Abstract
The title compound, C24H19ClN2, crystallizes with two independent molecules in the asymmetric unit. The prop-2-enyl substituents on the imidazole rings adopt similar conformations in the two molecules. The 4-and 5-substituted phenyl rings and the benzene ring make dihedral angles of 67.06 (8), 5.61 (8) and 41.09 (8)°, respectively, with the imadazole ring in one molecule and 71.53 (8), 28.85 (8) and 41.87 (8)°, respectively, in the other. The crystal structure features C—H⋯π interactions and weak π–π stacking interactions [centroid–centroid distances = 3.6937 (10) and 4.0232 (10) Å] between the chlorophenyl rings, which form a three-dimensional supramolecular structure.
Related literature
For pharmaceutical properties of imidazoles and imidazole-containing compounds, see, for example: Roman et al. (2007 ▶); Nanterment et al. (2004 ▶); Congiu et al. (2008 ▶); Venkatesan et al. (2008 ▶); Bhatnagar et al. (2011 ▶); Puratchikody & Doble (2007 ▶). For similar structures, see: Mohamed et al. (2013 ▶); Akkurt et al. (2013 ▶).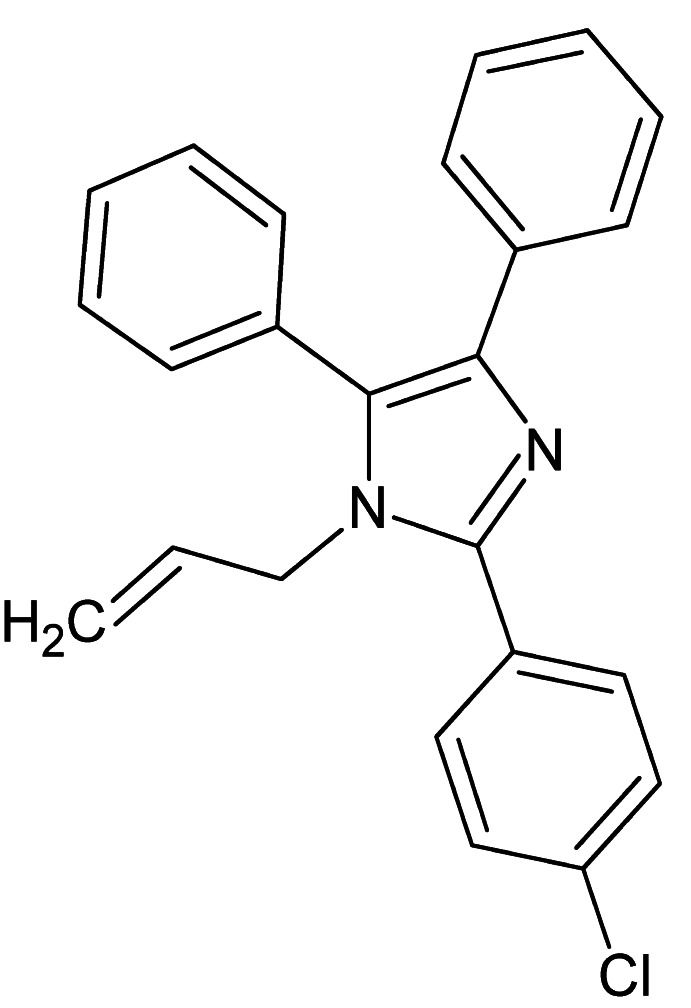
Experimental
Crystal data
C24H19ClN2
M r = 370.86
Triclinic,
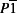
a = 10.0916 (7) Å
b = 13.1386 (9) Å
c = 15.6155 (10) Å
α = 72.924 (1)°
β = 86.849 (1)°
γ = 71.830 (1)°
V = 1879.0 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.21 mm−1
T = 100 K
0.57 × 0.33 × 0.28 mm
Data collection
Bruker SMART APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.919, T max = 0.942
20791 measured reflections
7718 independent reflections
6764 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.106
S = 1.07
7718 reflections
487 parameters
H-atom parameters constrained
Δρmax = 0.34 e Å−3
Δρmin = −0.23 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813012592/hg5314sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813012592/hg5314Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813012592/hg5314Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg2, Cg4 and Cg8 are the centroids of the C4–C9, C19–C24 and C43–C48 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5⋯Cg4i | 0.95 | 2.76 | 3.5968 (17) | 147 |
| C11—H11⋯Cg8ii | 0.95 | 2.83 | 3.5879 (19) | 137 |
| C33—H33⋯Cg2iii | 0.95 | 2.89 | 3.8217 (18) | 166 |
Symmetry codes: (i) 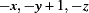 ; (ii)
; (ii) 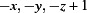 ; (iii)
; (iii) 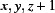 .
.
Acknowledgments
Manchester Metropolitan University, Erciyes University and Granada University are gratefully acknowledged for supporting this study. The authors also thank José Romero Garzón, Centro de Instrumentación Científica, Universidad de Granada, for the data collection.
supplementary crystallographic information
Comment
Imidazoles have been intensivley reported to serve as usefull building blocks for synthesis of diverse class of bioactive molecules. In addition imidazole comtaining compounds exhibited a wide spectrum of pharmaceutical properties such as pesticides, fungicides, antibacterial anti-inflammatory, anti-tubercular, anti-diabetic, antimalarial and antitumour (Roman et al., 2007; Nanterment et al., 2004; Congiu et al., 2008; Venkatesan et al., 2008; Bhatnagar et al., 2011; Puratchikody & Doble 2007). In this aspect and further to our study on synthesis of tetrasubstituted imidazoles as potential bioactive molecules, we herein report the crystal structure of the title compound.
As seen in the Fig. 1, the title compound contains two independent molecules, 1 (with Cl1) and 2 (with Cl2), in the asymmetric unit. The prop-2-ene substituents on the imidazole rings adopt similar conformations in molecules 1 and 2. The 4-and 5-substituted phenyl rings (C4–C9 and C19–C24) and the benzene ring (C13–C18) attached to the atom Cl1 makes dihedral angles of 67.06 (8), 5.61 (8) and 41.09 (8)°, respectively, with the imadazole ring (N1/N2/C1–C3) of molecule 1 and the corresponding angles are A/B = 71.53 (8), A/C = 28.85 (8) and A/D = 41.87 (8)°, respectively, in molecule 2; where A (N3/N4/C25–C27), B (C28–C33), C (C43–C48) and D (C37–C42). All bond lengths are normal and are comparable with those reported for the similar structures (Mohamed et al., 2013; Akkurt et al., 2013).
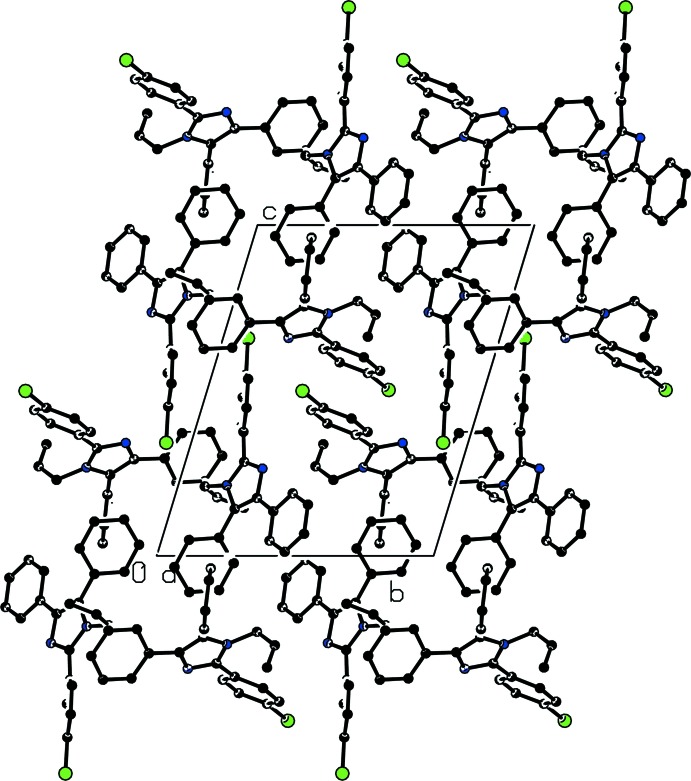
The crystal structure is stabilized by C—H···π interactions (Table 1) and weak π-π stacking interactions [Cg3···Cg3 (-x, -y, 1 - z) = 3.6937 (10) Å and Cg7···Cg7 (-x, 1 - y, 1 - z) = 4.0232 (10) Å; where Cg3 and Cg7 are the centroids of the C13–C18 and C37–C42 benzene rings respectively, which are attached to the Cl1 and Cl2 atoms]. Fig. 2 shows the packing diagram of (I) along the a axis.
Experimental
The title compound was prepared, according to our reported method (Mohamed et al., 2013) in 81% yield. Suitable single crystals were obtained by slow evaporation of a solution in ethanol, m.p. 305–307 K.
Refinement
All H atoms were placed in geometrically, with C—H = 0.95 and 0.99 Å, and refined as riding with Uiso(H) = 1.2 Ueq(C) of the parent atom.
Figures
Fig. 1.
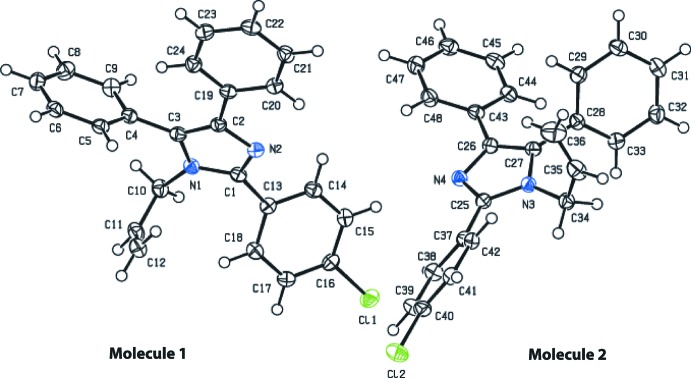
View of the two molecules of the title compound in the asymmetric unit with the atom numbering scheme. Displacement ellipsoids for non-H atoms are drawn at the 50% probability level.
Fig. 2.
A view of the packing diagram of the title compound viewing along the a axis
Crystal data
| C24H19ClN2 | Z = 4 |
| Mr = 370.86 | F(000) = 776 |
| Triclinic, P1 | Dx = 1.311 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.0916 (7) Å | Cell parameters from 9362 reflections |
| b = 13.1386 (9) Å | θ = 4.6–55.6° |
| c = 15.6155 (10) Å | µ = 0.21 mm−1 |
| α = 72.924 (1)° | T = 100 K |
| β = 86.849 (1)° | Prism, colourless |
| γ = 71.830 (1)° | 0.57 × 0.33 × 0.28 mm |
| V = 1879.0 (2) Å3 |
Data collection
| Bruker SMART APEX CCD area-detector diffractometer | 7718 independent reflections |
| Radiation source: sealed tube | 6764 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.023 |
| phi and ω scans | θmax = 26.5°, θmin = 1.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | h = −12→12 |
| Tmin = 0.919, Tmax = 0.942 | k = −16→16 |
| 20791 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.106 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0514P)2 + 0.7786P] where P = (Fo2 + 2Fc2)/3 |
| 7718 reflections | (Δ/σ)max = 0.001 |
| 487 parameters | Δρmax = 0.34 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | −0.20646 (4) | 0.09006 (3) | 0.65803 (2) | 0.0300 (1) | |
| N1 | −0.02163 (12) | 0.17529 (10) | 0.21321 (8) | 0.0200 (3) | |
| N2 | 0.06068 (12) | 0.28573 (10) | 0.26375 (8) | 0.0200 (3) | |
| C1 | −0.00889 (14) | 0.21232 (12) | 0.28437 (10) | 0.0197 (4) | |
| C2 | 0.09418 (14) | 0.29822 (11) | 0.17502 (9) | 0.0189 (4) | |
| C3 | 0.04422 (14) | 0.22994 (12) | 0.14208 (9) | 0.0196 (4) | |
| C4 | 0.05483 (15) | 0.20489 (12) | 0.05485 (10) | 0.0203 (4) | |
| C5 | −0.01427 (15) | 0.28619 (12) | −0.02220 (10) | 0.0218 (4) | |
| C6 | −0.00411 (15) | 0.26283 (13) | −0.10377 (10) | 0.0233 (4) | |
| C7 | 0.07436 (16) | 0.15777 (13) | −0.10974 (10) | 0.0259 (4) | |
| C8 | 0.14332 (17) | 0.07618 (13) | −0.03361 (11) | 0.0276 (4) | |
| C9 | 0.13453 (16) | 0.09918 (13) | 0.04827 (10) | 0.0249 (4) | |
| C10 | −0.09628 (16) | 0.09705 (13) | 0.20987 (10) | 0.0238 (4) | |
| C11 | −0.24612 (17) | 0.15345 (14) | 0.17687 (10) | 0.0277 (5) | |
| C12 | −0.30685 (17) | 0.26199 (14) | 0.14500 (11) | 0.0301 (5) | |
| C13 | −0.06083 (15) | 0.17650 (12) | 0.37484 (10) | 0.0202 (4) | |
| C14 | 0.02610 (16) | 0.16120 (12) | 0.44742 (10) | 0.0227 (4) | |
| C15 | −0.01711 (16) | 0.13543 (12) | 0.53460 (10) | 0.0237 (4) | |
| C16 | −0.14858 (16) | 0.12304 (12) | 0.54890 (10) | 0.0222 (4) | |
| C17 | −0.23644 (16) | 0.13630 (13) | 0.47904 (10) | 0.0239 (4) | |
| C18 | −0.19289 (16) | 0.16348 (13) | 0.39192 (10) | 0.0240 (4) | |
| C19 | 0.17971 (14) | 0.37187 (11) | 0.13467 (10) | 0.0195 (4) | |
| C20 | 0.22749 (15) | 0.42241 (12) | 0.18880 (10) | 0.0221 (4) | |
| C21 | 0.31189 (16) | 0.48977 (12) | 0.15487 (10) | 0.0238 (4) | |
| C22 | 0.35045 (15) | 0.50794 (12) | 0.06640 (11) | 0.0244 (4) | |
| C23 | 0.30485 (16) | 0.45768 (13) | 0.01221 (10) | 0.0249 (5) | |
| C24 | 0.22005 (15) | 0.39068 (12) | 0.04585 (10) | 0.0229 (4) | |
| Cl2 | −0.42972 (4) | 0.65508 (3) | 0.50155 (3) | 0.0334 (1) | |
| N3 | 0.19935 (12) | 0.35109 (10) | 0.73274 (8) | 0.0194 (3) | |
| N4 | 0.17457 (12) | 0.23629 (10) | 0.65869 (8) | 0.0205 (3) | |
| C25 | 0.11724 (15) | 0.33401 (12) | 0.67463 (9) | 0.0201 (4) | |
| C26 | 0.29983 (15) | 0.18823 (12) | 0.70807 (9) | 0.0197 (4) | |
| C27 | 0.31680 (14) | 0.25751 (12) | 0.75442 (9) | 0.0192 (4) | |
| C28 | 0.43246 (15) | 0.24556 (11) | 0.81453 (10) | 0.0195 (4) | |
| C29 | 0.56009 (16) | 0.25167 (13) | 0.77852 (10) | 0.0236 (4) | |
| C30 | 0.67230 (16) | 0.23572 (13) | 0.83402 (11) | 0.0261 (4) | |
| C31 | 0.65703 (16) | 0.21555 (13) | 0.92575 (11) | 0.0259 (5) | |
| C32 | 0.53069 (17) | 0.20896 (13) | 0.96205 (10) | 0.0269 (4) | |
| C33 | 0.41862 (16) | 0.22345 (13) | 0.90689 (10) | 0.0239 (4) | |
| C34 | 0.17099 (16) | 0.44544 (13) | 0.77037 (10) | 0.0243 (5) | |
| C35 | 0.25101 (17) | 0.52512 (13) | 0.72946 (12) | 0.0310 (5) | |
| C36 | 0.33377 (19) | 0.52130 (15) | 0.66240 (14) | 0.0399 (6) | |
| C37 | −0.01782 (15) | 0.41483 (12) | 0.63418 (9) | 0.0202 (4) | |
| C38 | −0.12790 (16) | 0.37477 (13) | 0.62550 (10) | 0.0246 (4) | |
| C39 | −0.25512 (16) | 0.44770 (13) | 0.58485 (11) | 0.0277 (5) | |
| C40 | −0.27089 (15) | 0.56125 (13) | 0.55261 (10) | 0.0245 (4) | |
| C41 | −0.16323 (16) | 0.60279 (13) | 0.55934 (10) | 0.0230 (4) | |
| C42 | −0.03662 (16) | 0.52952 (12) | 0.60013 (9) | 0.0218 (4) | |
| C43 | 0.39728 (15) | 0.07904 (12) | 0.70529 (10) | 0.0202 (4) | |
| C44 | 0.48893 (15) | 0.00912 (12) | 0.77813 (10) | 0.0222 (4) | |
| C45 | 0.58242 (16) | −0.09211 (13) | 0.77391 (11) | 0.0256 (4) | |
| C46 | 0.58688 (16) | −0.12606 (13) | 0.69738 (11) | 0.0273 (5) | |
| C47 | 0.49576 (17) | −0.05782 (13) | 0.62534 (11) | 0.0279 (5) | |
| C48 | 0.40109 (16) | 0.04347 (13) | 0.62921 (10) | 0.0251 (5) | |
| H5 | −0.06880 | 0.35820 | −0.01870 | 0.0260* | |
| H6 | −0.05110 | 0.31900 | −0.15580 | 0.0280* | |
| H7 | 0.08080 | 0.14180 | −0.16560 | 0.0310* | |
| H8 | 0.19690 | 0.00410 | −0.03750 | 0.0330* | |
| H9 | 0.18270 | 0.04300 | 0.10000 | 0.0300* | |
| H10A | −0.09340 | 0.04490 | 0.27070 | 0.0290* | |
| H10B | −0.04730 | 0.05210 | 0.17000 | 0.0290* | |
| H11 | −0.30190 | 0.10640 | 0.17930 | 0.0330* | |
| H12A | −0.25510 | 0.31230 | 0.14130 | 0.0360* | |
| H12B | −0.40240 | 0.29010 | 0.12560 | 0.0360* | |
| H14 | 0.11660 | 0.16870 | 0.43660 | 0.0270* | |
| H15 | 0.04190 | 0.12650 | 0.58340 | 0.0280* | |
| H17 | −0.32590 | 0.12690 | 0.49060 | 0.0290* | |
| H18 | −0.25320 | 0.17330 | 0.34350 | 0.0290* | |
| H20 | 0.20190 | 0.41050 | 0.24960 | 0.0260* | |
| H21 | 0.34330 | 0.52350 | 0.19250 | 0.0290* | |
| H22 | 0.40760 | 0.55440 | 0.04310 | 0.0290* | |
| H23 | 0.33170 | 0.46910 | −0.04830 | 0.0300* | |
| H24 | 0.18910 | 0.35720 | 0.00790 | 0.0270* | |
| H29 | 0.57050 | 0.26690 | 0.71560 | 0.0280* | |
| H30 | 0.75960 | 0.23860 | 0.80910 | 0.0310* | |
| H31 | 0.73310 | 0.20630 | 0.96360 | 0.0310* | |
| H32 | 0.52050 | 0.19450 | 1.02490 | 0.0320* | |
| H33 | 0.33240 | 0.21830 | 0.93230 | 0.0290* | |
| H34A | 0.19370 | 0.41560 | 0.83560 | 0.0290* | |
| H34B | 0.07000 | 0.48710 | 0.76200 | 0.0290* | |
| H35 | 0.24060 | 0.58450 | 0.75480 | 0.0370* | |
| H36A | 0.34770 | 0.46350 | 0.63470 | 0.0480* | |
| H36B | 0.38000 | 0.57630 | 0.64130 | 0.0480* | |
| H38 | −0.11570 | 0.29680 | 0.64760 | 0.0290* | |
| H39 | −0.32990 | 0.42020 | 0.57930 | 0.0330* | |
| H41 | −0.17570 | 0.68070 | 0.53630 | 0.0280* | |
| H42 | 0.03790 | 0.55760 | 0.60490 | 0.0260* | |
| H44 | 0.48700 | 0.03130 | 0.83100 | 0.0270* | |
| H45 | 0.64390 | −0.13850 | 0.82380 | 0.0310* | |
| H46 | 0.65140 | −0.19510 | 0.69440 | 0.0330* | |
| H47 | 0.49800 | −0.08050 | 0.57270 | 0.0330* | |
| H48 | 0.33850 | 0.08880 | 0.57960 | 0.0300* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0315 (2) | 0.0358 (2) | 0.0215 (2) | −0.0115 (2) | 0.0067 (2) | −0.0067 (2) |
| N1 | 0.0199 (6) | 0.0201 (6) | 0.0199 (6) | −0.0079 (5) | −0.0001 (5) | −0.0038 (5) |
| N2 | 0.0186 (6) | 0.0195 (6) | 0.0207 (6) | −0.0055 (5) | 0.0020 (5) | −0.0047 (5) |
| C1 | 0.0172 (7) | 0.0189 (7) | 0.0215 (7) | −0.0042 (5) | −0.0004 (5) | −0.0049 (5) |
| C2 | 0.0163 (7) | 0.0181 (7) | 0.0197 (7) | −0.0028 (5) | 0.0002 (5) | −0.0044 (5) |
| C3 | 0.0173 (7) | 0.0189 (7) | 0.0201 (7) | −0.0046 (5) | 0.0006 (5) | −0.0030 (5) |
| C4 | 0.0191 (7) | 0.0225 (7) | 0.0220 (7) | −0.0102 (6) | 0.0018 (5) | −0.0066 (6) |
| C5 | 0.0182 (7) | 0.0224 (7) | 0.0259 (8) | −0.0078 (6) | 0.0016 (6) | −0.0074 (6) |
| C6 | 0.0204 (7) | 0.0293 (8) | 0.0210 (7) | −0.0116 (6) | −0.0003 (6) | −0.0041 (6) |
| C7 | 0.0292 (8) | 0.0322 (8) | 0.0227 (7) | −0.0158 (7) | 0.0046 (6) | −0.0113 (6) |
| C8 | 0.0313 (8) | 0.0221 (7) | 0.0312 (8) | −0.0082 (6) | 0.0049 (7) | −0.0110 (6) |
| C9 | 0.0275 (8) | 0.0211 (7) | 0.0247 (8) | −0.0076 (6) | 0.0003 (6) | −0.0043 (6) |
| C10 | 0.0278 (8) | 0.0232 (7) | 0.0236 (7) | −0.0133 (6) | 0.0010 (6) | −0.0058 (6) |
| C11 | 0.0285 (8) | 0.0352 (9) | 0.0242 (8) | −0.0179 (7) | −0.0012 (6) | −0.0067 (7) |
| C12 | 0.0266 (8) | 0.0364 (9) | 0.0278 (8) | −0.0096 (7) | −0.0040 (6) | −0.0094 (7) |
| C13 | 0.0221 (7) | 0.0173 (7) | 0.0212 (7) | −0.0064 (6) | 0.0014 (6) | −0.0052 (5) |
| C14 | 0.0217 (7) | 0.0219 (7) | 0.0257 (8) | −0.0087 (6) | 0.0012 (6) | −0.0065 (6) |
| C15 | 0.0258 (8) | 0.0238 (7) | 0.0226 (7) | −0.0086 (6) | −0.0008 (6) | −0.0072 (6) |
| C16 | 0.0270 (8) | 0.0191 (7) | 0.0198 (7) | −0.0073 (6) | 0.0047 (6) | −0.0051 (6) |
| C17 | 0.0201 (7) | 0.0257 (8) | 0.0263 (8) | −0.0087 (6) | 0.0038 (6) | −0.0071 (6) |
| C18 | 0.0223 (7) | 0.0248 (7) | 0.0243 (8) | −0.0076 (6) | −0.0017 (6) | −0.0055 (6) |
| C19 | 0.0161 (7) | 0.0159 (6) | 0.0239 (7) | −0.0028 (5) | 0.0006 (5) | −0.0044 (5) |
| C20 | 0.0212 (7) | 0.0217 (7) | 0.0227 (7) | −0.0058 (6) | 0.0019 (6) | −0.0065 (6) |
| C21 | 0.0225 (7) | 0.0215 (7) | 0.0293 (8) | −0.0072 (6) | −0.0013 (6) | −0.0093 (6) |
| C22 | 0.0195 (7) | 0.0216 (7) | 0.0318 (8) | −0.0092 (6) | 0.0037 (6) | −0.0048 (6) |
| C23 | 0.0244 (8) | 0.0258 (8) | 0.0240 (8) | −0.0089 (6) | 0.0048 (6) | −0.0059 (6) |
| C24 | 0.0240 (8) | 0.0229 (7) | 0.0232 (7) | −0.0088 (6) | 0.0012 (6) | −0.0074 (6) |
| Cl2 | 0.0213 (2) | 0.0305 (2) | 0.0393 (2) | −0.0009 (2) | −0.0062 (2) | −0.0029 (2) |
| N3 | 0.0187 (6) | 0.0195 (6) | 0.0189 (6) | −0.0043 (5) | −0.0008 (5) | −0.0054 (5) |
| N4 | 0.0191 (6) | 0.0219 (6) | 0.0190 (6) | −0.0057 (5) | −0.0006 (5) | −0.0043 (5) |
| C25 | 0.0199 (7) | 0.0224 (7) | 0.0183 (7) | −0.0074 (6) | 0.0015 (5) | −0.0056 (6) |
| C26 | 0.0192 (7) | 0.0203 (7) | 0.0176 (7) | −0.0057 (6) | 0.0002 (5) | −0.0029 (5) |
| C27 | 0.0174 (7) | 0.0200 (7) | 0.0188 (7) | −0.0053 (5) | 0.0012 (5) | −0.0040 (5) |
| C28 | 0.0198 (7) | 0.0163 (7) | 0.0229 (7) | −0.0050 (5) | −0.0019 (5) | −0.0064 (5) |
| C29 | 0.0238 (8) | 0.0247 (7) | 0.0221 (7) | −0.0078 (6) | 0.0010 (6) | −0.0062 (6) |
| C30 | 0.0210 (7) | 0.0277 (8) | 0.0311 (8) | −0.0094 (6) | 0.0006 (6) | −0.0087 (6) |
| C31 | 0.0248 (8) | 0.0238 (8) | 0.0292 (8) | −0.0074 (6) | −0.0069 (6) | −0.0066 (6) |
| C32 | 0.0305 (8) | 0.0304 (8) | 0.0210 (7) | −0.0111 (7) | −0.0013 (6) | −0.0070 (6) |
| C33 | 0.0229 (8) | 0.0264 (8) | 0.0235 (7) | −0.0085 (6) | 0.0022 (6) | −0.0084 (6) |
| C34 | 0.0230 (8) | 0.0249 (8) | 0.0253 (8) | −0.0040 (6) | −0.0018 (6) | −0.0110 (6) |
| C35 | 0.0282 (8) | 0.0216 (8) | 0.0436 (10) | −0.0056 (6) | −0.0063 (7) | −0.0110 (7) |
| C36 | 0.0366 (10) | 0.0293 (9) | 0.0537 (12) | −0.0145 (8) | 0.0069 (8) | −0.0083 (8) |
| C37 | 0.0198 (7) | 0.0236 (7) | 0.0163 (7) | −0.0041 (6) | 0.0005 (5) | −0.0071 (6) |
| C38 | 0.0249 (8) | 0.0218 (7) | 0.0254 (8) | −0.0066 (6) | 0.0000 (6) | −0.0050 (6) |
| C39 | 0.0204 (8) | 0.0299 (8) | 0.0320 (8) | −0.0090 (6) | −0.0023 (6) | −0.0058 (7) |
| C40 | 0.0192 (7) | 0.0266 (8) | 0.0227 (7) | −0.0016 (6) | −0.0013 (6) | −0.0054 (6) |
| C41 | 0.0254 (8) | 0.0206 (7) | 0.0207 (7) | −0.0044 (6) | 0.0002 (6) | −0.0055 (6) |
| C42 | 0.0228 (7) | 0.0244 (7) | 0.0197 (7) | −0.0078 (6) | 0.0007 (6) | −0.0079 (6) |
| C43 | 0.0178 (7) | 0.0198 (7) | 0.0243 (7) | −0.0080 (6) | 0.0014 (6) | −0.0060 (6) |
| C44 | 0.0212 (7) | 0.0222 (7) | 0.0241 (7) | −0.0091 (6) | 0.0001 (6) | −0.0053 (6) |
| C45 | 0.0203 (7) | 0.0231 (7) | 0.0303 (8) | −0.0057 (6) | −0.0029 (6) | −0.0038 (6) |
| C46 | 0.0218 (8) | 0.0217 (7) | 0.0380 (9) | −0.0042 (6) | 0.0010 (6) | −0.0107 (7) |
| C47 | 0.0270 (8) | 0.0283 (8) | 0.0322 (8) | −0.0075 (7) | 0.0004 (6) | −0.0154 (7) |
| C48 | 0.0240 (8) | 0.0244 (8) | 0.0257 (8) | −0.0054 (6) | −0.0039 (6) | −0.0068 (6) |
Geometric parameters (Å, º)
| Cl1—C16 | 1.7458 (16) | C18—H18 | 0.9500 |
| Cl2—C40 | 1.7480 (17) | C20—H20 | 0.9500 |
| N1—C1 | 1.364 (2) | C21—H21 | 0.9500 |
| N1—C3 | 1.3925 (19) | C22—H22 | 0.9500 |
| N1—C10 | 1.465 (2) | C23—H23 | 0.9500 |
| N2—C2 | 1.3822 (18) | C24—H24 | 0.9500 |
| N2—C1 | 1.320 (2) | C25—C37 | 1.477 (2) |
| N3—C34 | 1.467 (2) | C26—C27 | 1.371 (2) |
| N3—C27 | 1.386 (2) | C26—C43 | 1.476 (2) |
| N3—C25 | 1.3706 (19) | C27—C28 | 1.479 (2) |
| N4—C25 | 1.324 (2) | C28—C29 | 1.393 (2) |
| N4—C26 | 1.384 (2) | C28—C33 | 1.393 (2) |
| C1—C13 | 1.475 (2) | C29—C30 | 1.389 (2) |
| C2—C19 | 1.477 (2) | C30—C31 | 1.387 (2) |
| C2—C3 | 1.380 (2) | C31—C32 | 1.384 (2) |
| C3—C4 | 1.483 (2) | C32—C33 | 1.391 (2) |
| C4—C5 | 1.396 (2) | C34—C35 | 1.495 (2) |
| C4—C9 | 1.402 (2) | C35—C36 | 1.306 (3) |
| C5—C6 | 1.387 (2) | C37—C38 | 1.397 (2) |
| C6—C7 | 1.388 (2) | C37—C42 | 1.397 (2) |
| C7—C8 | 1.387 (2) | C38—C39 | 1.391 (2) |
| C8—C9 | 1.390 (2) | C39—C40 | 1.387 (2) |
| C10—C11 | 1.505 (2) | C40—C41 | 1.380 (2) |
| C11—C12 | 1.315 (3) | C41—C42 | 1.388 (2) |
| C13—C14 | 1.400 (2) | C43—C44 | 1.402 (2) |
| C13—C18 | 1.397 (2) | C43—C48 | 1.393 (2) |
| C14—C15 | 1.384 (2) | C44—C45 | 1.388 (2) |
| C15—C16 | 1.385 (2) | C45—C46 | 1.387 (2) |
| C16—C17 | 1.380 (2) | C46—C47 | 1.386 (2) |
| C17—C18 | 1.385 (2) | C47—C48 | 1.391 (2) |
| C19—C20 | 1.401 (2) | C29—H29 | 0.9500 |
| C19—C24 | 1.396 (2) | C30—H30 | 0.9500 |
| C20—C21 | 1.391 (2) | C31—H31 | 0.9500 |
| C21—C22 | 1.386 (2) | C32—H32 | 0.9500 |
| C22—C23 | 1.387 (2) | C33—H33 | 0.9500 |
| C23—C24 | 1.390 (2) | C34—H34A | 0.9900 |
| C5—H5 | 0.9500 | C34—H34B | 0.9900 |
| C6—H6 | 0.9500 | C35—H35 | 0.9500 |
| C7—H7 | 0.9500 | C36—H36A | 0.9500 |
| C8—H8 | 0.9500 | C36—H36B | 0.9500 |
| C9—H9 | 0.9500 | C38—H38 | 0.9500 |
| C10—H10A | 0.9900 | C39—H39 | 0.9500 |
| C10—H10B | 0.9900 | C41—H41 | 0.9500 |
| C11—H11 | 0.9500 | C42—H42 | 0.9500 |
| C12—H12B | 0.9500 | C44—H44 | 0.9500 |
| C12—H12A | 0.9500 | C45—H45 | 0.9500 |
| C14—H14 | 0.9500 | C46—H46 | 0.9500 |
| C15—H15 | 0.9500 | C47—H47 | 0.9500 |
| C17—H17 | 0.9500 | C48—H48 | 0.9500 |
| C1—N1—C3 | 107.20 (12) | C24—C23—H23 | 120.00 |
| C1—N1—C10 | 126.91 (12) | C22—C23—H23 | 120.00 |
| C3—N1—C10 | 125.83 (12) | C19—C24—H24 | 120.00 |
| C1—N2—C2 | 106.30 (12) | C23—C24—H24 | 120.00 |
| C27—N3—C34 | 124.52 (12) | N3—C25—N4 | 111.56 (13) |
| C25—N3—C27 | 106.76 (12) | N3—C25—C37 | 124.29 (14) |
| C25—N3—C34 | 128.66 (13) | N4—C25—C37 | 124.15 (13) |
| C25—N4—C26 | 105.60 (12) | N4—C26—C27 | 110.18 (13) |
| N1—C1—C13 | 126.68 (14) | N4—C26—C43 | 121.86 (13) |
| N2—C1—C13 | 121.93 (14) | C27—C26—C43 | 127.93 (14) |
| N1—C1—N2 | 111.38 (13) | N3—C27—C26 | 105.91 (13) |
| N2—C2—C3 | 109.72 (12) | N3—C27—C28 | 123.01 (13) |
| N2—C2—C19 | 118.31 (13) | C26—C27—C28 | 131.07 (14) |
| C3—C2—C19 | 131.84 (13) | C27—C28—C29 | 119.28 (13) |
| N1—C3—C2 | 105.40 (12) | C27—C28—C33 | 121.48 (14) |
| N1—C3—C4 | 121.12 (13) | C29—C28—C33 | 119.18 (14) |
| C2—C3—C4 | 133.41 (13) | C28—C29—C30 | 120.42 (14) |
| C3—C4—C9 | 120.62 (13) | C29—C30—C31 | 120.05 (15) |
| C5—C4—C9 | 118.78 (14) | C30—C31—C32 | 119.84 (15) |
| C3—C4—C5 | 120.61 (14) | C31—C32—C33 | 120.30 (14) |
| C4—C5—C6 | 120.60 (15) | C28—C33—C32 | 120.20 (15) |
| C5—C6—C7 | 120.34 (14) | N3—C34—C35 | 114.07 (13) |
| C6—C7—C8 | 119.61 (14) | C34—C35—C36 | 126.56 (17) |
| C7—C8—C9 | 120.41 (16) | C25—C37—C38 | 119.00 (14) |
| C4—C9—C8 | 120.26 (15) | C25—C37—C42 | 121.96 (14) |
| N1—C10—C11 | 113.67 (14) | C38—C37—C42 | 118.96 (14) |
| C10—C11—C12 | 125.92 (17) | C37—C38—C39 | 120.79 (15) |
| C1—C13—C18 | 124.28 (14) | C38—C39—C40 | 118.82 (15) |
| C14—C13—C18 | 118.67 (14) | Cl2—C40—C39 | 119.90 (13) |
| C1—C13—C14 | 116.95 (14) | Cl2—C40—C41 | 118.58 (13) |
| C13—C14—C15 | 121.38 (15) | C39—C40—C41 | 121.53 (15) |
| C14—C15—C16 | 118.29 (15) | C40—C41—C42 | 119.34 (15) |
| Cl1—C16—C15 | 119.59 (12) | C37—C42—C41 | 120.57 (15) |
| C15—C16—C17 | 121.89 (14) | C26—C43—C44 | 121.28 (13) |
| Cl1—C16—C17 | 118.52 (13) | C26—C43—C48 | 120.42 (14) |
| C16—C17—C18 | 119.38 (15) | C44—C43—C48 | 118.30 (14) |
| C13—C18—C17 | 120.39 (15) | C43—C44—C45 | 120.76 (14) |
| C20—C19—C24 | 117.97 (14) | C44—C45—C46 | 120.53 (15) |
| C2—C19—C20 | 118.62 (13) | C45—C46—C47 | 119.06 (16) |
| C2—C19—C24 | 123.36 (13) | C46—C47—C48 | 120.79 (15) |
| C19—C20—C21 | 120.95 (14) | C43—C48—C47 | 120.55 (15) |
| C20—C21—C22 | 120.31 (14) | C28—C29—H29 | 120.00 |
| C21—C22—C23 | 119.37 (15) | C30—C29—H29 | 120.00 |
| C22—C23—C24 | 120.43 (14) | C29—C30—H30 | 120.00 |
| C19—C24—C23 | 120.96 (14) | C31—C30—H30 | 120.00 |
| C4—C5—H5 | 120.00 | C30—C31—H31 | 120.00 |
| C6—C5—H5 | 120.00 | C32—C31—H31 | 120.00 |
| C5—C6—H6 | 120.00 | C31—C32—H32 | 120.00 |
| C7—C6—H6 | 120.00 | C33—C32—H32 | 120.00 |
| C8—C7—H7 | 120.00 | C28—C33—H33 | 120.00 |
| C6—C7—H7 | 120.00 | C32—C33—H33 | 120.00 |
| C7—C8—H8 | 120.00 | N3—C34—H34A | 109.00 |
| C9—C8—H8 | 120.00 | N3—C34—H34B | 109.00 |
| C8—C9—H9 | 120.00 | C35—C34—H34A | 109.00 |
| C4—C9—H9 | 120.00 | C35—C34—H34B | 109.00 |
| C11—C10—H10A | 109.00 | H34A—C34—H34B | 108.00 |
| N1—C10—H10B | 109.00 | C34—C35—H35 | 117.00 |
| C11—C10—H10B | 109.00 | C36—C35—H35 | 117.00 |
| H10A—C10—H10B | 108.00 | C35—C36—H36A | 120.00 |
| N1—C10—H10A | 109.00 | C35—C36—H36B | 120.00 |
| C10—C11—H11 | 117.00 | H36A—C36—H36B | 120.00 |
| C12—C11—H11 | 117.00 | C37—C38—H38 | 120.00 |
| C11—C12—H12A | 120.00 | C39—C38—H38 | 120.00 |
| C11—C12—H12B | 120.00 | C38—C39—H39 | 121.00 |
| H12A—C12—H12B | 120.00 | C40—C39—H39 | 121.00 |
| C15—C14—H14 | 119.00 | C40—C41—H41 | 120.00 |
| C13—C14—H14 | 119.00 | C42—C41—H41 | 120.00 |
| C14—C15—H15 | 121.00 | C37—C42—H42 | 120.00 |
| C16—C15—H15 | 121.00 | C41—C42—H42 | 120.00 |
| C18—C17—H17 | 120.00 | C43—C44—H44 | 120.00 |
| C16—C17—H17 | 120.00 | C45—C44—H44 | 120.00 |
| C17—C18—H18 | 120.00 | C44—C45—H45 | 120.00 |
| C13—C18—H18 | 120.00 | C46—C45—H45 | 120.00 |
| C21—C20—H20 | 120.00 | C45—C46—H46 | 120.00 |
| C19—C20—H20 | 120.00 | C47—C46—H46 | 120.00 |
| C20—C21—H21 | 120.00 | C46—C47—H47 | 120.00 |
| C22—C21—H21 | 120.00 | C48—C47—H47 | 120.00 |
| C21—C22—H22 | 120.00 | C43—C48—H48 | 120.00 |
| C23—C22—H22 | 120.00 | C47—C48—H48 | 120.00 |
| C3—N1—C1—N2 | −0.36 (17) | C13—C14—C15—C16 | 1.1 (2) |
| C10—N1—C1—N2 | −177.56 (14) | C14—C15—C16—Cl1 | 179.79 (12) |
| C3—N1—C1—C13 | −179.04 (15) | C14—C15—C16—C17 | −0.4 (2) |
| C10—N1—C1—C13 | 3.8 (2) | C15—C16—C17—C18 | −0.4 (2) |
| C1—N1—C3—C4 | 177.32 (14) | Cl1—C16—C17—C18 | 179.41 (13) |
| C1—N1—C10—C11 | 91.50 (18) | C16—C17—C18—C13 | 0.5 (2) |
| C3—N1—C10—C11 | −85.20 (18) | C2—C19—C24—C23 | −177.66 (15) |
| C1—N1—C3—C2 | 0.06 (17) | C20—C19—C24—C23 | −0.1 (2) |
| C10—N1—C3—C4 | −5.4 (2) | C24—C19—C20—C21 | 0.4 (2) |
| C10—N1—C3—C2 | 177.30 (14) | C2—C19—C20—C21 | 178.03 (14) |
| C2—N2—C1—C13 | 179.25 (14) | C19—C20—C21—C22 | −0.1 (2) |
| C2—N2—C1—N1 | 0.49 (17) | C20—C21—C22—C23 | −0.4 (2) |
| C1—N2—C2—C3 | −0.45 (17) | C21—C22—C23—C24 | 0.7 (2) |
| C1—N2—C2—C19 | −176.82 (13) | C22—C23—C24—C19 | −0.4 (2) |
| C25—N3—C27—C28 | 178.62 (13) | N4—C25—C37—C42 | −136.27 (16) |
| C34—N3—C25—N4 | −176.98 (13) | N4—C25—C37—C38 | 40.4 (2) |
| C25—N3—C34—C35 | −104.98 (18) | N3—C25—C37—C38 | −140.18 (15) |
| C34—N3—C27—C28 | −4.0 (2) | N3—C25—C37—C42 | 43.2 (2) |
| C25—N3—C27—C26 | 0.04 (15) | N4—C26—C27—N3 | −0.33 (16) |
| C27—N3—C25—N4 | 0.27 (16) | C27—C26—C43—C44 | 29.4 (2) |
| C27—N3—C25—C37 | −179.26 (13) | N4—C26—C43—C48 | 28.1 (2) |
| C27—N3—C34—C35 | 78.23 (18) | C43—C26—C27—N3 | 177.88 (14) |
| C34—N3—C27—C26 | 177.43 (13) | C27—C26—C43—C48 | −149.97 (16) |
| C34—N3—C25—C37 | 3.5 (2) | N4—C26—C27—C28 | −178.74 (14) |
| C26—N4—C25—C37 | 179.07 (13) | N4—C26—C43—C44 | −152.57 (15) |
| C26—N4—C25—N3 | −0.46 (16) | C43—C26—C27—C28 | −0.5 (3) |
| C25—N4—C26—C27 | 0.49 (16) | N3—C27—C28—C33 | 74.0 (2) |
| C25—N4—C26—C43 | −177.85 (13) | N3—C27—C28—C29 | −108.89 (17) |
| N1—C1—C13—C14 | 139.66 (16) | C26—C27—C28—C29 | 69.3 (2) |
| N1—C1—C13—C18 | −44.2 (2) | C26—C27—C28—C33 | −107.87 (19) |
| N2—C1—C13—C18 | 137.26 (17) | C27—C28—C29—C30 | −177.11 (15) |
| N2—C1—C13—C14 | −38.9 (2) | C29—C28—C33—C32 | 0.7 (2) |
| C3—C2—C19—C24 | 4.5 (3) | C33—C28—C29—C30 | 0.1 (2) |
| N2—C2—C3—N1 | 0.23 (17) | C27—C28—C33—C32 | 177.86 (15) |
| N2—C2—C3—C4 | −176.54 (16) | C28—C29—C30—C31 | −1.2 (3) |
| C3—C2—C19—C20 | −173.05 (16) | C29—C30—C31—C32 | 1.4 (3) |
| N2—C2—C19—C20 | 2.4 (2) | C30—C31—C32—C33 | −0.6 (3) |
| N2—C2—C19—C24 | 179.91 (14) | C31—C32—C33—C28 | −0.5 (3) |
| C19—C2—C3—N1 | 175.95 (15) | N3—C34—C35—C36 | 4.3 (3) |
| C19—C2—C3—C4 | −0.8 (3) | C25—C37—C38—C39 | −177.66 (14) |
| N1—C3—C4—C9 | −65.7 (2) | C42—C37—C38—C39 | −0.9 (2) |
| C2—C3—C4—C5 | −69.3 (2) | C25—C37—C42—C41 | 177.44 (13) |
| N1—C3—C4—C5 | 114.35 (17) | C38—C37—C42—C41 | 0.8 (2) |
| C2—C3—C4—C9 | 110.7 (2) | C37—C38—C39—C40 | 0.3 (2) |
| C3—C4—C9—C8 | 179.65 (15) | C38—C39—C40—Cl2 | −179.77 (12) |
| C5—C4—C9—C8 | −0.4 (2) | C38—C39—C40—C41 | 0.6 (2) |
| C9—C4—C5—C6 | −0.1 (2) | Cl2—C40—C41—C42 | 179.66 (11) |
| C3—C4—C5—C6 | 179.87 (15) | C39—C40—C41—C42 | −0.7 (2) |
| C4—C5—C6—C7 | 0.4 (2) | C40—C41—C42—C37 | 0.0 (2) |
| C5—C6—C7—C8 | −0.3 (3) | C26—C43—C44—C45 | −178.42 (15) |
| C6—C7—C8—C9 | −0.2 (3) | C48—C43—C44—C45 | 1.0 (2) |
| C7—C8—C9—C4 | 0.5 (3) | C26—C43—C48—C47 | 178.02 (15) |
| N1—C10—C11—C12 | 6.4 (2) | C44—C43—C48—C47 | −1.4 (2) |
| C18—C13—C14—C15 | −1.0 (2) | C43—C44—C45—C46 | 0.0 (2) |
| C14—C13—C18—C17 | 0.2 (2) | C44—C45—C46—C47 | −0.5 (3) |
| C1—C13—C18—C17 | −175.91 (15) | C45—C46—C47—C48 | 0.1 (3) |
| C1—C13—C14—C15 | 175.37 (15) | C46—C47—C48—C43 | 0.9 (3) |
Hydrogen-bond geometry (Å, º)
Cg2, Cg4 and Cg8 are the centroids of the C4–C9, C19–C24 and C43–C48 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12A···N1 | 0.95 | 2.55 | 2.880 (2) | 100 |
| C20—H20···N2 | 0.95 | 2.44 | 2.804 (2) | 102 |
| C36—H36A···N3 | 0.95 | 2.56 | 2.887 (2) | 100 |
| C5—H5···Cg4i | 0.95 | 2.76 | 3.5968 (17) | 147 |
| C11—H11···Cg8ii | 0.95 | 2.83 | 3.5879 (19) | 137 |
| C33—H33···Cg2iii | 0.95 | 2.89 | 3.8217 (18) | 166 |
Symmetry codes: (i) −x, −y+1, −z; (ii) −x, −y, −z+1; (iii) x, y, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5314).
References
- Akkurt, M., Fronczek, F. R., Mohamed, S. K., Talybov, A. H., Marzouk, A. A. E. & Abdelhamid, A. A. (2013). Acta Cryst. E69, o527–o528. [DOI] [PMC free article] [PubMed]
- Bhatnagar, A., Sharma, P. K. & Kumar, N. (2011). Int. J. Pharm Tech. Res 3, 268–282.
- Bruker (2001). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Congiu, C., Cocco, M. T. & Onnis, V. (2008). Bioorg. Med. Chem. Lett. 18, 989–993. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Mohamed, S. K., Akkurt, M., Marzouk, A. A., Abbasov, V. M. & Gurbanov, A. V. (2013). Acta Cryst. E69, o474–o475. [DOI] [PMC free article] [PubMed]
- Nanterment, P. G., Barrow, J. C., Lindsley, S. R., Young, M., Mao, S., Carroll, S., Bailey, C., Bosserman, M., Colussi, D., McMasters, D. R., Vacca, J. P. & Selnick, H. G. (2004). Bioorg. Med. Chem. Lett. 14, 2141–2145. [DOI] [PubMed]
- Puratchikody, A. & Doble, M. (2007). Bioorg. Med. Chem. Lett. 15, 1083–1090. [DOI] [PubMed]
- Roman, G., Riley, J. G., Vlahakis, J. Z., Kinobe, R. T., Brien, J. F., Nakatsu, K. & Szarek, W. A. (2007). Bioorg. Med. Chem. 15, 3225–3234. [DOI] [PubMed]
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Venkatesan, A. M., Agarwal, A., Abe, T., Ushirogochi, H. O., Santos, D., Li, Z., Francisco, G., Lin, Y. I., Peterson, P. J., Yang, Y., Weiss, W. J., Shales, D. M. & Mansour, T. S. (2008). Bioorg. Med. Chem. 16, 1890–1902. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813012592/hg5314sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813012592/hg5314Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813012592/hg5314Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report