pcf081 [DOI] [PubMed] [Google Scholar]
- Ma Z., Baskin T. I., Brown K. M., Lynch J. P. (2003). Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol. 131 1381–1390 10.1104/pp.012161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor D. R., Deak K. I., Ingram P. A., Malamy J. E. (2008). Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20 2643–2660 10.1105/tpc.107.055475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes J. V., Garvin D. F., Wang Y., Sorrells M. E., Klein P. E., Schaffert R. E., et al. (2004). Comparative mapping of a major aluminum tolerance gene in Sorghum and other species in the Poaceae. Genetics 167 1905–1914 10.1534/genetics.103.023580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S., Tuteja N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444 139–158 10.1016/j.abb.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Malamy J. E. (2005). Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28 67–77 10.1111/j.1365-3040.2005.01306.x [DOI] [PubMed] [Google Scholar]
- Malamy J. E., Benfey P. N. (1997). Down and out in Arabidopsis: the formation of lateral roots. Trends Plant Sci. 2 390–396 10.1016/S1360-1385(97)90054-6 [DOI] [Google Scholar]
- Malamy J. E., Ryan K. S. (2001). Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 127 899–909 10.1104/pp.010406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C. Z., Yang L., Zheng B.-S., Wu Y.-R., Liu F.-Y., Wu P. (2004). Comparative mapping of QTLs for Al tolerance in rice and identification of positional Al-induced genes. J. Zhejiang Univ. Sci. 5 634–643 10.1631/jzus.2004.0634 [DOI] [PubMed] [Google Scholar]
- Maraschin F. D. S., Memelink J., Offringa R. (2009). Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 59 100–109 10.1111/j.1365-313X.2009.03854.x [DOI] [PubMed] [Google Scholar]
- Marin-Rodriguez M. C., Orchard J., Seymour G. B. (2002). Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53 2115–2119 10.1093/jxb/erf089 [DOI] [PubMed] [Google Scholar]
- Markmann K., Parniske M. (2009). Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci. 14 77–86 10.1016/j.tplants.2008.11.009 [DOI] [PubMed] [Google Scholar]
- Marsh J. F., Rakocevic A., Mitra R. M., Brocard L., Sun J., Eschstruth A., et al. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144 324–335 10.1104/pp.106.093021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Inoue E., Watanabe-Takahashi A., Yamaya T., Takahashi H. (2003). Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132 597–605 10.1104/pp.102.019802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Yamaya T., Takahashi H. (2004). Regulation of high-affinity sulfate transporters in plants: towards systematic analysis of sulphur signaling and regulation. J. Exp. Bot. 55 1843–1849 10.1093/jxb/erh175 [DOI] [PubMed] [Google Scholar]
- Matsumoto H. (2000). Cell biology of aluminum toxicity and tolerance in higher plants. Int. Rev. Cytol. 200 1–46 10.1016/S0074-7696(00)00001-2 [DOI] [PubMed] [Google Scholar]
- Matsumoto H., Senoo Y., Kasai M., Maeshima M. (1996). Response of the plant root to aluminum stress: analysis of the inhibition of the root elongation and changes in membrane function. J. Plant Res. 109 99–105 10.1007/BF02344294 [DOI] [Google Scholar]
- Mayzlish-Gati E., De-Cuyper C., Goormachtig S., Beeckman T., Vuylsteke M., Brewer P. B., et al. (2012). Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol. 160 1329–1341 10.1104/pp.112.202358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K. L., Bruskiewich R., Mackill D., Buell C. R., Leach J. E., Leung H. (2006). Sequencing multiple and diverse rice varieties. Connecting whole-genome variation with phenotypes. Plant Physiol. 141 26–31 10.1104/pp.106.077313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M., Murray J. (2000). The role and regulation of D-type cyclins in the plant cell cycle. Plant Mol. Biol. 43 621–633 10.1023/A:1006482115915 [DOI] [PubMed] [Google Scholar]
- Meng Y., Ma X., Chen D., Wu P., Chen M. (2010). MicroRNA-mediated signaling involved in plant root development. Biochem. Biophys. Res. Commun. 393 345–349 10.1016/j.bbrc.2010.01.129 [DOI] [PubMed] [Google Scholar]
- Menzel M. I., Oros-Peusquens A.-M., Pohlmeier A., Shah J., Schurr U., Schneider H. U. (2007). Comparing 1H-NMR imaging and relaxation mapping of German white asparagus from five different cultivation sites. J. Plant Nutr. Soil Sci. 170 24–38 10.1002/jpln.200625114 [DOI] [Google Scholar]
- Mergemann H., Sauter M. (2000). Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 124 609–614 10.1104/pp.124.2.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E., Mun J. H., Yeun L. H., Jayaraman D., Rouge P., Barre A., et al. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant-Microbe Interact. 20 912–921 10.1094/MPMI-20-8-0912. [DOI] [PubMed] [Google Scholar]
- Middleton P. H., Jakab J., Penmetsa R. V., Starker C. G., Doll J., Kalo P., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234 10.1105/tpc.106.048264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R., Gleason C., Edwards A. (2004). A Ca 2 calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. U.S.A. 101 4701–4705 10.1073/pnas.0400595101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A. S., Raghothama K. G., et al. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U.S.A. 102 7760–7765 10.1073/pnas.0500778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Kinoshita A., Fukuda H., Sawa S. (2009). Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem. J. Plant Res. 122 31–39 10.1007/s10265-008-0207-3 [DOI] [PubMed] [Google Scholar]
- Miyasaka S. C., Buta G. J., Howell R. K., Foy C. D. (1991). Mechanism of aluminum tolerance in snapbeans: root exudation of citric acid. Plant Physiol. 96 737–743 10.1104/pp.96.3.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka S. C., Kochian L. V., Shaff J. E., Foy C. D. (1989). Mechanisms of aluminum tolerance in wheat: an investigation of genotypic differences in rhizosphere pH, K, and H transport, and root-cell membrane potentials. Plant Physiol. 91 1188–1196 10.1104/pp.91.3.1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molas M. L., Kiss J. Z. (2008). PKS1 plays a role in red-light-based positive phototropism in roots. Plant Cell Environ. 31 842–849 10.1111/j.1365-3040.2008.01797.x [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno M. A., Van Norman J. M., Moreno A., Zhang J., Ahnert S. E., Benfey P. N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329 1306–1311 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M. T. (2010). Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 61 705–720 10.1146/annurev.arplant.043008.092042 [DOI] [PubMed] [Google Scholar]
- Moubayidin L., Di Mambro R., Sabatini S. (2009). Cytokinin–auxin crosstalk. Trends Plant Sci. 14 557–562 10.1016/j.tplants.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Mullen J. L., Hangarter R. P. (2003). Genetic analysis of the gravitropic set-point angle in lateral roots of Arabidopsis. Adv. Space Res. 31 2229–2236 10.1016/S0273-1177(03)00249-7 [DOI] [PubMed] [Google Scholar]
- Mullen J. L., Wolverton C., Ishikawa H., Hangarter R. P., Evans M. L. (2002). Spatial separation of light perception and growth response in maize root phototropism. Plant Cell Environ. 25 1191–1196 10.1046/j.1365-3040.2002.00899.x [DOI] [PubMed] [Google Scholar]
- Muller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453 1094–1097 10.1038/nature06943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 651–681 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Munos S., Cazettes C., Fizames C., Gaymard F., Tillard P., Lepetit M., et al. (2004). Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16 2433–2447 10.1105/tpc.104.024380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P., Canivenc G., Muller B. (2005). A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 138 2061–2074 10.1104/pp.105.060061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nap J., Bisseling T. (1990). The roots of nodulins. Physiol. Plant. 79 407–414 10.1111/j.1399-3054.1990.tb06760.x [DOI] [Google Scholar]
- Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgen R. (2003). Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 33 633–650 10.1046/j.1365-313X.2003.01657.x [DOI] [PubMed] [Google Scholar]
- Nishimura T., Nakano H., Hayashi K.-I., Niwa C., Koshiba T. (2009). Differential downward stream of auxin synthesized at the tip has a key role in gravitropic curvature via TIR1/AFBs-mediated auxin signaling pathways. Plant Cell Physiol. 50 1874–1885 10.1093/pcp/pcp129 [DOI] [PubMed] [Google Scholar]
- Noh B., Bandyopadhyay A., Peer W. A., Spalding E. P., Murphy A. S. (2003). Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423 999–1002 10.1038/nature01716 [DOI] [PubMed] [Google Scholar]
- Noh B., Murphy A. S., Spalding E. P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454 10.1105/tpc.010350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutman P. S. (1948). Physiological studies on nodule formation: I. The relation between nodulation and lateral root formation in red clover. Ann. Bot. 12 81–96 [Google Scholar]
- Ogawa M., Kay P., Wilson S., Swain S. M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21 216–233 10.1105/tpc.108.063768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama N., Takei K., Sakakibara H., Hayashi H., Yoneyama T., Fujiwara T. (2002). Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol. 43 1493–1501 10.1093/pcp/pcf183 [DOI] [PubMed] [Google Scholar]
- Okushima Y., Overvoorde P. J., Arima K., Alonso J. M., Chan A., Chang C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463 10.1105/tpc.104.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah B., Briere C., Becard G., Denarie J., Gough C. (2005). Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signaling pathway. Plant J. 44 195–207 10.1111/j.1365-313X.2005.02522.x [DOI] [PubMed] [Google Scholar]
- O’Malley R. C., Rodriguez F. I., Esch J. J., Binder B. M., O’Donnell P., Klee H. J., et al. (2005). Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 41 651–659 10.1111/j.1365-313X.2004.02331.x [DOI] [PubMed] [Google Scholar]
- Osmont K. S., Sibout R., Hardtke C. S. (2007). Hidden branches: developments in root system architecture. Annu. Rev. Plant Biol. 58 93–113 10.1146/annurev.arplant.58.032806.104006 [DOI] [PubMed] [Google Scholar]
- Overvoorde P., Fukaki H., Beeckman T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2 a001537 10.1101/cshperspect.a001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Villalobos D., Hardtke C. S. (2012). Natural genetic variation of root system architecture from Arabidopsis to Brachypodium: towards adaptive value. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 1552–1558 10.1098/rstb.2011.0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette A. J., Benfey P. N. (2005). Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 138 636–640 10.1104/pp.104.058362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek A., Singla-Pareek S. L., Sopory S. K., Grover A. (2007). “Analysis of salt stress-related transcriptome fingerprints from diverse plant species,” in Genomics-Assisted Crop Improvement Vol. 1 eds Varshney R., Tuberosa R. (Berlin: Springer) 267–287 [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbiosis. Nat.Rev. Microbiol 6 763–775 10.1038/nrmicro1987 [DOI] [PubMed] [Google Scholar]
- Parry G., Marchant A., May S., Swarup R., Swarup K., James N., et al. (2001). Quick on the uptake: characterization of a family of plant auxin influx carriers. J. Plant Growth Regul. 20 217–225 10.1007/s003440010030 [DOI] [Google Scholar]
- Paszkowski U., Boller T. (2002). The growth defect of lrt1, a maize mutant lacking lateral roots, can be complemented by symbiotic fungi or high phosphate nutrition. Planta 214 584–590 10.1007/s004250100642 [DOI] [PubMed] [Google Scholar]
- Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., et al. (2009a). Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14 399–408 10.1016/j.tplants.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Péret B., Larrieu A., Bennett M. J. (2009b). Lateral root emergence: a difficult birth. J. Exp. Bot. 60 3637–3643 10.1093/jxb/erp232 [DOI] [PubMed] [Google Scholar]
- Perez-Perez J. M. (2007). Hormone signaling and root development: an update on the latest Arabidopsis thaliana research. Funct. Plant Biol. 34 163–171 10.1071/FP06341 [DOI] [PubMed] [Google Scholar]
- Perez-Torres C.-A., Lopez-Bucio J., Cruz-Ramirez A., Ibarra-Laclette E., Dharmasiri W., Estelle M., et al. (2008). Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20 3258–3272 10.1105/tpc.108.058719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisova M., Klima P., Horak J., Valkova M., Malbeck J., Soucek P., et al. (2009). Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. U.S.A. 106 3609–3614 10.1073/pnas.0811539106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret J. S., Al-Belushi M. E., Deadman M. (2007). Non-destructive visualization and quantification of roots using computed tomography. Soil Biol. Biochem. 39 391–399 10.1016/j.soilbio.2006.07.018 [DOI] [Google Scholar]
- Perrin R. M., Young L.-S., Murthy U. M. N., Harrison B. R., Wang Y., Will J. L., et al. (2005). Gravity signal transduction in primary roots. Ann. Bot. 96 737–743 10.1093/aob/mci227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136 2675–2688 10.1242/dev.030353 [DOI] [PubMed] [Google Scholar]
- Pirozynski K. A., Malloch D. W. (1975). The origin of land plants: a matter of mycotrophism. Biosystems 6 153 10.1016/0303-2647(75)90023-4 [DOI] [PubMed] [Google Scholar]
- Provorov N. A. (2000). The population genetics of nodule bacteria. Zh. Obshch. Biol. 61 229–257 [PubMed] [Google Scholar]
- Puig J., Pauluzzi G., Guiderdoni E., Gantet P. (2012). Regulation of shoot and root development through mutual signaling. Mol. Plant 5 974–983 10.1093/mp/sss047 [DOI] [PubMed] [Google Scholar]
- Quintero F. J., Ohta M., Shi H., Zhu J.-K., Pardo J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99 9061–9066 10.1073/pnas.132092099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani Debi B., Chhun T., Taketa S., Tsurumi S., Xia K., Miyao A., et al. (2005). Defects in root development and gravity response in the aem1 mutant of rice are associated with reduced auxin efflux. J. Plant Physiol. 162 678–685 10.1016/j.jplph.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Rebouillat J., Dievart A., Verdeil J. L., Escoute J., Giese G., Breitler J. C., et al. (2009). Molecular genetics of rice root development. Rice 2 15–34 10.1007/s12284-008-9016-5 [DOI] [Google Scholar]
- Reed J. W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6 420–425 10.1016/S1360-1385(01)02042-8 [DOI] [PubMed] [Google Scholar]
- Relic B., Talmont F., Kopcinska J., Golinowski W., Prome J. C., Broughton W. J. (1993). Biological activity of Rhizobium sp. NGR234 nod-factors on Macroptilium atropurpureum. Mol. Plant Microbe Interact. 6 764–774 10.1094/MPMI-6-764 [DOI] [PubMed] [Google Scholar]
- Remans T., Nacry P., Pervent M., Filleur S., Diatloff E., Mounier E., et al. (2006a). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U.S.A. 103 19206–19211 10.1073/pnas.0605275103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T., Nacry P., Prevent M., Girin T., Tillard P., Lepetit M., et al. (2006b). A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 140 909–921 10.1104/pp.105.075721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M., Svistoonoff S., Loudet O., Nussaume L., Desnos T. (2006). Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ. 29 115–125 10.1111/j.1365-3040.2005.01405.x [DOI] [PubMed] [Google Scholar]
- Richter S., Anders N., Wolters H., Beckmann H., Thomann A., Heinrich R., et al. (2010). Role of the GNOM gene in Arabidopsis apical-basal patterning – from mutant phenotype to cellular mechanism of protein action. Eur. J. Cell Biol. 89 138–144 10.1016/j.ejcb.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Barrueco C., De Castro F. B. (1973). Cytokinin-induced pseudonodules on Alnus glutinosa. Physiol. Plant. 29 277–280 10.1111/j.1399-3054.1973.tb03107.x [DOI] [Google Scholar]
- Ruppel N. J., Hangarter R. P., Kiss J. Z. (2001). Red-light-induced positive phototropism in Arabidopsis roots. Planta 212 424–430 10.1007/s004250000410 [DOI] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B.-H., Matsumoto T. K., et al. (2001). AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc. Natl. Acad. Sci. U.S.A. 98 14150–14155 10.1073/pnas.241501798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C., Kohlen W., Charnikhova T., Van Zeijl A., Van Bezouwen L., De Ruijter N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155 721–734 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K., Simaskova M., Duclercq J., Petrasek J., Zazimalova E., Simon S., et al. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. U.S.A. 106 4284–4289 10.1073/pnas.0900060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P. R., Ditomaso J. M., Kochian L. V. (1993). Aluminium toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. J. Exp. Bot. 44 437–446 10.1093/jxb/44.2.437 [DOI] [Google Scholar]
- Saab I. N., Sharp R. E., Pritchard J., Voetberg G. S. (1990). Increased endogenous abscisic acid maintains primary root growth and inhibits shoot growth of maize seedlings at low water potentials. Plant Physiol. 93 1329–1336 10.1104/pp.93.4.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., et al. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472 10.1016/S0092-8674(00)81535-4 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17 354 10.1101/gad.252503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. (2000). Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr. Opin. Plant Biol. 3 188–195 10.1016/S1369-5266(00)80064-3 [DOI] [PubMed] [Google Scholar]
- Saito K., Yoshikawa M., Yano K., Miwa H., Uchida H., Asamizu E., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19 610–624 10.1105/tpc.106.046938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Calderon L., Lopez-Bucio J., Chacon-Lopez A., Cruz-Ramirez A., Nieto-Jacobo F., Dubrovsky J. G., et al. (2005). Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 46 174–184 10.1093/pcp/pci011 [DOI] [PubMed] [Google Scholar]
- Sano T., Nagata T. (2002). The possible involvement of a phosphate-induced transcription factor encoded by Phi-2 gene from Tobacco in ABA-signaling pathways. Plant Cell Physiol. 43 12–20 10.1093/pcp/pcf002 [DOI] [PubMed] [Google Scholar]
- Scheres B., McKhann H. I., Van Den Berg C. (1996). Roots redefined: anatomical and genetic analysis of root development. Plant Physiol. 111 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W., Schikora A. (2001). Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 125 2078–2084 10.1104/pp.125.4.2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmohl N., Pilling J., Fisahn J., Horst W. J. (2000). Pectin methylesterase modulates aluminium sensitivity in Zea mays and Solanum tuberosum. Physiol. Plant. 109 419–427 10.1034/j.1399-3054.2000.100408.x [DOI] [Google Scholar]
- Schüßler A., Schwarzott D., Walker C. (2001). A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105 1413 10.1017/S0953756201005196 [DOI] [Google Scholar]
- Schulze J., Temple G., Temple S. J., Beschow H., Vance C. P. (2006). Nitrogen fixation by white lupin under phosphorus deficiency. Ann. Bot. 98 731–740 10.1093/aob/mcl154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane M., Lambers H. (2005). Cluster roots: a curiosity in context. Plant Soil 274 101–125 10.1007/s11104-004-2725-7 [DOI] [Google Scholar]
- Sharp R. E., Silk W. K., Hsiao T. C. (1988). Growth of the maize primary root at low water potentials: I. Spatial distribution of expansive growth. Plant Physiol. 87 50–57 10.1104/pp.87.1.50PMCid:35153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidhar H. E., Henry A., Hardy B. (2012). Methodologies for Root Drought Studies in Rice. Los Baños: International Rice Research Institute, 65 p [Google Scholar]
- Sheng M., Tang M., Chen H., Yang B., Zhang F., Huang Y. (2008). Influence of arbuscular mycorrhizae on photosynthesis and water status of maize plants under salt stress. Mycorrhiza 18 287–296 10.1007/s00572-008-0180-7 [DOI] [PubMed] [Google Scholar]
- Shilev S., Sancho E. D., Benlloch-Gonzalez M. (2010). Rhizospheric bacteria alleviate salt-produced stress in sunflower. J. Environ. Manage. 95 S37–S41 10.1016/j.jenvman.2010.07.019 [DOI] [PubMed] [Google Scholar]
- Shimizu A., Yanagihara S., Kawasaki S., Ikehashi H. (2004). Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theor. Appl. Genet. 109 1361–1368 10.1007/s00122-004-1751-4 [DOI] [PubMed] [Google Scholar]
- Signora L., De Smet I., Foyer C. H., Zhang H. (2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28 655–662 10.1046/j.1365-313x.2001.01185.x [DOI] [PubMed] [Google Scholar]
- Simon L., Bousquet J., Levesque R. C., Lalonde M. (1993). Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 363 67–69 10.1038/363067a0 [DOI] [Google Scholar]
- Sivaguru M., Ezaki B., He Z.-H., Tong H., Osawa H., Baluska F., et al. (2000). Aluminum-induced 1 → 3-beta-D-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol. 124 991–1006 10.1104/pp.124.3.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M., Horst W. J. (1998). The distal part of the transition zone is the most aluminum-sensitive apical root zone of Maize. Plant Physiol. 116 155–163 10.1104/pp.116.1.155 [DOI] [Google Scholar]
- Sivaguru M., Horst W. J., Eticha D., Matsumoto H. (2006). Aluminum inhibits apoplastic flow of high-molecular weight solutes in root apices of Zea mays L. J. Plant Nutr. Soil Sci. 169 679–690 10.1002/jpln.200620603 [DOI] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debelle F., Gough C., Bisseling T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 10.1126/science.1111025 [DOI] [PubMed] [Google Scholar]
- Snapp S. S., Shennan C. (1992). Effects of salinity on root growth and death dynamics of tomato, Lycopersicon esculentum Mill. New Phytol. 121 71–79 10.1111/j.1469-8137.1992.tb01094.x [DOI] [PubMed] [Google Scholar]
- Soni R. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7 85–103 10.1105/tpc.7.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreevidya V. S., Hernandez-Oane R. J., Gyaneshwar P., Lara-Flores M., Ladha J. K., Reddy P. M. (2010). Changes in auxin distribution patterns during lateral root development in rice. Plant Sci. 178 531–538 10.1016/j.plantsci.2010.03.004 [DOI] [Google Scholar]
- Steffens B., Sauter M. (2005). Epidermal cell death in rice is regulated by ethylene, gibberellin, and abscisic acid. Plant Physiol. 139 713–721 10.1104/pp.105.064469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B., Sauter M. (2009). Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 21 184–196 10.1105/tpc.108.061887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B., Wang J., Sauter M. (2006). Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 223 604–612 10.1007/s00425-005-0111-1 [DOI] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C. L., Paris S., et al. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318 10.1126/science.286.5438.316 [DOI] [PubMed] [Google Scholar]
- Strabala T., O’Donnell P. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR. Plant Physiol. 140 1331–1344 10.1104/pp.105.075515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack D., Fester T., Hause B., Schliemann W., Walter M. H. (2003). Arbuscular mycorrhiza: biological, chemical, and molecular aspects. J. Chem. Ecol. 29 1955–1979 10.1023/A:1025695032113 [DOI] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., et al. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962 10.1038/nature00841 [DOI] [PubMed] [Google Scholar]
- Sun J., Xu Y., Ye S., Jiang H., Chen Q., Liu F., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21 1495–1511 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Tian Q.-Y., Chen J., Zhang W.-H. (2010). Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 61 347–356 10.1093/jxb/erp306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suralta R. R., Inukai Y., Yamauchi A. (2008). Genotypic variations in responses of lateral root development to transient moisture stresses in rice cultivars. Plant Prod. Sci. 11 324–335 10.1626/pps.11.324 [DOI] [Google Scholar]
- Svistoonoff S., Sy M.-O., Diagne N., Barker D. G., Bogusz D., Franche C. (2010). Infection-specific activation of the Medicago truncatula Enod11 early nodulin gene promoter during actinorhizal root nodulation. Mol. Plant Microbe Interact. 23 740–747 10.1094/MPMI-23-6-0740 [DOI] [PubMed] [Google Scholar]
- Swarup K., Benkova E., Swarup R., Casimiro I., Péret B., Yang Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10 946–954 10.1038/ncb1754 [DOI] [PubMed] [Google Scholar]
- Swarup R., Kramer E. M., Perry P., Knox K., Ottoline Leyser H. M., Haseloff J., et al. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7 1057–1065 10.1038/ncb1316 [DOI] [PubMed] [Google Scholar]
- Swensen S. M. (1996). The evolution of actinorhizal symbioses: evidence for multiple origins of the symbiotic association. Am. J. Bot. 83 1503–1512 10.2307/2446104 [DOI] [Google Scholar]
- Takahashi H. (2010). Regulation of sulfate transport and assimilation in plants. Int. Rev. Cell Mol. Biol. 281 129–159 10.1016/S1937-6448(10)81004-4 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Noji M., Saito K. (1999). Molecular regulation and engineering of sulfur transport and assimilation. Tanpakushitsu Kakusan Koso 44 2291–2298 [PubMed] [Google Scholar]
- Takahashi H., Watanabe-Takahashi A., Smith F. W., Blake-Kalff M., Hawkesford M. J., Saito K. (2000). The roles of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J. 23 171–182 10.1046/j.1365-313x.2000.00768.x [DOI] [PubMed] [Google Scholar]
- Taramino G., Sauer M., Stauffer J. L., Multani D., Niu X., Sakai H., et al. (2007). The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 50 649–659 10.1111/j.1365-313X.2007.03075.x [DOI] [PubMed] [Google Scholar]
- Tester M., Davenport R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91 503–527 10.1093/aob/mcg058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi C. A., Delatorre C. A., Lahner B., Salt D. E., Abel S. (2004). Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 37 801–814 10.1111/j.1365-313X.2004.02005.x [DOI] [PubMed] [Google Scholar]
- Ticconi C. A., Lucero R. D., Sakhonwasee S., Adamson A. W., Creff A., Nussaume L., et al. (2009). ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. U.S.A. 106 14174–14179 10.1073/pnas.0901778106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich H. (1977). Vergleichend morphologische Untersuchungen zur Identität der Gramineen-Primärwurzel. Flora 166 415–421 [Google Scholar]
- Tirichine L., Sandal N., Madsen L. H., Radutoiu S., Albrektsen A. S., Sato S., et al. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107 10.1126/science.1132397 [DOI] [PubMed] [Google Scholar]
- Tirlapur U. K., Konig K. (1999). Technical advance: near-infrared femtosecond laser pulses as a novel non-invasive means for dye-permeation and 3D imaging of localised dye-coupling in the Arabidopsis root meristem. Plant J. 20 363–370 10.1046/j.1365-313X.1999.t01-1-00603.x [DOI] [PubMed] [Google Scholar]
- Tracy S. R., Roberts J. A., Black C. R., McNeill A., Davidson R., Mooney S. J. (2010). The X-factor: visualizing undisturbed root architecture in soils using X-ray computed tomography. J. Exp. Bot. 61 311–313 10.1093/jxb/erp386 [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A., Theologis A. (2004). Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136 2982–3000 10.1104/pp.104.049999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C.-W., Zhao K., Wright M. H., Ali M. L., Jung J., Kimball J., et al. (2010). Development of a research platform for dissecting phenotype-genotype associations in rice (Oryza spp.). Rice 3 205–217 10.1007/s12284-010-9056-5 [DOI] [Google Scholar]
- Ueda M., Koshino-Kimura Y., Okada K. (2005). Stepwise understanding of root development. Curr. Opin. Plant Biol. 8 71–76 10.1016/j.pbi.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Uexküll H. R., Mutert E. (1995). Global extent, development and economic impact of acid soils. Plant Soil 171 1–15 10.1007/BF00009558 [DOI] [Google Scholar]
- Umehara M., Hanada A., Magome H., Takeda-Kamiya N., Yamaguchi S. (2010). Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 51 1118–1126 10.1093/pcp/pcq084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N., Kim E. J., Rubio G., Yamaguchi T., Muto S., Tsuboi A., et al. (2000). The Arabidopsis HKT1 gene homolog mediates inward Na(+) currents in Xenopus laevis oocytes and Na(+) uptake in Saccharomyces cerevisiae. Plant Physiol. 122 1249–1259 10.1104/pp.122.4.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D. P. S., Hu C.-A., Zhang M. (1992). Root nodule development: origin, function and regulation of nodulin genes. Physiol. Plant. 85 253–265 10.1111/j.1399-3054.1992.tb04730.x [DOI] [Google Scholar]
- Vernie T., Moreau S., de Billy F., Plet J., Combier J.-P., Rogers C., et al. (2008). EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20 2696–2713 10.1105/tpc.108.059857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierheilig H., Bago B., Lerat S., Piche Y. (2002). Shoot-produced, light-dependent factors are partially involved in the expression of the arbuscular mycorrhizal (AM) status of AM host and non-host plants. J. Plant Nutr. 165 21–25 10.1002/1522-2624(200202)165:1 [DOI] [Google Scholar]
- Vilella A. J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. (2009). EnsemblCompara genetrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19 327–335 10.1101/gr.073585.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P., Ivanov I. I., Filleur S., Gan Y., Remans T., Forde B. G. (2006). Nitrogen regulation of root branching. Ann. Bot. 97 875–881 10.1093/aob/mcj601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. R., Hu H., Wand G.-H., Li J., Chen J.-Y., Wu P. (2009). Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol. Plant 2 823–831 10.1093/mp/ssp023 [DOI] [PubMed] [Google Scholar]
- Wang J.-W., Wang L.-J., Mao Y.-B., Cai W.-J., Xue H.-W., Chen X.-Y. (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17 2204–2216 10.1105/tpc.105.033076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegers B. S., Cheer A. Y., Silk W. K. (2009). Modeling the hydraulics of root growth in three dimensions with phloem water sources. Plant Physiol. 150 2092–103 10.1104/pp.109.138198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson L. C., Ribrioux S. P. C. P., Fitter A. H., Leyser H. M. O. (2001). Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 126 875–882 10.1104/pp.126.2.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woll K., Borsuk L. A., Stransky H., Nettleton D., Schnable P. S., Hochholdinger F. (2005). Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol. 139 1255–1267 10.1104/pp.105.067330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lewis D. R., Spalding E. P. (2007). Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19 1826–1837 10.1105/tpc.106.048777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 3024–3036 10.1101/gad.852200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., et al. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442 705–708 10.1038/nature04920 [DOI] [PubMed] [Google Scholar]
- Xu M., Zhu L., Shou H., Wu P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46 1674–1681 10.1093/pcp/pci183 [DOI] [PubMed] [Google Scholar]
- Xue R., Zhang B. (2007). Increased endogenous methyl jasmonate altered leaf and root development in transgenic soybean plants. J. Genet. Genomics 34 339–346 10.1016/S1673-8527(07)60036-8 [DOI] [PubMed] [Google Scholar]
- Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., et al. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42 1017–1023 10.1093/pcp/pce127 [DOI] [PubMed] [Google Scholar]
- Yamaji N., Huang C. F., Nagao S., Yano M., Sato Y., Nagamura Y., et al. (2009). A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21 3339–3349 10.1105/tpc.109.070771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Shah T., Warburton M., Buckler E. S., McMullen M. D., Crouch J. (2009). Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 4:e8451. 10.1371/journal.pone.0008451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Yoshida S., Müller J., Singh S., Banba M., Vickers K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. 105 20540–20545 10.1073/pnas.0811417106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Wu Z., Zhou J., Du L., Guo L., Wu Y., et al. (2005). OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 138 2087–2096 10.1104/pp.105.063115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., Nishitani K. (2004). Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol. 45 1111–1121 10.1093/pcp/pch151 [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Takeuchi Y., Sekimoto H. (2007a). Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225 1031–1038 10.1007/s00425-006-0410-1 [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Kusumoto D. (2007b). Nitrogen deficiency as well as phosphorus deficiency in Sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular. Planta 227 125–132 10.1007/s00425-007-0600-5 [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Sekimoto H. (2008). Strigolactones, host recognition signals for root parasitic plants and arbuscular mycorrhizal fungi, from Fabaceae plants. New Phytol. 179 484–494 10.1111/j.1469-8137.2008.02462.x [DOI] [PubMed] [Google Scholar]
- Yoshida S., Parniske M. (2005). Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J. Biol. Chem. 280 9203–9209 10.1074/jbc.M411665200 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N., Takahashi H., Smith F. W., Yamaya T., Saito K. (2002). Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 29 465–473 10.1046/j.0960-7412.2001.01231.x [DOI] [PubMed] [Google Scholar]
- Young L. M., Evans M. L. (1996). Patterns of auxin and abscisic acid movement in the tips of gravistimulated primary roots of maize. Plant Growth Regul. 20 253–258 10.1007/BF00043315 [DOI] [PubMed] [Google Scholar]
- Yun H., Joo S., Park C., Kim S. (2009). Effects of brassinolide and IAA on ethylene production and elongation in maize primary roots. J. Plant Biol. 52 268–274 10.1007/s12374-009-9032-z [DOI] [Google Scholar]
- Zeng H.-Q., Zhu Y.-Y., Bao Y., Shen Q.-R., Guo K., Huang S.-Q., et al. (2010). Relationship between the development of tomato lateral roots and expression of miR164, NAC1 under P deficiency. Acta Metall. Sin. 16 166–171 [Google Scholar]
- Zhang H., Forde B. G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 407–409 10.1126/science.279.5349.407 [DOI] [PubMed] [Google Scholar]
- Zhang H., Jennings A., Barlow P. W., Forde B. (1999). Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. U.S.A. 96 6529–6534 10.1073/pnas.96.11.6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.-Q., Todhunter R. J., Tiwari H. K., Gore M. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42 355–360 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L., Zhou D.-X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21 736–748 10.1105/tpc.108.061655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant. Biol. 53 247–273 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K., Liu J., Xiong L. (1998). Genetic analysis of salt tolerance in Arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell 10 1181–1191 10.1105/tpc.10.7.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidan I., Azaizeh H., Neumann P. M. (1990). Does salinity reduce growth in maize root epidermal cells by inhibiting their capacity for cell wall acidification? Plant Physiol. 93 7–11 10.1104/pp.93.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]