Abstract
In recent years, the role of epigenetic phenomenon, such as methylation, in mediating vulnerability to behavioral illness has become increasingly appreciated. One prominent locus at which epigenetic phenomena are thought to be in play is the Monoamine Oxidase A (MAOA) locus. In order to examine the role of methylation at this locus, we performed quantitative methylation analysis across the promoter region of this gene in lymphoblast lines derived from 191 subjects participating in the Iowa Adoption Studies (IAS). We analyzed the resulting data with respect to genotype and lifetime symptom counts for the more common major behavioral disorders in the IAS, antisocial personality disorder (ASPD), and substance use disorders (alcohol (AD) and nicotine dependence (ND)). We found that methylation status was significantly associated with lifetime symptom counts for ND (p<0.001) and AD (p<0.008) in women, but not men. Furthermore, a trend was found for women homozygous for the 3,3 allele to have a higher degree of overall methylation than women homozygous for the 4,4 allele (p<0.10). We conclude that methylation of MAOA may play a significant role in common psychiatric illness and that further examination of epigenetic processes at this locus is in order.
INTRODUCTION
Over the past several years, it has become increasingly evident that gene-environment interactions (GxE) and residual gene-environment correlations (rGE) have a prominent role in the etiology of most common behavioral illnesses [1]. However, the exact processes underlying these interactions and the extent of their relative contributions are unclear. At the molecular level, epigenetic phenomena such as DNA methylation and histone modification are thought to contribute to these processes. Unfortunately, empirical data to support this hypothesis at behaviorally relevant loci have been scarce.
Two candidate loci at which epigenetic phenomena may participate in GxE, rGE or E effects are the Serotonin Transporter (SLC6A4) and Monoamine Oxidase A (MAOA). The protein products of both of these two loci play prominent roles in regulating serotonergic and monoaminergic transmission, respectively. These moderating roles have come under increasing scrutiny due to recent studies which have demonstrated prominent GxE effects for depression at SLC6A4 [2] and for aggression at MAOA [3, 4]. Hence, there is a great deal of curiosity as to the mechanisms through which E or GxE effects could influence biological processes at these loci.
One mechanism through which GxE or E effects could become manifest at the molecular level is through altering relevant gene expression through methylation of gene promoters in response to environmental stressors. In our initial study of the relationship between promoter methylation and behavioral phenomena, we conducted quantitative methylation analyses of the SLC6A4 associated promoter CpG island and demonstrated that methylation of this promoter is both sex dependent and associated with increased vulnerability to major depression. However, whether there is a similar promoter associated CpG island at MAOA, and if it exists, whether its methylation has behavioral consequences is unclear.
Two types of disorders that could potentially be influenced by methylation induced changes in MAOA activity are Antisocial Personality Disorder (ASPD) and substance use disorders (SUD). Already, genetic variation in a variable nucleotide repeat (VNTR) located immediately upstream of the MAOA minimal central promoter has been associated with different vulnerability to ASPD [5-8] and two forms of SUD; alcohol dependence (AD) [6-9] and nicotine dependence (ND) [10]. Therefore, given the prior reports of GxE effects at this locus with respect to ASPD [4], it is reasonable to hypothesize that epigenetic processes, such as methylation, which influence MAOA activity may also be a factor with respect to these disorders at the MAOA locus.
In this report, using a set of similar techniques to our prior methylation and gene expression analyses of SCL6A4 [11] and the resources of the Iowa Adoption Studies (IAS), a large longitudinal adoption study focusing on the role of GxE effects in SUD [12], we examined the relationship of MAOA genotype and methylation to SUD and ASPD.
METHODS
The procedures used in the IAS have been described in detail elsewhere [12]. Briefly, the IAS is a case and control adoption study of G, E and GxE effects in SUD and ASPD. This study, founded by Remi Cadoret, contrasts the outcomes of 475 adoptees from the State of Iowa who are at high biological risk for SUD or ASPD (i.e., one of their biological parents was severely affected) with those of 475 adoptees who were not at biological risk for either SUD or ASPD. After birth, each of these adoptees was randomly placed in an adoptive home. Since their inception in the study, the adoptees and their adoptive environments have been serially assessed. The subjects included in this pilot study were the first 95 males and 96 females to participate in this wave of the study. The overall study design and all procedures described in this communication were approved by the University of Iowa Institutional Review Board.
Briefly, the behavioral and biological material used in these studies were obtained from subjects who participated in the last two waves of the Iowa Adoptions Studies (IAS). In both of these waves, each subject was interviewed with a version of the Semi Structured Assessment for the Genetics of Alcoholism, Version 2 (SSAGA-II) [13]. In addition, in the latest round of the study, phlebotomy was performed on each of the participants. Symptom counts and categorical diagnoses for each of the disorders (ASPD, AD, ND) were derived from SSAGA-II data using the individual dependence or personality disorder criteria from DSM-IV [14], with the highest total symptom count from these two interviews being defined as the lifetime symptom count.
RNA and DNA used in the studies were derived from lymphoblast cell lines using biomaterial contributed by the participants. These lymphoblast cell lines were prepared using standard EBV transfection techniques from the specimens contributed by the study participants [15]. Total RNA was prepared from lymphoblast using a Midi RNA purification kit from Invitrogen (Carlsbad, CA) according to the manufacturer’s instructions. DNA was prepared from lymphoblast cell pellets using cold protein precipitation [16].
PCR amplification of the MAOA variable nucleotide repeat (VNTR) polymorphism was conducted using the method of Sabol and colleagues [17]. The resulting PCR products were electrophoresed on a 6% non-denaturing polyacrylamide gel and imaged using silver staining [18]. The resulting alleles were compared to internal standards and the genotypes were called by two individuals blind to affected status.
RTPCR was conducted as previously described [11, 19]. Briefly, RNA was reverse transcribed using an Applied Biosystems cDNA archiving kit (Foster City, CA). Then, 12.5 ng aliquots of cDNA were robotically dispensed and RTPCR performed using reagents from Applied Biosystems including primer-probe sets for MAOA (Hs 00165140) and the endogenous control loci GAPDH (from the GAPDH Control kit) and LDHA (Hs 00855332).
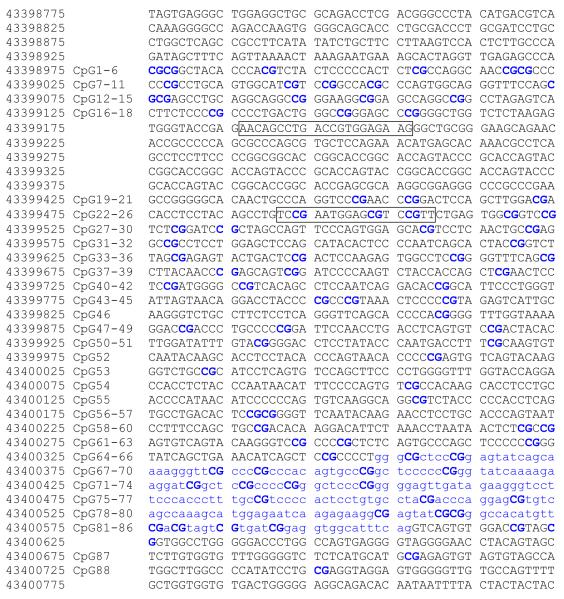
The existence, location, size and sequence of the MAOA CpG islands were determined using the default browser settings of the University of California Genome Browser (UCSC) website (www.genome.ucsc.edu). The sequences for these islands are freely available from the website or from the authors on request.
Quantitative methylation analyses for each of the samples at these CpG residues were conducted by Sequenom Inc. (San Diego, CA) as previously described [20]. First, aliquots of purified DNA were treated using bisulfite modification [21]. Treatment of DNA with bisulfite deaminates unmethylated cytosine residues to uracil . Since uracil base pairs with adenosine, thymidines are incorporated into subsequent DNA strands in the place of unmethylated cytosine residues during subsequent PCR amplifications. Next, contigs covering the CpG islands (see Figure 1) were PCR amplified. Because of the size of the region, the CpG enriched regions were PCR amplified in four separate reactions. The primers for each of those PCR amplifications are as follows: Amplicon A (from BP 43398925 to 43399181):F- TTAAAGAATGAAAGTATTAGGTTGAGAGTT and R-ATACCCACTCTTAAAAACCAACCCC; Amplicon B (from BP 43399430 to 43399858): F-GGGTGTTGAATTTTGAGGAGAAG and R-AAAACACAACTACCCAAATCCC; Amplicon C (from BP 43400453 to 43400805) : F-GGGGAGTTGATAGAAGGGTTTTTTTTAT and R-TATATCTACCTCCCCCAATCACACC and Amplicon D (from BP 43400486 to 43400035): F-AAAGGGTGGGAAGGATTTTTTTATTAATT and R-CATCCTCAATATCCAACTTCCCCTA using standard touchdown PCR conditions [22]. Please see supplementary Table I for the exact details as to primer sequence, position and the sequence covered by each primer set. Methylation ratios for each of the CpG residues (methyl CpG/total CpG) were then determined using a MassARRAY™ mass spectrometer using proprietary peak picking and spectra interpretation tools [23, 24].
Figure 1.
The sequence and structure of the MAOA promoter region. The first CpG island begins at bp 43398975 and contains 18 CpG residues. A second CpG island begins at bp 43399493 and contains 70 CpG residues. The position of each of the CpG residues is noted in the figure. The first exon of MAOA is denoted by small blue letters and is wholly contained within the second island. The positions of the primers used to amplify the MAOA VNTR are denoted by boxed letters. The transcription start site (TSS) is at bp43400353 between CpG residues 64 and 65.
The data were analyzed using the JMP (version 7; SAS Institute, Cary, SC) using Pearson’s correlation coefficients, regression [analysis of variance (ANOVA) and ordinal logistic regression (OLR)] or Chi-square testing as indicated in the text [25]. All tests were two-tailed and all analyses were conducted by gender.
RESULTS
The characteristics of the IAS subjects who contributed the biomaterials to this study are given in Table I. In total, 96 female and 95 male subjects provided biomaterials for the study. The male subjects were significantly older than the female subjects (t-test, p<0.002) and had a significantly higher symptom count for ASPD (Chi-Square, p<0.001).
Table I.
Demographic and Clinical Characteristics of the IAS Subjects
| Male | Female | |
|---|---|---|
| N | 95 | 96 |
| Age (years ± SD) | 42.4 ± 8.5 | 38.8 ± 6.8 |
| Ethnicity | ||
| White | 87 | 91 |
| African American | 5 | 2 |
| White of Hispanic Origin | 2 | 1 |
| Other | 1 | 2 |
| DSM IV Symptom Counts | ||||||
|---|---|---|---|---|---|---|
| ASPD | AD | ND | ||||
| # Symptoms | M | F | M | F | M | F |
| 0 | 18 | 41 | 35 | 49 | 47 | 50 |
| 1 | 26 | 30 | 25 | 25 | 4 | 6 |
| 2 | 21 | 9 | 16 | 13 | 10 | 7 |
| 3 | 7 | 7 | 11 | 3 | 15 | 8 |
| 4 | 10 | 5 | 2 | 2 | 6 | 14 |
| 5 | 9 | 3 | 3 | 3 | 8 | 8 |
| 6 | 4 | 3 | 2 | 0 | 3 | 3 |
| 7 | 0 | 0 | 1 | 0 | 2 | 0 |
The MAOA VNTR genotypes for the subjects are given in Table II. The testing for Hardy Weinberg equilibrium in the female subjects was unremarkable.
Table II.
MAOA VNTR Genotype.
| Genotype | Female Subjects | Male Subjects* |
|---|---|---|
| 2, 2 | 0 | 1 |
| 2, 4 | 1 | - |
| 3, 3 | 18 | 34 |
| 3, 4 | 41 | - |
| 3, 5 | 1 | - |
| 3.5, 3.5 | 0 | 1 |
| 3.5, 4 | 1 | - |
| 4, 4 | 31 | 59 |
| 4, 5 | 3 | - |
Male subjects are hemizygous with respect to this X-chromosome locus.
Sequence analysis of MAOA demonstrated the presence of two CpG islands in the gene (Figure 1). The first island, stretching from bp 43398975 to bp 43399158, contains 18 CpG residues and is approximately 1200 bp upstream of the transcription start site for MAOA. The second CpG island begins at bp 43399493 and contains 70 CpG residues. Exon 1 of MAOA is wholly contained within the CpG island with the transcription start site (TSS) for the gene occurring between CpG residues 64 and 65. The MAOA VNTR is found between the two CpG islands.
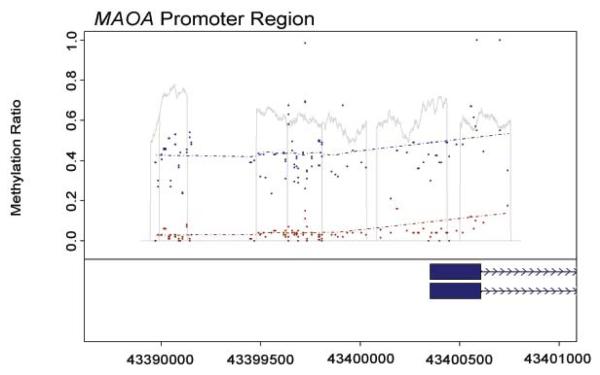
The average methylation ratio at each of these residues is shown in Figure 2. As the figure demonstrates, females have consistently higher methylation ratios at each CpG residue than males (who are hemizygous for this gene). The exact average at each of these residues for each gender is given in supplementary Table III. Please note that secondary to methodological limitations with respect to the ability of the mass spectrograph to resolve individual residues, the values for CpG residues, 1-2, 5-7, 11-12, 19-20, 30-31, 43-44, 55-57, 67-68, 72-73, and 79-80 are shown as aggregates.
Figure 2.
The average methylation ratios (methyl CpG/total CpG) at each CpG residue for each sex. The bp position on the X chromosome is given on the X axis and corresponds to the position of each of the residues in Figure 1. The average values for female subjects are depicted by blue squares, while the average values for males are depicted by red circles. The position of MAOA exon 1 is denoted by the box with the direction of transcription being indicated by the line with arrows.
The interrelationships of MAOA methylation between individual residues for each gender are also shown in supplementary Table III. As the table shows, the correlation between methylation is higher between residues in the smaller 5′ CpG island than it is in between residues in the larger CpG island that encompasses Exon 1. Of particular potential interest, methylation of the two residues immediately flanking the TSS, CpG 64 and 65, is poorly correlated with methylation throughout the rest of the island. However, methylation at the residues CpG 58-63 and CpG 66-70 is highly inter-correlated.
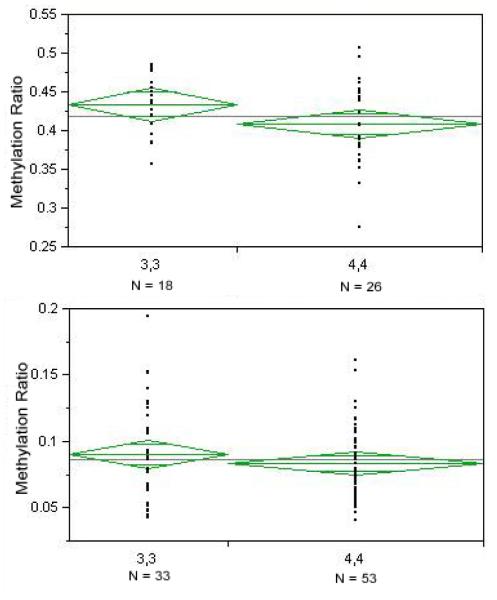
In order to test the hypothesis that MAOA genotype influences the amount of methylation, we analyzed the relationship of average methylation to genotype at the VNTR (Figure 3) for each gender. There was a trend for female 3,3 homozygotes to have a higher average methylation than female 4,4 homozygotes (43.3% ± 3.8 vs 40.9% ± 5.2; p<0.10). There was no significant difference between males hemizygous for the 3 repeat allele as compared to those with the 4 allele although the arithmetic difference was in the same direction (9.0 ± 3.7 vs 8.3 ± 2.6; p<0.32).
Figure 3.
The relationship of MAOA VNTR genotype to methylation in females (above) and males (below). There was a trend for association for female 3,3 homozygotes to have higher average methylation (methyl CpG/total CpG) than female 4,4 homozygotes (43.3% ± 3.8 vs 40.9% ± 5.2; p<0.10). There was no significant difference between males hemizygous for the 3 repeat allele as compared to those with the 4 allele although the arithmetic difference was in the same direction (9.0 ± 3.7 vs 8.3 ± 2.6; p<0.32).
We then analyzed the relationship between symptom counts for ASPD, AD and ND with average methylation for each gender using ordinal regression analysis. There was no relationship between ASPD and overall methylation for neither men (OLR, p<0.37).or women (OLR, p>0.70). There also were not any significant relationships between average methylation and AD (OLR, p<0.23) and ND (OLR, p<0.68) in male subjects. However, there were strong relationships between average overall methylation and symptom counts for AD (OLR, p<0.008) and ND (OLR, p<0.002) in female subjects (see supplementary Figure IV).
In order to identify the residues driving the strong correlations between overall methylation and symptom counts for AD and ND in women, we analyzed the relationship between methylation at individual CpG residues and symptom counts. With respect to former, methylation at CpG residues 27, 38, 41, and 48 were nominally significantly associated (p<0.05 before correction for multiple comparisons) with AD symptom count in female subjects. With respect to the latter, methylation at CpG residues 18, 42, 48, 52, 64, 65, 67-68, 69, and 77 were nominally associated (p<0.05 before correction for multiple comparisons) with ND symptom counts. The complete results of the analyses for each of the four disorders examined are given in supplementary Table IV.
Finally, in an attempt to discern whether gene expression was correlated with MAOA genotype or methylation, we attempted to measure MAOA gene expression using our previously described techniques [11, 22, 26]. Unfortunately, despite several attempts, we could not reliably detect MAOA gene expression.
DISCUSSION
In summary, we report that MAOA methylation is associated with ND and AD in women, but not men. In addition, we did not find a significant relationship between ASPD and CpG methylation in men nor women. Finally, there was a trend for MAOA genotype to be associated with methylation in women.
Before these data are discussed, several limitations of the study should be noted. First, the methylation studies were conducted using lymphoblast DNA. Although we and others have demonstrated that lymphoblast cells are reliable surrogates for their CNS counterparts, they are still not neurons [26-29]. Second, none of the values reported in the Results section are corrected for multiple comparisons. Given the high correlation between the clinical syndromes examined and between methylation at individual CpG residues, we felt it would be too difficult to objectively arrive at a correction term for these comparisons. Therefore, as per our prior communication with regard to SLC6A4, we caution the reader to be aware of the inevitable false positives that will result because of this lack of correction. Third, these methylation values are assessed at only one point in time, making it impossible to tell if they reflect change from a prior unmethylated or less methylated state. Likewise, no direction of effect can be inferred. It may be that the association is the result of methylation leading to substance use or the reverse may better describe the direction of effect. Finally, the IAS subjects used in this study come from an epidemiologically sound but high-risk population that is largely White. Therefore, generalizations to other populations should be done judiciously though we carefully examined for the effects of ethnicity and did not find any effects of ethnicity on these results.
The results with respect to ND are perhaps the most compelling. Review of the animal model literature shows that MAOA knockout mice exhibit impaired nicotine preference but have normal responses to other novel stimuli [30]. Furthermore, treatment of rats with the monoamine oxidase inhibitor phenelzine enhances the discriminant stimulus effect of nicotine [31] and increases nicotine self administration [32, 33]. Review of the literature with respect to humans, reveals that the targeting of neurotransmitter systems regulated by MAOA, using agents such as reboxetine, a selective norepinephrine reuptake inhibitor, or bupropion, which targets the dopaminergic system, have been shown to be clinically effective in the treatment of ND [34-36]. Finally, platelet MAOA activity is reduced in smokers [37].
This evidence is made even more compelling by closer inspection and consideration of the MAOA methylation data with respect to ND in the female subjects. The control of transcription initiation is one of the major mechanisms through which cells regulate gene expression [38]. Hence, the TSS is a frequent target of epigenetic modifications including methylation and histone modification [39, 40] . Therefore, if the current findings are meaningful, we would expect any significant changes in ND association MAOA methylation to preferentially affect the MAOA TSS. This is indeed what is observed with a strong clustering of CpG residues that are either nominally significantly associated or with a trend for association (p<0.10) surrounding the TSS (supplementary Table IV).
In contrast, the differential methylation associated with AD in women is found proximal to the TSS. Whereas this differential methylation may also be functional, a concise understanding of how it may affect gene expression is not clear at the current time. However, given the strong a priori support for the involvement of MAOA in AD [6-9], the current findings are invigorating and suggest a need for further examination in an independent group of subjects.
It is important to note that our primary outcome measure with respect to methylation in this study was overall methylation, not individual CpG residue methylation. This is because we did not have firm hypotheses as to which CpG residues might be most important in this pilot study. If only a subset of the CpG residues are differentially methylated in association with substance use, this decision may have decreased our sensitivity. In order to eliminate this problem for others wishing to confirm or extend our work, we have provided the average methylation and the p-values for their association in the supplementary material (Supplementary Table IV).
In contrast to our prior work in which we documented the relationship between promoter methylation and gene expression at the serotonin transporter (SLC6A4), we were unable to reliably detect MAOA expression using RTPCR because of the low expression of this transcript in these cell lines (please see the gene expression files from [26]). Although the low expression may be secondary to a number of factors such as the presence or absence of transcriptional factors not related to the methylation, this limits these findings.
It is important to note that ND was the most common syndrome in this group of subjects. Therefore, the failure to show differential methylation effects for AD may be a function of the greater power with respect to ND. Given the positive results discussed above with respect to ND and the biological plausibility of these regulatory changes, further studies with larger number of subjects are clearly indicated for each of these syndromes.
It will be essential for these findings to be confirmed in actual catecholaminergic neurons before they can be generally accepted. In particular, examination of methylation of MAOA in human post-mortem tissue would be an excellent test of these findings.
A critical question is why significant effects are observed in females, but not males. There are several possible reasons. First, the effect size may simply be lower in males. Second, the epigenetic regulation of MAOA may be different in hemizygous males than in dizygous females. Thirdly, twin studies suggest that the genetic factors affecting ND may differ between men and women [41]. Fourth, it may be that ASPD contributes to ND and AD despite being unrelated to MAOA methylation for both men and women. Because ASPD is more common in men it may obscure associations for men to a greater degree than it does for women. Finally, it may be that substance use produces different and more pronounced methylation signatures for females than for males.
Surprisingly, we did not find any relationship between MAOA methylation and ASPD. Nor did we find a significant relationship between genotype and methylation. Once again, this may simply be a function of low power. At the same time, these findings do not preclude specific GxE effects on ASPD at this locus because we did not examine the relationship of environmental factors hypothesized to elicit such effects, such as maltreatment [4], in this study. We plan to revisit this topic when we have measured methylation in a greater number of IAS subjects.
In summary, we report that methylation of the MAOA promoter is associated with ND and AD in females. We suggest that further examination of these findings is in order.
Supplementary Material
Acknowledgments
This work was generously supported by a grant to Dr. Philibert (DA 015789). The authors wish to express their gratitude to the late Dr. Remi Cadoret, founder of the Iowa Adoption Studies, whose pioneering efforts led to these studies. The authors also thank Pamela Wernett for her careful proofreading, Bill Adams for his aid in preparing the graphics and Sequenom Inc. for their prompt, precise work.
REFERENCES
- 1.Jaffee SR, Price TS. Gene-environment correlations: a review of the evidence and implications for prevention of mental illness. Mol Psychiatry. 2007;12(5):432–442. doi: 10.1038/sj.mp.4001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caspi A, Sugden K, Moffitt TE, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–9. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 3.Kim-Cohen J, Caspi A, Taylor A, et al. MAOA, maltreatment, and gene-environment interaction predicting children’s mental health: new evidence and a meta-analysis. Mol Psychiatry. 2006;11(10):903–913. doi: 10.1038/sj.mp.4001851. [DOI] [PubMed] [Google Scholar]
- 4.Caspi A, McClay J, Moffitt TE, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297(5582):851–4. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 5.Koller G, Bondy B, Preuss UW, Bottlender M, Soyka M. No association between a polymorphism in the promoter region of the MAOA gene with antisocial personality traits in alcoholics. Alcohol Alcohol. 2003;38(1):31–4. doi: 10.1093/alcalc/agg003. [DOI] [PubMed] [Google Scholar]
- 6.Samochowiec J, Lesch KP, Rottmann M, et al. Association of a regulatory polymorphism in the promoter region of the monoamine oxidase A gene with antisocial alcoholism. Psychiatry Res. 1999;86(1):67–72. doi: 10.1016/s0165-1781(99)00020-7. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt LG, Sander T, Kuhn S, et al. Different allele distribution of a regulatory MAOA gene promoter polymorphism in antisocial and anxious-depressive alcoholics. J Neural Transm. 2000;107(6):681–9. doi: 10.1007/s007020070069. [DOI] [PubMed] [Google Scholar]
- 8.Contini V, Marques FZ, Garcia CE, Hutz MH, Bau CH. MAOA-uVNTR polymorphism in a Brazilian sample: further support for the association with impulsive behaviors and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2006;141(3):305–8. doi: 10.1002/ajmg.b.30290. [DOI] [PubMed] [Google Scholar]
- 9.Saito T, Lachman HM, Diaz L, et al. Analysis of monoamine oxidase A (MAOA) promoter polymorphism in Finnish male alcoholics. Psychiatry Res. 2002;109(2):113–9. doi: 10.1016/s0165-1781(02)00013-6. [DOI] [PubMed] [Google Scholar]
- 10.Jin Y, Chen D, Hu Y, et al. Association between monoamine oxidase gene polymorphisms and smoking behaviour in Chinese males. Int J Neuropsychopharmacol. 2006;9(5):557–64. doi: 10.1017/S1461145705006218. [DOI] [PubMed] [Google Scholar]
- 11.Philibert RA, Sandhu H, Hollenbeck N, et al. The Relationship of 5HTT Methylation and Genotype on mRNA Expression and Liability to Major Depression and Alcohol Dependence in Subjects from the Iowa Adoption Studies. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2007 doi: 10.1002/ajmg.b.30657. epub ahead of publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yates WR, Cadoret RJ, Troughton E, Stewart MA. An adoption study of DSM-IIIR alcohol and drug dependence severity. Drug and Alcohol Dependence. 1996;41(1):9. doi: 10.1016/0376-8716(96)01221-5. [DOI] [PubMed] [Google Scholar]
- 13.Bucholz KK, Cadoret R, Cloninger CR, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–58. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 14.Association AP. Diagnostic and Statistical Manual of Mental Disorder. Fourth Edition American Psychiatric Association; Washington D.C.: 1994. [Google Scholar]
- 15.Klaus GGB. Lymphocytes: A practical Approach. IRL Press; Oxford: 1987. pp. 149–162. [Google Scholar]
- 16.Lahiri DK, Nurnberger JI., Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Research. 1991;19(19):5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103(3):273–9. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- 18.Merril CR, Pratt ME. A silver stain for the rapid quantitative detection of proteins or nucleic acids on membranes or thin layer plates. Analytical Biochemistry. 1986;156(1):96–110. doi: 10.1016/0003-2697(86)90160-0. [DOI] [PubMed] [Google Scholar]
- 19.Bradley SL, Dodelzon K, Sandhu HK, Philibert RA. Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. Am J Med Genet B Neuropsychiatr Genet. 2005;136(1):58–61. doi: 10.1002/ajmg.b.30185. [DOI] [PubMed] [Google Scholar]
- 20.Philibert RA, King BH, Winfield S, et al. Association of an X-chromosome dodecamer insertional variant allele with mental retardation. [erratum appears in Mol Psychiatry 1999 Mar;4(2):197.] Molecular Psychiatry. 1998;3(4):303–9. doi: 10.1038/sj.mp.4000442. [DOI] [PubMed] [Google Scholar]
- 21.Frommer M, McDonald LE, Millar DS, et al. A Genomic Sequencing Protocol that Yields a Positive Display of 5-Methylcytosine Residues in Individual DNA Strands. Proceedings of the National Academy of Sciences. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philibert R, Madan A, Andersen A, et al. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet. 2007;144(1):101–5. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]
- 23.Ehrich M, Nelson MR, Stanssens P, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–90. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 2007;35(5):e29. doi: 10.1093/nar/gkl1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleiss JL. Statistical Methods for Rates and Proportions. 2nd ed John Wiley & Sons Inc; New York, NY: 1981. [Google Scholar]
- 26.Philibert RA, Ryu GY, Yoon JG, et al. Transcriptional profiling of subjects from the Iowa adoption studies. Am J Med Genet B Neuropsychiatr Genet. 2007;144(5):683–90. doi: 10.1002/ajmg.b.30512. [DOI] [PubMed] [Google Scholar]
- 27.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philibert RA, Crowe R, Ryu GY, et al. Transcriptional profiling of lymphoblast lines from subjects with panic disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144(5):674–82. doi: 10.1002/ajmg.b.30502. [DOI] [PubMed] [Google Scholar]
- 29.Johnston CM, Lovell FL, Leongamornlert DA, et al. Large-Scale Population Study of Human Cell Lines Indicates that Dosage Compensation Is Virtually Complete. PLoS Genetics. 2008;4(1):e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agatsuma S, Lee M, Zhu H, et al. Monoamine oxidase A knockout mice exhibit impaired nicotine preference but normal responses to novel stimuli. Hum. Mol. Genet. 2006;15(18):2721–2731. doi: 10.1093/hmg/ddl206. [DOI] [PubMed] [Google Scholar]
- 31.Wooters TE, Bardo MT. The monoamine oxidase inhibitor phenelzine enhances the discriminative stimulus effect of nicotine in rats. Behav Pharmacol. 2007;18(7):601–8. doi: 10.1097/FBP.0b013e3282eff0d5. [DOI] [PubMed] [Google Scholar]
- 32.Villegier AS, Lotfipour S, McQuown SC, Belluzzi JD, Leslie FM. Tranylcypromine enhancement of nicotine self-administration. Neuropharmacology. 2007;52(6):1415–25. doi: 10.1016/j.neuropharm.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Guillem K, Vouillac C, Azar MR, et al. Monoamine oxidase A rather than monoamine oxidase B inhibition increases nicotine reinforcement in rats. Eur J Neurosci. 2006;24(12):3532–40. doi: 10.1111/j.1460-9568.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- 34.Miller DK, Wong EHF, Chesnut MD, Dwoskin LP. Reboxetine: Functional Inhibition of Monoamine Transporters and Nicotinic Acetylcholine Receptors. J Pharmacol Exp Ther. 2002;302(2):687–695. doi: 10.1124/jpet.302.2.687. [DOI] [PubMed] [Google Scholar]
- 35.George T, Weinberger A. Monoamine Oxidase Inhibition for Tobacco Pharmacotherapy. Mol Ther. 2007 doi: 10.1038/sj.clpt.6100474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David SP, Brown RA, Papandonatos GD, et al. Pharmacogenetic clinical trial of sustained-release bupropion for smoking cessation. Nicotine & Tobacco Research. 2007;9(8):821–833. doi: 10.1080/14622200701382033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berlin I, Anthenelli RM. Monoamine oxidases and tobacco smoking. Int J Neuropsychopharmacol. 2001;4(1):33–42. doi: 10.1017/S1461145701002188. [DOI] [PubMed] [Google Scholar]
- 38.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424(6945):147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 39.Kawaji H, Frith M, Katayama S, et al. Dynamic usage of transcription start sites within core promoters. Genome Biology. 2006;7(12):R118. doi: 10.1186/gb-2006-7-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang G, Lin JC, Wei V, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101(19):7357–62. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lessov CN, Martin NG, Statham DJ, et al. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychological Medicine. 2004;34(05):865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.