Abstract
In the title molecular salt, C6H11N3O2 2+·2C6H2N3O7 −·2H2O, the histidine molecule exists as a histidinium dication, being protonated at the N atom of the imidazole ring. The charges are balanced by two picrate anions and the compound crystallizes as a dihydrate. In the crystal, the components are linked via N—H⋯O and O—H⋯O hydrogen bonds and weak C—H⋯O interactions, forming a three-dimensional supermolecular structure.
Related literature
For the role of hydrogen bonding in the construction of supramolecular structures, see: Braga et al. (2004 ▶); Harrowfield et al. (1995 ▶). For picrates of biologically important molecules, see: Harrison et al. (2007 ▶); Swamy et al. (2007 ▶); Bibal et al. (2003 ▶); Olsher et al. (1996 ▶). For bond angles in related structures, see: Yang et al. (2001 ▶).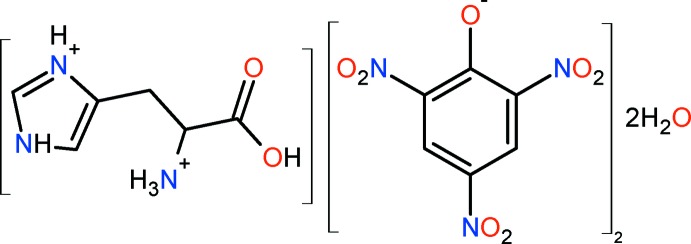
Experimental
Crystal data
C6H11N3O2 2+·2C6H2N3O7 −·2H2O
M r = 649.42
Monoclinic,
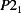
a = 6.6060 (4) Å
b = 25.7003 (13) Å
c = 7.9627 (5) Å
β = 107.532 (7)°
V = 1289.08 (13) Å3
Z = 2
Mo Kα radiation
μ = 0.15 mm−1
T = 293 K
0.20 × 0.15 × 0.10 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.970, T max = 0.985
5817 measured reflections
2982 independent reflections
2560 reflections with I > 2σ(I)
R int = 0.020
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.088
S = 1.09
2982 reflections
439 parameters
7 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.21 e Å−3
Δρmin = −0.18 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2 and SAINT (Bruker, 2004 ▶); data reduction: SAINT and XPREP (Bruker, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813013949/su2602sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813013949/su2602Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813013949/su2602Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯O10 | 0.89 | 2.19 | 2.909 (3) | 138 |
| N1—H1A⋯O12 | 0.89 | 2.11 | 2.841 (4) | 139 |
| N1—H1B⋯O18W | 0.89 | 1.85 | 2.700 (5) | 158 |
| N1—H1C⋯O15i | 0.89 | 2.16 | 3.007 (4) | 159 |
| N2—H2B⋯O10 | 0.91 (5) | 1.86 (5) | 2.709 (4) | 154 (4) |
| N2—H2B⋯O16 | 0.91 (5) | 2.49 (4) | 3.125 (4) | 128 (3) |
| N3—H3⋯O9ii | 0.86 | 2.56 | 2.992 (5) | 112 |
| N3—H3⋯O14ii | 0.86 | 2.25 | 3.077 (4) | 160 |
| O2—H2⋯O3iii | 0.82 | 1.86 | 2.657 (3) | 165 |
| O17W—H17A⋯O5iv | 0.84 (2) | 2.27 (3) | 3.082 (4) | 162 (9) |
| O17W—H17B⋯O3 | 0.84 (2) | 2.14 (8) | 2.864 (4) | 144 (12) |
| O18W—H18A⋯O17W v | 0.83 (2) | 1.83 (2) | 2.664 (5) | 176 (5) |
| O18W—H18B⋯O7 | 0.83 (2) | 2.32 (4) | 3.005 (5) | 140 (6) |
| C3—H3B⋯O10 | 0.97 | 2.59 | 3.210 (4) | 122 |
| C9—H9⋯O8vi | 0.93 | 2.40 | 3.177 (4) | 141 |
| C17—H17⋯O11iv | 0.93 | 2.36 | 3.177 (3) | 147 |
Symmetry codes: (i) 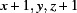 ; (ii)
; (ii) 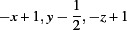 ; (iii)
; (iii) 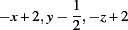 ; (iv)
; (iv) 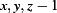 ; (v)
; (v) 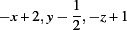 ; (vi)
; (vi) 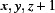 .
.
Acknowledgments
MS thanks the UGC Networking Centre, School of Chemistry, University of Hyderabad, India, for the award of a Visiting Research Fellowship to use the facilities at the School, which the authors also thank for access to the X-ray diffraction equipment.
supplementary crystallographic information
Comment
Intermolecular and inter-ionic hydrogen bonding interactions, which are not only the strongest of the noncovalent interactions but also highly directional, play an important role in constructing supramolecular structures (Braga et al., 2004). Picrate is generally used as an accompanying ion in many systems involving extraction and transport of metal ions to improve the extractability (Bibal et al., 2003). Picrate interacts as a monodentate, bidentate and tridentate ligand (Olsher et al., 1996). Furthermore, picrate is a penta-dentate ligand when it coordinates with cation by chelating pairs of oxygen atoms from p-nitro groups of adjacent picrates, and with successive cations linking the array into a two or three-dimensional network (Harrowfield et al., 1995) and picrates of biologically important molecules (Harrison et al., 2007; Swamy et al., 2007). We have prepared a new picrate of L-Histidinium hydrate and its crystal structure is reported herein.
The asymmetric unit of the title compound, Fig. 1, contains an L-histidinium cation, two picrate anions and two water molecules. The histidine molecule exists as an histidinium ion due to the protonation at the N atom of the imidazole ring. The charges are equilibrated by two picrate anions and crystallizes as a dihydrate. The imidazole ring (N2/N3/C4/C5/C6) makes a dihedral angle of 5.0 (2) and 4.9 (2)° with the benzene rings (C7—C12 and C13—C18), respectively, of the picrate anions.
The picrate anions adopt the keto form with C7—O3 and C13—O10 bond distance of 1.261 (4) and 1.249 (4) Å, C7—C8, C7—C12, C13—C14 and C13—C18 bond distance of 1.445 (4), 1.444 (4), 1.451 (4) and 1.450 (4) Å, respectively, which is longer than the other C—C bond lengths (between 1.374 (5) to 1.460 (4) Å) in the benzene ring. The bond angles C12—C7—C8 and C14—C13—C18 is 111.3 (3) and 110.6 (3)°, respectively, which is the case in some picrate complexes, while the corresponding bond angle of picric acid is 116.4 (5)° [Yang et al., 2001]. In the picrate anion the depronated phenolate oxygen atom deviates slightly from the plane of the benzene ring (torsion angle O3-C7-C8-C9 = -175.4 (3) ° and O10-C13-C14-C15 = 178.2 (3)°). The twist angles between the benzene rings (C7—C12 and C13—C18) and the ortho nitro groups (N4, N6, N7 and N9) are 41.5 (2), 32.4 (2), 32.2 (2) and 34.8 (2), respectively. The para-positioned nitro groups are twisted by 3.4 (2)° (N5) and 5.8 (2)° (N8), and are most likely influenced by a weak hydrogen bond interaction (O18—H18B···O7). The picrate ions are stacked head-to-tail, presumably as a result of charge-transfer interactions.
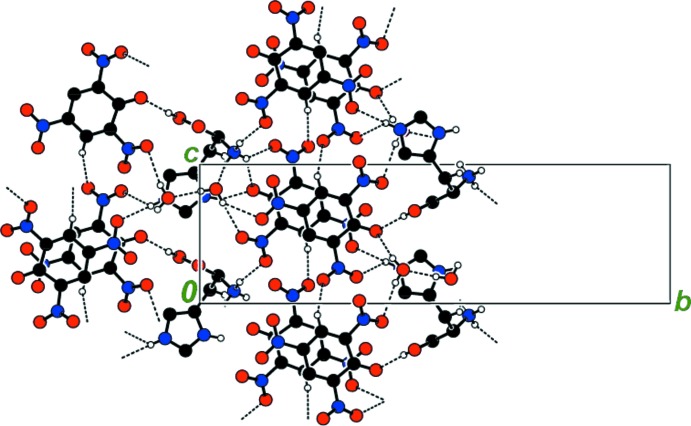
In the crystal the cation, the picrate anions and the water molecules of crystallization are involved in N—H···O and O—H···O hydrogen bonds and week C—H···O interactions, to form a three-dimensional supramolecular network (Table 1 and Fig. 2).
Experimental
1:2 stoichiometric proportions of analar grades L-histidine and picric acid (E-Merck) were dissolved in a triply distilled water and ethanol mixture and the two solutions were thoroughly mixed together using mechanical stirrer for about three hours. The clear yellow solution obtained was filtered off to get the crude material. The material was re-dissolved in a water-ethanol solvent mixture and kept aside without any mechanical movement for crystal growth in a dust free environment. Bright yellowish crystals that formed in 5 days were collected carefully from the mother liquor. Several re-crystallizations were done to get ultra pure crystals. The yield in the reaction was ca. 60%. Analysis calc. for C18H18N9O18 : C: 33.34, H: 2.79, N: 19.44 %; Found: C: 33.21, H: 3.74, N: 19.60%.
Refinement
The water molecule H-atoms, the methine (CH) H atom, and the CH and one NH H atom of the imidazole ring, were located in a difference Fourier map and freely refined. The OH, the NH3, one NH H atom of the imidazole ring, and the CH~2~ H atoms were positioned geometrically and refined using a riding model: O-H = 0.82 Å, N—H = 0.89 Å (NH3), N—H = 0.86 Å, C—H = 0.97 Å for CH2 H atoms, with Uiso(H) = 1.5Ueq(O,N) for the OH and NH3 H atoms and = 1.2Ueq(N,C) for other H atoms.
Figures
Fig. 1.
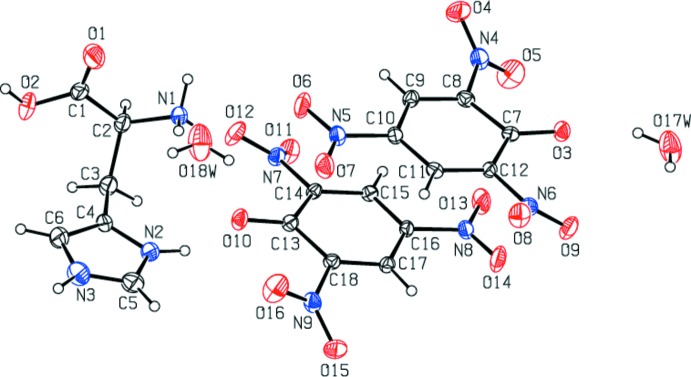
View of the molecular structure of the title compound, showing the atom labelling. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
A partial view along the a axisof the crystal packing of the title compound. Dashed lines indicate N—H···O and O—H···O hydrogen bonds and weak C—H···O interactions (see Table 1 for details).
Crystal data
| C6H11N3O22+·2C6H2N3O7−·2H2O | F(000) = 668 |
| Mr = 649.42 | Dx = 1.673 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2yb | Cell parameters from 5817 reflections |
| a = 6.6060 (4) Å | θ = 2.4–31.1° |
| b = 25.7003 (13) Å | µ = 0.15 mm−1 |
| c = 7.9627 (5) Å | T = 293 K |
| β = 107.532 (7)° | Square, yellow |
| V = 1289.08 (13) Å3 | 0.20 × 0.15 × 0.10 mm |
| Z = 2 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 2982 independent reflections |
| Radiation source: fine-focus sealed tube | 2560 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.020 |
| ω and φ scans | θmax = 28.9°, θmin = 2.8° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | h = −8→8 |
| Tmin = 0.970, Tmax = 0.985 | k = −26→34 |
| 5817 measured reflections | l = −10→9 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.039 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.088 | w = 1/[σ2(Fo2) + (0.0341P)2 + 0.2411P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max < 0.001 |
| 2982 reflections | Δρmax = 0.21 e Å−3 |
| 439 parameters | Δρmin = −0.18 e Å−3 |
| 7 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0095 (12) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 1.1170 (5) | −0.00275 (12) | 1.2736 (5) | 0.0774 (11) | |
| O2 | 0.8469 (5) | −0.04330 (11) | 1.3342 (4) | 0.0566 (8) | |
| H2 | 0.9346 | −0.0664 | 1.3719 | 0.085* | |
| O3 | 0.9237 (4) | 0.37442 (9) | 0.5194 (3) | 0.0366 (6) | |
| O4 | 1.1711 (5) | 0.33930 (12) | 1.0182 (3) | 0.0618 (9) | |
| O5 | 0.9113 (6) | 0.38681 (13) | 0.8677 (4) | 0.0659 (9) | |
| O6 | 1.0105 (5) | 0.15561 (11) | 0.8787 (3) | 0.0527 (7) | |
| O7 | 0.9511 (5) | 0.13208 (10) | 0.6075 (3) | 0.0507 (7) | |
| O8 | 0.9119 (4) | 0.26246 (10) | 0.1693 (3) | 0.0456 (6) | |
| O9 | 0.7415 (5) | 0.33288 (10) | 0.1914 (3) | 0.0489 (7) | |
| O10 | 0.5060 (4) | 0.11319 (9) | 0.8159 (3) | 0.0379 (6) | |
| O11 | 0.4555 (4) | 0.23048 (10) | 1.1305 (3) | 0.0443 (6) | |
| O12 | 0.6326 (5) | 0.16004 (10) | 1.1346 (3) | 0.0505 (7) | |
| O13 | 0.4413 (5) | 0.35254 (9) | 0.6731 (4) | 0.0529 (7) | |
| O14 | 0.3963 (5) | 0.32377 (10) | 0.4098 (3) | 0.0522 (7) | |
| O15 | 0.2242 (5) | 0.14022 (12) | 0.3056 (3) | 0.0565 (8) | |
| O16 | 0.4623 (6) | 0.09005 (11) | 0.4743 (4) | 0.0668 (9) | |
| N1 | 0.8717 (5) | 0.07237 (10) | 1.0875 (4) | 0.0354 (6) | |
| H1A | 0.7843 | 0.0987 | 1.0431 | 0.053* | |
| H1B | 0.8912 | 0.0535 | 1.0000 | 0.053* | |
| H1C | 0.9959 | 0.0848 | 1.1535 | 0.053* | |
| N2 | 0.4952 (5) | 0.00857 (12) | 0.7727 (4) | 0.0380 (7) | |
| N3 | 0.4987 (6) | −0.07402 (13) | 0.7471 (5) | 0.0590 (10) | |
| H3 | 0.4956 | −0.1045 | 0.7020 | 0.071* | |
| N4 | 1.0229 (5) | 0.34810 (12) | 0.8865 (4) | 0.0388 (7) | |
| N5 | 0.9722 (4) | 0.16635 (11) | 0.7214 (4) | 0.0353 (7) | |
| N6 | 0.8457 (5) | 0.29442 (11) | 0.2552 (3) | 0.0322 (6) | |
| N7 | 0.5317 (4) | 0.19734 (11) | 1.0568 (3) | 0.0312 (6) | |
| N8 | 0.4235 (4) | 0.31696 (11) | 0.5679 (4) | 0.0315 (6) | |
| N9 | 0.3649 (5) | 0.13122 (11) | 0.4434 (3) | 0.0364 (7) | |
| C1 | 0.9350 (6) | −0.00520 (14) | 1.2718 (5) | 0.0423 (9) | |
| C2 | 0.7771 (6) | 0.03902 (13) | 1.1981 (4) | 0.0344 (7) | |
| C3 | 0.5503 (5) | 0.02145 (14) | 1.0983 (4) | 0.0365 (8) | |
| H3A | 0.4943 | 0.0022 | 1.1793 | 0.044* | |
| H3B | 0.4629 | 0.0522 | 1.0624 | 0.044* | |
| C4 | 0.5280 (5) | −0.01146 (13) | 0.9395 (4) | 0.0331 (7) | |
| C5 | 0.4757 (6) | −0.03046 (16) | 0.6596 (5) | 0.0493 (10) | |
| C6 | 0.5283 (7) | −0.06396 (16) | 0.9212 (5) | 0.0504 (10) | |
| C7 | 0.9258 (5) | 0.32737 (12) | 0.5650 (4) | 0.0256 (6) | |
| C8 | 0.9775 (5) | 0.30948 (13) | 0.7450 (4) | 0.0283 (7) | |
| C9 | 0.9939 (5) | 0.25863 (13) | 0.7963 (4) | 0.0275 (7) | |
| H9 | 1.0308 | 0.2500 | 0.9151 | 0.033* | |
| C10 | 0.9549 (5) | 0.22013 (12) | 0.6693 (4) | 0.0265 (6) | |
| C11 | 0.9070 (4) | 0.23282 (12) | 0.4924 (4) | 0.0259 (6) | |
| H11 | 0.8856 | 0.2068 | 0.4077 | 0.031* | |
| C12 | 0.8915 (5) | 0.28421 (11) | 0.4435 (4) | 0.0235 (6) | |
| C13 | 0.4825 (5) | 0.15872 (12) | 0.7585 (4) | 0.0260 (6) | |
| C14 | 0.5012 (5) | 0.20440 (12) | 0.8692 (4) | 0.0232 (6) | |
| C15 | 0.4816 (5) | 0.25485 (12) | 0.8099 (4) | 0.0246 (6) | |
| H15 | 0.4977 | 0.2825 | 0.8884 | 0.030* | |
| C16 | 0.4375 (5) | 0.26384 (12) | 0.6310 (4) | 0.0237 (6) | |
| C17 | 0.3974 (4) | 0.22294 (12) | 0.5116 (4) | 0.0247 (6) | |
| H17 | 0.3562 | 0.2293 | 0.3910 | 0.030* | |
| C18 | 0.4195 (5) | 0.17329 (12) | 0.5740 (4) | 0.0249 (6) | |
| O17W | 1.0107 (7) | 0.43238 (13) | 0.2418 (5) | 0.0724 (10) | |
| O18W | 1.0051 (8) | 0.03423 (14) | 0.8230 (5) | 0.0779 (11) | |
| H17A | 1.006 (14) | 0.415 (3) | 0.150 (7) | 0.18 (4)* | |
| H17B | 1.04 (2) | 0.411 (3) | 0.325 (9) | 0.31 (7)* | |
| H18A | 1.001 (9) | 0.0022 (8) | 0.808 (7) | 0.086 (19)* | |
| H18B | 0.977 (15) | 0.049 (2) | 0.726 (5) | 0.19 (4)* | |
| H2A | 0.763 (6) | 0.0623 (16) | 1.296 (5) | 0.039 (10)* | |
| H6 | 0.539 (7) | −0.0916 (19) | 0.999 (6) | 0.064 (13)* | |
| H5 | 0.455 (7) | −0.029 (2) | 0.536 (6) | 0.068 (13)* | |
| H2B | 0.479 (7) | 0.0432 (19) | 0.751 (5) | 0.053 (12)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0441 (17) | 0.047 (2) | 0.123 (3) | 0.0016 (14) | −0.0029 (17) | 0.0338 (19) |
| O2 | 0.0723 (19) | 0.0389 (16) | 0.0567 (16) | 0.0113 (15) | 0.0166 (14) | 0.0245 (13) |
| O3 | 0.0544 (15) | 0.0211 (12) | 0.0300 (11) | −0.0025 (11) | 0.0064 (10) | −0.0010 (9) |
| O4 | 0.083 (2) | 0.0564 (19) | 0.0295 (13) | −0.0121 (17) | −0.0084 (13) | −0.0061 (13) |
| O5 | 0.096 (2) | 0.0489 (19) | 0.0517 (16) | 0.0160 (18) | 0.0213 (15) | −0.0173 (14) |
| O6 | 0.0750 (19) | 0.0426 (16) | 0.0386 (14) | −0.0024 (14) | 0.0143 (13) | 0.0187 (12) |
| O7 | 0.0717 (19) | 0.0268 (14) | 0.0524 (16) | −0.0034 (14) | 0.0168 (13) | 0.0001 (13) |
| O8 | 0.0684 (17) | 0.0436 (16) | 0.0298 (12) | −0.0007 (14) | 0.0225 (12) | −0.0067 (11) |
| O9 | 0.0775 (19) | 0.0295 (13) | 0.0292 (12) | 0.0062 (13) | 0.0000 (12) | 0.0058 (10) |
| O10 | 0.0591 (16) | 0.0204 (12) | 0.0299 (12) | 0.0018 (11) | 0.0070 (11) | 0.0016 (9) |
| O11 | 0.0660 (16) | 0.0427 (15) | 0.0298 (12) | 0.0072 (13) | 0.0229 (12) | −0.0033 (11) |
| O12 | 0.080 (2) | 0.0350 (14) | 0.0269 (12) | 0.0154 (14) | 0.0007 (12) | 0.0059 (11) |
| O13 | 0.079 (2) | 0.0174 (13) | 0.0575 (16) | 0.0002 (13) | 0.0135 (14) | −0.0002 (12) |
| O14 | 0.081 (2) | 0.0376 (15) | 0.0368 (13) | 0.0005 (15) | 0.0155 (13) | 0.0172 (12) |
| O15 | 0.0706 (19) | 0.0514 (18) | 0.0320 (13) | −0.0160 (15) | −0.0078 (12) | −0.0063 (12) |
| O16 | 0.112 (3) | 0.0364 (17) | 0.0446 (16) | 0.0147 (18) | 0.0118 (17) | −0.0133 (13) |
| N1 | 0.0395 (15) | 0.0194 (14) | 0.0404 (16) | −0.0002 (12) | 0.0017 (12) | 0.0029 (11) |
| N2 | 0.0445 (17) | 0.0244 (15) | 0.0380 (16) | −0.0015 (14) | 0.0019 (13) | −0.0004 (13) |
| N3 | 0.082 (3) | 0.0247 (17) | 0.058 (2) | −0.0058 (18) | 0.0021 (18) | −0.0120 (15) |
| N4 | 0.0573 (19) | 0.0330 (18) | 0.0281 (14) | −0.0092 (15) | 0.0158 (13) | −0.0041 (12) |
| N5 | 0.0356 (15) | 0.0281 (16) | 0.0416 (16) | −0.0027 (13) | 0.0110 (12) | 0.0070 (13) |
| N6 | 0.0444 (16) | 0.0279 (15) | 0.0219 (13) | −0.0078 (13) | 0.0066 (11) | −0.0031 (11) |
| N7 | 0.0428 (16) | 0.0261 (15) | 0.0220 (13) | −0.0012 (13) | 0.0056 (11) | −0.0016 (11) |
| N8 | 0.0286 (14) | 0.0255 (15) | 0.0399 (15) | −0.0017 (12) | 0.0096 (11) | 0.0060 (12) |
| N9 | 0.0547 (18) | 0.0253 (15) | 0.0290 (14) | −0.0073 (14) | 0.0122 (13) | −0.0055 (12) |
| C1 | 0.048 (2) | 0.0270 (19) | 0.0382 (19) | −0.0013 (17) | −0.0072 (16) | 0.0034 (15) |
| C2 | 0.0470 (19) | 0.0246 (17) | 0.0274 (15) | 0.0004 (15) | 0.0048 (14) | 0.0022 (13) |
| C3 | 0.0412 (18) | 0.0329 (18) | 0.0341 (17) | 0.0030 (16) | 0.0097 (14) | 0.0045 (14) |
| C4 | 0.0338 (16) | 0.0230 (16) | 0.0377 (17) | −0.0032 (14) | 0.0037 (13) | 0.0027 (14) |
| C5 | 0.058 (2) | 0.038 (2) | 0.043 (2) | −0.0062 (19) | 0.0018 (18) | −0.0087 (18) |
| C6 | 0.064 (3) | 0.031 (2) | 0.045 (2) | −0.0083 (19) | 0.0001 (19) | 0.0019 (18) |
| C7 | 0.0269 (15) | 0.0258 (16) | 0.0221 (14) | −0.0025 (13) | 0.0046 (11) | −0.0039 (12) |
| C8 | 0.0311 (16) | 0.0290 (17) | 0.0248 (15) | −0.0051 (14) | 0.0081 (12) | −0.0064 (13) |
| C9 | 0.0295 (15) | 0.0314 (18) | 0.0224 (14) | −0.0003 (14) | 0.0090 (12) | 0.0034 (13) |
| C10 | 0.0251 (14) | 0.0234 (16) | 0.0313 (15) | −0.0005 (13) | 0.0088 (11) | 0.0038 (12) |
| C11 | 0.0254 (14) | 0.0262 (16) | 0.0263 (14) | −0.0033 (13) | 0.0082 (11) | −0.0041 (12) |
| C12 | 0.0288 (15) | 0.0187 (14) | 0.0213 (14) | −0.0009 (12) | 0.0051 (11) | −0.0007 (11) |
| C13 | 0.0275 (15) | 0.0248 (16) | 0.0251 (15) | 0.0014 (13) | 0.0070 (12) | −0.0005 (13) |
| C14 | 0.0267 (14) | 0.0246 (16) | 0.0176 (14) | 0.0022 (13) | 0.0057 (11) | 0.0006 (11) |
| C15 | 0.0256 (14) | 0.0233 (15) | 0.0239 (14) | 0.0018 (13) | 0.0057 (11) | −0.0023 (12) |
| C16 | 0.0229 (14) | 0.0202 (15) | 0.0276 (15) | 0.0015 (12) | 0.0072 (11) | 0.0048 (12) |
| C17 | 0.0254 (14) | 0.0274 (17) | 0.0215 (14) | −0.0005 (13) | 0.0074 (11) | 0.0027 (12) |
| C18 | 0.0270 (15) | 0.0252 (16) | 0.0215 (15) | −0.0004 (13) | 0.0058 (12) | −0.0030 (11) |
| O17W | 0.126 (3) | 0.0397 (18) | 0.0615 (19) | −0.0171 (19) | 0.043 (2) | −0.0111 (15) |
| O18W | 0.125 (3) | 0.043 (2) | 0.085 (3) | 0.009 (2) | 0.060 (2) | 0.0001 (18) |
Geometric parameters (Å, º)
| O1—C1 | 1.199 (5) | N8—C16 | 1.448 (4) |
| O2—C1 | 1.311 (5) | N9—C18 | 1.468 (4) |
| O2—H2 | 0.8200 | C1—C2 | 1.534 (5) |
| O3—C7 | 1.261 (4) | C2—C3 | 1.537 (5) |
| O4—N4 | 1.220 (4) | C2—H2A | 1.01 (4) |
| O5—N4 | 1.220 (4) | C3—C4 | 1.491 (5) |
| O6—N5 | 1.232 (4) | C3—H3A | 0.9700 |
| O7—N5 | 1.242 (4) | C3—H3B | 0.9700 |
| O8—N6 | 1.230 (4) | C4—C6 | 1.357 (5) |
| O9—N6 | 1.224 (4) | C5—H5 | 0.96 (5) |
| O10—C13 | 1.249 (4) | C6—H6 | 0.93 (5) |
| O11—N7 | 1.225 (3) | C7—C12 | 1.444 (4) |
| O12—N7 | 1.224 (4) | C7—C8 | 1.445 (4) |
| O13—N8 | 1.222 (4) | C8—C9 | 1.363 (5) |
| O14—N8 | 1.230 (4) | C9—C10 | 1.383 (4) |
| O15—N9 | 1.228 (4) | C9—H9 | 0.9300 |
| O16—N9 | 1.224 (4) | C10—C11 | 1.387 (4) |
| N1—C2 | 1.494 (4) | C11—C12 | 1.372 (4) |
| N1—H1A | 0.8900 | C11—H11 | 0.9300 |
| N1—H1B | 0.8900 | C13—C14 | 1.451 (4) |
| N1—H1C | 0.8900 | C13—C18 | 1.450 (4) |
| N2—C5 | 1.328 (5) | C14—C15 | 1.373 (4) |
| N2—C4 | 1.380 (4) | C15—C16 | 1.385 (4) |
| N2—H2B | 0.91 (5) | C15—H15 | 0.9300 |
| N3—C5 | 1.303 (5) | C16—C17 | 1.388 (4) |
| N3—C6 | 1.365 (5) | C17—C18 | 1.361 (4) |
| N3—H3 | 0.8600 | C17—H17 | 0.9300 |
| N4—C8 | 1.463 (4) | O17W—H17A | 0.84 (2) |
| N5—C10 | 1.438 (4) | O17W—H17B | 0.84 (2) |
| N6—C12 | 1.461 (4) | O18W—H18A | 0.83 (2) |
| N7—C14 | 1.457 (4) | O18W—H18B | 0.83 (2) |
| C1—O2—H2 | 109.5 | C6—C4—N2 | 105.8 (3) |
| C2—N1—H1A | 109.5 | C6—C4—C3 | 130.7 (3) |
| C2—N1—H1B | 109.5 | N2—C4—C3 | 123.5 (3) |
| H1A—N1—H1B | 109.5 | N3—C5—N2 | 108.3 (4) |
| C2—N1—H1C | 109.5 | N3—C5—H5 | 124 (3) |
| H1A—N1—H1C | 109.5 | N2—C5—H5 | 128 (3) |
| H1B—N1—H1C | 109.5 | C4—C6—N3 | 107.0 (4) |
| C5—N2—C4 | 109.0 (3) | C4—C6—H6 | 134 (3) |
| C5—N2—H2B | 129 (3) | N3—C6—H6 | 119 (3) |
| C4—N2—H2B | 121 (3) | O3—C7—C12 | 123.9 (3) |
| C5—N3—C6 | 109.8 (3) | O3—C7—C8 | 124.7 (3) |
| C5—N3—H3 | 125.1 | C12—C7—C8 | 111.3 (3) |
| C6—N3—H3 | 125.1 | C9—C8—C7 | 125.1 (3) |
| O5—N4—O4 | 123.7 (3) | C9—C8—N4 | 116.1 (3) |
| O5—N4—C8 | 118.8 (3) | C7—C8—N4 | 118.7 (3) |
| O4—N4—C8 | 117.5 (3) | C8—C9—C10 | 119.1 (3) |
| O6—N5—O7 | 121.8 (3) | C8—C9—H9 | 120.4 |
| O7—N5—O7 | 0.0 (3) | C10—C9—H9 | 120.4 |
| O6—N5—C10 | 118.9 (3) | C9—C10—C11 | 120.7 (3) |
| O7—N5—C10 | 119.3 (3) | C9—C10—N5 | 119.7 (3) |
| O9—N6—O8 | 123.9 (3) | C11—C10—N5 | 119.6 (3) |
| O9—N6—C12 | 119.2 (3) | C12—C11—C10 | 119.3 (3) |
| O8—N6—C12 | 116.9 (3) | C12—C11—H11 | 120.3 |
| O12—N7—O11 | 122.8 (3) | C10—C11—H11 | 120.3 |
| O12—N7—C14 | 120.2 (3) | C11—C12—C7 | 124.5 (3) |
| O11—N7—C14 | 117.0 (3) | C11—C12—N6 | 116.0 (3) |
| O13—N8—O14 | 123.4 (3) | C7—C12—N6 | 119.4 (3) |
| O13—N8—C16 | 119.0 (3) | O10—C13—C14 | 123.9 (3) |
| O14—N8—C16 | 117.6 (3) | O10—C13—C18 | 125.4 (3) |
| O16—N9—O15 | 123.6 (3) | C14—C13—C18 | 110.6 (3) |
| O16—N9—C18 | 119.5 (3) | C15—C14—C13 | 125.0 (2) |
| O15—N9—C18 | 116.9 (3) | C15—C14—N7 | 116.1 (3) |
| O1—C1—O2 | 126.3 (4) | C13—C14—N7 | 118.8 (3) |
| O1—C1—C2 | 121.9 (3) | C14—C15—C16 | 118.7 (3) |
| O2—C1—C2 | 111.7 (3) | C14—C15—H15 | 120.6 |
| N1—C2—C1 | 107.1 (3) | C16—C15—H15 | 120.6 |
| N1—C2—C3 | 112.3 (3) | C15—C16—C17 | 121.1 (3) |
| C1—C2—C3 | 115.1 (3) | C15—C16—N8 | 119.1 (3) |
| N1—C2—H2A | 105 (2) | C17—C16—N8 | 119.8 (3) |
| C1—C2—H2A | 111 (2) | C18—C17—C16 | 118.8 (3) |
| C3—C2—H2A | 106 (2) | C18—C17—H17 | 120.6 |
| C4—C3—C2 | 115.9 (3) | C16—C17—H17 | 120.6 |
| C4—C3—H3A | 108.3 | C17—C18—C13 | 125.4 (3) |
| C2—C3—H3A | 108.3 | C17—C18—N9 | 117.1 (2) |
| C4—C3—H3B | 108.3 | C13—C18—N9 | 117.5 (3) |
| C2—C3—H3B | 108.3 | H17A—O17W—H17B | 106 (3) |
| H3A—C3—H3B | 107.4 | H18A—O18W—H18B | 109 (3) |
| O1—C1—C2—N1 | 17.5 (5) | C8—C7—C12—C11 | −0.3 (4) |
| O2—C1—C2—N1 | −164.4 (3) | O3—C7—C12—N6 | −1.7 (5) |
| O1—C1—C2—C3 | 143.1 (4) | C8—C7—C12—N6 | −177.6 (3) |
| O2—C1—C2—C3 | −38.8 (4) | O9—N6—C12—C11 | 148.0 (3) |
| N1—C2—C3—C4 | 62.2 (4) | O8—N6—C12—C11 | −30.3 (4) |
| C1—C2—C3—C4 | −60.7 (4) | O9—N6—C12—C7 | −34.5 (4) |
| C5—N2—C4—C6 | 0.3 (4) | O8—N6—C12—C7 | 147.2 (3) |
| C5—N2—C4—C3 | −178.1 (3) | O10—C13—C14—C15 | 178.2 (3) |
| C2—C3—C4—C6 | 92.6 (5) | C18—C13—C14—C15 | −5.6 (4) |
| C2—C3—C4—N2 | −89.5 (4) | O10—C13—C14—N7 | −5.3 (5) |
| C6—N3—C5—N2 | 1.7 (5) | C18—C13—C14—N7 | 170.9 (2) |
| C4—N2—C5—N3 | −1.2 (5) | O12—N7—C14—C15 | −150.2 (3) |
| N2—C4—C6—N3 | 0.7 (4) | O11—N7—C14—C15 | 28.7 (4) |
| C3—C4—C6—N3 | 179.0 (4) | O12—N7—C14—C13 | 33.0 (4) |
| C5—N3—C6—C4 | −1.5 (5) | O11—N7—C14—C13 | −148.1 (3) |
| O3—C7—C8—C9 | −175.4 (3) | C13—C14—C15—C16 | 1.0 (5) |
| C12—C7—C8—C9 | 0.4 (4) | N7—C14—C15—C16 | −175.5 (2) |
| O3—C7—C8—N4 | 2.6 (5) | C14—C15—C16—C17 | 4.9 (4) |
| C12—C7—C8—N4 | 178.4 (3) | C14—C15—C16—N8 | −178.0 (3) |
| O5—N4—C8—C9 | −139.3 (3) | O13—N8—C16—C15 | −3.4 (4) |
| O4—N4—C8—C9 | 40.2 (4) | O14—N8—C16—C15 | 175.9 (3) |
| O5—N4—C8—C7 | 42.6 (5) | O13—N8—C16—C17 | 173.8 (3) |
| O4—N4—C8—C7 | −138.0 (3) | O14—N8—C16—C17 | −6.9 (4) |
| C7—C8—C9—C10 | −1.4 (5) | C15—C16—C17—C18 | −5.4 (4) |
| N4—C8—C9—C10 | −179.4 (3) | N8—C16—C17—C18 | 177.5 (3) |
| C8—C9—C10—C11 | 2.2 (4) | C16—C17—C18—C13 | 0.1 (5) |
| C8—C9—C10—N5 | 179.8 (3) | C16—C17—C18—N9 | 176.9 (3) |
| O6—N5—C10—C9 | 3.9 (4) | O10—C13—C18—C17 | −178.8 (3) |
| O7—N5—C10—C9 | −175.1 (3) | C14—C13—C18—C17 | 5.1 (4) |
| O6—N5—C10—C11 | −178.5 (3) | O10—C13—C18—N9 | 4.4 (5) |
| O7—N5—C10—C11 | 2.5 (4) | C14—C13—C18—N9 | −171.8 (3) |
| C9—C10—C11—C12 | −2.2 (4) | O16—N9—C18—C17 | 147.2 (3) |
| N5—C10—C11—C12 | −179.7 (3) | O15—N9—C18—C17 | −31.3 (4) |
| C10—C11—C12—C7 | 1.2 (5) | O16—N9—C18—C13 | −35.7 (4) |
| C10—C11—C12—N6 | 178.6 (2) | O15—N9—C18—C13 | 145.8 (3) |
| O3—C7—C12—C11 | 175.6 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O10 | 0.89 | 2.19 | 2.909 (3) | 138 |
| N1—H1A···O12 | 0.89 | 2.11 | 2.841 (4) | 139 |
| N1—H1B···O18W | 0.89 | 1.85 | 2.700 (5) | 158 |
| N1—H1C···O15i | 0.89 | 2.16 | 3.007 (4) | 159 |
| N2—H2B···O10 | 0.91 (5) | 1.86 (5) | 2.709 (4) | 154 (4) |
| N2—H2B···O16 | 0.91 (5) | 2.49 (4) | 3.125 (4) | 128 (3) |
| N3—H3···O9ii | 0.86 | 2.56 | 2.992 (5) | 112 |
| N3—H3···O14ii | 0.86 | 2.25 | 3.077 (4) | 160 |
| O2—H2···O3iii | 0.82 | 1.86 | 2.657 (3) | 165 |
| O17W—H17A···O5iv | 0.84 (2) | 2.27 (3) | 3.082 (4) | 162 (9) |
| O17W—H17B···O3 | 0.84 (2) | 2.14 (8) | 2.864 (4) | 144 (12) |
| O18W—H18A···O17Wv | 0.83 (2) | 1.83 (2) | 2.664 (5) | 176 (5) |
| O18W—H18B···O7 | 0.83 (2) | 2.32 (4) | 3.005 (5) | 140 (6) |
| C3—H3B···O10 | 0.97 | 2.59 | 3.210 (4) | 122 |
| C9—H9···O8vi | 0.93 | 2.40 | 3.177 (4) | 141 |
| C17—H17···O11iv | 0.93 | 2.36 | 3.177 (3) | 147 |
Symmetry codes: (i) x+1, y, z+1; (ii) −x+1, y−1/2, −z+1; (iii) −x+2, y−1/2, −z+2; (iv) x, y, z−1; (v) −x+2, y−1/2, −z+1; (vi) x, y, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2602).
References
- Bibal, B., Declercq, J. P., Dutasta, J. P., Tinant, B. & Valade, A. G. (2003). Tetrahedron, 59, 5849–5854.
- Braga, D., Maini, L., Polito, M. & Grepioni, F. (2004). Struct. Bond. 111, 1–32.
- Bruker (2004). APEX2, SAINT and XPREP Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Harrison, W. T. A., Bindya, S., Ashok, M. A., Yathirajan, H. S. & Narayana, B. (2007). Acta Cryst. E63, o3143.
- Harrowfield, J. M., Skelton, B. W. & White, A. H. (1995). Aust. J. Chem. 48, 1311–1331.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Olsher, U., Feinberg, H., Frolow, F. & Shoham, G. (1996). Pure Appl. Chem. 68, 1195–1199.
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Swamy, M. T., Ashok, M. A., Yathirajan, H. S., Narayana, B. & Bolte, M. (2007). Acta Cryst. E63, o4919.
- Yang, L., Zhang, T. L., Feng, C. G., Zhang, J. G. & Yu, K. B. (2001). Energ. Mater. 9, 37–39.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813013949/su2602sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813013949/su2602Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813013949/su2602Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report