Abstract

Understanding potential health risks is a significant challenge due to the large numbers of diverse chemicals with poorly characterized exposures and mechanisms of toxicities. The present study analyzes 976 chemicals (including failed pharmaceuticals, alternative plasticizers, food additives, and pesticides) in Phases I and II of the U.S. EPA’s ToxCast project across 331 cell-free enzymatic and ligand-binding high-throughput screening (HTS) assays. Half-maximal activity concentrations (AC50) were identified for 729 chemicals in 256 assays (7,135 chemical–assay pairs). Some of the most commonly affected assays were CYPs (CYP2C9 and CYP2C19), transporters (mitochondrial TSPO, norepinephrine, and dopaminergic), and GPCRs (aminergic). Heavy metals, surfactants, and dithiocarbamate fungicides showed promiscuous but distinctly different patterns of activity, whereas many of the pharmaceutical compounds showed promiscuous activity across GPCRs. Literature analysis confirmed >50% of the activities for the most potent chemical–assay pairs (54) but also revealed 10 missed interactions. Twenty-two chemicals with known estrogenic activity were correctly identified for the majority (77%), missing only the weaker interactions. In many cases, novel findings for previously unreported chemical–target combinations clustered with known chemical–target interactions. Results from this large inventory of chemical–biological interactions can inform read-across methods as well as link potential targets to molecular initiating events in adverse outcome pathways for diverse toxicities.
1. Introduction
Evaluating the safety and hazard of chemicals for potential human health and environmental effects is undergoing a major transformation.1 This 21st century toxicology paradigm has emerged from the limitations of the current paradigm in regard to cost, time, and throughput, as well as the development of modern biological tools. These tools can probe chemical–biological interactions at fundamental levels, focusing on the molecular and cellular pathways that are targets of chemical disruption.2 In this manner, we can begin to understand mechanisms of chemical toxicity that may invoke disease or health effect end points. A more mechanistic understanding will help elucidate common pathways of toxicity and susceptibilities underlying human-relevant outcomes.
Toxicity information is limited or absent for tens of thousands of compounds potentially entering the environment.1,3,4 Even for pharmaceuticals designed with a particular biological activity in mind, there is little public information about unexpected toxicities or adverse responses that may be initiated by off-target binding to nuclear receptors, G-protein-coupled receptors, and receptor tyrosine kinases, or by a myriad of events upstream or downstream to receptor engagement.5,6 Evaluating the untested chemicals through the current safety assessment paradigm is limited in throughput, cost, time, and mechanistic revelation. As such, high-throughput screening (HTS) of in vitro chemical–target interactions across chemicals, including pharmaceuticals and chemicals of known and unknown toxicities through a broad range of biochemical assays will help describe the chemical–assay space for which there has been no information to date. Our broader hypothesis is that biochemical HTS, when combined with the diverse assays within the ToxCast portfolio, provides an anchor for predictive signatures and mechanistic pathways leading to toxicity. With further analysis, these types of screens may help identify novel initial molecular events potentially associated with pathways of toxicity7 and inform systems modeling efforts aimed at characterizing adverse outcome pathways.8−16
EPA’s ToxCast project and the federal Tox21 collaboration are generating HTS data and building modeling approaches to identify and characterize biological pathways of toxicity.2,3,17 This approach employs a large, structurally diverse chemical library to probe a wide spectrum of biological targets and cell-based activities, which enables grouping and prioritizing of chemicals based on their in vitro activity profiles, as well as deeper exploration of system biology relationships linking biological activities to in vivo toxicology. Further applications of this approach have the potential to enhance and refine structure, metabolism, or presumed mode of action-based read-across methods,18 as well as to identify potential targets for molecular initiating events in adverse outcome pathways for diverse toxicities. These approaches can be applied to testing prioritization, hazard and safety assessment workflows, design of green alternative chemicals, or screening for adverse effects for drug development processes.
ToxCast Phase I screened 310 unique compounds, mainly food-use pesticides with rich in vivo data profiles in 467 biochemical or cell-based assays from 9 assay technologies.3,8−16 Despite the somewhat limited chemical diversity of this initial test library, pesticidal compounds were found to have sufficiently rich bioactivity profiles, in vitro and in vivo, to yield promising new insights and initial predictive models. To expand on these efforts, ToxCast Phase II has significantly broadened the scope and diversity of the chemical library with 667 newly added chemicals, selected based on factors such as production volume, potential for human exposure, possible hazards to human and ecological populations, and availability of in vivo toxicity data. Some of these chemicals have known biological activities, whereas most have limited toxicity data as compared to Phase I or have not been previously characterized. Specifically, mammalian biological data, including toxicity, are publically unavailable for the majority of these chemicals. The Phase II chemical set includes phthalates, alternative plasticizers, antimicrobials, pesticides, food additives, toxicity reference compounds, and 111 failed drugs donated by six major pharmaceutical companies.
Previously, we reported an analysis of the original ToxCast 310 Phase I chemical set (PhaseI_v1) in a biochemical screen consisting of 292 protein targets (note: ToxCast Phase I originally reported results for 309 unique chemicals. Upon further review, 2 compounds previously labeled as separately sourced replicates were found to be distinct stereoisomers, yielding a revised total of 310 unique chemicals).9 The assays were selected from a commercial NovaScreen panel (NVS) for preclinical drug development. These assays measure chemical binding to nuclear receptors, G-protein-coupled receptors (GPCR), transporters, and ion channels, and enzymatic inhibition or activation for a range of proteins including kinases, phosphatases, CYP450s, proteases, and histone deacetylases. These results are augmented in the present study with newly reported NVS results for the ToxCast Phase II chemical set bringing the chemical inventory to 976 unique chemicals, spanning considerably greater chemical structure diversity and use categories, and with the potential to probe a greater number of biological pathways than Phase I compounds.
This report provides the first critical analysis of findings for 976 chemicals from ToxCast Phase I and II profiled across 331 assays within the NVS panel. The 331 assays evaluated here are currently all of the cell-free assays in ToxCast Phase II. These comprise most of the cell-free assays run in Phase I, with the addition of one new assay (hCAR_Agonist).9 The rationale for assay selection was addressed in the Phase I article, and the reduced number of assays in the present study reflect the elimination of assays shown to have lesser value based on redundancy (i.e., a subset of cytochrome P450s, CYPs) and first-generation predictive models. We focused on relative specificity toward different assay types and their sensitivity to different chemical classes, extending the Phase I study of 310 chemicals. In addition, assays and chemicals were analyzed for similarity based on the chemical–assay activities. Chemical activity similarities were further characterized by enriched chemical structure fragments. Results demonstrate that this large, in vitro chemical–target inventory contains potential novel interactions for many unknown chemical–target combinations, as well as off-site targets for some well-characterized compounds.
2. Experimental Procedures
2.1. Chemical Library
Phases I and II of the ToxCast chemical library considered in this study contain 1020 diverse compound samples, consisting of 976 unique structures, and 44 replicate samples for quality control purposes. These chemicals more specifically correspond to the ToxCast PhaseI_v1 and Phase IIa,b chemical libraries. The rationale for chemical selection was based on several criteria, including chemical nominations within the EPA and other federal agencies (e.g., National Toxicology Program/National Institutes for Environmental Health Sciences; National Center for Advancing Translational Sciences/National Institutes of Health; U.S. Food and Drug Administration); international organizations such as the Organisation for Economic Co-operation and Development; and other stakeholder groups. The chemical use groups for the nominated chemicals include (but are not limited to) marketed drugs with known target activities and over 100 failed (terminated) pharmaceuticals donated by several pharmaceutical companies; phthalates and alternative plasticizers; antimicrobials; pesticides; and food additives. Other selection criteria included DMSO solubility and compound commercial availability and affordability. A tabular listing and Structure Data Format (SDF) file of the complete ToxCast chemical library, of which the 976 compound library is a subset, is available for download at http://www.epa.gov/ncct/dsstox/sdf_toxcst.html.
2.2. Chemical Quality Control (QC)
Chemical information associated with the ToxCast library (i.e., chemical names, CAS RN, and substance description) was quality reviewed and structure-annotated within the U.S. EPA’s DSSTox project (http://www.epa.gov/ncct/dsstox/). Chemical samples were commercially procured, diluted in dimethyl sulfoxide (DMSO) to a stock concentration of 20 mM, and plated by Evotec (formerly BioFocus DPI, South San Francisco, CA). Analytical QC for the Phase I chemical inventory consisted of high-throughput liquid and gas chromatography mass spectrometry determination of sample purity, parent mass, and sample stability in DMSO over time (http://www.epa.gov/ncct/toxcast/chemicals.html). Similar analytical QC methods are being applied to analyzing the Phase II portion of the current ToxCast library in association with the Tox21 project and will be made publically available upon completion. Additional information about these chemicals can be found by querying the DSSTox_GSID, from Supporting Information, Table S2, in the publically available DSSTox Structure Browser (http://epa.gov/dsstox_structurebrowser/), which provides additional structure-based link-outs to information in EPA’s ACToR database (http://actor.epa.gov) and in PubChem (http://pubchem.ncbi.nlm.nih.gov/).
2.3. Assay Description
The NVS assays, developed and run by Caliper, a PerkinElmer company (Hanover, MD), consisted of 331 assays that detect whether a test chemical alters the binding of ligands to receptors (131) or inhibits enzymatic activity (100). The 100 inhibitory enzymatic assays were also assessed for enzymatic activation, resulting in an additional 100 assays. Details on individual assays, catalog numbers, quality assurance methods, and literature references can be obtained at www.caliperls.com/products/contract-research/. Supporting Information, Table S1 contains a list of all assays, their technical specifications, and active links to specific assay protocols. The assay distribution by protein family consists of the binding format: 77 G-protein coupled receptors (GPCR), including 32 aminergic GPCRs; 19 nuclear receptors, 10 from subfamily 1 and 9 from subfamily 3 (steroid); 7 ion channels; 13 ligand-gated ion channels (LGIC), including 9 cys-loop and 4 ionotropic glutamate receptors; 11 transporters; and 3 other binding assays (other); and enzymatic inhibition (and activation) format, 37 kinases; 19 phosphatases; 15 proteases; 3 cholinesterases; 10 CYPs; and 17 other enzymes including histone deacetylases, phosphodiesterases, monoamine oxidases, and cyclooxygenases. Most assays use human (240) or rat (60) targets; the remaining assays use bovine (10), guinea pig (10), sheep (4), mouse (3), rabbit (2), mixed rodent (1 rat/mouse), and boar (1) targets. Assays were assigned to assay categories (Table 1) for further analysis.
Table 1. Biochemical Activity Profiles by Assay Categorya.
| AC50sd |
|||||
|---|---|---|---|---|---|
| assay category | assaysb | activesc | actives %c | ≤10 μM | ≤1 μM |
| activator: cholinesterase | 3 | 1 | 0.03 | 0 | 0 |
| activator: CYP | 10 | 10 | 0.10 | 10 | 7 |
| activator: kinase | 37 | 32 | 0.09 | 16 | 7 |
| activator: other enzyme | 16 | 2 | 0.01 | 1 | 0 |
| activator: phosphatase | 19 | 27 | 0.15 | 9 | 1 |
| activator: protease | 15 | 5 | 0.03 | 2 | 0 |
| cholinesterase | 3 | 151 | 5.16 | 50 | 15 |
| CYP | 10 | 843 | 8.64 | 450 | 129 |
| GPCR (aminergic) | 32 | 1579 | 5.06 | 540 | 148 |
| GPCR (other) | 45 | 1175 | 2.68 | 287 | 55 |
| ion channel | 7 | 226 | 3.31 | 83 | 17 |
| kinase | 37 | 277 | 0.77 | 49 | 10 |
| LGIC (cys loop) | 9 | 109 | 1.24 | 35 | 8 |
| LGIC (ionotropic glutamate) | 4 | 28 | 0.72 | 1 | 0 |
| nuclear receptor (subfamily 1) | 10 | 282 | 2.89 | 90 | 41 |
| nuclear receptor (subfamily 3) | 9 | 393 | 4.47 | 144 | 52 |
| other | 3 | 111 | 3.79 | 36 | 15 |
| other enzyme | 17 | 484 | 2.92 | 105 | 25 |
| phosphatase | 19 | 262 | 1.41 | 69 | 19 |
| protease | 15 | 351 | 2.40 | 81 | 14 |
| transporter | 11 | 787 | 7.33 | 271 | 61 |
| total | 331 | 7135 | 53.19 | 2329 | 624 |
Twenty-one assay categories are defined in the Experimental Procedures section.
Number of assays in each category.
Actives based on the number of AC50 values recorded within an assay category; active fraction (active %) indicates the percentage of theoretical maximum.
Number of actives recorded ≤10 μM and ≤1 μM, respectively.
2.4. Screening Strategy
The screening strategy for Phase II follows the same protocol as that used with the Phase I chemicals.9 For Phase II, all chemicals were initially screened at a single concentration in duplicate (161,700 chemical–assay pairs in duplicate). A single concentration of 10 μM was used for CYP assays and 25 μM for all other assays. Assay–chemical combinations were defined as active in the primary screening if they met a predefined threshold of the mean assay signal differing by at least 30% from the vehicle (DMSO) control signal or the mean assay signal varying by a minimum of 2.0 median absolute deviations from the median (MAD2). From these criteria, 14,961 active chemical–assay combinations were conducted in concentration–response along with 25 inactive chemical–assay combinations (14,986 total chemical–assay pairs ordered, 9% of original single concentration pairs). Each chemical was run in 8 serial dilutions using half-log spacing descending from a top concentration of 50–0.023 μM (or 20 μM–0.009 μM for CYP assays). Consistent with Phase I, lower chemical concentrations were used for CYP assays due to increased sensitivity to inhibition, as expected for enzymes active against xenobiotic chemicals.9
2.5. AC50 Calculation
The raw data (percent activity versus positive control) were received by the EPA and processed within a common custom workflow for all assays within the ToxCast project.19 All Phase I and II data were subjected to custom curve-fitting algorithms for processing and computing AC50 values (chemical concentration at which 50% of maximum activity is achieved). These algorithms were developed in the open source R language,20,21 similar to the analysis used for the Phase I chemicals.9 The code and source data are available from the authors upon request. The R scripts utilized a four parameter Hill function to fit the data and included the following assumptions: all concentration responses were assumed to be monotonic; outliers were discounted when there was a monotonic curve; a variable slope was allowed; and negative inhibition (activation) was allowed for enzymatic assays. The asymptotes of the curve were constrained to be between −10 and 10% activity (lower) and 100 and 110% activity (upper), to allow for consistent extrapolation of the AC50 across assay–chemical combinations. AC50 extrapolations were allowed at concentrations 3-fold lower than the lowest concentration tested and 3-fold higher than the highest concentration tested to capture potential activity outside of the tested concentrations. Additional criteria for generating AC50 values included Hill curves r2 coefficient of determination values ≥0.6, p value ≤0.01 (from a test of significant difference between the top and bottom of the curve fit), and Emax (maximum activity) ≥ 30% baseline activity. To capture activity when a dose–response curve was nonmonotonic or could not be calculated from these criteria, a special case AC50 was assigned to the first concentration when a chemical inhibited an assay by at least 50% at any two consecutive concentrations. Specifically, there were 189 chemical–assay pair special cases, which can be seen from the curve fits provided in the Supporting Information. When chemical–assay data did not meet these requirements, a dose–response curve was not generated, and the chemical was deemed inactive in the assay. In cases where a chemical was tested in both Phases I and II, data were not combined to calculate AC50s due to potential variation during temporally separate experimentation; thus, the lowest AC50 value was taken. AC50 values were transformed to −log(AC50/1000000) to reflect potency. Inactive chemical–assay pairs were set at 1000000 μM (1M) before transformation to ensure a potency of zero.
2.6. Clustering
Transformed data (i.e., −log(AC50/1000000) were clustered with Partek Discovery Suite 6.6 software (Partek, Inc., St. Louis, MO). Chemical–assay unsupervised hierarchical clustering was performed using Euclidean distance as the similarity metric and Ward’s method for assembling clusters. Assay–assay and chemical–chemical similarity matrices were built using Pearson’s correlation as the similarity metric and Ward’s method for assembling clusters.
2.7. Enrichment Score
In order to identify the assays affected by chemicals clustered together in the chemical–chemical similarity matrix, we developed an assay category enrichment score (ES) for each chemical. The ES scores were calculated for each chemical over the 21 assay categories, showing which categories the chemical is preferentially affecting. Initial ESc, ac = avg(AC50)c, ac/avg (AC50)c, ac_rest, where c is a given chemical [1,967], ac is a given assay category [1,21], and ac_rest is all other assay categories not associated with the given ac. For example, if the ac was GPCR_aminergic, then ac_rest would be all assays except GPCR_aminergic and GPCR_other. Each ESc,ac within an ac is normalized by the maximum ES, i.e., final ESc, ac = initial ESc, ac /maximum(ES)C, ac, where C is all chemicals.
2.8. Chemical Structure Enrichment
Univariate analyses between chemical–assay categories and chemical structure fragments were used to determine chemical structure fragment assay category associations. Chemical–assay category ES scores were calculated as in 2.7 (above), but using only AC50s ≤10 μM (i.e., calculated AC50s >10 μM were assigned an AC50 of zero) to reduce potential nonspecific interactions. Chemical structure fingerprints were determined for all chemicals. A fingerprint is a bit string indicating whether a chemical structure contains a particular chemical fragment. The fragment libraries used are the FP3, FP4, and MACCS fingerprint sets available with Open Babel,22 PaDEL,23 and PubChem (http://pubchem.ncbi.nlm.nih.gov) fingerprint sets. All fragments are described as SMARTS strings. Fingerprinting was carried out using Open Babel and comparing the fragment SMARTS strings to the chemical SMILES strings. In total, there were 6655 fragments used, but only 2950 fragments were found in at least one chemical structure, and 1907 fragments were found in 5 or more structures. For each fragment–assay pair, a 2 × 2 table was built (have fragment, yes or no vs active in assay, yes or no), and standard statistical metrics were calculated. All univariate associations were used where there were at least 5 true positives (cases where a chemical had a fragment and was active in an assay), and the positive predictive value (PPV) was greater than 0.5. These associations were plotted in Cytoscape: An Open Source Platform for Complex Network Analysis and Visualization (www.cytoscape.org).24 Chemical structures were visualized through MarvinSketch viewer (ChemAxon Ltd.) using the U.S. EPA ACToR Web site (http://actor.epa.gov).
3. Results
3.1. Data Overview
Since data analyzed in the present study are a compilation of two temporally distinct phases of testing, Phase I and Phase II, assay performance was evaluated using chemical replicates between phases and within phases, on AC50s up to the highest dose tested. For the 11 chemicals replicated across Phases I and II, there was a 95% concordance over 3045 chemical–assay pairs (2907 concordant, including 100 positives and 2807 negatives). Nine chemicals replicated within Phase II showed 93% concordance over 2079 chemical–assay pairs (1941 concordant, including 27 positives and 1914 negatives). Discrepancies of AC50 activities above 10 μM accounted for 74% of the discordances. All subsequent results described herein aggregate results for the 44 total replicates of 11 Phase I compounds and thus consider the set of 976 unique chemicals among the 1020 chemical sample inventory.
Results are summarized in Table 1 for 976 unique chemical actives tested against the 331 biochemical assay portfolio. Active chemical–assay pairs refer to a recorded AC50 value listed in Supporting Information, Table S2. Overall, 729 of 976 chemicals (75%) were active in at least one assay, and 256 of 331 assays (77%) had at least one active chemical. This yielded 7,135 active chemical–assay pairs out of a maximum of 323,056 possible. The assay categories most affected include GPCR (aminergic), GPCR (other), and CYP categories, with 1579, 1175, and 843 actives, respectively. Limiting the AC50s to a threshold of 10 μM or 1 μM gave 2329 and 624 chemical–assay actives, respectively. The GPCR (aminergic) category had the greatest number of actives at ≤1 μM (148). A chemical affected 10 assays on average, ranging from 0 (274 chemicals) to 90 (1 chemical). An assay was affected by 28 chemicals on average, ranging from 0 (75 assays) to 264 (1 assay). There were 75 assays not affected by any chemical tested, of which 70 are activator assays measuring increased assay activity.
Chemicals and assays were assessed for their promiscuity and potency (micromolar AC50 value). The 20 most promiscuous chemicals are listed in Table 2. They represented a broad range of chemical categories, including heavy metal chemicals (phenylmercuric acetate, mercuric chloride, tributyltin methacrylate, and tributyltin chloride), surfactants (sodium dodecylbenzenesulfonate, perfluorooctane sulfonic acid, dodecylbenzene sulfonate triethanolamine (1:1)), dithiocarbamate fungicides (mancozeb, maneb, and metiram zinc), and a variety of pharmaceuticals. The 20 most promiscuous assays are listed in Table 3. They represent a broad range of assay categories, including transporters (translocator protein (PBR/TSPO), dopamine, norepinephrine), CYPs (CYP1A2, CYP2B6, CYP2C19, CYP2C9), GPCR (aminergic, 5HT7 and DRD1), GPCR (other, opiate κ and opiate μ), nuclear receptor (subfamily 3, AR and GR), nuclear receptor (subfamily 1, pregnane X receptor (PXR)), ion channel (NaCh), other enzyme (MAOAC), protease (BACE), and other (SIGMA receptor).
Table 2. Top 20 Most Promiscuous Chemicalsa.
| AC50s |
||||
|---|---|---|---|---|
| chemical name | total | ≤10 μM | ≤1 μM | |
| 1 | phenylmercuric acetate | 90 | 47 | 20 |
| 2 | mancozeb | 88 | 41 | 13 |
| 3 | gentian violet | 86 | 51 | 5 |
| 4 | sodium dodecylbenzenesulfonate | 82 | 19 | 0 |
| 5 | tributyltin methacrylate | 79 | 48 | 12 |
| 6 | tributyltin chloride | 77 | 45 | 9 |
| 7 | mercuric chloride | 73 | 45 | 14 |
| 8 | perfluorooctane sulfonic acid | 72 | 13 | 2 |
| 9 | {4-[3-(aminomethyl)phenyl]piperidin-1-yl}{5-[(2-fluorophenyl)ethynyl]furan-2-yl}methanone (pharma) | 71 | 25 | 4 |
| 10 | dodecylbenzene sulfonate triethanolamine (1:1) | 66 | 7 | 1 |
| 11 | SSR241586 (pharma) | 66 | 30 | 8 |
| 12 | emamectin benzoate | 65 | 14 | 2 |
| 13 | {4-[5-(aminomethyl)-2-fluorophenyl]piperidin-1-yl}(4-bromo-3-methyl-5-propoxythiophen-2-yl)methanone hydrochloride (pharma) | 64 | 19 | 2 |
| 14 | (1R)-1-[(ethoxycarbonyl)oxy]ethyl 1-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methyl}-2-{[1-(propan-2-yl)piperidin-4-yl]carbamoyl}-1H-indole-5-carboxylate hydrochloride(pharma) | 63 | 29 | 2 |
| 15 | maneb | 62 | 31 | 16 |
| 16 | SSR150106 (pharma) | 62 | 41 | 13 |
| 17 | didecyl dimethyl ammonium chloride | 62 | 30 | 2 |
| 18 | zamifenacin (pharma) | 60 | 27 | 11 |
| 19 | SSR125047 (pharma) | 59 | 16 | 3 |
| 20 | metiram | 56 | 16 | 4 |
Columns indicate the top 20 most promiscuous chemicals, the total number of calculated AC50s, and the number of AC50s at ≤10 μM and ≤1 μM, respectively. The number of AC50s corresponds to the number of assays affected by a given chemical at the AC50 value constraint.
Table 3. Top 20 Most Promiscuous Assaysa.
| AC50s |
||||
|---|---|---|---|---|
| assay target | assay category | total | ≤10 μM | ≤1 μM |
| hCYP2C19 | CYP | 264 | 144 | 53 |
| hCYP2C9 | CYP | 152 | 81 | 19 |
| rPBR | transporter | 147 | 62 | 18 |
| hPXR | nuclear receptor (subfamily 1) | 140 | 73 | 35 |
| hNET | transporter | 136 | 48 | 13 |
| hPBR | transporter | 117 | 36 | 5 |
| hDAT | transporter | 117 | 45 | 7 |
| hCYP1A2 | CYP | 108 | 60 | 16 |
| gDAT | transporter | 98 | 26 | 4 |
| h5HT7 | GPCR (aminergic) | 96 | 35 | 13 |
| hGR | nuclear receptor (subfamily 3) | 96 | 35 | 6 |
| hOpiate_mu | GPCR (other) | 92 | 27 | 5 |
| hDRD1 | GPCR (aminergic) | 89 | 36 | 9 |
| rNaCh_site2 | ion channel | 87 | 37 | 13 |
| hCYP2B6 | CYP | 81 | 43 | 16 |
| gSIGMA_NonSelective | other | 80 | 31 | 13 |
| gOpiateK | GPCR (other) | 75 | 18 | 4 |
| rMAOAC | other enzyme | 73 | 15 | 6 |
| hAR | nuclear receptor (subfamily 3) | 73 | 33 | 8 |
| hBACE | protease | 73 | 28 | 3 |
Columns indicate the topmost promiscuous assay targets (corresponding to one assay), the respective assay category, the total number of calculated AC50s, and the number of AC50s affecting the assay at ≤10 μM and ≤1 μM. The number of AC50s corresponds to the number of chemicals affecting a given assay at the AC50 value constraint.
Fifty-four chemicals had AC50s at the lowest concentration tested for one or more assays (LCT ∼0.009 μM for CYPs and 0.023 μM for non-CYPs), indicating high potency for these chemical–assay pairs. For many of these chemicals, high potency was observed for two or more assays of the same or related category (e.g., CP-471358 and CP-544439 inhibition of matrix metalloproteinases MMP2, MMP9, and MMP13; chlorpromazine inhibition of GPCR ligand binding activity for DRD1, DRD2s, 5HT2C, Adra1A, Adr1B, and H1 and inhibition of NET transporter binding activity) (Table 4). More than half of the compounds listed in Table 4 have literature references to substantiate these observed chemical–target interactions. Results from the data overview indicate that both broad and specific biochemical activities are detected across 323,056 possible chemical–assay pairs in ToxCast Phases I and II.
Table 4. Potent Chemical–Assay Pairs with AC50s at the Lowest Dose Testeda.
| AC50s |
||||
|---|---|---|---|---|
| chemical name | total | LCTb | assay target(s) | refs |
| chlorpromazine hydrochloride | 55 | 7 | TR_hNET, GPCR_hDRD1/2s, p5HT2C, hH1, rAdra1A/1B | (55−58) |
| haloperidol | 42 | 6 | OR_gSIGMA_NonSelective, GPCR_hDRD1/2s/4.4, GPCR_rAdra1_NonSelective, GPCR_bDR_NonSelective | (59−61) |
| trelanserin (SL650472 pharma) | 24 | 6 | GPCR_h5HT2A/7, p5HT2C, r5HT_NonSelective, GPCR_hDRD1/4.4 | (62) |
| 17β-estradiol | 9 | 5 | NR_hAR, bPR, mERa, hER, bER | (63,64) |
| 17α-ethinylestradiol | 24 | 4 | NR_hAR, mERa, hER, bER | (65) |
| CP-471358 pharma | 6 | 3 | ENZ_hMMP13/2/9 | (66) |
| CP-544439 pharma | 10 | 3 | ENZ_hMMP13/2/9 | (67) |
| diethylstilbestrol | 31 | 3 | NR_mERa, hER, bER | (68) |
| 2,2-bis(4-hydroxyphenyl)-1,1,1-trichloroethane | 36 | 3 | NR_mERa, bER, hCAR_Antagonist | (69) |
| zamifenacin (pharma) | 60 | 3 | GPCR_gMPeripheral_NonSelective, hM3/5 | (70,71) |
| flufenacet | 10 | 2 | MP_rPBR, NR_hPXR | |
| maneb | 62 | 2 | ENZ_hPTPN9/4 | |
| methadone hydrochloride | 37 | 2 | GPCR_rOpiate_NonSelective/Na | (72) |
| progesterone | 11 | 2 | NR_hAR, NR_bPR | (73) |
| SR144190 pharma | 24 | 2 | GPCR_hNK2, NR_hPXR | (74) |
| tetraconazole | 24 | 1 | ADME_hCYP2C19 | |
| diniconazole | 5 | 1 | ADME_hCYP2C19 | |
| flufenpyr-ethyl | 2 | 1 | ADME_hCYP4F12_Activator | |
| tannic acid | 26 | 1 | ENZ_hGSK3b | (75) |
| chlorpyrifos oxon | 14 | 1 | ENZ_hES | (76) |
| mancozeb | 88 | 1 | ENZ_hDUSP3 | |
| CP-105696 pharma | 5 | 1 | GPCR_gLTB4 | (77) |
| diphenhydramine hydrochloride | 41 | 1 | GPCR_hH1 | (78) |
| elzasonan (CP-448187 pharma) | 5 | 1 | GPCR_hAdrb2 | |
| emamectin benzoate | 65 | 1 | GPCR_hNTS | |
| disodium (5S,6R,7R)-5-(1,3-benzodioxol-5-yl)-2-butyl-7-{2-[(2S)-2-carboxylatopropyl]-4-methoxyphenyl}-6,7-dihydro-5H-cyclopenta[b]pyridine-6-carboxylate (pharma) | 7 | 1 | GPCR_hETA | |
| PHA-00543613 pharma | 12 | 1 | LGIC_rNNR_BungSens | (79) |
| CP-457920 pharma | 2 | 1 | LGIC_bGABARa5 | (80) |
| 2,6-difluoro-5-[3-(2-hydroxypropan-2-yl)imidazo[1,2-b][1,2,4]triazin-7-yl]biphenyl-2-carbonitrile (pharma) | 4 | 1 | LGIC_bGABARa5 | (81) |
| sodium 3-(4-{[1-(3-ethoxyphenyl)-2-(4-methylphenyl)-1H-imidazol-4-yl]carbonyl}piperazin-1-yl)naphthalene-1-carboxylate (pharma) | 31 | 1 | GPCR_mCCKAPeripheral | (82) |
| CP-122721 pharma | 44 | 1 | GPCR_rNK1 | (83) |
| SAR102779 pharma | 43 | 1 | GPCR_hNK2 | (84) |
| SSR241586 pharma | 66 | 1 | GPCR_rNK3 | (85) |
| SAR150640 pharma | 53 | 1 | GPCR_hDRD2s | (86) |
| SSR103800 pharma | 33 | 1 | GPCR_gOpiateK | |
| enadoline (pharma) | 6 | 1 | GPCR_gOpiateK | (87) |
| SSR125047 pharma | 59 | 1 | OR_gSIGMA_NonSelective | (88) |
| spiroxamine | 18 | 1 | OR_gSIGMA_NonSelective | |
| 4-(3-{[4-(2-methyl-1H-imidazol-1-yl)phenyl]sulfanyl}phenyl)tetrahydro-2H-pyran-4-carboxamide methanesulfonate (pharma) | 19 | 1 | TR_gDAT | |
| 1-[2-(3,4-dichlorophenoxy)-5-fluorophenyl]-N-methylmethanamine (pharma) | 11 | 1 | TR_rSERT | (89) |
| UK-416244 pharma | 43 | 1 | TR_rSERT | (90) |
| 6-{2-[4-(1,2-benzothiazol-3-yl)piperazin-1-yl]ethyl}-4,4,8-trimethyl-3,4-dihydroquinolin-2(1H)-one methanesulfonate (pharma) | 9 | 1 | GPCR_p5HT2C | |
| SB243213A pharma | 8 | 1 | GPCR_p5HT2C | (91) |
| volinanserin (pharma) | 41 | 1 | GPCR_h5HT2A | (92) |
| bisphenol B | 31 | 1 | NR_mERa | (93) |
| 5,5-diphenylhydantoin | 2 | 1 | NR_hPXR | (94) |
| fenamiphos | 5 | 1 | NR_hPXR | |
| imazamox | 2 | 1 | NR_hPXR | |
| napropamide | 14 | 1 | NR_hPXR | |
| pyraclostrobin | 9 | 1 | NR_hPXR | |
| sodium hexyldecyl sulfate | 29 | 1 | NR_hPXR | |
| sorbitan | 7 | 1 | NR_hPXR | |
| PK11195 | 21 | 1 | MP_hPBR | (95) |
| zoxamide | 2 | 1 | MP_rPBR | |
Columns indicate chemicals with AC50s at the lowest dose tested, the total number of calculated AC50s for the given chemical, the number of AC50s at the lowest dose (i.e., number of assays affected at the lowest dose), the assay target(s) with AC50 values at the lowest dose tested, and the literature evidence of the chemical–target interaction, if applicable.
LCT: lowest concentration tested.
The literature evidence also revealed missed chemical–target interactions for 10 of the 54 chemicals. Missed known chemical–target interactions included (1) chemicals that missed one assay within the same category (haloperidol affected all 5HT receptors, except h5HT2A; zamifenacin affected all muscarinic receptors, except hM1); (2) assays with species orthologues which were not affected (CP-105696 inhibited the guinea pig but missed the human leukotriene assay, hLTB4_BLT1; PHA-00554613 inhibited the rat but missed the human neonicotinoid receptor, hNNR_NBungSens assay; and UK-416244 inhibited the rat but missed the human serotonergic transporter, hSERT); (3) specific chemical inhibitors which missed the nonspecific assays (CP-457920 and 2,6-difluoro-5-[3-(2-hydroxypropan-2-yl)imidazo[1,2-b][1,2,4]triazin-7-yl]biphenyl-2-carbonitrile inhibited bGABARa5 but not bGABAR_Agonist or rGABAR_NonSelective); (4) chemicals that inhibited one or no assays in a category (elzasonan a 5HT1b antagonist did not inhibit any 5HT assays; emamectin benzoate is known to bind to mammalian GABA receptors, albeit at a lower affinity than in insects, but it only inhibited 1/5 GABA assays; 5,5-diphenylhydantoin is a substrate and inhibitor for many CYPs, although it did not inhibit any CYPs in this screen).
Evaluating this list for false negatives (missing chemical–target interactions) is limited by publically available information. To evaluate false negatives and true positives (where AC50s were calculated for known chemical–target interactions) in a systematic way, we chose well-known chemical–target interactions in a specific set of assays. The EPA’s Endocrine Disruptor Screening Program (EDSP) has an estrogen receptor reference chemical list of known estrogenic compounds of varying potency and nonestrogenic compounds with categories of potency defined as follows: strong actives exhibit an effective concentration at 50% maximal activity (EC50) of <10–11 M; moderate 10–9 to 10–8 M; weak 10–7 to 10–6 M, and very weak >10–5 M (http://www.regulations.gov/#!documentDetail;D=EPA-HQ-OPP-2012-0818-0017). Twenty-two chemicals in common were evaluated in the bER, hER, and mERa assays. We found that all inactives (6), strong actives (3), and antagonists (1) were correctly identified. None of the very weak (3) chemicals were correctly identified, but a majority of the weak positives (7 of 9) were in at least two of the assays. Together, these findings indicate that these data are robust with respect to correctly identifying activity within an assay category for weak to strong inhibitors (77%, 17 of 22) correct using two assays); however, limitations are also realized with respect to potentially identifying very weak inhibitors.
3.2. Hierarchical Clustering
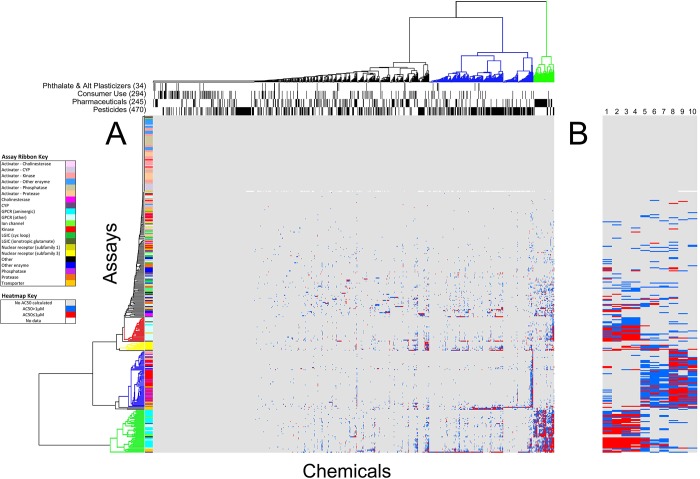
To determine how the chemical–assay space was structured, we performed hierarchical clustering based on chemical–assay potency (Figure 1). Clustering by potency ordered the chemical–assay space into sectors ranging from no activity to high activity. For some clusters, it was possible to annotate by assay category or chemical class (Figure 1A). Figure 1B shows the clustering for 10 of the most promiscuous chemicals including heavy metal chemicals, surfactants, and dithiocarbamate fungicides. Distinct effects can be seen among these chemical classes, indicating some degree of specificity. Four functional categories listed across the top of the hierarchical cluster (pesticides, pharmaceuticals, consumer use chemicals, and phthalates and alternative plasticizers) give a diverse representation of the chemicals evaluated in this data set (Figure 1A). These data show varying degrees of sensitivity and specificity as will be discussed in section 3.3.
Figure 1.
Hierarchical clustering of 976 chemicals by 331 biochemical assays. Hierarchical clustering was based on chemical–assay potency as measured by the inverse log of AC50s (in micromolar) divided by 1,000,000 (−log(AC50/1000000)). Actives with AC50 values ≤1 μM are colored red and others blue. Gray indicates inactive and white not tested in Phase I (hCAR_Agonist assay). (A) Left ribbon indicates clusters of 21 assay categories (see Assay Ribbon Key). Left colored tree structure indicates relatively homogeneous assay category clustering including GPCR (other), red; nuclear receptor (subfamily 3), yellow; kinases and phosphatases, blue; GPCR (aminergic), green. Top bars indicate clusters of 4 chemical use groups. Top colored tree structure indicates highly promiscuous, mostly pharmaceuticals (green) and promiscuous, mostly pesticides (blue). (B) Details for 10 of the most promiscuous chemicals listed in Table 2. Heavy metals (1-phenylmercuric acetate, 2-mercuric chloride, 3-tributyltin methacrylate, 4-tributyltin chloride); surfactants (5-sodium dodecylbenzenesulfonate, 6-perfluorooctane sulfonic acid, and 7-dodecylbenzene sulfonate triethanolamine (1:1)); and dithiocarbamate fungicides (8-mancozeb, 9-maneb, and 10-metiram).
Prominent assay clusters containing relatively homogeneous assay categories included GPCR (other), nuclear receptor (subfamily 3), kinase and phosphatase, and GPCR (aminergic) (Figure 1A). The clustering was clearly following at least some of the biology of the constituent chemical–assay pairs. For example, the GPCR aminergic cluster (green) included several adrenergic GPCRs (rAdra1A/B, rAdra1/2_NonSelective, hAdrb1/2, hAdra2A/C, and rmAdra2B) and corresponding transporters (hNET) and also included several dopaminergic GPCRs (hDRD1, hDRD2s, hDRD4.4, and bDR_NonSelective) and their corresponding transporters (hDAT and gDAT). This implies a degree of consistency and specificity to the chemical biology of the perturbation. The kinase and phosphatase cluster (blue) was highly enriched with these assays and also included a phosphodiesterase (PDE4A1) and class-III HDAC (SIRT2). The largest nuclear receptor cluster (yellow) contained 8 assays (386 chemical–assay actives) all within subfamily 3, the steroid receptors (GR, AR, PR, and ER). Many of these assays were also among the most promiscuous (see Table 3).
3.3. Chemical Class Clusters
3.3.1. Pesticides
Pesticide active compounds, a main component of the ToxCast Phase I testing due to the availability of in vivo data, are designed to be biologically active in target species (i.e., insects, weeds, and fungi), while producing minimal effects on human biology. A diverse set of 470 pesticides in the combined Phase I and Phase II library considered here affected a large number of assays (236, 71%). Roughly 20% (105) affected no assays, whereas 41% (195) were included among the most promiscuous clusters (indicated by the top green and blue colored tree structures (Figure 1A) containing a total of 315 chemicals). The top active assays affected by pesticides included human CYP2C19, CYP2C9, CYP1A2, and CYP2B6 enzymatic inhibition assays (165, 79, 74, and 55 chemicals, respectively), human PXR binding assay (116), and rat and human TSPO binding assays (104 and 74). Examples of chemical–assay actives are highlighted for their known chemical–target interactions. Carbofuran, an insecticide known to inhibit cholinesterase, inhibited only 3 assays: butyrylcholinesterase and, at submicromolar AC50 concentrations, the human and rat acetylcholinesterase enzymatic assays. Picoxystrobin, a strobilurin fungicide that inhibits fungal respiration, affected only 4 assays, including the human CYP2C9 and CYP2C19 enzyme assays, as well as the human TSPO and, at submicromolar AC50 concentrations, the rat TSPO enzymatic assay. Two other members of the strobilurin class of fungicides, described in the previous Phase I analysis,9 also affected a small number of assays (3–9), including TSPO. Hence, for a number of examples, the activity displayed in this biochemical platform for selected pesticides reflected bioactivities associated with their known pesticidal modes of action and/or potentially related targets.
3.3.2. Pharmaceuticals
Pharmaceuticals (245), including the 111 donated compounds, are intended for biologic activity like pesticides but toward mammalian biology. The pharmaceuticals are also more likely to have a defined target. There were 36 pharmaceuticals that did not produce an AC50 for any of the target assays tested here; however, 37 pharmaceuticals clustered tightly by inhibition of GPCR binding activities (Figure 1A).
Several examples could be found to indicate chemical–assay actives for known chemical–target interactions. For example, volinanserin, a serotonin receptor antagonist,25 inhibited 41 different assays, the majority of which were GPCRs and transporters (78%), including the serotonergic system (affecting 9 out of 11 total serotonergic assays) (see Supporting Information, Table S2). Several examples could be found to indicate chemical–assay actives for unknown chemical–target interactions. For example, anthralin is a topical antiproliferative, anti-inflammatory agent used to treat psoriasis;26 however, the mechanisms of action are unknown. Anthralin inhibited 22 assays, almost exclusively enzymes, including caspases 1–5 and 8 (where the inflammatory caspase 5 activity was inhibited at submicromolar AC50 concentrations), MMPs 1/2/3/7/13, and cyclooxygenases PTGS1/COX1 and PTGS2/COX2 (see Supporting Information, Table S2). These data indicate that the biochemical activity profiles of pharmaceuticals provide information on potential novel effects in addition to their known activities, giving possible leads for further targeted studies.
3.3.3. Consumer Use Chemicals
Consumer use chemicals tested here (171), exclusive of pesticides and pharmaceuticals that were described above, showed spotted activity based on the number of targets affected across the assay space (42% of this chemical class) or, in some cases, were highly promiscuous (9% of this chemical class) (Figure 1A). Example chemicals in this class include food additives and ingredients in sunscreens, cosmetics, soaps, and shampoos. Of the chemicals showing activity (100), the median number of assays affected per chemical was 2, with a range of 1 to 72 assays. Consider caffeine as an example. Caffeine is a known adenosine receptor antagonist. Caffeine and the structurally related methylxanthine, theophylline, inhibited all three adenosine receptor binding assays (hAdo2a, hAdo1, and nonselective bAdo receptor). Interestingly, adenosine receptor binding activities were also inhibited by the 2′-deoxyadenosine analogue, cladribine, an effect recently reported.27 The 13 perfluorinated compounds, which are industrial and consumer use products (e.g., surfactants and nonstick applications), showed a wide range of chemical-specific activity (median 6 assays). For example, 1,1,2,2-tetrahydroperfluoro-1-decanol inhibited none of the assays, whereas perfluorooctane sulfonic acid (PFOS) inhibited 72 assays. Across all perfluorinated compounds, the most commonly affected assay (inhibited by 10 of 13 chemicals) was the leukotriene B4 (GPCR_gLTB4). Other leukotriene assays (GPCR_hLTB4_BLT1, gLTD4) were inhibited by 30% (4) of the perfluorinated compounds. Although these data indicate that most chemicals labeled for consumer use in the ToxCast library, excluding pesticides, were generally inactive in the biochemical HTS platform, there are case examples where some broad use categories are active against particular assay categories.
3.3.4. Phthalate Plasticizers and Alternatives
There is evidence that phthalate plasticizers are endocrine active.28 Alternatives to phthalate plasticizers have been proposed to eliminate this undesired effect;29 however, many alternatives have not undergone rigorous testing.30 Thirty-four plasticizers, including 7 phthalates and 27 potential alternatives, were tested here. Overall, 12 plasticizers were inactive across all 331 assays (see Figure 1A). These included 1 phthalate and 11 nonphthalate alternatives. Of the remaining 22 compounds, 15 were active in 1 to 2 assays (3 phthalates and 12 nonphthalates), and 7 were active in 3 to 6 assays (3 phthalates and 4 nonphthalates). The most frequent assay targets for these compounds were CYPs (CYP2C19 and CYP1A2) and TSPO (human and rat). Only two plasticizers inhibited any nuclear receptor binding and only at higher concentrations (DEHP inhibited hGR at an AC50 = 95 μM, and diethyl succinate inhibited rMR at an AC50 = 49 μM; see Supporting Information, Table S2). Limited phthalate and estrogen receptor binding is not surprising, given that half-maximal inhibitory concentrations for binding have been reported to be 1000 μM.31 No statistical difference (p > 0.05 from t test) between the responses of the phthalate and nonphthalate plasticizers to these particular assays could be concluded.
3.4. Assay and Chemical Similarity Matrices
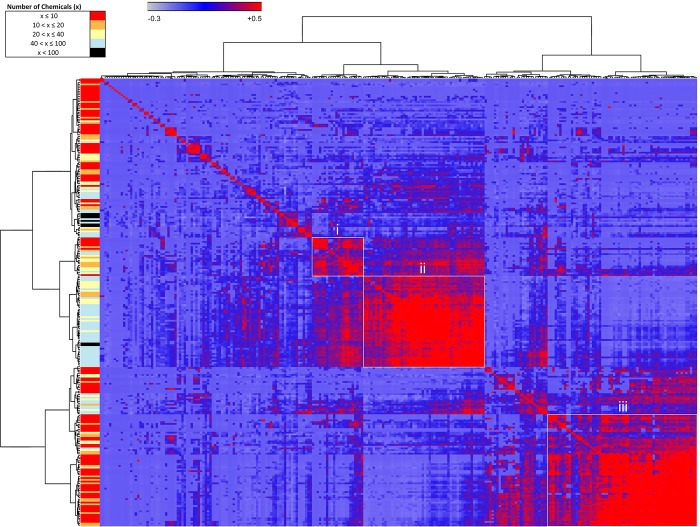
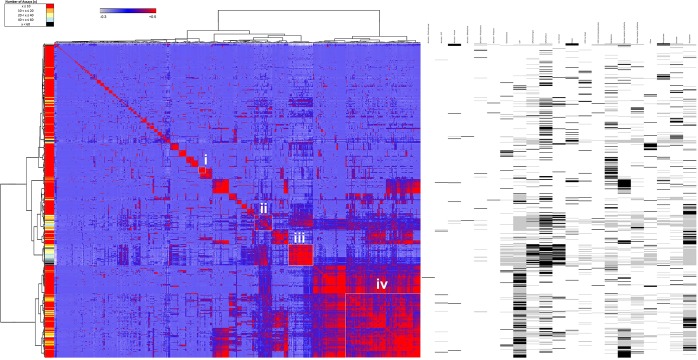
To evaluate the similarities among assays (Figure 2) and chemicals (Figure 3), we performed Pearson’s correlation based on chemical–assay activities. Assays or chemicals that are most associated together cluster as blocks along the diagonal. Nonoverlapping clusters of assays (Figure 2) or chemicals (Figure 3) with positive Pearson’s correlations are shown as examples. The left-hand ribbon indicates the number of chemicals active in an assay (Figure 2) or assays active for a given chemical (Figure 3). A caveat of this correlation model is that the size of a cluster may be driven by one to two assays in the chemical similarity matrix or one or two chemicals in the assay similarity matrix.
Figure 2.
Assay–assay similarity matrix clustered based on chemical–assay profiles. Pearson correlations (−0.3 to 0.5) indicated the strength of associations, which were visualized in the heatmap from blue to red, respectively. Similar assays clustered along the diagonal have high associations. Selected clusters (i, ii, and iii) are shown as examples and listed in Table 5. The left color bar indicates the number of chemicals affecting a given assay.
Figure 3.
Chemical–chemical similarity matrix clustered based on chemical–assay profiles. Pearson correlations (−0.3 to 0.5) indicated the strength of associations, which were visualized in the heatmap from blue to red, respectively. Similar chemicals clustered along the diagonal have high associations. Selected clusters (i, ii, iii, and iv) are shown as examples and listed in Table 6. The left color bar indicates the number of assays affected by a given chemical. The right panel indicates the assay enrichment scores (defined in the Experimental Procedures section) for each chemical and can be used to describe the chemicals in the clusters.
Table 5. Known Estrogenic and Nonestrogenic Compoundsa.
| AC50s
(μM) |
||||
|---|---|---|---|---|
| relative potency | chemical name | bER | hER | mERa |
| inactive | atrazine | 0 | 0 | 0 |
| inactive | linuron | 0 | 0 | 0 |
| inactive | haloperidol | 0 | 0 | 0 |
| inactive | phenobarbital sodium salt | 0 | 0 | 0 |
| inactive | progesterone | 0 | 0 | 0 |
| inactive | ketoconazole | 0 | 0 | 0 |
| inactive summary | 100% | 100% | 100% | |
| very weak | ethylparaben | 0 | 0 | 0 |
| very weak | methoxychlor | 0 | 0 | 0 |
| very weak | butyl benzyl phthalate | 0 | 0 | 0 |
| very weak summary | 0% | 0% | 0% | |
| weak | 4-(1,1,3,3-tetramethylbutyl)phenol | 33.000 | 7.200 | 8.200 |
| weak | kepone | 0 | 0 | 0 |
| weak | genistein | 0.130 | 0.032 | 0.130 |
| weak | 4-cumylphenol | 16.000 | 0 | 12.000 |
| weak | bisphenol B | 0.430 | 0.300 | 0.023 |
| weak | o,p-DDT | 0 | 0 | 0 |
| weak | bisphenol A | 0.630 | 0.820 | 1.100 |
| weak | 4-nonylphenol, branched | 33.000 | 20.000 | 5.600 |
| weak | butylparaben | 56.999 | 17.000 | 23.000 |
| weak summary | 78% | 67% | 78% | |
| strong | 17β-estradiol | 0.023 | 0.023 | 0.023 |
| strong | diethylstilbestrol | 0.023 | 0.023 | 0.023 |
| strong | 17α-ethinylestradiol | 0.023 | 0.023 | 0.023 |
| strong summary | 100% | 100% | 100% | |
| antagonist | tamoxifen | 0.100 | 0.330 | 0.200 |
| antagonist summary | 100% | 100% | 100% | |
The chemicals are listed along with their relative potency for the estrogen receptor (as noted within the EPA’s Endocrine Disruptor Screening Program) and calculated AC50 values for the three estrogen assays. The summary for each potency category indicates the percent of chemical–AC50 combinations that were correctly identified in the HTS.
3.4.1. Evaluation of Assay Similarities
Assay–assay similarity was performed over the 256 active assays (Figure 2). The basis for highly similar clusters (Pearson’s correlation ≥0.65) in some cases was associated with a specific response to relatively few chemicals, such as the case with the human caspases 1–3 (active for 5–9 chemicals) and histone deacetylases 3 and 6 (active for 5–8 chemicals). In other cases, high-similarity clustering was driven by a large number of chemicals causing activity in related assays, such as that seen with TSPOs (inhibited by 117–147 chemicals), acetylcholinesterases (41–46 chemicals), and estrogen receptors (18–30 chemicals). Three other clusters are highlighted in Figure 2 and Table 6 to demonstrate the composition of a cluster that aggregated due to chemical–assay response similarity. Cluster i holds 22 assays perturbed by 2 to 64 chemicals. These were mainly GPCR_other assays (18). Two subclusters (nonoverlapping clusters with Pearson’s correlation ≥0.65) within Cluster i each contain peptide GPCRs, including glutamate, vasoactive intestinal peptide, endothelin, and angiotensin II GPCRs, as well as the neuropeptide Y GPCRs. Cluster ii involves 52 assays active for 16 to 136 chemicals. The assays were mainly within the GPCR_aminergic (32), GPCR_other (7), and transporter (6) categories. One subcluster contained rhodopsin-like biogenic amine GPCRs, including histamine, muscarinic acetylcholine, adrenergic, serotonergic, and dopaminergic GPCRs, another containing three opiate GPCRs, and the third containing two ion channels. Cluster iii involves 41 assays active for 2 to 41 chemicals. The assays represented were mainly within the kinase (21) and phosphatase (14) categories. Both subclusters included relatively equal numbers of kinases and class I cysteine-based protein tyrosine phosphatases; however, the kinases were distinct for each subcluster with tyrosine and CAM kinases in one and mainly tyrosine kinases in the other. The assay similarities revealed clusters of targets known to be similar (e.g., GPCRs), as well as varied or dissimilar targets. These findings demonstrate a biological basis to clustering of chemicals in the assay similarity matrix.
Table 6. Assay–Assay Similaritiesa.
| clustersb | assay categoriesc | assay targets |
|---|---|---|
| Cluster i (22) | ||
| subcluster1 | GPCR (other) (4) | rmMGluR5, rVIP (nonselective), hETB, bAT2 |
| subcluster2 | GPCR (other) (2) | hNPY2, bNPY (nonselective) |
| Cluster ii (52) | ||
| subcluster1 | GPCR (aminergic) (18) | hH1, gH2, hM1–5, gMperipheral, rAdra1A&B, rAdra1&2(nonselective), hAdra2A, rmAdra2B, hAdrb2, r5HT(nonselective), h5HT2A, bDR(nonselective) |
| GPCR (other) (3) | gOpiateK, rOpiate Non-Selective | |
| ion channel (2) | rNaCh, rCaBTZCHL | |
| subcluster2 | GPCR (aminergic) (7) | hDRD1, hDRD2s, hDRD4.4, h5HT5A, h5HT7, hAdra2C, hAdrb1 |
| GPCR (other) (2) | hOpiateD1 & Mu | |
| Cluster iii (41) | ||
| subcluster1 | activator: kinase (4) | hNEK2, hFyn, hIGF1R, hSRC |
| kinase (3) | hMAPKAP2, hMAPK3, hPAK4 | |
| activator: phosphatase (1) | hPPM1A | |
| phosphatase (4) | hPTPN4, 9, 14, hACP1 | |
| subcluster2 | kinase (9) | hRAF1, hCSF1R, hEGFR, hMAPK1, hTie2, hMsk1, hVEGFR2&3, hAurA |
| phosphatase (7) | hPTPN1, 2, 6, 11, 12, hPTPRB, hPTEN | |
Three clusters were identified through assay similarity analysis for further analysis.
Subclusters were identified from clusters along the diagonal with ≥65% similarity. Numbers in parentheses indicate the number of assays within a cluster.
Numbers in parentheses indicate the number of assays within an assay category.
3.4.2. Evaluation of Chemical Similarities
Clustering by chemical–chemical similarity was performed on 729 active chemicals (Figure 3). There are 55 such clusters (>65% similar) based on a small number of assays (<4) that range in size from 2 to 17 chemicals. Some examples include a cluster of 17 chemicals active in only the CYP2C19 assay and a cluster of 14 chemicals active in only the human PXR assay. Other clusters are driven by a few promiscuous chemicals, such as the cluster containing maneb, mancozeb, and metiram, and another cluster containing surfactants such as sodium dodecyl sulfate and PFOS. Smaller clusters with chemicals active in a limited number of assays revealed specificity within the data set: α-cyclodextrin, 17β-estradiol, and genistein (active in 5–9 assays) inhibited the three estrogen assays; two compounds CP-544439 and CP-471358 (active in 6–7 assays) inhibited MMPs 1/2/3/9/13, as well as the leukotriene B4 GPCR; rifampicin and tert-butylhydroquinone (active in 6–7 assays) inhibited PTGS1/COX1 and PTGS2/COX2 enzymatic activities, as well as CYP2C9 and CYPC19; and eight chemicals (caffeine, theophylline, cladribine, cyanazine, triflusulfuron-methyl, and prometryn, active in 3–9 assays) inhibited three human adenosine A receptors A1.
Four other clusters are highlighted in Figure 3 and Table 7 to demonstrate the substructure of a cluster that aggregated due to chemical–assay response similarity. To describe the clustered chemicals in terms of activity in the 21 assay categories, we used an enrichment score for each chemical (Figure 3, right panel). This chemical–assay category score indicates specificity of a chemical to affect assays within a given assay category over other categories. Cluster i has 29 chemicals active in 1 to 19 assays, enriched for Other enzyme. The subclusters include1-[cis-1-(3-ethoxyphenyl)-4-methylcyclohexyl]-4-phenylpiperazine methanesulfonate (1:1) (pharma), coumarin, DTBP, pioglitazone hydrochlorine, a number of nitrotoluene and phenol compounds, Michler’s ketone, dichlobenil, and 3-({(3R,4R)-6-[(5-fluoro-1,3-benzothiazol-2-yl)methoxy]-4-hydroxy-3,4-dihydro-2H-chromen-3-yl}methyl)benzoic acid (pharma). Cluster ii includes 36 chemicals active in 7 to 65 assays, mostly enriched with GPCR (other) and ion channel assays, and includes five other assay enrichment categories. Four subclusters include two chemicals in each group. Subcluster1 includes 4-dodecylbenzenesulfonic acid and docusate sodium and is mostly enriched for the GPCR (other) assay category, whereas the other three subclusters are enriched for the nuclear receptor (subfamily3) category and includes subcluster2, with 4,4′-sulfonylbis[2-(prop-2-en-1-yl)phenol] and clorophene; subcluster3, with bisphenol A and B; and subcluster4, with progesterone and androstenedione. Cluster iii involves 49 chemicals active in 12 to 90 assays, mostly enriched for GPCRs (both categories) and ion channels, as well as five others. Five subclusters were identified, including two subclusters with the most promiscuous chemicals, tributyltin chloride and tributyltin methacrylate, and phenylmercuric acetate and mercuric chloride. The other three clusters include at least half of the pharmaceuticals, most likely in the GPCR assay cluster from Figure 2. Cluster iv involves 149 chemicals active in 2 to 48 assays, enriched for CYPs, transporters, and nuclear receptor (subfamily 1). Many subfamilies were identified, but six were listed and contain mostly pesticides. The other subfamilies not listed were almost exclusively pesticides from Phase I of testing and discussed previously.9 The chemical similarities revealed known assay clusters (e.g., for bisphenols or heavy metal compounds) and previously unreported clusters (e.g., for pharmaceuticals). These findings demonstrate a structural basis to clustering of assays in the chemical similarity matrix.
Table 7. Chemical–Chemical Similaritiesa.
| clustersb | chemical namec |
|---|---|
| Cluster i (29) | |
| enriched assay group | other enzyme (29) |
| subcluster1 | 1-[cis-1-(3-ethoxyphenyl)-4-methylcyclohexyl]-4-phenylpiperazine methanesulfonate (1:1) (pharma); coumarin; DTBP; pioglitazone hydrochloride |
| subcluster2 | 2,4-DNT; 2,3-DNT; 2,6-DNT; 2,4,5-trichlorophenol; DNB; Michlers ketone; 4-nitrophenol; dichlobenil; 4-chloro-3-methylphenol; 3-({(3R,4R)-6-[(5-fluoro-1,3-benzothiazol-2-yl)methoxy]-4-hydroxy-3,4-dihydro-2H-chromen-3-yl}methyl)benzoic acid (pharma) |
| Cluster ii (36) | |
| enriched assay group | GPCR (other) (33), ion channel (26), CYP (33), GPCR (aminergic) (32), nuclear receptor (subfamily 3) (31), transporter (33), other enzyme (22) |
| subcluster1 | 4-dodecylbenzenesulfonic acid; docusate sodium |
| subcluster2 | 4,4′-sulfonylbis[2-(prop-2-en-1-yl)phenol]; clorophene |
| subcluster3 | bisphenol A; bisphenol B |
| subcluster4 | androstenedione; progesterone |
| Cluster iii (49) | |
| enriched assay group | GPCR (aminergic) (49), GPCR (other) (47), ion channel (47), transporter (46), cholinesterase (38), CYP (36), other (34), other enzyme (27) |
| subcluster1 | 1,3-diphenylguanidine; spiroxamine; CP-728663 pharma; 1,3-dichloro-6,7,8,9,10,12-hexahydroazepino[2,1-b]quinazoline hydrochloride (1:1)(pharma) |
| subcluster2 | tributyltin chloride; tributyltin methacrylate |
| subcluster3 | methadone hydrochloride; volinanserin(pharma); haloperidol; diphenhydramine hydrochloride; SSR125047 pharma; dodecyltrimethylammonium chloride; pentamidine isethionate; AVE8488 pharma; SR271425 pharma; SB236057A pharma; GSK163929B pharma; SSR240612 pharma; UK-416244 pharma; SSR241586 pharma; SAR102779 pharma; SAR150640 pharma; chlorpromazine hydrochloride; SSR150106 pharma; zamifenacin (pharma) |
| subcluster4 | phenylmercuric acetate; mercuric chloride |
| subcluster5 | amiodarone hydrochloride; (1R)-1-[(ethoxycarbonyl)oxy]ethyl 1-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methyl}-2-{[1-(propan-2-yl)piperidin-4-yl]carbamoyl}-1H-indole-5-carboxylate hydrochloride (pharma); didecyldimethylammonium chloride; {4-[5-(aminomethyl)-2-f luorophenyl]piperidin-1-yl}(4-bromo-3-methyl-5-propoxythiophen-2-yl)methanone hydrochloride (pharma); 5-(benzylsulfonyl)-2-{[2-(dimethylamino)ethyl](ethyl)amino}-N,N-diethyl-4-(4-phenylpiperidin-1-yl)benzamide (pharma); gentian violet; {4-[3-(aminomethyl)phenyl]piperidin-1-yl}{5-[(2-fluorophenyl)ethynyl]furan-2-yl}methanone (pharma) |
| Cluster iv (149) | |
| enriched assay group | CYP (149), transporter (111), nuclear receptor (subfamily 1) (81) |
| subcluster1 | rifampicin; tert-Butylhydroquinone |
| subcluster2 | decane; clotrimazole; cyproconazole; fenbuconazole |
| subcluster3 | 3,3′,5,5′-tetrabromobisphenol A; methoxychlor; paclobutrazol; triticonazole; fipronil; dimethoxane |
| subcluster4 | 2,5-ditert-butylbenzene-1,4-diol; Phenothiazine |
| subcluster5 | CP-456773 pharma; O-ethyl O-(4-nitrophenyl) phenylphosphonothioate |
| subcluster6 | metolachlor; chlorpyrifos-methyl; 2-(4-fluorophenoxy)-N-[4-(2-hydroxypropan-2-yl)benzyl]pyridine-3-carboxamide (pharma); DEHP; acetochlor; trifloxystrobin; fenpropathrin; disulfiram; chlorobenzilate; benodanil; 4,4′-methylenebis(2-methylaniline); 6-chloro-2-phenyl-8,8a-dihydro-3aH-indeno[1,2-d][1,3]thiazol-3a-ol (pharma); butyl benzyl phthalate; 7,12-dimethylbenz(a)anthracene; dibutyl phthalate; triphenyl phosphate; dipropylene glycol dibenzoate; norflurazon; carfentrazone-ethyl; rotenone; 3-pyridinecarboxamide, 2-(2,1,3-benzoxadiazol-5-yloxy)-N-[[4-(1-hydroxy-1-methylethyl)phenyl]methyl]-(pharma); picoxystrobin; fenoxycarb |
Four clusters were identified through chemical similarity analysis for further analysis. The chemicals within each cluster were enriched for a number of assay categories, with the most associated in bold. Numbers in parentheses indicate the number or chemicals within the cluster and associated with the assay category.
Subclusters were identified from clusters along the diagonal with ≥65% similarity.
Chemicals within each subcluster are listed.
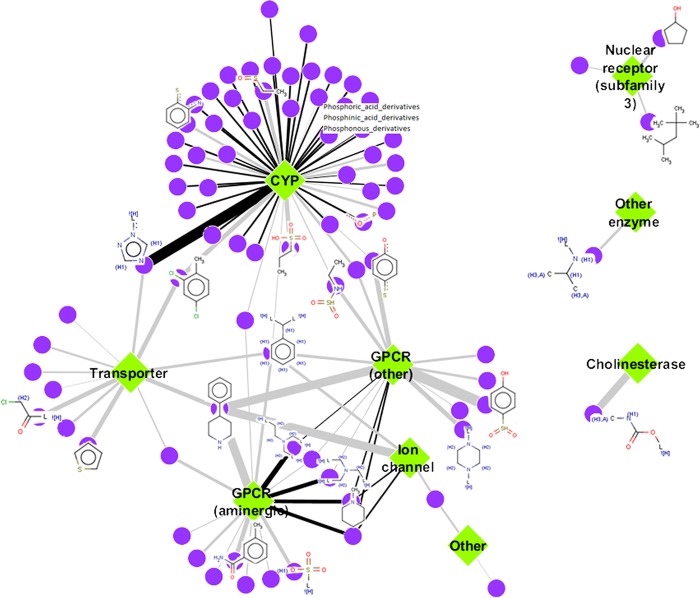
3.5. Common Chemical Structure Features Enriched for Specific Assay Categories
From the chemical activity similarity cluster, enriched assay categories can be identified. In turn, evaluating similar chemical structure fragments enriched in the assay categories can help describe the types of chemicals that are clustering together (Figure 4). Univariate analyses linking chemical structure fragments to assay categories (based on AC50s ≥10 μM) revealed 107 associations for 84 unique chemical fragments and 9 assay categories. CYPs had the most associated chemical fragments, with a majority being subfragments involving phosphoric acid, phosphinic acid, and phosphonous derivatives; however, 1,2,4-triazole had the highest PPV. A benzene ring with two constituents and 4-phenylpiperidine were structures shared by the CYPs, transporters, GPCRs, and ion channel categories. Additionally, variations of the benzene ring were seen throughout the associations. Three assay categories that did not share chemical structures with other categories included NR_subfamily3_steroid associated with cyclopentanol and 2,2,4-trimethylpentane; the other enzyme category associated with isopropylamine; and cholinesterase associated with N-methylcarbamate. Overall, the chemical fragments from chemicals in this data set that are enriched for affecting an assay category are generally unique, but overlap is observed.
Figure 4.
Chemical fragment–assay category associations. Univariate associations between chemical-structure fragments and enrichment scores for chemical–assay categories were performed to identify chemical fragments in chemicals specifically enriched for affecting an assay category. Chemical fragment–assay categories with true positives ≥5 and positive predictive value (PPV) >0.5 are visualized. Purple nodes are the chemical fragments, and green diamonds are the assay categories. Example chemical fragment structures are shown for the associations with the highest PPV (indicated by increasing edge thickness). Numbers of true positives are indicated by edge color from gray (5–9) to black (10–26).
4. Discussion
The data set analyzed here profiles 976 structurally and categorically diverse chemicals in the ToxCast library across 331 biological assays. These data were derived from two temporally separated testing phases: (1) 310 unique chemicals analyzed across 292 assays from the original PhaseI_v1 of ToxCast, reanalyzed using an improved algorithm for automated curve-fitting across 230 assays retained in Phase II; and (2) new data generated for 676 unique structures (including 9 Phase I compounds in replicate) in Phase II analyzed across 231 assays. In total, the combined set provides bioactivity data for 976 chemicals (Phase I, II) profiled across 331 assays (231 assays plus 100 analyzed for activation) for ligand–receptor binding and enzymatic activities. Patterns derived from these profiles can only be interpreted within the context of the specific assays covered here and do not imply in vivo toxicity or lack thereof since the biochemical screen does not consider kinetics and metabolism (ADME) and intra/inter-cellular signaling. Results from this study extend the previous analysis9 to a broader chemical landscape, revealing both major and minor patterns for groups of chemicals and biochemical assays.
Roughly a quarter of the 976 compounds tested showed no demonstrable activity (AC50) in any of the assays used herein, including many consumer use chemicals, phthalates, and green alternatives. The remaining compounds tested here showed specific or promiscuous activities. Clustering revealed coarse relationships by chemical use category, chemical structure, and/or biological target. For example, most pesticidal actives and pharmaceuticals could be resolved by target signatures, AChE and nuclear receptors on the one hand versus GPCRs and transporters on the other. A similar degree of promiscuity ascribed to the Phase I chemical library9 was also noted here for the broader chemical landscape. Perhaps surprisingly, 7 of the 20 most promiscuous compounds were pharmaceuticals. This promiscuity, directed across the GPCR assays, partly reflected the large contingent of GPCR assays included in this screen but also suggests a potential for off-target effects. Inherent chemical factors such as surfactant or detergent properties and metal-based or metal-chelating properties could also account for promiscuity. In particular, the polymeric dithiocarbamate pesticides affected multiple kinase and phosphatase assays that require divalent cations (magnesium or manganese) and/or have active site cysteine residues sensitive to oxidation. This specific effect was noted previously for maneb, mancozeb, and metiram,9 consistent with a mechanism of action associated with the inhibition of metal-dependent and sulfhydryl enzymes,32,33 and seems to be specific to these compounds across the broader chemical landscape tested here. Lastly, compounds containing tin (e.g., tributyltin) and mercury (e.g., phenylmercuric acetate) broadly disrupted GPCR ligand-binding activities but not kinase or phosphatase activities. These examples suggest a systematic basis to the promiscuity and a degree of specificity to the biological target or assay conditions. Such systematic activity may be reflected in characteristic responses in vivo. For example, the promiscuous GPCR activity of the mercury-containing compounds is consistent with the neuropsychiatric effects seen in mercury poisoning, such as Minamata Disease or Mad Hatter Syndrome, whose symptoms include delirium and hallucinations, ataxia, numbness in hands and feet, general muscle weakness, narrowing of the field of vision, and damage to hearing and speech.34,35
Biochemical profiling also identified previously unreported chemical–target interactions that may suggest potential modes of action or off-target effects. Anthralin, for example, is a topical drug used for treatment of psoriasis, but its mechanism of action is unknown. Consistent with the biology of psoriasis as an autoimmune skin disease,36 anthralin specifically inhibited the activity of several protein targets that may play a role in inflammatory pathways (caspases, MMPs, and COX1 and 2). Tannic acid, which is known for inhibiting inflammatory processes,37 also inhibited these protein targets. UK-416244, a selective serotonin reuptake inhibitor (SSRI) (http://www.chemspider.com/Chemical-Structure.8123181.html), inhibited the rat serotonin transporter (AC50 of 0.023 μM) as expected but also showed moderate activity on GPCRs including the human and rat adrenergic receptors (AC50s of 0.29–12 μM), which are known off-targets for SSRIs.5 The most common target of the perfluorinated compounds was the leukotriene B4 GPCR. Although this is not a known interaction, the interaction is plausible since leukotrienes are fatty acid signaling molecules, and perfluorinated compounds are hydrophobic and lipophilic. In addition, both chemicals and proteins are suggested players in immune function and may give clues to off-target effects.38,39
Pharmaceuticals, of which 245 are included in this study, serve not only as an anchor for known interactions and off-target effects but also as a kernel for clustering environmental chemicals by potency, efficacy, and specificity. A similarity matrix clustering chemical compounds by their assay targets classified two triazine herbicides (cyanazine and prometryn) with caffeine, theophylline, and cladribine based on a relatively selective effect on human adenosine receptors. The latter three compounds are known (or suspected) to disrupt pathways in adenosine signaling or metabolism and may serve to anchor the potential activity of cyanazine or prometryn to adenosine signaling.
Co-clustering of biologically related assays is also useful in discovering potentially unknown interactions. Consider, for example, assay clusters for biogenic amine GPCRs and peptide GPCRs from the assay by assay similarity matrix. A number of chemical compounds preferentially affected these two GPCR subfamilies. In addition, the biogenic amine GPCRs had a closer relationship with the disruption of opiate GPCRs, transporters for norepinephrine, serotonin, and dopamine, and sodium or calcium channels. Consideration to the broader range of biochemical targets may be informative data to train structure models for predicting chemical–target interactions. The major and minor patterns of associations from the biochemical profiling can be used in conjunction with (or to inform) computational approaches, such as the structure–activity relationship or similarity ensemble approach analyses.5,40,41
Additional analyses for discovering unknown interactions may come from the enrichment of common chemical fragments in association with specific or multiple assay targets. These chemical fragment associations suggest that if a new chemical contains this chemical fragment, it has a greater likelihood of affecting a given assay category and could provide clues governing chemical interactions for a specific or multiple assay targets. For example, phenol-containing compounds are well known to interact with the estrogen receptor (within the steroid subfamily 3 of nuclear receptors),42 but chemicals containing the phenol fragment were also associated with promiscuous inhibition of CYPs, thereby not specifically associating with the steroid subfamily 3 category. Similarly, organophosphates are known cholinesterase inhibitors for some insecticides, and chemicals containing this fragment were also associated with the inhibition of various CYPs and not specifically with cholinesterase inhibition. The cyclopentanol and 2,2,4-trimethylpentane fragments are inherent features of but not robust drivers of chemicals predicted to inhibit steroid receptor binding.43 The gross association of fragments with assay enrichment in some cases and not others may provide a departure point for building more specific structure–activity relationship models, i.e., determining chemical property or feature modifiers that could potentially distinguish more specific activity subclasses within the fragment-containing clusters. However, examples of fragment–assay associations consistent with a priori knowledge include the association of an N-methylcarbamate fragment of some insecticides to anticholinesterase activity44 and the isopropylamine fragment to some inhibitors of monoamine oxidases.45 Beyond feature enrichment for certain chemical clusters (chemotyping), this biochemical HTS, together with nonbiochemical ToxCast assays and structure similarity, can serve as a broader resource for mapping structure–activity relationships to biological profiles and in vivo toxicities. The full range of the biochemical HTS could be used, for example, to test the hypothesis that structural analogues and/or chemicals sharing a common mechanistic category show sufficiently similar signals and whether or not such response signatures are specific enough to delineate a unique read-across grouping. Comparisons have been made between the Phase I chemical–assay effects and adverse outcomes, where a considerable amount of in vivo data is available.3,46,47 A challenge will be to address the clustering of Phase I and Phase II chemicals by biological response given the lack of in vivo data for most of the Phase II chemicals.
Biochemical inhibition of CYP activity may indicate a potential for the chemical to disrupt intermediary metabolism in vivo or alternatively that the xenobiotic may be preferentially metabolized in vivo. Other specific interactions, either unrelated to cellular mechanisms or without obvious biological significance may be detected in the biochemical screen. For example, the soluble dietary fiber α-cyclodextrin inhibited all three estrogen assays (hER, mERa, and bER) and clustered with the known ER ligands estradiol and genistein. This interaction may be similar to that known for β-cyclodextrin through clathrate formation to sequester estrogen in a lattice.48 Cyclodextrins are relatively nontoxic, however, due to their inability to penetrate lipophilic membranes.49 Although these cell-free data may not always point to a biologically relevant process, the specificity of activity within a complex chemical–assay matrix may, nevertheless, provide useful information about the chemical compounds in their interaction with specific protein targets.
Since these chemicals were tested at or below 50 μM, it may be difficult to extrapolate the current AC50 concentrations to in vivo activity since the concentrations are high relative to what might be achieved during in vivo exposure.50 However, a number of chemical–assay pairs showed an AC50 at or below 1 μM, which would seem attainable at least in high-dose testing animal studies. Along these lines, negative compounds that exhibit adverse outcomes in vivo could have been missed in the cell-free biochemical screen due to (a) bioavailability resulting in a xenobiotic metabolite not in the ToxCast chemical inventory; (b) the biological target not being represented, e.g., the 331 assays do not cover all possible targets; (c) the 50 μM concentration not being high enough; or (d) the fact that the chemical–assay pair activity was not observed in the single screen due to assay variability, compound instability, or assay interference and hence not followed up by concentration–response. Furthermore, the concentration–response data is being made public with this article for user interpretation (http://actor.epa.gov). These points indicate areas for future research, specifically performing more extensive ADME analyses, consulting other cell-based assays for target validation and further expansion of the assay set, and identifying bioactivity signatures of broad chemical classes for which we could then query the toxicological space.
Moving forward, this large data set contributes significantly toward advancing new a regulatory paradigm through predictive toxicity and a better understanding of mechanisms.2 The biochemical HTS screen offers preliminary evidence for chemical targets in a cell or tissue that, when combined with information from the literature or targeted studies, indicates potential pathways of toxicity. Predictive signatures developed from combinations of assays may be linked to an adverse outcome pathway.3,46,47,51,52 Efforts are underway to incorporate chemical features into these types of analyses to identify potential structural alerts that in combination with the HTS assays may be more predictive. These types of data and alerts can potentially provide biological predictions about chemicals with little to no biological information. In addition, in silico models are incorporating this information with cell–cell and tissue level behavior to predict outcomes of changing dose and time within a developing organ, without the need to pick up a pipet.53
Overall, these results expand the ToxCast database to include a large and more structurally diverse set of chemicals. They also contribute to a novel framework for describing the interaction of environmental chemicals with important biochemical targets. By broadly surveying both the chemical landscape and biological target space, patterns of biochemical activity have been identified which can help focus future research toward potential chemical–biological interactions that may result in adverse outcomes in vivo. The use of a quantitative approach in determining the potency of chemicals against each biological target, utilizing a diverse chemical library containing many reference chemicals with known activities as chemical probes, provides the context to better understand the potential for hazard of those chemicals with limited toxicity information. As these interactions become better defined through methods such as incorporation into adverse outcome pathways,7,54 these in vitro screening methods may ultimately serve as an efficient means of avoiding unwanted biological activity early in the chemical design and development process.
Acknowledgments
We thank Mr. John Southerland for excellent management of EPA contracts with Caliper Discovery Alliances and Services, A PerkinElmer company and Evotec. Special thanks to the entire ToxCast team and pharmaceutical partners for their helpful insights. We also thank Dr. Scott Auerbach (NIEHS/NTP), Dr. Steve Simmons (EPA/NHEERL), and Dr. Mark Miller (EPA/OSP) for their valuable comments.
Glossary
Abbreviations
- AC50
half-maximal activity concentration
- CYP
cytochrome P450
- DMSO
dimethyl sulfoxide
- DNT
dinitrotoluene
- DTBP di-tert-butyl peroxide
- ES enrichment score
- GPCR
G-protein-coupled receptor
- HTS high-throughput screening
- LCT
lowest concentration tested
- LGIC
ligand-gated ion channel
- μM
micromolar
- mM
millimolar
- M
molar
- MAD2
2.0 median absolute deviations from the median
- MMP
metalloproteinase
- NVS
NovaScreen panel
- p
probability
- PFOS
perfluorooctane sulfonic acid
- PPV
positive predictive value
- PXR
pregnane X receptor
- QC
quality control
- SDF
Structure Data Format
- SSRI
selective serotonin reuptake inhibitor
- TSPO
translocator protein
Supporting Information Available
All assays and assay descriptions; miniSOPs and curve fit figures linked to Table S1; and matrix of all chemical–assay AC50 values, in order as displayed in Figure 1. This material is available free of charge via the Internet at http://pubs.acs.org.
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- NRC (2007) Toxicity Testing in the 21st Century: A Vision and a Strategy, National Academies Press, Washington, D.C. [Google Scholar]
- Kavlock R.; Chandler K.; Houck K.; Hunter S.; Judson R.; Kleinstreuer N.; Knudsen T.; Martin M.; Padilla S.; Reif D.; Richard A.; Rotroff D.; Sipes N.; Dix D. (2012) Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Judson R. S.; Houck K. A.; Kavlock R. J.; Knudsen T. B.; Martin M. T.; Mortensen H. M.; Reif D. M.; Rotroff D. M.; Shah I.; Richard A. M.; Dix D. J. (2010) In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ. Health Perspect. 118, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC (1984) Toxicity Testing: Strategies to Determine Needs and Priorities, National Academies Press, Washington, D.C. [PubMed] [Google Scholar]
- Keiser M. J.; Setola V.; Irwin J. J.; Laggner C.; Abbas A. I.; Hufeisen S. J.; Jensen N. H.; Kuijer M. B.; Matos R. C.; Tran T. B.; Whaley R.; Glennon R. A.; Hert J.; Thomas K. L.; Edwards D. D.; Shoichet B. K.; Roth B. L. (2009) Predicting new molecular targets for known drugs. Nature 462, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.; Xie L.; Bourne P. E. (2011) Structure-based systems biology for analyzing off-target binding. Curr. Opin. Struct. Biol. 21, 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankley G. T.; Bennett R. S.; Erickson R. J.; Hoff D. J.; Hornung M. W.; Johnson R. D.; Mount D. R.; Nichols J. W.; Russom C. L.; Schmieder P. K.; Serrrano J. A.; Tietge J. E.; Villeneuve D. L. (2010) Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Martin M. T.; Dix D. J.; Judson R. S.; Kavlock R. J.; Reif D. M.; Richard A. M.; Rotroff D. M.; Romanov S.; Medvedev A.; Poltoratskaya N.; Gambarian M.; Moeser M.; Makarov S. S.; Houck K. A. (2010) Impact of environmental chemicals on key transcription regulators and correlation to toxicity end points within EPA’s ToxCast program. Chem. Res. Toxicol. 23, 578–590. [DOI] [PubMed] [Google Scholar]
- Knudsen T. B.; Houck K. A.; Sipes N. S.; Singh A. V.; Judson R. S.; Martin M. T.; Weissman A.; Kleinstreuer N. C.; Mortensen H. M.; Reif D. M.; Rabinowitz J. R.; Setzer R. W.; Richard A. M.; Dix D. J.; Kavlock R. J. (2011) Activity profiles of 309 ToxCast chemicals evaluated across 292 biochemical targets. Toxicology 282, 1–15. [DOI] [PubMed] [Google Scholar]
- Berg E. L.; Kunkel E. J.; Hytopoulos E.; Plavec I. (2006) Characterization of compound mechanisms and secondary activities by BioMAP analysis. J. Pharmacol. Toxicol. Methods 53, 67–74. [DOI] [PubMed] [Google Scholar]
- Giuliano K. A.; Johnston P. A.; Gough A.; Taylor D. L. (2006) Systems cell biology based on high-content screening. Methods Enzymol. 414, 601–619. [DOI] [PubMed] [Google Scholar]
- Rotroff D. M.; Beam A. L.; Dix D. J.; Farmer A.; Freeman K. M.; Houck K. A.; Judson R. S.; LeCluyse E. L.; Martin M. T.; Reif D. M.; Ferguson S. S. (2010) Xenobiotic-metabolizing enzyme and transporter gene expression in primary cultures of human hepatocytes modulated by ToxCast chemicals. J. Toxicol. Environ. Health, Part B 13, 329–346. [DOI] [PubMed] [Google Scholar]
- Chandler K. J.; Barrier M.; Jeffay S.; Nichols H. P.; Kleinstreuer N. C.; Singh A. V.; Reif D. M.; Sipes N. S.; Judson R. S.; Dix D. J.; Kavlock R.; Hunter E. S. III; Knudsen T. B. (2011) Evaluation of 309 environmental chemicals using a mouse embryonic stem cell adherent cell differentiation and cytotoxicity assay. PLoS One 6, e18540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.; Xia M.; Cho M. H.; Sakamuru S.; Shinn P.; Houck K. A.; Dix D. J.; Judson R. S.; Witt K. L.; Kavlock R. J.; Tice R. R.; Austin C. P. (2011) Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ. Health Perspect. 119, 1142–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J.; Auld D. S.; Jadhav A.; Johnson R. L.; Simeonov A.; Yasgar A.; Zheng W.; Austin C. P. (2006) Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U.S.A. 103, 11473–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. J.; Huang R.; Austin C. P.; Xia M. (2010) The future of toxicity testing: a focus on in vitro methods using a quantitative high-throughput screening platform. Drug Discovery Today 15, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. S.; Gray G. M.; Bucher J. R. (2008) Toxicology. Transforming environmental health protection. Science 319, 906–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth A. P.; Lapenna S.; Serafimova R. (2013) QSAR and metabolic assessment tools in the assessment of genotoxicity. Methods Mol. Biol. 930, 125–162. [DOI] [PubMed] [Google Scholar]
- Judson R. S.; Martin M. T.; Egeghy P.; Gangwal S.; Reif D. M.; Kothiya P.; Wolf M.; Cathey T.; Transue T.; Smith D.; Vail J.; Frame A.; Mosher S.; Cohen Hubal E. A.; Richard A. M. (2012) Aggregating data for computational toxicology applications: The U.S. Environmental Protection Agency (EPA) Aggregated Computational Toxicology Resource (ACToR) System. Int. J. Mol. Sci. 13, 1805–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R.; Gentleman R. (1996) R: A Language for Data Analysis and Graphics. J. Comp. Graph. Stat. 5, 299–314. [Google Scholar]
- R Development Core Team (2012) R: A Language and Environment for Statistical Computing, ISBN 3-900051-07-0, R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org. [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. (2011) Open Babel: An open chemical toolbox. J. Cheminform. 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap C. W. (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32, 1466–1474. [DOI] [PubMed] [Google Scholar]
- Cline M. S.; Smoot M.; Cerami E.; Kuchinsky A.; Landys N.; Workman C.; Christmas R.; Avila-Campilo I.; Creech M.; Gross B.; Hanspers K.; Isserlin R.; Kelley R.; Killcoyne S.; Lotia S.; Maere S.; Morris J.; Ono K.; Pavlovic V.; Pico A. R.; Vailaya A.; Wang P. L.; Adler A.; Conklin B. R.; Hood L.; Kuiper M.; Sander C.; Schmulevich I.; Schwikowski B.; Warner G. J.; Ideker T.; Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C. J.; Fadayel G. M.; Sullivan C. K.; Taylor V. L. (1992) 5-HT2 receptors exert a state-dependent regulation of dopaminergic function: studies with MDL 100,907 and the amphetamine analogue, 3,4-methylenedioxymethamphetamine. Eur. J. Pharmacol. 223, 65–74. [DOI] [PubMed] [Google Scholar]
- Jiaravuthisan M. M.; Sasseville D.; Vender R. B.; Murphy F.; Muhn C. Y. (2007) Psoriasis of the nail: anatomy, pathology, clinical presentation, and a review of the literature on therapy. J. Am. Acad. Dermatol. 57, 1–27. [DOI] [PubMed] [Google Scholar]
- Jensen K.; Johnson L. A.; Jacobson P. A.; Kachler S.; Kirstein M. N.; Lamba J.; Klotz K. N. (2012) Cytotoxic purine nucleoside analogues bind to A1, A2A, and A3 adenosine receptors. Naunyn-Schmiedeberg’s Arch. Pharmacol. 385, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witorsch R. J.; Thomas J. A. (2010) Personal care products and endocrine disruption: A critical review of the literature. Crit. Rev. Toxicol. 40(Suppl 3), 1–30. [DOI] [PubMed] [Google Scholar]
- Tickner J. A.; Schettler T.; Guidotti T.; McCally M.; Rossi M. (2001) Health risks posed by use of Di-2-ethylhexyl phthalate (DEHP) in PVC medical devices: a critical review. Am. J. Ind. Med. 39, 100–111. [DOI] [PubMed] [Google Scholar]
- U.S. Consumer Product Safety Commission by Versar, Inc. and Syracuse Research Corporation (SRC) (2010) Review of exposure and toxicity data for phthalate substitutes, www.cpsc.gov/about/cpsia/phthalsub.pdf (accessed Jan 4, 2012).
- Blair R. M.; Fang H.; Branham W. S.; Hass B. S.; Dial S. L.; Moland C. L.; Tong W.; Shi L.; Perkins R.; Sheehan D. M. (2000) The estrogen receptor relative binding affinities of 188 natural and xenochemicals: structural diversity of ligands. Toxicol. Sci. 54, 138–153. [DOI] [PubMed] [Google Scholar]
- Dubois K. P.; Raymund A. B.; Hietbrink B. E. (1961) Inhibitory action of dithiocarbamates on enzymes of animal tissues. Toxicol. Appl. Pharmacol. 3, 236–255. [DOI] [PubMed] [Google Scholar]
- Miller D. B. (1982) Neurotoxicity of the pesticidal carbamates. Neurobehav. Toxicol. Teratol. 4, 779–787. [PubMed] [Google Scholar]
- O’Carroll R. E.; Masterton G.; Dougall N.; Ebmeier K. P.; Goodwin G. M. (1995) The neuropsychiatric sequelae of mercury poisoning. The Mad Hatter’s disease revisited. Br. J. Psychiatry 167, 95–98. [DOI] [PubMed] [Google Scholar]
- Harada M. (1995) Minamata disease: methylmercury poisoning in Japan caused by environmental pollution. Crit. Rev. Toxicol. 25, 1–24. [DOI] [PubMed] [Google Scholar]
- Haroon M.; Fitzgerald O. (2012) Pathogenetic overview of psoriatic disease. J. Rheumatol., Suppl. 89, 7–10. [DOI] [PubMed] [Google Scholar]
- Chen X.; Beutler J. A.; McCloud T. G.; Loehfelm A.; Yang L.; Dong H. F.; Chertov O. Y.; Salcedo R.; Oppenheim J. J.; Howard O. M. (2003) Tannic acid is an inhibitor of CXCL12 (SDF-1alpha)/CXCR4 with antiangiogenic activity. Clin. Cancer Res. 9, 3115–3123. [PubMed] [Google Scholar]
- Di Gennaro A.; Haeggstrom J. Z. (2012) The leukotrienes: immune-modulating lipid mediators of disease. Adv. Immunol. 116, 51–92. [DOI] [PubMed] [Google Scholar]
- Keil D. E.; Mehlmann T.; Butterworth L.; Peden-Adams M. M. (2008) Gestational exposure to perfluorooctane sulfonate suppresses immune function in B6C3F1 mice. Toxicol. Sci. 103, 77–85. [DOI] [PubMed] [Google Scholar]
- Lounkine E.; Keiser M. J.; Whitebread S.; Mikhailov D.; Hamon J.; Jenkins J. L.; Lavan P.; Weber E.; Doak A. K.; Cote S.; Shoichet B. K.; Urban L. (2012) Large-scale prediction and testing of drug activity on side-effect targets. Nature 486, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini G. V.; Shapland R. H.; van Hoorn W. P.; Mason J. S.; Hopkins A. L. (2006) Global mapping of pharmacological space. Nat. Biotechnol. 24, 805–815. [DOI] [PubMed] [Google Scholar]
- Duax W. L.; Griffin J. F.; Weeks C. M.; Wawrzak Z. (1988) The mechanism of action of steroid antagonists: insights from crystallographic studies. J. Steroid Biochem. 31, 481–492. [DOI] [PubMed] [Google Scholar]
- Tabira Y.; Nakai M.; Asai D.; Yakabe Y.; Tahara Y.; Shinmyozu T.; Noguchi M.; Takatsuki M.; Shimohigashi Y. (1999) Structural requirements of para-alkylphenols to bind to estrogen receptor. Eur. J. Biochem. 262, 240–245. [DOI] [PubMed] [Google Scholar]
- Reigart J. R., and Roberts J. R. (1999) N-Methyl Carbamate Insecticides, in Recognition and Management of Pesticide Poisoning (Langner G., Ed.) pp 48–54, U.S. Environmental Protection Agency, Washington, D.C. [Google Scholar]
- Vilches-Herrera M.; Miranda-Sepulveda J.; Rebolledo-Fuentes M.; Fierro A.; Luhr S.; Iturriaga-Vasquez P.; Cassels B. K.; Reyes-Parada M. (2009) Naphthylisopropylamine and N-benzylamphetamine derivatives as monoamine oxidase inhibitors. Bioorg. Med. Chem. 17, 2452–2460. [DOI] [PubMed] [Google Scholar]
- Sipes N. S.; Martin M. T.; Reif D. M.; Kleinstreuer N. C.; Judson R. S.; Singh A. V.; Chandler K. J.; Dix D. J.; Kavlock R. J.; Knudsen T. B. (2011) Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data. Toxicol. Sci. 124, 109–127. [DOI] [PubMed] [Google Scholar]
- Martin M. T.; Knudsen T. B.; Reif D. M.; Houck K. A.; Judson R. S.; Kavlock R. J.; Dix D. J. (2011) Predictive model of rat reproductive toxicity from ToxCast high throughput screening. Biol. Reprod. 85, 327–339. [DOI] [PubMed] [Google Scholar]
- Oishi K.; Toyao K.; Kawano Y. (2008) Suppression of estrogenic activity of 17-beta-estradiol by beta-cyclodextrin. Chemosphere 73, 1788–1792. [DOI] [PubMed] [Google Scholar]
- Del Valle E. M. M. (2004) Cyclodextrins and their uses: a review. Process Biochem. (Oxford, U. K.) 39, 1033–1046. [Google Scholar]
- Wetmore B. A.; Wambaugh J. F.; Ferguson S. S.; Sochaski M. A.; Rotroff D. M.; Freeman K.; Clewell H. J. III; Dix D. J.; Andersen M. E.; Houck K. A.; Allen B.; Judson R. S.; Singh R.; Kavlock R. J.; Richard A. M.; Thomas R. S. (2012) Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol. Sci. 125, 157–174. [DOI] [PubMed] [Google Scholar]
- Dix D. J.; Houck K. A.; Judson R. S.; Kleinstreuer N. C.; Knudsen T. B.; Martin M. T.; Reif D. M.; Richard A. M.; Shah I.; Sipes N. S.; Kavlock R. J. (2012) Incorporating biological, chemical, and toxicological knowledge into predictive models of toxicity. Toxicol. Sci. 130, 440–441. [DOI] [PubMed] [Google Scholar]; author reply 442-443.
- Knudsen T. B.; Kleinstreuer N. C. (2011) Disruption of embryonic vascular development in predictive toxicology. Birth Defects Res., Part C 93, 312–323. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer N.; Dix D.; Rountree M.; Baker N.; Sipes N.; Reif D.; Spencer R.; Knudsen T. (2013) A computational model predicting disruption of blood vessel development. PLoS Comput. Biol. 9, e1002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer N. C., and Knudsen T. B. (2011) Predictive Modeling and Computational Toxicology, in Developmental and Reproductive Toxicology: A Practical Approach (Hood R. D., Ed.) pp 578–591, Informa Healthcare Books, London, U.K. [Google Scholar]
- Hals P. A.; Hall H.; Dahl S. G. (1988) Muscarinic cholinergic and histamine H1 receptor binding of phenothiazine drug metabolites. Life Sci. 43, 405–412. [DOI] [PubMed] [Google Scholar]
- Huerta-Bahena J.; Villalobos-Molina R.; Garcia-Sainz J. A. (1983) Trifluoperazine and chlorpromazine antagonize alpha 1- but not alpha2- adrenergic effects. Mol. Pharmacol. 23, 67–70. [PubMed] [Google Scholar]
- Trichard C.; Paillere-Martinot M. L.; Attar-Levy D.; Recassens C.; Monnet F.; Martinot J. L. (1998) Binding of antipsychotic drugs to cortical 5-HT2A receptors: a PET study of chlorpromazine, clozapine, and amisulpride in schizophrenic patients. Am. J. Psychiatry 155, 505–508. [DOI] [PubMed] [Google Scholar]
- York D. H. (1972) Dopamine receptor blockade--a central action of chlorpromazine on striatal neurones. Brain Res. 37, 91–99. [DOI] [PubMed] [Google Scholar]
- Arnt J.; Skarsfeldt T. (1998) Do novel antipsychotics have similar pharmacological characteristics? A review of the evidence. Neuropsychopharmacology 18, 63–101. [DOI] [PubMed] [Google Scholar]
- Tam S. W.; Cook L. (1984) Sigma opiates and certain antipsychotic drugs mutually inhibit (+)-[3H] SKF 10,047 and [3H]haloperidol binding in guinea pig brain membranes. Proc. Natl. Acad. Sci. U.S.A. 81, 5618–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppurainen H.; Kuikka J. T.; Viinamaki H.; Husso M.; Tiihonen J. (2009) Dopamine D2/3 receptor binding potential and occupancy in midbrain and temporal cortex by haloperidol, olanzapine and clozapine. Psychiatry Clin. Neurosci. 63, 529–537. [DOI] [PubMed] [Google Scholar]
- McCort G.; Hoornaert C.; Aletru M.; Denys C.; Duclos O.; Cadilhac C.; Guilpain E.; Dellac G.; Janiak P.; Galzin A. M.; Delahaye M.; Guilbert F.; O’Connor S. (2001) Synthesis and SAR of 3- and 4-substituted quinolin-2-ones: discovery of mixed 5-HT(1B)/5-HT(2A) receptor antagonists. Bioorg. Med. Chem. 9, 2129–2137. [DOI] [PubMed] [Google Scholar]
- LaFrate A. L.; Carlson K. E.; Katzenellenbogen J. A. (2009) Steroidal bivalent ligands for the estrogen receptor: design, synthesis, characterization and binding affinities. Bioorg. Med. Chem. 17, 3528–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S.; Miyamoto H.; Shima H.; Chang C. (1998) From estrogen to androgen receptor: a new pathway for sex hormones in prostate. Proc. Natl. Acad. Sci. U.S.A. 95, 5527–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider C. V.; Hartig P. C.; Cardon M. C.; Wilson V. S. (2009) Comparison of chemical binding to recombinant fathead minnow and human estrogen receptors alpha in whole cell and cell-free binding assays. Environ. Toxicol. Chem. 28, 2175–2181. [DOI] [PubMed] [Google Scholar]
- Planting A.; van der Gaast A.; Schoffski P.; Bartkowski M.; Verheij C.; Noe D.; Ferrante K.; Verweij J. (2005) A phase I and pharmacologic study of the matrix metalloproteinase inhibitor CP-471,358 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 55, 136–142. [DOI] [PubMed] [Google Scholar]
- Dalvie D.; Cosker T.; Boyden T.; Zhou S.; Schroeder C.; Potchoiba M. J. (2008) Metabolism distribution and excretion of a matrix metalloproteinase-13 inhibitor, 4-[4-(4-fluorophenoxy)-benzenesulfonylamino]tetrahydropyran-4-carboxylic acid hydroxyamide (CP-544439), in rats and dogs: assessment of the metabolic profile of CP-544439 in plasma and urine of humans. Drug Metab. Dispos. 36, 1869–1883. [DOI] [PubMed] [Google Scholar]
- Shiau A. K.; Barstad D.; Loria P. M.; Cheng L.; Kushner P. J.; Agard D. A.; Greene G. L. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937. [DOI] [PubMed] [Google Scholar]
- Gaido K. W.; Leonard L. S.; Maness S. C.; Hall J. M.; McDonnell D. P.; Saville B.; Safe S. (1999) Differential interaction of the methoxychlor metabolite 2,2-bis-(p-hydroxyphenyl)-1,1,1-trichloroethane with estrogen receptors alpha and beta. Endocrinology 140, 5746–5753. [DOI] [PubMed] [Google Scholar]
- Watson N.; Reddy H.; Stefanich E.; Eglen R. M. (1995) Characterization of the interaction of zamifenacin at muscarinic receptors in vitro. Eur. J. Pharmacol. 285, 135–142. [DOI] [PubMed] [Google Scholar]
- Watson N.; Daniels D. V.; Ford A. P.; Eglen R. M.; Hegde S. S. (1999) Comparative pharmacology of recombinant human M3 and M5 muscarinic receptors expressed in CHO-K1 cells. Br. J. Pharmacol. 127, 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak G. W.; Snowman A. M.; Snyder S. H. (1975) Selective enhancement of [3H]opiate agonist binding by divalent cations. Mol. Pharmacol. 11, 735–744. [PubMed] [Google Scholar]
- Williams S. P.; Sigler P. B. (1998) Atomic structure of progesterone complexed with its receptor. Nature 393, 392–396. [DOI] [PubMed] [Google Scholar]
- Emonds-Alt X.; Advenier C.; Cognon C.; Croci T.; Daoui S.; Ducoux J. P.; Landi M.; Naline E.; Neliat G.; Poncelet M.; Proietto V.; Van Broeck D.; Vilain P.; Soubrie P.; Le Fur G.; Maffrand J. P.; Breliere J. C. (1997) Biochemical and pharmacological activities of SR 144190, a new potent non-peptide tachykinin NK2 receptor antagonist. Neuropeptides 31, 449–458. [DOI] [PubMed] [Google Scholar]
- Ishizaki H.; Yamada S.; Yanagiguchi K.; Koyama Z.; Ikeda T.; Hayashi Y. (2009) Pre-treatment with tannic acid inhibits the intracellular IL-8 production by chitosan in a human oral epithelial cancer cell line. Oral Med. Pathol. 13, 135–141. [Google Scholar]
- Amitai G.; Moorad D.; Adani R.; Doctor B. P. (1998) Inhibition of acetylcholinesterase and butyrylcholinesterase by chlorpyrifos-oxon. Biochem. Pharmacol. 56, 293–299. [DOI] [PubMed] [Google Scholar]
- Showell H. J.; Pettipher E. R.; Cheng J. B.; Breslow R.; Conklyn M. J.; Farrell C. A.; Hingorani G. P.; Salter E. D.; Hackman B. C.; Wimberly D. J.; et al. (1995) The in vitro and in vivo pharmacologic activity of the potent and selective leukotriene B4 receptor antagonist CP-105696. J. Pharmacol. Exp. Ther. 273, 176–184. [PubMed] [Google Scholar]
- Butler S.; Hall F. R.; Sanford H. N. (1950) An investigation of the mode of action of benadryl as an antihistaminic. J. Lab. Clin. Med. 36, 806. [PubMed] [Google Scholar]
- Rago B.; Liu J.; Tan B.; Holliman C. (2011) Application of the dried spot sampling technique for rat cerebrospinal fluid sample collection and analysis. J. Pharm. Biomed. Anal. 55, 1201–1207. [DOI] [PubMed] [Google Scholar]
- Bednar M.; Chieffo C.; Morse T.; Duplin M.; Gibbs M. (2004) Plasma and cerebrospinal fluid (CSF) pharmacokinetics of CP-457,920, a selective alpha 5 GABA-A receptor inverse agonist in young, healthy volunteers. Clin. Pharmacol. Ther. 75, 90. [Google Scholar]
- Gauthier D. R. Jr.; Limanto J.; Devine P. N.; Desmond R. A.; Szumigala R. H. Jr.; Foster B. S.; Volante R. P. (2005) Palladium-catalyzed regioselective arylation of imidazo[1,2-b][1,2,4]triazine: synthesis of an alpha 2/3-selective GABA agonist. J. Org. Chem. 70, 5938–5945. [DOI] [PubMed] [Google Scholar]
- Zhu C.; Hansen A. R.; Bateman T.; Chen Z.; Holt T. G.; Hubert J. A.; Karanam B. V.; Lee S. J.; Pan J.; Qian S.; Reddy V. B.; Reitman M. L.; Strack A. M.; Tong V.; Weingarth D. T.; Wolff M. S.; MacNeil D. J.; Weber A. E.; Duffy J. L.; Edmondson S. D. (2008) Discovery of imidazole carboxamides as potent and selective CCK1R agonists. Bioorg. Med. Chem. Lett. 18, 4393–4396. [DOI] [PubMed] [Google Scholar]
- McLean S.; Ganong A.; Seymour P. A.; Bryce D. K.; Crawford R. T.; Morrone J.; Reynolds L. S.; Schmidt A. W.; Zorn S.; Watson J.; Fossa A.; DePasquale M.; Rosen T.; Nagahisa A.; Tsuchiya M.; Heym J. (1996) Characterization of CP-122,721; a nonpeptide antagonist of the neurokinin NK1 receptor. J. Pharmacol. Exp. Ther. 277, 900–908. [PubMed] [Google Scholar]
- Kelly J. (2009) Appendices, in Principles of CNS Drug Development: From Test Tube to Patient pp 287–293, John Wiley & Sons, Ltd., Chichester, U.K. [Google Scholar]
- Metro T. X.; Cochi A.; Gomez Pardo D.; Cossy J. (2011) Asymmetric synthesis of an antagonist of neurokinin receptors: SSR 241586. J. Org. Chem. 76, 2594–2602. [DOI] [PubMed] [Google Scholar]
- Croci T.; Cecchi R.; Marini P.; Rouget C.; Viviani N.; Germain G.; Guagnini F.; Fradin Y.; Descamps L.; Pascal M.; Advenier C.; Breuiller-Fouche M.; Leroy M. J.; Bardou M. (2007) In vitro and in vivo pharmacological characterization of ethyl-4-[trans-4-[((2S)-2-hydroxy-3-[4-hydroxy-3[(methylsulfonyl)amino]-phenoxy]p ropyl) amino]cyclohexyl]benzoate hydrochloride (SAR150640), a new potent and selective human beta3-adrenoceptor agonist for the treatment of preterm labor. J. Pharmacol. Exp. Ther. 321, 1118–1126. [DOI] [PubMed] [Google Scholar]
- Walsh S. L.; Strain E. C.; Abreu M. E.; Bigelow G. E. (2001) Enadoline, a selective kappa opioid agonist: comparison with butorphanol and hydromorphone in humans. Psychopharmacology (Berlin, Ger.) 157, 151–162. [DOI] [PubMed] [Google Scholar]
- Gray J. A.; Roth B. L. (2007) The pipeline and future of drug development in schizophrenia. Mol. Psychiatry 12, 904–922. [DOI] [PubMed] [Google Scholar]
- Mente S.; Gallaschun R.; Schmidt A.; Lebel L.; Vanase-Frawley M.; Fliri A. (2008) Quantitative structure-activity relationship of phenoxyphenyl-methanamine compounds with 5HT2A, SERT, and hERG activities. Bioorg. Med. Chem. Lett. 18, 6088–6092. [DOI] [PubMed] [Google Scholar]
- Middleton D. S.; Andrews M.; Glossop P.; Gymer G.; Hepworth D.; Jessiman A.; Johnson P. S.; MacKenny M.; Stobie A.; Tang K.; Morgan P.; Jones B. (2008) Designing rapid onset selective serotonin re-uptake inhibitors. Part 3: Site-directed metabolism as a strategy to avoid active circulating metabolites: structure-activity relationships of (thioalkyl)phenoxy benzylamines. Bioorg. Med. Chem. Lett. 18, 5303–5306. [DOI] [PubMed] [Google Scholar]
- Bromidge S. M.; Dabbs S.; Davies D. T.; Davies S.; Duckworth D. M.; Forbes I. T.; Gaster L. M.; Ham P.; Jones G. E.; King F. D.; Mulholland K. R.; Saunders D. V.; Wyman P. A.; Blaney F. E.; Clarke S. E.; Blackburn T. P.; Holland V.; Kennett G. A.; Lightowler S.; Middlemiss D. N.; Trail B.; Riley G. J.; Wood M. D. (2000) Biarylcarbamoylindolines are novel and selective 5-HT(2C) receptor inverse agonists: identification of 5-methyl-1-[[2-[(2-methyl-3-pyridyl)oxy]- 5-pyridyl]carbamoyl]-6-trifluoromethylindoline (SB-243213) as a potential antidepressant/anxiolytic agent. J. Med. Chem. 43, 1123–1134. [DOI] [PubMed] [Google Scholar]
- Knight A. R.; Misra A.; Quirk K.; Benwell K.; Revell D.; Kennett G.; Bickerdike M. (2004) Pharmacological characterisation of the agonist radioligand binding site of 5-HT(2A), 5-HT(2B) and 5-HT(2C) receptors. Naunyn-Schmiedeberg’s Arch. Pharmacol. 370, 114–123. [DOI] [PubMed] [Google Scholar]
- Ashcroft F. J.; Newberg J. Y.; Jones E. D.; Mikic I.; Mancini M. A. (2011) High content imaging-based assay to classify estrogen receptor-alpha ligands based on defined mechanistic outcomes. Gene 477, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G.; Cunningham M.; Kim S.; Burn T.; Lin J.; Sinz M.; Hamilton G.; Rizzo C.; Jolley S.; Gilbert D.; Downey A.; Mudra D.; Graham R.; Carroll K.; Xie J.; Madan A.; Parkinson A.; Christ D.; Selling B.; LeCluyse E.; Gan L. S. (2002) CYP3A4 induction by drugs: correlation between a pregnane X receptor reporter gene assay and CYP3A4 expression in human hepatocytes. Drug Metab. Dispos. 30, 795–804. [DOI] [PubMed] [Google Scholar]
- Le Fur G.; Vaucher N.; Perrier M. L.; Flamier A.; Benavides J.; Renault C.; Dubroeucq M. C.; Gueremy C.; Uzan A. (1983) Differentiation between two ligands for peripheral benzodiazepine binding sites, [3H]RO5–4864 and [3H]PK 11195, by thermodynamic studies. Life Sci. 33, 449–457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.