Abstract
In the title compound, C24H20N2, one of the ring C atoms and one of the ring N atoms are disordered over two sets of sites in a 0.615 (3):0.385 (3) ratio. The two parts of the disordered imidazole ring adopt an envelope conformation, with the undisordered ring N atom as the flap, displaced by −0.118 (6) and 0.226 (7) Å, respectively, in the two disorder components from the plane through the other ring atoms. The crystal structure features C—H⋯N hydrogen bonds and C—H⋯π interactions, which lead to the formation of infinite chains along [010].
Related literature
For the biological significance of imidazole derivatives, see, for example: Kumar (2010 ▶); Castaño et al. (2008 ▶); Banfi et al. (2006 ▶); Bogle et al. (1994 ▶). For the synthesis and the structures of similar imidazoles, see: Mohamed et al. (2013a
▶,b
▶); Akkurt et al. (2013 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).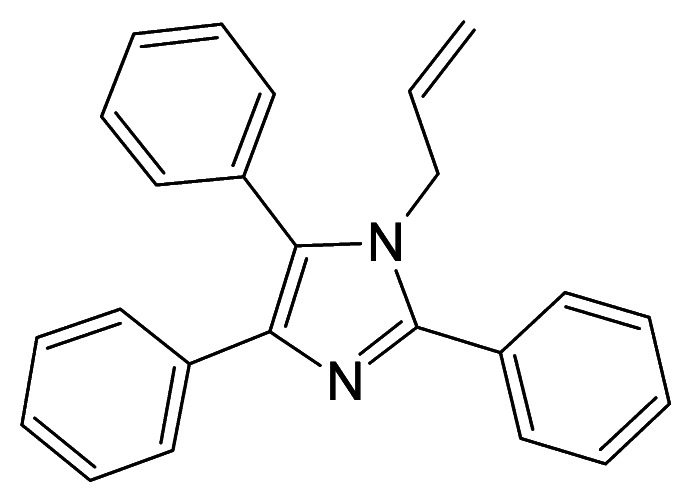
Experimental
Crystal data
C24H20N2
M r = 336.42
Monoclinic,
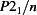
a = 10.362 (3) Å
b = 8.938 (2) Å
c = 19.387 (5) Å
β = 90.340 (5)°
V = 1795.5 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.07 mm−1
T = 100 K
0.14 × 0.14 × 0.003 mm
Data collection
Bruker SMART APEX CCD area-detector diffractometer
Absorption correction: refined from ΔF (XABS2; Parkin et al., 1995 ▶) T min = 0.990, T max = 1.000
17127 measured reflections
3168 independent reflections
2442 reflections with I > 2σ(I)
R int = 0.046
Refinement
R[F 2 > 2σ(F 2)] = 0.062
wR(F 2) = 0.183
S = 1.04
3168 reflections
299 parameters
12 restraints
H-atom parameters constrained
Δρmax = 0.49 e Å−3
Δρmin = −0.29 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813014104/zp2005sup1.cif
Structure factors: contains datablock(s) shelxl. DOI: 10.1107/S1600536813014104/zp2005Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813014104/zp2005Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg3 and Cg4 are the centroids of the C4–C9 and C10–C15 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C17A—H17A⋯N1i | 0.95 | 2.45 | 3.246 (8) | 141 |
| C21A—H21A⋯Cg3ii | 0.95 | 2.98 | 3.888 (5) | 160 |
| C21B—H21B⋯Cg4i | 0.95 | 2.99 | 3.914 (4) | 163 |
Symmetry codes: (i) 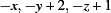 ; (ii)
; (ii) 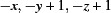 .
.
Acknowledgments
Manchester Metropolitan University, Erciyes University and Granada University are gratefully acknowledged for supporting this study. The authors also thank José Romero Garzón, Centro de Instrumentación Científica, Universidad de Granada, for the data collection.
supplementary crystallographic information
Comment
The substituted imidazole derivatives are valuable in treatment of many systemic microbial infections and exhibit different types of pharmacological and biological activities (Kumar 2010). A number of substituted imidazoles, including clotrimazole, are selective inhibitors of nitric oxide synthase, which makes them interesting drug targets in inflammation, neurodegenerative diseases and tumors of the nervous system (Bogle et al., 1994; Castaño et al., 2008). Imidazoles also belong to the class of azole antifungals (Banfi et al., 2006), which includes 1-vinyl imidazole, ketoconazole, miconazole, and clotrimazole. In this aspect we have prepared a series of tetrasubstituted imidazoles including the title compound as potential bio-active precursors. Herein, we report the single-crystal X-ray structure of 2,4,5-Triphenyl-1-(prop-2-en-1-yl)-1H-imidazole (I).
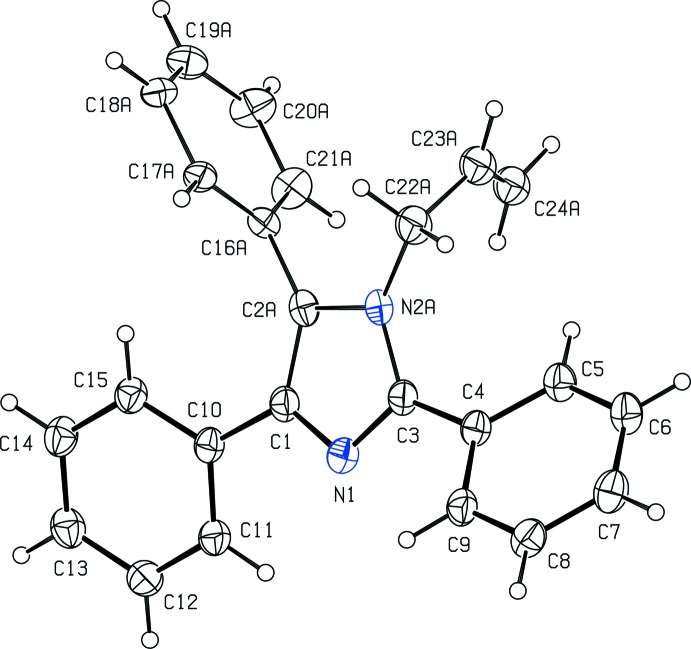
In the title compound (I), (Fig. 1), the two parts of the disordered imidazole ring adopt an envelope conformation [the puckering parameters (Cremer & Pople, 1975) are Q(2) = 0.076 (3) Å, φ(2) = 357 (4) ° for (N1/N2A/C1/C2A/C3), and Q(2) = 0.146 (4) Å, φ(2) = 180 (3) ° for (N1/N2B/C1/C2B/C3)]. The phenyl rings (C4–C9, C10–C15, C16A–C21A and C16B–C21B) makes dihedral angles of 35.91 (7), 18.14 (17), 85.0 (2) and 87.8 (2)°, respectively, with the mean plane of the imadazole ring (N1/N2A/C1/C2A/C3) and the corresponding angles are 18.8 (3), 35.3 (2), 85.7 (3) and 83.6 (3)°, respectively, for (N1/N2B/C1/C2B/C3). The bond lengths in (I) are within normal ranges and are comparable with those reported for the similar structures (Mohamed et al., 2013a,b; Akkurt et al., 2013).
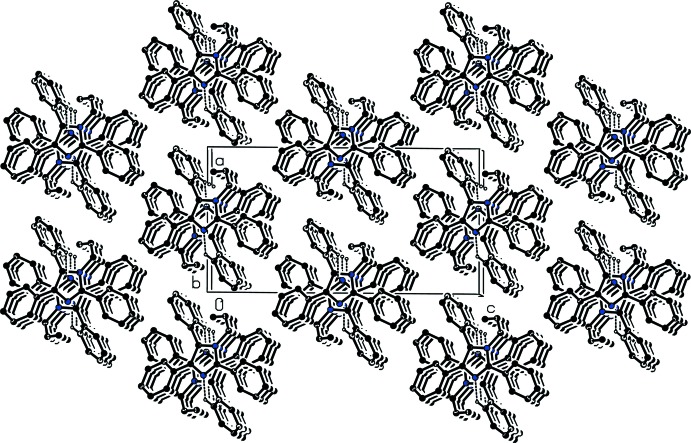
In the crystal structure, molecules are linked by intermolecular C—H···N hydrogen bonds (Table 1, Fig. 2). In addition, C—H···π interactions contribute to the stabilization of the crystal packing.
Experimental
The title compound was synthesized according to our reported method (Mohamed et al. 2013a) in 83% yield. Colourless plates suitable for X-ray analyses were obtained by slow evaporation of a solution of (I) in ethanol, m.p. 377–379 K.
Refinement
All H atoms were placed in geometrically, with C—H = 0.95 and 0.99 Å, and refined as riding with Uiso(H) = 1.2 Ueq(C) of the parent atom. The carbon (C2) and nitrogen (N2) atoms which are adjacent at the imidazole ring and the phenyl (C16–C21) and propane (C22—C24) groups which attached to them respectively, are disordered over two sites (with the suffixes A and B) with an occupancy ratio of 0.615 (3):0.385 (3). The atoms of the disordered propane groups were set to equal each other by an EADP instruction. The disordered phenyl ring (C16B–C21B) was constrained to a rigid hexagon with the AFIX 66 instruction, and for the other atoms of disorder the SIMU and DELU instructions were used in the refinement procedure.
Figures
Fig. 1.
View of the major component of the disordered title compound with the atom numbering scheme. Displacement ellipsoids for non-H atoms are drawn at the 30% probability level.
Fig. 2.
The packing diagram of the title compound viewing along the b axis. The hydrogen atoms not involved in hydrogen bonding and the minor component of the disorder have been omitted for clarity.
Crystal data
| C24H20N2 | F(000) = 712 |
| Mr = 336.42 | Dx = 1.245 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 2698 reflections |
| a = 10.362 (3) Å | θ = 2.2–21.5° |
| b = 8.938 (2) Å | µ = 0.07 mm−1 |
| c = 19.387 (5) Å | T = 100 K |
| β = 90.340 (5)° | Plate, colourless |
| V = 1795.5 (8) Å3 | 0.14 × 0.14 × 0.003 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX CCD area-detector diffractometer | 3168 independent reflections |
| Radiation source: sealed tube | 2442 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.046 |
| phi and ω scans | θmax = 25.0°, θmin = 2.1° |
| Absorption correction: part of the refinement model (ΔF) (XABS2; Parkin et al., 1995) | h = −12→12 |
| Tmin = 0.990, Tmax = 1.000 | k = −10→10 |
| 17127 measured reflections | l = −23→23 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.062 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.183 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0987P)2 + 0.7593P] where P = (Fo2 + 2Fc2)/3 |
| 3168 reflections | (Δ/σ)max < 0.001 |
| 299 parameters | Δρmax = 0.49 e Å−3 |
| 12 restraints | Δρmin = −0.29 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | −0.08400 (18) | 0.74686 (19) | 0.49897 (9) | 0.0410 (6) | |
| N2A | 0.1170 (5) | 0.7384 (5) | 0.5420 (2) | 0.0353 (16) | 0.615 (3) |
| C1 | −0.0060 (2) | 0.8077 (2) | 0.45077 (12) | 0.0403 (7) | |
| C2A | 0.1231 (6) | 0.8099 (6) | 0.4794 (3) | 0.0360 (17) | 0.615 (3) |
| C3 | −0.0081 (2) | 0.6952 (2) | 0.54940 (11) | 0.0402 (7) | |
| C4 | −0.0590 (2) | 0.6243 (2) | 0.61210 (11) | 0.0394 (7) | |
| C5 | 0.0089 (2) | 0.5150 (2) | 0.64866 (11) | 0.0437 (7) | |
| C6 | −0.0425 (2) | 0.4520 (3) | 0.70782 (12) | 0.0486 (8) | |
| C7 | −0.1627 (3) | 0.4953 (3) | 0.73117 (12) | 0.0515 (8) | |
| C8 | −0.2316 (2) | 0.6021 (3) | 0.69493 (13) | 0.0489 (8) | |
| C9 | −0.1804 (2) | 0.6654 (3) | 0.63597 (13) | 0.0454 (8) | |
| C10 | −0.0566 (2) | 0.8751 (2) | 0.38715 (11) | 0.0390 (7) | |
| C11 | −0.1781 (2) | 0.8326 (3) | 0.36242 (12) | 0.0441 (8) | |
| C12 | −0.2281 (2) | 0.8935 (3) | 0.30231 (12) | 0.0482 (8) | |
| C13 | −0.1588 (2) | 0.9992 (3) | 0.26605 (12) | 0.0496 (8) | |
| C14 | −0.0392 (2) | 1.0439 (3) | 0.29035 (12) | 0.0494 (8) | |
| C15 | 0.0118 (2) | 0.9824 (2) | 0.35016 (11) | 0.0429 (7) | |
| C16A | 0.2459 (4) | 0.8604 (4) | 0.44996 (18) | 0.0334 (11) | 0.615 (3) |
| C17A | 0.2941 (6) | 0.9947 (9) | 0.4667 (4) | 0.0396 (16) | 0.615 (3) |
| C18A | 0.4150 (3) | 1.0459 (4) | 0.43689 (18) | 0.0403 (11) | 0.615 (3) |
| C19A | 0.4771 (4) | 0.9553 (5) | 0.39306 (19) | 0.0470 (14) | 0.615 (3) |
| C20A | 0.4300 (4) | 0.8172 (5) | 0.3761 (2) | 0.0543 (14) | 0.615 (3) |
| C21A | 0.3143 (5) | 0.7684 (5) | 0.4041 (2) | 0.0500 (16) | 0.615 (3) |
| C22A | 0.2230 (4) | 0.7339 (5) | 0.5920 (2) | 0.0505 (10) | 0.615 (3) |
| C23A | 0.3217 (4) | 0.6142 (5) | 0.5810 (2) | 0.0505 (10) | 0.615 (3) |
| C24A | 0.3129 (6) | 0.5098 (6) | 0.5380 (2) | 0.0505 (10) | 0.615 (3) |
| C24B | 0.3213 (17) | 0.999 (2) | 0.4606 (8) | 0.060 (2) | 0.385 (3) |
| C17B | 0.2961 (4) | 0.5016 (5) | 0.5314 (2) | 0.085 (4) | 0.385 (3) |
| C18B | 0.4132 (4) | 0.4561 (4) | 0.5598 (2) | 0.0480 (19) | 0.385 (3) |
| C19B | 0.4774 (3) | 0.5477 (5) | 0.6067 (2) | 0.058 (2) | 0.385 (3) |
| C20B | 0.4245 (4) | 0.6848 (5) | 0.6252 (2) | 0.063 (3) | 0.385 (3) |
| N2B | 0.1125 (8) | 0.7634 (8) | 0.4574 (4) | 0.039 (2) | 0.385 (3) |
| C2B | 0.1184 (10) | 0.6892 (9) | 0.5210 (5) | 0.038 (3) | 0.385 (3) |
| C16B | 0.2432 (3) | 0.6387 (5) | 0.5499 (2) | 0.0393 (19) | 0.385 (3) |
| C23B | 0.3136 (8) | 0.8876 (10) | 0.4214 (4) | 0.060 (2) | 0.385 (3) |
| C21B | 0.3074 (4) | 0.7303 (4) | 0.5968 (2) | 0.059 (3) | 0.385 (3) |
| C22B | 0.2197 (8) | 0.7681 (9) | 0.4076 (4) | 0.060 (2) | 0.385 (3) |
| H6 | 0.00520 | 0.37850 | 0.73250 | 0.0580* | |
| H7 | −0.19750 | 0.45210 | 0.77180 | 0.0620* | |
| H8 | −0.31430 | 0.63210 | 0.71050 | 0.0590* | |
| H9 | −0.22900 | 0.73820 | 0.61130 | 0.0540* | |
| H5 | 0.09120 | 0.48350 | 0.63290 | 0.0530* | |
| H22B | 0.26730 | 0.83200 | 0.59130 | 0.0610* | 0.615 (3) |
| H23A | 0.39730 | 0.61750 | 0.60890 | 0.0610* | 0.615 (3) |
| H24A | 0.23890 | 0.50240 | 0.50900 | 0.0610* | 0.615 (3) |
| H24B | 0.38020 | 0.43820 | 0.53430 | 0.0610* | 0.615 (3) |
| H11 | −0.22710 | 0.76080 | 0.38720 | 0.0530* | |
| H12 | −0.31050 | 0.86250 | 0.28590 | 0.0580* | |
| H13 | −0.19300 | 1.04080 | 0.22470 | 0.0600* | |
| H14 | 0.00840 | 1.11730 | 0.26590 | 0.0590* | |
| H15 | 0.09440 | 1.01370 | 0.36610 | 0.0510* | |
| H17A | 0.24920 | 1.05690 | 0.49830 | 0.0480* | 0.615 (3) |
| H18A | 0.44920 | 1.14150 | 0.44820 | 0.0490* | 0.615 (3) |
| H19A | 0.55590 | 0.98800 | 0.37330 | 0.0560* | 0.615 (3) |
| H20A | 0.47620 | 0.75470 | 0.34530 | 0.0650* | 0.615 (3) |
| H21A | 0.28130 | 0.67260 | 0.39230 | 0.0600* | 0.615 (3) |
| H22A | 0.18590 | 0.72080 | 0.63850 | 0.0610* | 0.615 (3) |
| H17B | 0.25230 | 0.43900 | 0.49930 | 0.1020* | 0.385 (3) |
| H18B | 0.44940 | 0.36250 | 0.54720 | 0.0570* | 0.385 (3) |
| H19B | 0.55740 | 0.51670 | 0.62610 | 0.0700* | 0.385 (3) |
| H20B | 0.46830 | 0.74740 | 0.65720 | 0.0760* | 0.385 (3) |
| H21B | 0.27130 | 0.82390 | 0.60940 | 0.0710* | 0.385 (3) |
| H22C | 0.26510 | 0.67060 | 0.40850 | 0.0730* | 0.385 (3) |
| H22D | 0.18350 | 0.78200 | 0.36070 | 0.0730* | 0.385 (3) |
| H23B | 0.38800 | 0.87800 | 0.39330 | 0.0730* | 0.385 (3) |
| H24C | 0.25300 | 1.02150 | 0.49140 | 0.0730* | 0.385 (3) |
| H24D | 0.39560 | 1.06170 | 0.45950 | 0.0730* | 0.385 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0465 (10) | 0.0351 (10) | 0.0414 (11) | −0.0013 (8) | 0.0027 (9) | 0.0000 (8) |
| N2A | 0.046 (2) | 0.028 (3) | 0.032 (3) | 0.004 (2) | −0.0003 (18) | 0.0005 (16) |
| C1 | 0.0441 (13) | 0.0320 (12) | 0.0449 (13) | 0.0033 (9) | 0.0037 (10) | 0.0038 (9) |
| C2A | 0.049 (3) | 0.029 (3) | 0.030 (3) | 0.005 (2) | −0.003 (2) | −0.0054 (18) |
| C3 | 0.0446 (13) | 0.0327 (12) | 0.0432 (13) | 0.0008 (9) | 0.0022 (10) | 0.0040 (9) |
| C4 | 0.0460 (13) | 0.0309 (11) | 0.0413 (12) | −0.0001 (9) | 0.0022 (10) | −0.0008 (9) |
| C5 | 0.0513 (14) | 0.0391 (12) | 0.0409 (12) | 0.0048 (10) | 0.0069 (10) | 0.0014 (10) |
| C6 | 0.0609 (16) | 0.0417 (13) | 0.0433 (13) | 0.0056 (11) | 0.0074 (11) | 0.0063 (10) |
| C7 | 0.0666 (16) | 0.0457 (14) | 0.0422 (13) | −0.0032 (12) | 0.0094 (12) | 0.0021 (11) |
| C8 | 0.0518 (14) | 0.0391 (13) | 0.0559 (15) | −0.0005 (11) | 0.0143 (12) | −0.0039 (11) |
| C9 | 0.0502 (14) | 0.0319 (12) | 0.0540 (14) | 0.0001 (10) | 0.0025 (11) | 0.0034 (10) |
| C10 | 0.0455 (13) | 0.0305 (11) | 0.0411 (12) | 0.0009 (9) | 0.0034 (10) | −0.0004 (9) |
| C11 | 0.0456 (13) | 0.0334 (12) | 0.0533 (14) | 0.0013 (10) | 0.0012 (11) | −0.0001 (10) |
| C12 | 0.0486 (14) | 0.0419 (13) | 0.0539 (14) | −0.0003 (11) | −0.0080 (11) | −0.0054 (11) |
| C13 | 0.0610 (15) | 0.0465 (14) | 0.0413 (13) | 0.0024 (12) | −0.0080 (11) | 0.0011 (11) |
| C14 | 0.0612 (16) | 0.0450 (14) | 0.0418 (13) | −0.0069 (12) | −0.0030 (11) | 0.0060 (11) |
| C15 | 0.0500 (13) | 0.0384 (12) | 0.0401 (12) | −0.0043 (10) | −0.0033 (10) | 0.0009 (10) |
| C16A | 0.0321 (19) | 0.041 (2) | 0.0270 (18) | 0.0065 (16) | 0.0013 (15) | −0.0007 (15) |
| C17A | 0.030 (3) | 0.038 (2) | 0.051 (3) | 0.005 (2) | 0.010 (2) | 0.000 (2) |
| C18A | 0.0308 (19) | 0.050 (2) | 0.040 (2) | −0.0044 (17) | 0.0001 (15) | −0.0003 (17) |
| C19A | 0.041 (2) | 0.059 (3) | 0.041 (2) | 0.0007 (18) | −0.0045 (16) | −0.0068 (18) |
| C20A | 0.050 (2) | 0.063 (3) | 0.050 (2) | 0.000 (2) | 0.0207 (19) | −0.020 (2) |
| C21A | 0.064 (3) | 0.048 (3) | 0.038 (2) | −0.006 (2) | 0.003 (2) | −0.0151 (18) |
| C22A | 0.0504 (16) | 0.061 (2) | 0.0401 (14) | 0.0030 (14) | 0.0057 (13) | 0.0122 (12) |
| C23A | 0.0504 (16) | 0.061 (2) | 0.0401 (14) | 0.0030 (14) | 0.0057 (13) | 0.0122 (12) |
| C24A | 0.0504 (16) | 0.061 (2) | 0.0401 (14) | 0.0030 (14) | 0.0057 (13) | 0.0122 (12) |
| C24B | 0.060 (4) | 0.073 (4) | 0.048 (3) | −0.007 (3) | −0.011 (3) | 0.010 (2) |
| C17B | 0.080 (7) | 0.040 (5) | 0.136 (10) | −0.013 (5) | 0.001 (7) | −0.001 (5) |
| C18B | 0.033 (3) | 0.066 (4) | 0.045 (3) | −0.002 (3) | −0.002 (3) | 0.004 (3) |
| C19B | 0.060 (4) | 0.055 (4) | 0.060 (4) | −0.009 (3) | 0.029 (3) | −0.015 (3) |
| C20B | 0.058 (4) | 0.069 (5) | 0.062 (4) | −0.004 (4) | −0.031 (4) | −0.018 (4) |
| N2B | 0.053 (4) | 0.036 (4) | 0.029 (4) | −0.008 (3) | 0.001 (3) | 0.003 (3) |
| C2B | 0.052 (4) | 0.024 (5) | 0.039 (5) | −0.005 (4) | −0.004 (4) | 0.000 (3) |
| C16B | 0.037 (3) | 0.050 (4) | 0.031 (3) | −0.014 (3) | 0.003 (3) | −0.008 (3) |
| C23B | 0.060 (4) | 0.073 (4) | 0.048 (3) | −0.007 (3) | −0.011 (3) | 0.010 (2) |
| C21B | 0.085 (6) | 0.052 (4) | 0.040 (4) | 0.007 (4) | 0.004 (4) | −0.018 (3) |
| C22B | 0.060 (4) | 0.073 (4) | 0.048 (3) | −0.007 (3) | −0.011 (3) | 0.010 (2) |
Geometric parameters (Å, º)
| N1—C1 | 1.353 (3) | C20A—C21A | 1.389 (6) |
| N1—C3 | 1.334 (3) | C20B—C21B | 1.390 (6) |
| N2A—C2A | 1.373 (7) | C22A—C23A | 1.496 (6) |
| N2A—C3 | 1.361 (6) | C22B—C23B | 1.468 (12) |
| N2A—C22A | 1.461 (6) | C23A—C24A | 1.254 (6) |
| N2B—C2B | 1.401 (12) | C23B—C24B | 1.255 (19) |
| N2B—C22B | 1.477 (11) | C5—H5 | 0.9500 |
| N2B—C1 | 1.296 (8) | C6—H6 | 0.9500 |
| C1—C2A | 1.445 (7) | C7—H7 | 0.9500 |
| C1—C10 | 1.467 (3) | C8—H8 | 0.9500 |
| C2A—C16A | 1.469 (7) | C9—H9 | 0.9500 |
| C2B—C3 | 1.426 (10) | C11—H11 | 0.9500 |
| C2B—C16B | 1.477 (11) | C12—H12 | 0.9500 |
| C3—C4 | 1.471 (3) | C13—H13 | 0.9500 |
| C4—C5 | 1.395 (3) | C14—H14 | 0.9500 |
| C4—C9 | 1.392 (3) | C15—H15 | 0.9500 |
| C5—C6 | 1.387 (3) | C17A—H17A | 0.9500 |
| C6—C7 | 1.383 (4) | C17B—H17B | 0.9500 |
| C7—C8 | 1.382 (4) | C18A—H18A | 0.9500 |
| C8—C9 | 1.384 (4) | C18B—H18B | 0.9500 |
| C10—C11 | 1.397 (3) | C19A—H19A | 0.9500 |
| C10—C15 | 1.394 (3) | C19B—H19B | 0.9500 |
| C11—C12 | 1.384 (3) | C20A—H20A | 0.9500 |
| C12—C13 | 1.381 (3) | C20B—H20B | 0.9500 |
| C13—C14 | 1.382 (3) | C21A—H21A | 0.9500 |
| C14—C15 | 1.385 (3) | C21B—H21B | 0.9500 |
| C16A—C17A | 1.339 (9) | C22A—H22A | 0.9900 |
| C16A—C21A | 1.406 (6) | C22A—H22B | 0.9900 |
| C16B—C17B | 1.390 (6) | C22B—H22C | 0.9900 |
| C16B—C21B | 1.390 (6) | C22B—H22D | 0.9900 |
| C17A—C18A | 1.457 (7) | C23A—H23A | 0.9500 |
| C17B—C18B | 1.390 (6) | C23B—H23B | 0.9500 |
| C18A—C19A | 1.341 (5) | C24A—H24A | 0.9500 |
| C18B—C19B | 1.390 (6) | C24A—H24B | 0.9500 |
| C19A—C20A | 1.367 (6) | C24B—H24C | 0.9500 |
| C19B—C20B | 1.390 (6) | C24B—H24D | 0.9500 |
| C1—N1—C3 | 107.06 (18) | C6—C5—H5 | 120.00 |
| C2A—N2A—C3 | 105.9 (4) | C5—C6—H6 | 120.00 |
| C2A—N2A—C22A | 124.1 (5) | C7—C6—H6 | 120.00 |
| C3—N2A—C22A | 129.4 (3) | C6—C7—H7 | 120.00 |
| C2B—N2B—C22B | 124.0 (8) | C8—C7—H7 | 120.00 |
| C1—N2B—C2B | 105.6 (7) | C7—C8—H8 | 120.00 |
| C1—N2B—C22B | 130.0 (7) | C9—C8—H8 | 120.00 |
| N1—C1—C2A | 107.2 (3) | C4—C9—H9 | 119.00 |
| N2B—C1—C10 | 122.9 (4) | C8—C9—H9 | 119.00 |
| N1—C1—C10 | 122.26 (19) | C10—C11—H11 | 120.00 |
| N1—C1—N2B | 112.2 (4) | C12—C11—H11 | 120.00 |
| C2A—C1—C10 | 130.0 (3) | C11—C12—H12 | 120.00 |
| N2A—C2A—C1 | 106.6 (5) | C13—C12—H12 | 120.00 |
| N2A—C2A—C16A | 122.1 (5) | C12—C13—H13 | 120.00 |
| C1—C2A—C16A | 131.1 (4) | C14—C13—H13 | 120.00 |
| N2B—C2B—C3 | 106.7 (7) | C13—C14—H14 | 120.00 |
| C3—C2B—C16B | 132.1 (7) | C15—C14—H14 | 120.00 |
| N2B—C2B—C16B | 120.8 (8) | C10—C15—H15 | 120.00 |
| C2B—C3—C4 | 129.6 (4) | C14—C15—H15 | 120.00 |
| N1—C3—C4 | 122.82 (19) | C16A—C17A—H17A | 120.00 |
| N1—C3—C2B | 105.6 (4) | C18A—C17A—H17A | 120.00 |
| N1—C3—N2A | 112.5 (2) | C16B—C17B—H17B | 120.00 |
| N2A—C3—C4 | 123.8 (2) | C18B—C17B—H17B | 120.00 |
| C5—C4—C9 | 118.0 (2) | C17A—C18A—H18A | 121.00 |
| C3—C4—C5 | 122.63 (19) | C19A—C18A—H18A | 121.00 |
| C3—C4—C9 | 119.34 (19) | C17B—C18B—H18B | 120.00 |
| C4—C5—C6 | 120.6 (2) | C19B—C18B—H18B | 120.00 |
| C5—C6—C7 | 120.5 (2) | C18A—C19A—H19A | 119.00 |
| C6—C7—C8 | 119.4 (2) | C20A—C19A—H19A | 119.00 |
| C7—C8—C9 | 120.2 (2) | C18B—C19B—H19B | 120.00 |
| C4—C9—C8 | 121.2 (2) | C20B—C19B—H19B | 120.00 |
| C11—C10—C15 | 118.1 (2) | C19A—C20A—H20A | 120.00 |
| C1—C10—C11 | 119.59 (19) | C21A—C20A—H20A | 120.00 |
| C1—C10—C15 | 122.34 (19) | C19B—C20B—H20B | 120.00 |
| C10—C11—C12 | 120.9 (2) | C21B—C20B—H20B | 120.00 |
| C11—C12—C13 | 120.3 (2) | C16A—C21A—H21A | 120.00 |
| C12—C13—C14 | 119.5 (2) | C20A—C21A—H21A | 120.00 |
| C13—C14—C15 | 120.5 (2) | C20B—C21B—H21B | 120.00 |
| C10—C15—C14 | 120.7 (2) | C16B—C21B—H21B | 120.00 |
| C17A—C16A—C21A | 119.3 (5) | N2A—C22A—H22B | 108.00 |
| C2A—C16A—C21A | 120.5 (4) | N2A—C22A—H22A | 108.00 |
| C2A—C16A—C17A | 120.3 (4) | H22A—C22A—H22B | 107.00 |
| C2B—C16B—C17B | 121.1 (5) | C23A—C22A—H22A | 108.00 |
| C17B—C16B—C21B | 120.0 (3) | C23A—C22A—H22B | 108.00 |
| C2B—C16B—C21B | 118.9 (5) | N2B—C22B—H22D | 109.00 |
| C16A—C17A—C18A | 120.4 (6) | H22C—C22B—H22D | 108.00 |
| C16B—C17B—C18B | 120.0 (4) | C23B—C22B—H22C | 109.00 |
| C17A—C18A—C19A | 118.6 (4) | C23B—C22B—H22D | 109.00 |
| C17B—C18B—C19B | 120.0 (4) | N2B—C22B—H22C | 109.00 |
| C18A—C19A—C20A | 121.7 (4) | C22A—C23A—H23A | 117.00 |
| C18B—C19B—C20B | 120.0 (3) | C24A—C23A—H23A | 117.00 |
| C19A—C20A—C21A | 119.8 (4) | C22B—C23B—H23B | 112.00 |
| C19B—C20B—C21B | 120.0 (4) | C24B—C23B—H23B | 112.00 |
| C16A—C21A—C20A | 120.2 (4) | H24A—C24A—H24B | 120.00 |
| C16B—C21B—C20B | 120.0 (4) | C23A—C24A—H24A | 120.00 |
| N2A—C22A—C23A | 115.9 (4) | C23A—C24A—H24B | 120.00 |
| N2B—C22B—C23B | 113.7 (7) | C23B—C24B—H24C | 120.00 |
| C22A—C23A—C24A | 125.4 (4) | C23B—C24B—H24D | 120.00 |
| C22B—C23B—C24B | 136.7 (11) | H24C—C24B—H24D | 120.00 |
| C4—C5—H5 | 120.00 | ||
| C3—N1—C1—C2A | −8.2 (3) | N1—C3—C4—C5 | 150.5 (2) |
| C3—N1—C1—C10 | 179.56 (17) | N2A—C3—C4—C9 | 139.5 (3) |
| C1—N1—C3—N2A | 9.1 (3) | C9—C4—C5—C6 | −1.4 (3) |
| C1—N1—C3—C4 | 178.39 (17) | C3—C4—C5—C6 | 179.5 (2) |
| C3—N2A—C22A—C23A | 105.6 (5) | C5—C4—C9—C8 | 1.2 (3) |
| C2A—N2A—C3—N1 | −6.0 (4) | C3—C4—C9—C8 | −179.6 (2) |
| C22A—N2A—C3—N1 | 165.4 (4) | C4—C5—C6—C7 | 0.7 (3) |
| C2A—N2A—C3—C4 | −175.2 (3) | C5—C6—C7—C8 | 0.1 (4) |
| C22A—N2A—C3—C4 | −3.8 (6) | C6—C7—C8—C9 | −0.2 (4) |
| C3—N2A—C2A—C16A | −174.0 (4) | C7—C8—C9—C4 | −0.4 (4) |
| C3—N2A—C2A—C1 | 0.6 (5) | C1—C10—C11—C12 | −179.6 (2) |
| C22A—N2A—C2A—C1 | −171.4 (4) | C11—C10—C15—C14 | −0.5 (3) |
| C2A—N2A—C22A—C23A | −84.4 (6) | C1—C10—C15—C14 | −179.8 (2) |
| C22A—N2A—C2A—C16A | 14.0 (8) | C15—C10—C11—C12 | 1.1 (3) |
| N1—C1—C10—C11 | −23.1 (3) | C10—C11—C12—C13 | −0.8 (4) |
| N1—C1—C2A—N2A | 4.7 (4) | C11—C12—C13—C14 | −0.1 (4) |
| C2A—C1—C10—C15 | −14.0 (4) | C12—C13—C14—C15 | 0.7 (4) |
| C2A—C1—C10—C11 | 166.6 (3) | C13—C14—C15—C10 | −0.4 (3) |
| N1—C1—C2A—C16A | 178.7 (5) | C2A—C16A—C17A—C18A | −179.2 (5) |
| C10—C1—C2A—N2A | 176.1 (3) | C17A—C16A—C21A—C20A | −0.6 (7) |
| C10—C1—C2A—C16A | −9.9 (7) | C21A—C16A—C17A—C18A | 1.0 (8) |
| N1—C1—C10—C15 | 156.25 (19) | C2A—C16A—C21A—C20A | 179.6 (4) |
| N2A—C2A—C16A—C17A | −86.4 (7) | C16A—C17A—C18A—C19A | −0.6 (8) |
| N2A—C2A—C16A—C21A | 93.4 (6) | C17A—C18A—C19A—C20A | −0.2 (6) |
| C1—C2A—C16A—C21A | −79.8 (6) | C18A—C19A—C20A—C21A | 0.6 (6) |
| C1—C2A—C16A—C17A | 100.4 (7) | C19A—C20A—C21A—C16A | −0.2 (6) |
| N1—C3—C4—C9 | −28.6 (3) | N2A—C22A—C23A—C24A | −8.4 (7) |
| N2A—C3—C4—C5 | −41.4 (4) |
Hydrogen-bond geometry (Å, º)
Cg3 and Cg4 are the centroids of the C4–C9 and C10–C15 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C17A—H17A···N1i | 0.95 | 2.45 | 3.246 (8) | 141 |
| C24A—H24A···N2A | 0.95 | 2.54 | 2.882 (8) | 101 |
| C21A—H21A···Cg3ii | 0.95 | 2.98 | 3.888 (5) | 160 |
| C21B—H21B···Cg4i | 0.95 | 2.99 | 3.914 (4) | 163 |
Symmetry codes: (i) −x, −y+2, −z+1; (ii) −x, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZP2005).
References
- Akkurt, M., Fronczek, F. R., Mohamed, S. K., Talybov, A. H., Marzouk, A. A. E. & Abdelhamid, A. A. (2013). Acta Cryst. E69, o527–o528. [DOI] [PMC free article] [PubMed]
- Banfi, E., Scialino, G., Zampieri, D., Mamolo, M. G., Vio, L., Ferrone, M., Maurizio Fermeglia, M., Paneni, M. S. & Sabrina Pric, S. (2006). J. Antimicrob. Chemother. 58, 76–84. [DOI] [PubMed]
- Bogle, R. G., Whitley, G. S., Soo, S. C., Johnstone, A. P. & Vallance, P. (1994). Br. J. Pharmacol. 111, 1257–1261. [DOI] [PMC free article] [PubMed]
- Bruker (2001). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Castaño, T., Encinas, A., Pérez, C., Castro, A., Campillo, N. E. & Gil, C. (2008). Bioorg. Med. Chem. 16, 6193–6206. [DOI] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kumar, J. R. (2010). Pharmacophore, 1, 167–177.
- Mohamed, S. K., Akkurt, M., Marzouk, A. A., Abbasov, V. M. & Gurbanov, A. V. (2013a). Acta Cryst. E69, o474–o475. [DOI] [PMC free article] [PubMed]
- Mohamed, S. K., Akkurt, M., Marzouk, A. A. E., Santoyo-Gonzalez, F. & Elremaily, M. A. A. (2013b). Acta Cryst. E69, o875–o876. [DOI] [PMC free article] [PubMed]
- Parkin, S., Moezzi, B. & Hope, H. (1995). J. Appl. Cryst. 28, 53–56.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813014104/zp2005sup1.cif
Structure factors: contains datablock(s) shelxl. DOI: 10.1107/S1600536813014104/zp2005Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813014104/zp2005Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report