Abstract
Heparin is a highly sulfated polysaccharide which serves biologically relevant roles as an anticoagulant and anti-cancer agent. While it is well known that modification of heparin’s sulfation pattern can drastically influence its ability to bind growth factors and other extracellular molecules, very little is known about the cellular uptake of heparin and the role sulfation patterns serve in affecting its internalization. In this study, we chemically synthesized several fluorescently-labeled heparins consisting of a variety of sulfation patterns. These polysaccharides were thoroughly characterized using anion exchange chromatography and size exclusion chromatography. Subsequently, we utilized flow cytometry and confocal imaging to show that sulfation patterns differentially affect the amount of heparin uptake in multiple cell types. This study provides the first comprehensive analysis of the effect of sulfation pattern on the cellular internalization of heparin or heparan sulfate like polysaccharides. The results of this study expand current knowledge regarding heparin internalization and provide insights into developing more effective heparin-based drug conjugates for applications in intracellular drug delivery.
Keywords: Heparin, Cellular Uptake, Internalization, Nucleus localization, Heparan Sulfate, Heparosan
Introduction
Heparin is a complex, biocompatible, biodegradable, and water-soluble glycosaminoglycan that is commonly found within mast cell granules. While its biological role is unclear, heparin is utilized clinically for its anti-coagulant properties. As an agonist of anti-thrombin, heparin is an effective treatment against deep-vein thrombosis and pulmonary emboli.[1] Recent studies have also shown that heparin has potent anti-cancer properties including an ability to hinder cancer invasion, metastasis, and tumor-derived angiogenesis.[2–4]
It is well known that altering heparin’s sulfation pattern can affect its biochemical properties. N-desulfation of heparin, removal of 2-O sulfate groups from iduronic acid residues, and removal of 6-O sulfate groups from glucosamine residues within heparin can inhibit heparin-FGF interactions.[5–7] Additionally, N-sulfate and 3-O sulfate groups are critical to heparin’s anti-coagulant activity.[8, 9]
Several recent publications have utilized covalently conjugated heparin-based drug delivery vehicles (DDV) to deliver anti-cancer molecules such as paclitaxel and litocholate.[10, 11] Conjugation to heparin provides additional therapeutic value because both the DDV as well as the drug prevent cancer progression. However, it is still unclear how altering heparin’s sulfation patterns can affect its cellular internalization, localization, and efficacy as a DDV. Previously, researchers have identified heparin scavenger receptors, however these receptors have not yet been isolated and their substrate specificities remain unknown. [12–14]
In this article, we chemically modify heparin and heparosan, a heparin precursor isolated from E. Coli K5, to show that modification of heparin’s sulfation pattern leads to increased cellular uptake – providing hints to define the ligand specificities of heparin receptors in cells. These exciting results provide new insight into heparin/heparan sulfate biology and the design of more effective heparin-conjugates for drug delivery.
Materials and Methods
Materials
HT-29 colon cancer cells and BXPC-3 pancreatic cancer cells were provided by Dr. Scott Kuwada (University of Hawaii). U87-Mg glioma cells were obtained from Dr. Randy Jensen (University of Utah). Hog mucosal heparin was obtained from Ming Han Chemicals (Oakland, CA). K1 CHO cells were obtained from the ATCC. DEAE-Sepharose gel was purchased from Amersham Biosciences. The analytical grade strong anion exchange column, size exclusion column, and weak anion exchange columns were obtained from Dionex, and Tosoh Biosciences, respectively. Disaccharide standards for strong anion exchange were obtained from Iduron Inc (Manchester, UK). Heparitinase I, II and III from flavobacterium heparinum were expressed as previously described.[15] Cell culture reagents were from Invitrogen Inc. Internalization inhibitors Chlorpromazine (CPZ), Filipin (FIL), Dynasore (DYN), 5-(N-Ethyl-N-isopropyl) amiloride (EIPA), and all other reagents and solvents were from Sigma-Aldrich.
Synthesis of Modified Heparins (M. Heps)
Briefly, Heparosan (NA), N-sulfo heparosan (NS), completely desulfated heparin (CDSHep), completely desulfated N-resulfated heparin (CDSNS), and 2-O desulfated heparin (2ODS) were synthesized as described in literature.[16–19] After extensive dialysis, each substrate was digested with a cocktail of heparitinase I, II, and III and subjected to disaccharide analysis by strong anion exchange chromatography.[20] More specifically, the substrates were prepared as described in the following sections.
Heparosan (NA)
Heparosan capsular polysaccharide was first isolated and purified from E.Coli K5 as previously described in literature.[16] The resulting polysaccharide was then further purified by dialysis against running water through a 3000 MWCO membrane for 3 days. After complete lyophilization, the product was weighed and characterized through anion exchange chromatography as described in the supplementary material.
N-Sulfated Heparosan (NS)
As described in literature, N-sulfated heparosan was prepared by N-deacetylation of heparosan followed by N-sulfation.[17] N-deacetylation was carried out by treating 1 g of heparosan with 2.5 M NaOH in water at 55 °C overnight. Next, N-deacetylated heparosan was neutralized to pH 7.0 and treated with 2.5 g each of NaCO3 and triethylamine-sulfur trioxide complex and stirred for 24 h at 48 °C. The pH of this reaction was maintained below pH 10 by addition of HCl. Subsequently, an additional 2.5 g each of NaCO3 and triethylamine-sulftur trioxide complex was added and the reaction was stirred for an additional 24 h. The resulting polysaccharide was dialyzed, lyophilized, and chemically characterized in a similar manner to NA.
Completely desulfated Heparin (CDSHep)
CDSHep was prepared by utilizing published protocols.[18] The pyridinium salt of heparin was first synthesized by passing a solution of 1 g of heparin in water through a column packed with Amberlite cation exchange resin. The resulting eluant was collected on ice, adjusted to pH 9 with pyridine, stirred for 30 minutes, and then concentrated in a rotary evaporator. 100 mg of the pyridinium salt of heparin was then completely desulfated by stirring overnight at 100 °C in a 10 ml mixture of 9:1 DMSO:Methanol. The resulting polysaccharide was dialyzed, lyophilized, and characterized as stated before.
Completely desulfated N-resulfated Heparin (CDSNS)
To synthesize CDSNS, CDSHep was subjected to N-sulfation as previously described for NS preparation.
2-O Desulfated Heparin (2ODS)
According to a previously published protocol, 10 mg of heparin was mixed with 1 mg of NaBH4 in 10 ml of 0.4 N NaOH.[19] This mixture was then frozen in a −80 refrigerator and lyophilized to dryness. The resulting crusty yellow solid was subsequently redissolved in water and neutralized to pH 7 with acetic acid. This polymer was then dialyzed for 3 days, lyophilized, and characterized as stated before.
Fluoresceinamine conjugation to M. Heps
First, a stock of 100 mg of fluoresceinamine (FA) was dissolved in a 1 ml mixture of 3:2:1 DMSO:Acetonitrile:Acetone. Additionally, a stock containing 22 mg of 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) in 1 ml of water was created. Next, 100 mg of each M. Hep substrate was dissolved in 1 ml of water. 300 µl of the FA stock was added to the M. Hep along with 300 µl of the EDC stock. This mixture was stirred overnight at room temperature and subsequently dialyzed and lyophilized. Utilizing FA modified heparins, the molecular weight of FA-M. Heps was analyzed by size exclusion HPLC as described previously.[20] The charge density of each substrate was analyzed by weak anion-exchange HPLC as described previously.[20]
Cell Treatment with FA-M. Heps
Approximately 50,000 cells were trypsinized and added into wells of a 96 well plate with DMEM containing 10% FBS and 1% Penicillin/Streptomycin (P/S). After approximately 16 hrs cells had were adherent and the media was replaced with HAMS F-12 containing 10% FBS and 1% P/S. To these wells, 200 µg of each FA-M. Hep was added and cells were incubated for 6 hrs in a humidified cell culture incubator.
Fluorescence assisted cell sorting
After treatment with FA-M. Heps, cells were resuspended in trypsin without phenol red. Trypsin was neutralized with HAMS F-12 media containing 10% FBS, and cell suspensions were analyzed on a FACScan instrument (Becton Dickenson Immunocytometry Systems, Mountain View, CA) with computer-aid from CellQuant software. A minimum of 5000 gated events were captured for each sample and used for comparison purposes.
Confocal Imaging
For confocal imaging, approximately 50,000 cells were grown on glass coverslips within 35 mm cell culture dishes with 1 ml media. Subsequently, 200 µg of FA-M. Hep conjugates were added and cells were allowed to internalize M. Heps for 16 hrs in a humidified incubator. Subsequently, the media was removed and cells were washed twice with PBS. 500 µl of 4% paraformaldehyde solution was then added to the cells and cells were maintained for 10 mins at room temperature. Next, cells were washed twice with PBS and stained with DAPI and Rhodamine Phalloidin for 10 mins each. After incubation with cellular stains, cells were mounted onto microscope slides and imaged with an FV1000-XY Confocal Olympus IX81 microscope with a 60X oil immersion lens.
Results
The central goal of this study is to determine whether modulation of sulfation pattern affects the cellular internalization of heparin. To achieve this goal, a library of floresceinamine-conjugated M. Heps was synthesized (Fig. 1) and extensively characterized via analysis of sulfation pattern, charge density, and size (Table S1 and Fig. S1). A variety of cells were incubated with each M. Hep and FACS was utilized to analyze the total amount of conjugate that was internalized after 6 hours (Fig. 2). Next, the cellular localization of each conjugate was assessed using confocal microscopy in two different cell lines – HT-29 colon cancer cells and U87Mg glioma cells (Figs. 3 and 4). The rate and mechanism of uptake of each conjugate was also assessed using FACS (Figs. 5 and 6).
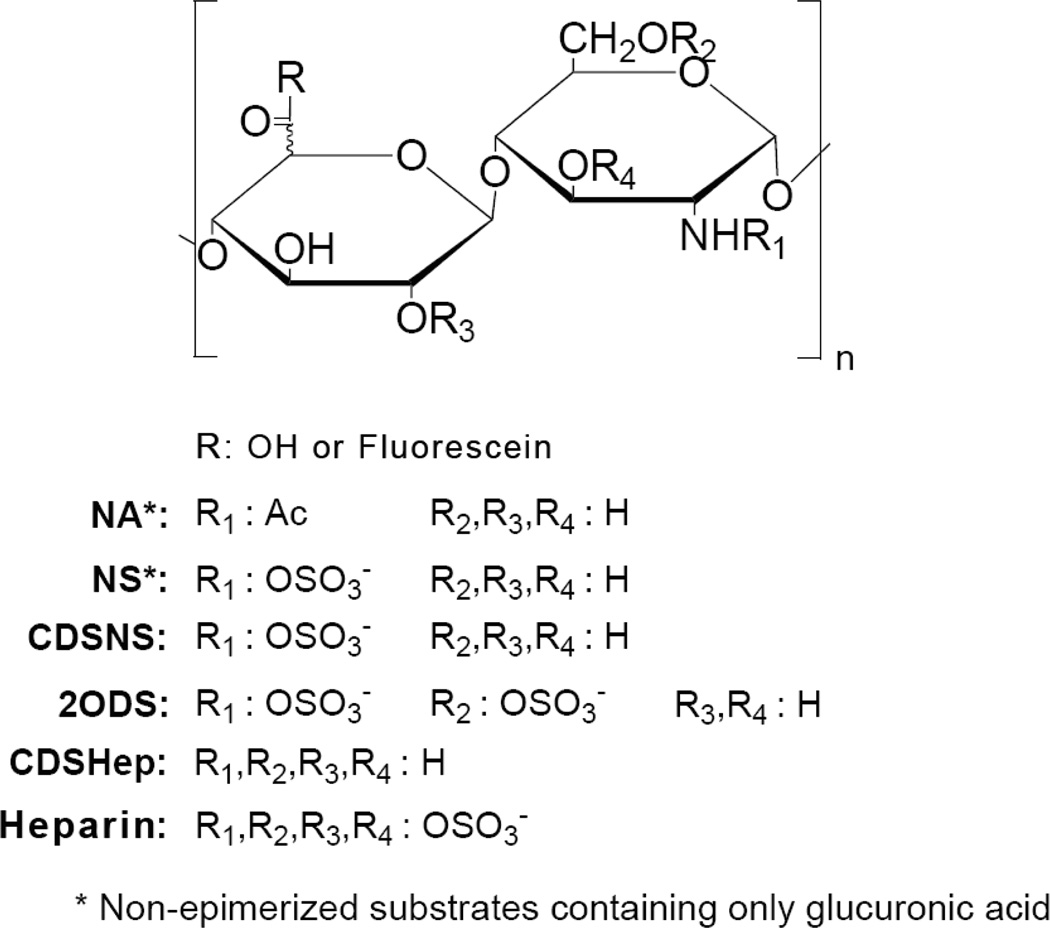
Figure 1.
Structures of modified heparins prepared in this study.
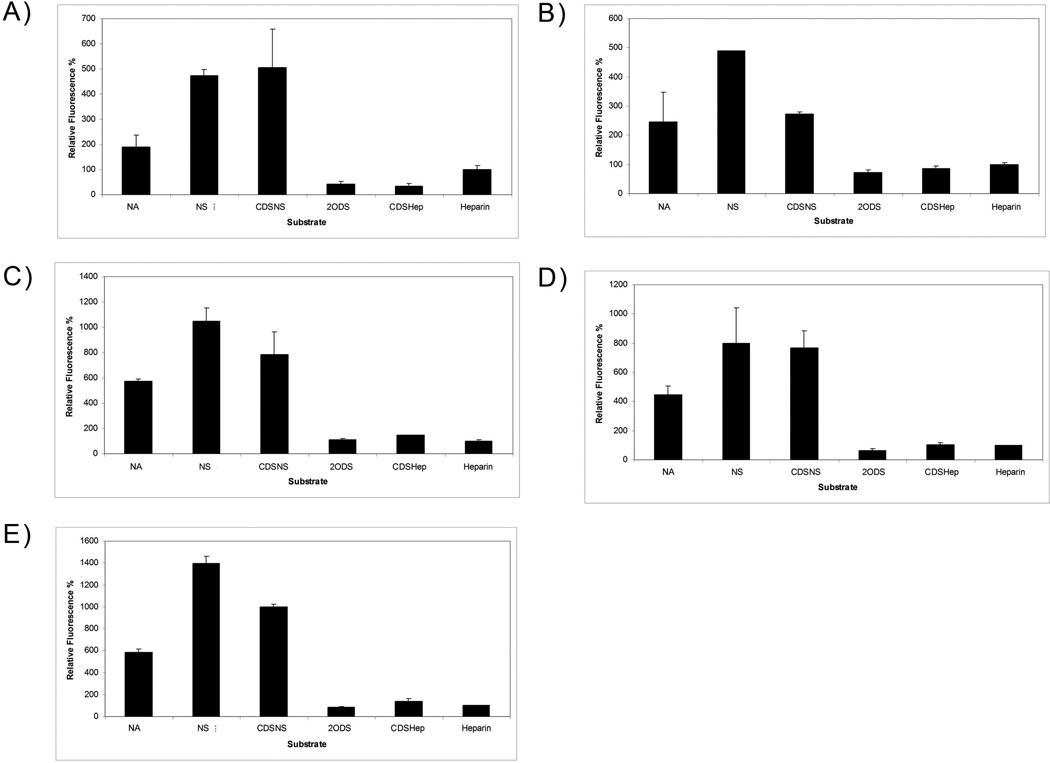
Figure 2.
Sulfation patterns determine the total amount of cellular uptake of M. Heps at 6 hours. The different panels indicate M. Hep uptake in a) BLMVEC, b) K1 CHO, c) BXPC-3, d) HT-29, and e) U87 Mg cells. Values are normalized against heparin and show that M. Heps such as NA, NS, and CDSNS show enhanced cell uptake relative to heparin. Internalization of M. Heps was determined by fluorescence assisted cell sorting analysis as described in the experimental section. Cellular auto fluorescence at the settings used was minimal.
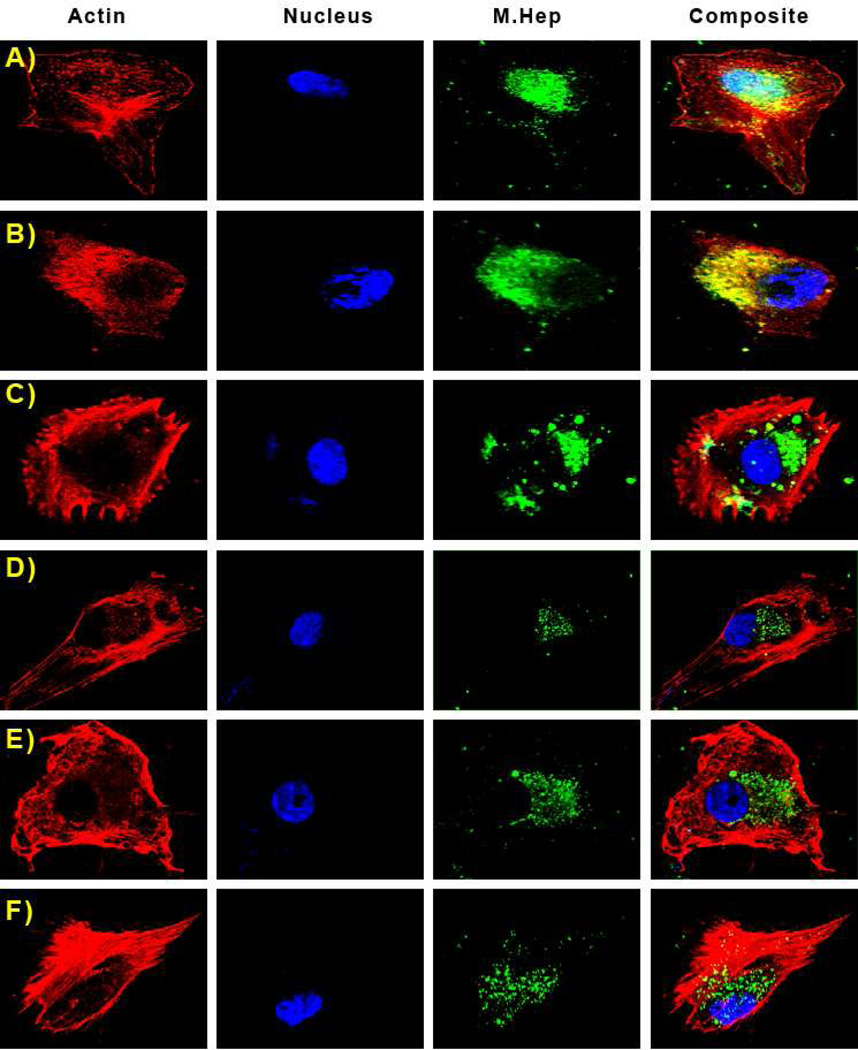
Figure 3.
Sulfation patterns determine the cellular localization of M. Heps in U87 Mg cells. Panels in this image are fluorescence from Rhodamine Phalloidin (actin), DAPI (nucleus), Fluoresceinamine (M. Heps), and an overlay of all fluorophores. Representative substrates are: A) NA, B) NS, C) CDSNS, D) 2ODS, E) CDSHep, F) Heparin. It is evident that NA polymers colocalize with DAPI in the nucleus of U87 Mg cells.
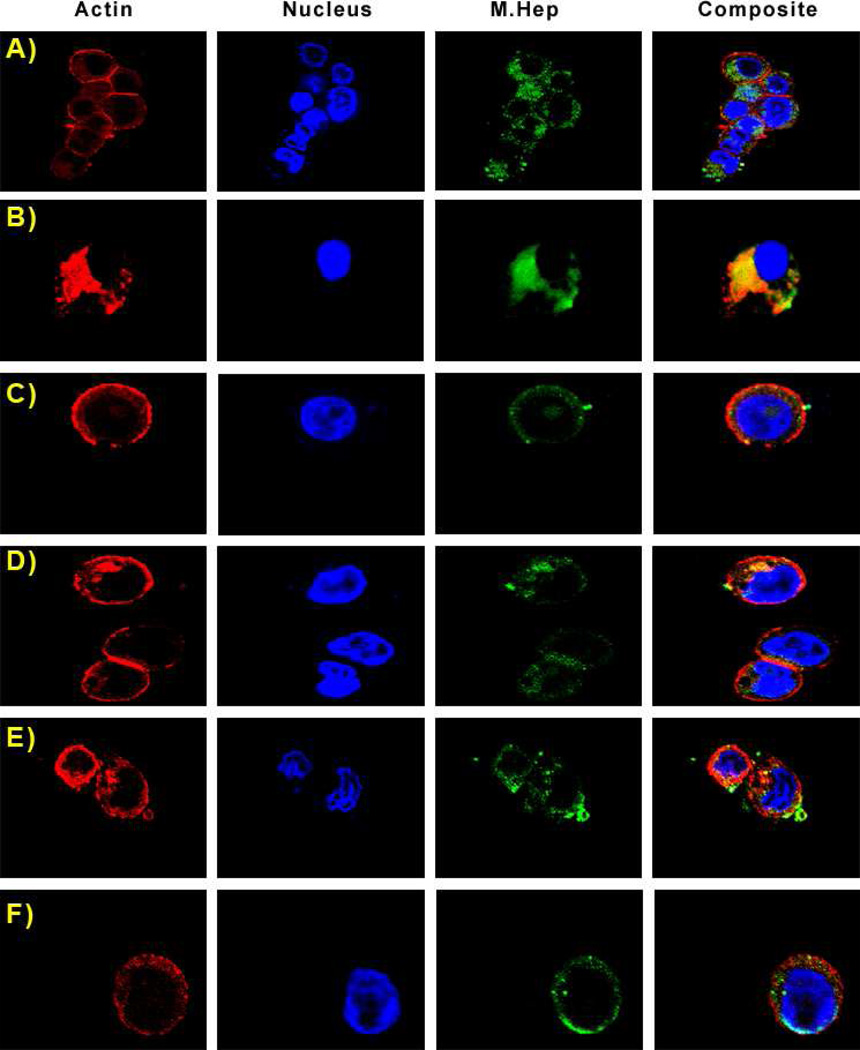
Figure 4.
Sulfation patterns determine the cellular localization of M. Heps, however no nuclear localization is visible for any substrate in HT-29 cells. Panels in this image are fluorescence from Rhodamine Phalloidin (actin), DAPI (nucleus), Fluoresceinamine (M. Heps), and an overlay of all fluorophores. Representative substrates are: A) NA, B) NS, C) CDSNS, D) 2ODS, E) CDSHep, F) Heparin.
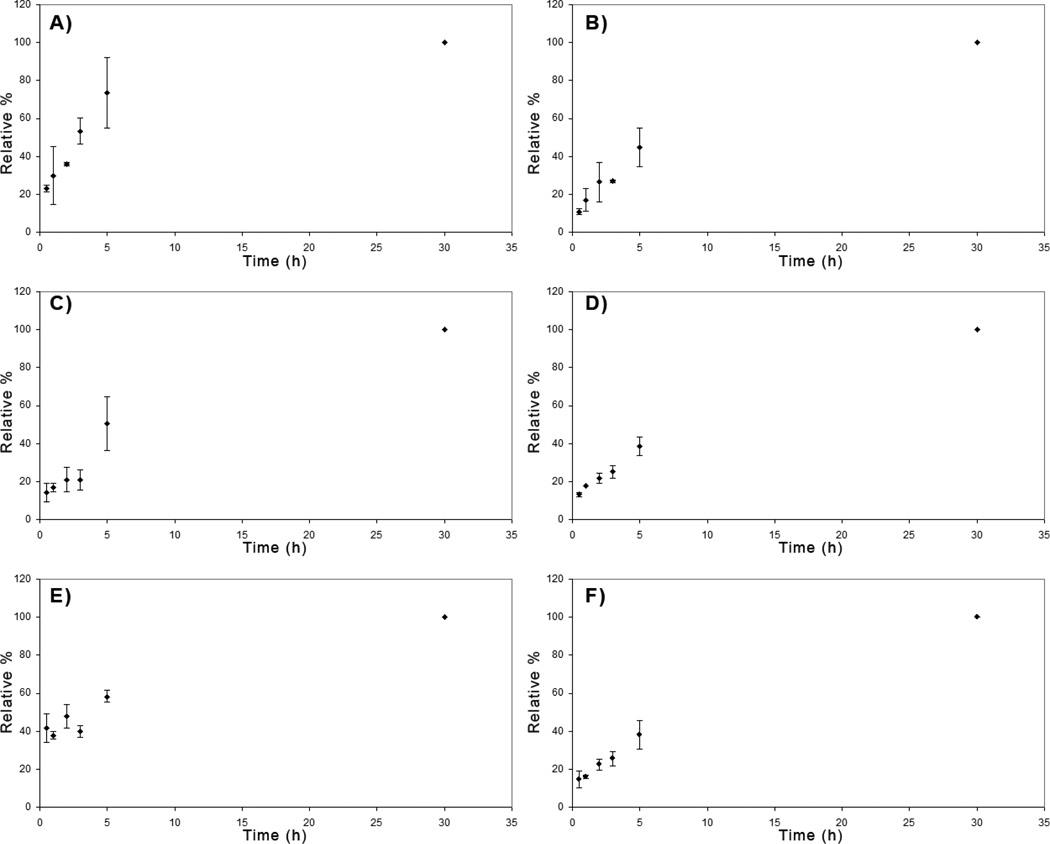
Figure 5.
Sulfation patterns determine rate of entry of M. Heps into HT-29 colon cancer cells. Values are normalized to the 30 hour time points for each substrate. Representative panels indicate the time-dependent internalization of: A) NA, B) NS, C) CDSNS, D) 2ODS, E) CDSHep, and F) Heparin
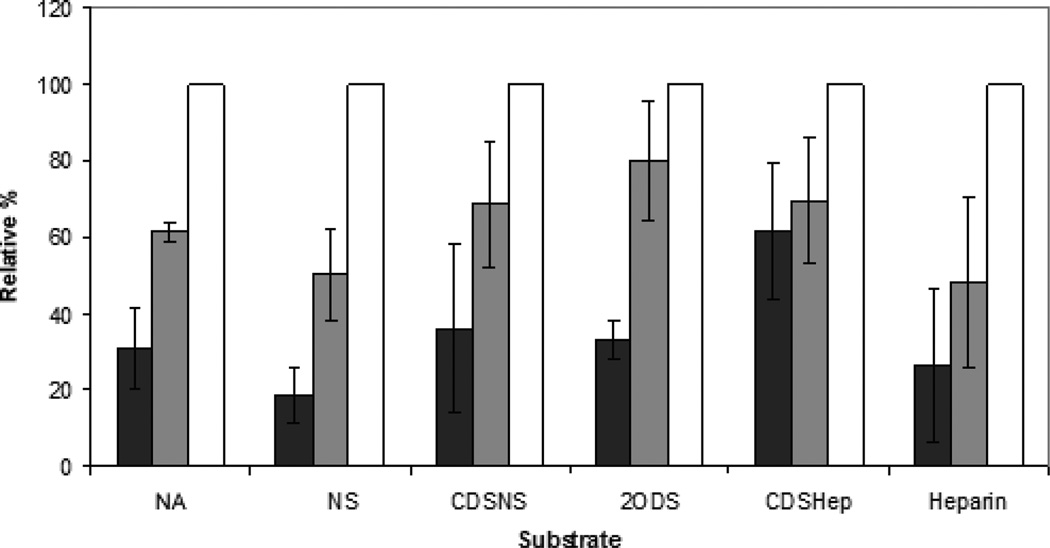
Figure 6.
Sulfation pattern has little effect on the concentration dependence of internalization. Representative bars indicate the relative fluorescence measured by FACS when 20 µg (■), 100 µg ( ), or 200 µg (□) of M.Heps were incubated with cells for 24 hours. Values are normalized to the total substrate internalized after 24 hours when 200 ▪g of the respective substrate are added.
), or 200 µg (□) of M.Heps were incubated with cells for 24 hours. Values are normalized to the total substrate internalized after 24 hours when 200 ▪g of the respective substrate are added.
Sulfation pattern affects the internalization of M. Heps into multiple cell types
To determine the effects of sulfation pattern on internalization, experiments were conducted to test the relative internalization of several M. Heps in the following cell lines (Fig. 2): bovine lung microvascular endothelial cells (BLMVEC), chinese hamster ovary K1 cells (CHO K1), BXPC-3 human pancreatic cancer cells, HT-29 human colorectal cancer cells, and U87Mg human glioma cells.
All cells were incubated with fluoresceinamine-conjugated M. Heps for 6 hours and subsequently subjected to analysis by FACS (Fig. 2). Relative to heparin, M. Hep substrates showed drastically altered internalization into different cell types. NA, NS, and CDSNS fluoresceinamine conjugates were internalized into all cell types significantly more than heparin. However, a partially positively charged polymer, CDSHep, was not internalized to the same extent as NA, NS, or CDSNS in all the cell lines tested. Furthermore, NS, not NA, was internalized to the greatest extent in all cell types tested; thus, these results indicate that uptake may be receptor-mediated and that the receptor recognizes and internalizes NS more than other M. Heps tested.
M. Heps are found throughout cellular bodies
To examine the effect of sulfation patterns on cellular localization, U87 Mg and HT-29 cells were incubated with M. Hep-FA conjugates overnight and localization was analyzed by confocal microscopy (Figs. 3 and 4). Rhodamine Phalloidin was utilized as a red dye to identify cellular actin and DAPI was utilized to label the cellular nuclei. Z-Slices were chosen so as to minimize colocalization of the actin stain with the nuclear stain.
Confocal images indicate that NS, CDSNS, 2ODS, CDSHep, and heparin all co-localize with cellular actin in U87 MG cells, and are found throughout cell bodies. Interestingly, NA co-localizes with DAPI, showing that this substrate may enter the nuclei of U87 MG cells. However, in HT-29 cells, all the M. Heps are found in cell bodies – thus indicating that nuclear localization may be both sulfation pattern-dependent and cell-specific.
Sulfation patterns affect the rate of internalization of M.Heps into cells
In addition to the extent of internalization, the effects of sulfation pattern on the rate of substrate internalization into HT-29 cells was analyzed by incubating equal amounts of each substrate with cells for various time points (Fig. 5). Compared to the amount of each substrate internalized after 30 hours, NA and CDSHep saturated the fastest. NS, CDSNS, and 2ODS saturated at an intermediate rate while heparin saturated at a slower rate. Cellular recognition of GAG sulfation patterns probably determines the rate of saturation.
In contrast, sulfation patterns do not determine the concentration dependence of internalization (Fig. 6). Only CDSHep showed a significant departure from Heparin by saturating at lower concentrations than all other polymers tested. This is most likely due to its amine functionality, as NA and other substrates did not show similar concentration dependence.
Discussion
In recent years, heparin-based conjugates have shown promise in preclinical studies as drug delivery vehicles. One of the reasons for their efficacy is because heparin-drug conjugates are able to attack cancer cells using multiple pathways – both the drug and the DDV are able to mitigate tumor progression.[21–24] However, the role of sulfation patterns in the cellular uptake of heparin is largely unknown. In this article, chemically modified heparosan from E. Coli K5 as well as chemically modified heparin are utilized to show that sulfation patterns determine heparin cellular uptake into several cell types. This knowledge inspires new designs of chemically modified heparin-drug conjugates that are favorable for drug delivery but lack heparin’s inherent drawbacks such as bleeding complications and heparin induced thrombocytopenia. Additionally, the results of this study further provide hints to illuminate the ligand specificities of elusive heparin scavenger receptors.
Previous studies have found that modification of sulfation pattern can alter the biological properties of heparin. Controlling the amount of 2-O, 3-O, and 6-O sulfation can drastically affect heparin’s ability to bind ligands. [5, 8, 9, 25] To test our hypothesis that sulfation patterns affect cellular internalization and the effectiveness of heparin as a DDV, we designed a library of heparins to represent a diverse group of polymers with different sulfation patterns and densities (Fig. 1). CDSHep, was the only substrate determined to have free amine groups and hence it had the least negative charge density. Unaltered heparin had the highest negative charge while other substrates had intermediate charge density. We also specifically designed molecules derived from heparosan that were non-epimerized and those derived from heparin which contained high iduronic acid content. Analysis by size exclusion chromatography revealed that only minor differences in size and polydispersity exist among the different substrates (Table S1 and Fig. S1).
After structural characterization, a variety of cell types were treated with M. Heps and analyzed via FACS to determine the uptake of each M. Hep (Fig. 2). The experiment was designed to include both tumorigenic as well as non-tumorigenic cell types. As hypothesized, modifying the sulfation patterns of heparin and heparosan significantly altered cellular internalization. One would expect that CDSHep, the most positively charged polymer, would be internalized by cells to the greatest extent. However, NA, NS, and CDSNS accumulated inside the cell to a much larger extent than CDSHep, 2ODS, and heparin in all cell types tested. This indicates that sulfation pattern, not charge density, determines cellular internalization of M. Heps. These results also give a glimpse into the substrate specificities of elusive heparin uptake receptors.[12–14] While researchers have not yet isolated these receptors, it is clear that sulfation pattern greatly affects cellular uptake of heparin and that these receptors prefer N-sulfo heparosan to any of the other M. Heps tested. Further evidence that heparin uptake may be receptor driven is determined by the concentration- dependence of the internalization of M.Heps (Fig 6). If internalization was a purely diffusion-driven process then the concentration dependence of internalization would be linear with concentration. However, it is evident that incubating cells with five times and ten times more substrate does not lead to a linear dose dependent increase in internalization.
Next, the cellular localization of M. Heps was sought after to determine the effects of sulfation pattern on sub-cellular targeting. Previously, it was found that GAGs and proteoglycans such as Syndecan-1 and Glypican-1 can enter the nucleus.[26–30] Additionally, anti-proliferative GAGs primed by xylosides can enter cellular nuclei and modulate histone 3 acetylation and cellular growth.[31] Heparin-poly-β-amino ester complexes have also been found to modulate nuclear transcription factors.[32] However, researchers have not yet identified if sulfation patterns affect the nuclear entry of these PGs and GAGs. Therefore, we examined the cellular localization of M. Heps tagged with fluoresceinamine by utilizing confocal microscopy (Figs. 3 and 4). The majority of M. Heps were found throughout cell bodies and excluded from the nucleus in both HT-29 and U87Mg cells. However, in U87Mg, NA was found to co-localize with DAPI. Based on these results, it may be possible to deduce that sulfation patterns can affect nuclear entry of GAGs. It is unlikely that charge density was responsible for nuclear entry as CDSHep did not enter the nuclei. Additionally, none of the epimerized substrates were visible in the nucleus suggesting that chain flexibility may affect nuclear entry as well. However, additional biochemical proof will be necessary to further understand the differential localization of heparin and heparin-like polymers.
In conclusion, this research presents the first comprehensive evidence that sulfation pattern, not charge density, determines heparin/heparan sulfate cellular uptake into several cell types. While the M. Heps are found throughout cells, nuclear localization of these glycosaminoglycans may be both sulfation pattern and cell-type dependent. The results of this study have broad implications in cell biology, heparan sulfate biochemistry, and drug delivery vehicle design. However, several new questions now surface: Why do NS and NA polymers enter cells to a greater extent than heparin? Do NA or NA/NS domains of heparan sulfate promote cellular uptake and nuclear localization of Syndecans and Glypicans? Additionally, would developing NA-, NS-, or CDSNS- based DDV yield better tumor-targeting capability than heparin-based DDV? In-depth analysis of nuclear localization and in vivo drug targeting is necessary to answer these intriguing questions.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH grants P01HL107152 and R01GM075168 to B.K and by the NIH fellowship F31CA168198 to K.R.
ABBREVIATIONS
- M. Hep
Modified heparin
- FGF
fibroblast growth factor
- DDV
drug delivery vehicle
- CPZ
Chlorpromazine
- FIL
Filipin
- DYN
dynasore
- EIPA
5-(N-Ethyl-N-isopropyl) amiloride
- FA
fluoresceinamine
- NA
Heparosan
- NS
N-Sulfo heparosan
- CDSHep
Completely desulfated heparin
- CDSNS
Completely desulfated N-resulfated heparin
- 2ODS
2-O desulfated heparin
- HS
heparan sulfate
Footnotes
Supporting Information. Charge and Size of M. Heps. Effect of treatment with multiple uptake inhibitors. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Hull R, Delmore T, Genton E, Hirsh J, Gent M, Sackett D, McLoughlin D, Armstrong P. Warfarin sodium versus low-dose heparin in the long-term treatment of venous thrombosis. N Engl J Med. 1979;301(16):855–858. doi: 10.1056/NEJM197910183011602. [DOI] [PubMed] [Google Scholar]
- 2.Borsig L. Antimetastatic activities of modified heparins: selectin inhibition by heparin attenuates metastasis. Semin Thromb Hemost. 2007;33(5):540–546. doi: 10.1055/s-2007-982086. [DOI] [PubMed] [Google Scholar]
- 3.Vlodavsky I, Mohsen M, Lider O, Svahn CM, Ekre HP, Vigoda M, Ishai-Michaeli R, Peretz T. Inhibition of tumor metastasis by heparanase inhibiting species of heparin. Invasion Metastasis. 1994;14(1–6):290–302. [PubMed] [Google Scholar]
- 4.Folkman J, Langer R, Linhardt RJ, Haudenschild C, Taylor S. Angiogenesis inhibition and tumor regression caused by heparin or a heparin fragment in the presence of cortisone. Science. 1983;221(4612):719–725. doi: 10.1126/science.6192498. [DOI] [PubMed] [Google Scholar]
- 5.Guimond S, Maccarana M, Olwin BB, Lindahl U, Rapraeger AC. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2, and FGF-4. J Biol Chem. 1993;268(32):23906–23914. [PubMed] [Google Scholar]
- 6.Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, Claesson-Welsh L. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem. 2000;275(32):24653–24660. doi: 10.1074/jbc.M908930199. [DOI] [PubMed] [Google Scholar]
- 7.Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem. 2008;283(16):10366–10376. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- 8.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of 3-O- and 6-O-sulfated glucosamine residues in the heparin-induced conformational change in antithrombin III. Biochemistry. 1987;26(20):6454–6461. doi: 10.1021/bi00394a024. [DOI] [PubMed] [Google Scholar]
- 9.Danishefsky I, Ahrens M, Klein S. Effect of heparin modification on its activity in enhancing the inhibition of thrombin by antithrombin III. Biochim Biophys Acta. 1977;498(1):215–222. doi: 10.1016/0304-4165(77)90101-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Wang Y, Xiang J, Yao K. Target-specific cellular uptake of taxol-loaded heparin-PEG-folate nanoparticles. Biomacromolecules. 2010;11(12):3531–3538. doi: 10.1021/bm101013s. [DOI] [PubMed] [Google Scholar]
- 11.Yu MK, Lee DY, Kim YS, Park K, Park SA, Son DH, Lee GY, Nam JH, Kim SY, Kim IS, Park RW, Byun Y. Antiangiogenic and apoptotic properties of a novel amphiphilic folate-heparin-lithocholate derivative having cellular internality for cancer therapy. Pharm Res. 2007;24(4):705–714. doi: 10.1007/s11095-006-9190-3. [DOI] [PubMed] [Google Scholar]
- 12.Falcone DJ. Heparin stimulation of plasminogen activator secretion by macrophage-like cell line RAW264.7: role of the scavenger receptor. J Cell Physiol. 1989;140(2):219–226. doi: 10.1002/jcp.1041400205. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe J, Haba M, Urano K, Yuasa H. Uptake mechanism of fractioned [(3)H]heparin in isolated rat kupffer cells: involvement of scavenger receptors. Biol Pharm Bull. 1996;19(4):581–586. doi: 10.1248/bpb.19.581. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe J, Muranishi H, Haba M, Yuasa H. Uptake of fluorescein isothiocyanate (FITC)-fractionated heparin by rat parenchymal hepatocytes in primary culture. Biol Pharm Bull. 1993;16(9):939–941. doi: 10.1248/bpb.16.939. [DOI] [PubMed] [Google Scholar]
- 15.Babu P, Kuberan B. Fluorescent-tagged heparan sulfate precursor oligosaccharides to probe the enzymatic action of heparitinase I. Anal Biochem. 2009;396(1):124–132. doi: 10.1016/j.ab.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuberan B, Lech M, Zhang L, Wu ZL, Beeler DL, Rosenberg RD. Analysis of heparan sulfate oligosaccharides with ion pair-reverse phase capillary high performance liquid chromatography-microelectrospray ionization time-of-flight mass spectrometry. J Am Chem Soc. 2002;124(29):8707–8718. doi: 10.1021/ja0178867. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd AG, Embery G, Fowler LJ. Studies on heparin degradation. I. Preparation of (35 S) sulphamate derivatives for studies on heparin degrading enzymes of mammalian origin. Biochem Pharmacol. 1971;20(3):637–648. doi: 10.1016/0006-2952(71)90150-x. [DOI] [PubMed] [Google Scholar]
- 18.Nagasawa K, Inoue Y, Kamata T. Solvolytic desulfation of glycosaminoglycuronan sulfates with dimethyl sulfoxide containing water or methanol. Carbohydr Res. 1977;58(1):47–55. doi: 10.1016/s0008-6215(00)83402-3. [DOI] [PubMed] [Google Scholar]
- 19.Ishihara M, Kariya Y, Kikuchi H, Minamisawa T, Yoshida K. Importance of 2-O-sulfate groups of uronate residues in heparin for activation of FGF-1 and FGF-2. J Biochem. 1997;121(2):345–349. doi: 10.1093/oxfordjournals.jbchem.a021593. [DOI] [PubMed] [Google Scholar]
- 20.Victor XV, Nguyen TK, Ethirajan M, Tran VM, Nguyen KV, Kuberan B. Investigating the elusive mechanism of glycosaminoglycan biosynthesis. J Biol Chem. 2009;284(38):25842–25853. doi: 10.1074/jbc.M109.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vannucchi S, Pasquali F, Chiarugi VP, Ruggiero M. Heparin inhibits A431 cell growth independently of serum and EGF mitogenic signalling. FEBS Lett. 1991;281(1–2):141–144. doi: 10.1016/0014-5793(91)80378-g. [DOI] [PubMed] [Google Scholar]
- 22.Norrby K. Heparin and angiogenesis: a low-molecular-weight fraction inhibits and a high-molecular-weight fraction stimulates angiogenesis systemically. Haemostasis. 1993;23(Suppl 1):141–149. doi: 10.1159/000216923. [DOI] [PubMed] [Google Scholar]
- 23.Folkman J, Weisz PB, Joullie MM, Li WW, Ewing WR. Control of angiogenesis with synthetic heparin substitutes. Science. 1989;243(4897):1490–1493. doi: 10.1126/science.2467380. [DOI] [PubMed] [Google Scholar]
- 24.Engelberg H. Actions of heparin that may affect the malignant process. Cancer. 1999;85(2):257–272. doi: 10.1002/(sici)1097-0142(19990115)85:2<257::aid-cncr1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Krauel K, Hackbarth C, Furll B, Greinacher A. Heparin-induced thrombocytopenia: in vitro studies on the interaction of dabigatran, rivaroxaban, and low-sulfated heparin, with platelet factor 4 and anti-PF4/heparin antibodies. Blood. 2012;119(5):1248–1255. doi: 10.1182/blood-2011-05-353391. [DOI] [PubMed] [Google Scholar]
- 26.Ishihara M, Fedarko NS, Conrad HE. Transport of heparan sulfate into the nuclei of hepatocytes. J Biol Chem. 1986;261(29):13575–13580. [PubMed] [Google Scholar]
- 27.Richardson TP, Trinkaus-Randall V, Nugent MA. Regulation of heparan sulfate proteoglycan nuclear localization by fibronectin. J Cell Sci. 2001;114(Pt 9):1613–1623. doi: 10.1242/jcs.114.9.1613. [DOI] [PubMed] [Google Scholar]
- 28.Hsia E, Richardson TP, Nugent MA. Nuclear localization of basic fibroblast growth factor is mediated by heparan sulfate proteoglycans through protein kinase C signaling. J Cell Biochem. 2003;88(6):1214–1225. doi: 10.1002/jcb.10470. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Sanderson RD. Heparanase regulates levels of syndecan-1 in the nucleus. PLoS One. 2009;4(3):e4947. doi: 10.1371/journal.pone.0004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Y, Haring M, Roughley PJ, Margolis RK, Margolis RU. Glypican and biglycan in the nuclei of neurons and glioma cells: presence of functional nuclear localization signals and dynamic changes in glypican during the cell cycle. J Cell Biol. 1997;139(4):851–864. doi: 10.1083/jcb.139.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson U, Johnsson R, Fransson LA, Ellervik U, Mani K. Attenuation of tumor growth by formation of antiproliferative glycosaminoglycans correlates with low acetylation of histone H3. Cancer Res. 2010;70(9):3771–3779. doi: 10.1158/0008-5472.CAN-09-4331. [DOI] [PubMed] [Google Scholar]
- 32.Berry D, Lynn DM, Sasisekharan R, Langer R. Poly(beta-amino ester)s promote cellular uptake of heparin and cancer cell death. Chem Biol. 2004;11(4):487–498. doi: 10.1016/j.chembiol.2004.03.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.