ABSTRACT
The human JC polyomavirus (JCPyV) is the causative agent of the fatal, demyelinating disease progressive multifocal leukoencephalopathy (PML). The Mad-1 prototype strain of JCPyV uses the glycan lactoseries tetrasaccharide c (LSTc) and serotonin receptor 5-HT2A to attach to and enter into host cells, respectively. Specific residues in the viral capsid protein VP1 are responsible for direct interactions with the α2,6-linked sialic acid of LSTc. Viral isolates from individuals with PML often contain mutations in the sialic acid-binding pocket of VP1 that are hypothesized to arise from positive selection. We reconstituted these mutations in the Mad-1 strain of JCPyV and found that they were not capable of growth. The mutations were then introduced into recombinant VP1 and reconstituted as pentamers in order to conduct binding studies and structural analyses. VP1 pentamers carrying PML-associated mutations were not capable of binding to permissive cells. High-resolution structure determination revealed that these pentamers are well folded but no longer bind to LSTc due to steric clashes in the sialic acid-binding site. Reconstitution of the mutations into JCPyV pseudoviruses allowed us to directly quantify the infectivity of the mutants in several cell lines. The JCPyV pseudoviruses with PML-associated mutations were not infectious, nor were they able to engage sialic acid as measured by hemagglutination of human red blood cells. These results demonstrate that viruses from PML patients with single point mutations in VP1 disrupt binding to sialic acid motifs and render these viruses noninfectious.
IMPORTANCE
Infection with human JC polyomavirus (JCPyV) is common and asymptomatic in healthy individuals, but during immunosuppression, JCPyV can spread from the kidney to the central nervous system (CNS) and cause a fatal, demyelinating disease, progressive multifocal leukoencephalopathy (PML). Individuals infected with HIV, those who have AIDS, or those receiving immunomodulatory therapies for autoimmune diseases are at serious risk for PML. Recent reports have demonstrated that viral isolates from PML patients often have distinct changes within the major capsid protein. Our structural-functional approach highlights that these mutations result in abolished engagement of the carbohydrate receptor motif LSTc that is necessary for infection. Viruses with PML-associated mutations are not infectious in glial cells, suggesting that they may play an alternative role in PML pathogenesis.
Introduction
The human JC polyomavirus (JCPyV) is an icosahedral, nonenveloped double-stranded DNA (dsDNA) virus and a member of the Polyomaviridae family (1). JCPyV infects approximately 50% of the population, and the infection is asymptomatic in healthy individuals (2, 3). Viral spread likely occurs via a fecal-oral route, as JCPyV is shed in the urine of healthy individuals (4) and can be detected in untreated wastewater (5–7). The site of initial infection is thought to be the stromal cells of the tonsils (8), followed by a persistent infection in the kidney (9) and in B lymphocytes of the bone marrow (10–12). In healthy individuals, JCPyV remains in the kidney, but in immunosuppressed individuals, JCPyV can spread to the central nervous system (CNS) (10, 13–15) and infect astrocytes and oligodendrocytes (16, 17). Oligodendrocytes produce myelin, and astrocytes are critical to the process of myelination in the CNS (18–20). JCPyV infection of astrocytes and cytolytic destruction of the oligodendrocytes cause the fatal, demyelinating disease progressive multifocal leukoencephalopathy (PML) (21, 22). PML is a devastating disease that can result in fatality within 3 months to 1 year of symptom onset if untreated (23). PML affects approximately 3 to 5% of HIV-1-positive individuals, is considered an AIDS-defining illness, and is one of the most common CNS-related diseases in AIDS (22). Since 2005, the incidence of PML has risen in individuals receiving immunomodulatory therapies for autoimmune diseases (24). In particular, individuals with multiple sclerosis (MS) who are receiving the biological therapy natalizumab have a 1:500 chance of developing PML (25, 26). Natalizumab is an anti-VLA-4 (α4β1 integrin) antibody that blocks extravasation of VLA-4+ T and B lymphocytes to the brain, where they normally bind to endothelial cells (27). Therefore, while this treatment prevents the movement of lymphocytes to the brain, thus protecting the brain of an MS patient from attack, the lack of immune surveillance can also result in increased spread of JCPyV to the brain and thus increase the chances of developing PML (25).
The mechanisms of JCPyV spread to the CNS and infection of glial cells are not well understood, although spread is thought to occur via a hematogenous route, possibly involving B lymphocytes (28–30). In addition, it is well documented that JCPyV undergoes certain polymorphic changes within the host that render it neurotropic. The nonpathogenic form of virus that resides in the kidney is referred to as the archetype strain (Cy) and can be detected in the urine of healthy individuals (4, 31–33). JCPyV undergoes rearrangements in the noncoding control region (NCCR), which contains the viral origin of replication and sequences that serve as binding sites for transcription factors necessary for transcription of viral early and late genes (34–38). These rearrangements include duplication of enhancer elements to convert the virus to the neuropathogenic form (34, 39, 40). Viruses found in the cerebral spinal fluid (CSF), brain tissue, and blood but not in the urine contain NCCR rearrangements and are referred to as PML-type strains (22). Mad-1 is the laboratory prototype strain of the PML-type strain that was originally isolated from the brain of a PML patient and contains a canonical 98-bp tandem repeat in the NCCR (35). While NCCR rearrangements are necessary for JCPyV growth in the CNS, the incidence of PML is relatively low, given the rates of seropositivity (22). Thus, it is likely that other viral, cellular, and individual host factors play a role in PML pathogenesis.
Recently, a number of studies have reported that viral isolates from PML patients also contain mutations in the viral capsid VP1 protein. VP1 is a pentameric protein that interacts with neighboring VP1 pentamers through C-terminal extensions linking together 72 pentamers to form the viral capsid. VP1 serves as the viral attachment protein and mediates direct interactions with cell surface receptors (41). Initial studies to define the receptors for JCPyV infection revealed that JCPyV utilizes sialic acid (42, 43) and the 5-HT2A receptor (44). The sialic acid component of the receptor was thought to be an α2,3- or α2,6-linked sialic acid (45). Our laboratory demonstrated that the presence of α2,6-linked sialic acid correlates with JCPyV infection of cells and tissues in the host, including B lymphocytes, kidney, and the glial cells astrocytes and oligodendrocytes (46). Moreover, JCPyV infection of B lymphocytes, kidney, and glial cells is mediated by sialic acid (42, 45, 47, 48). We identified the specific sialic acid receptor motif for the Mad-1 strain of JCPyV as lactoseries tetrasaccharide c (LSTc), which terminates in α2,6-linked sialic acid (41). The high-resolution crystal structure of JCPyV VP1 pentamers in complex with LSTc revealed that the protein specifically binds to the terminal α2,6-linked sialic acid and engages LSTc in a unique L-shaped conformation. Mutation of VP1 residues that contact LSTc leads to a severe defect in viral growth in glial cells (41). Interestingly, viruses isolated from the blood and CSF of individuals with PML exhibit mutations in these VP1 residues, including L54F, S266F, and S268F/Y (49–53), which are located in the sialic acid-binding pocket of VP1 (41). However, these mutations have never been found in the urine of healthy individuals, nor are they present in the urine of individuals with PML, in whom viral isolates with PML-associated mutations are found in the CSF and blood. This suggests that these mutations can arise within the JCPyV-infected individual (50–53). Analysis of viral isolates from urine, blood, and CSF from a single patient infected with a single JCPyV genotype supports this hypothesis. While virus isolated from the blood and CSF of this individual carried PML-associated mutations in VP1 (L54F, N264S, and S266F), these mutations were not present in virus isolated from the urine (53). Mutations in VP1 sialic acid-binding sites arise with high frequency, in as many as 80 to 90% of the viral isolates, indicating that the majority of the isolates from individuals with PML exhibit mutations in one or more of these sites in VP1 (50, 52–55). The role of some of the PML-associated mutations has been analyzed as virus-like particles (VLPs) in the JCPyV genotype 3 background. VLPs with PML-associated mutations have an altered capacity to engage sialic acid-containing receptors, as VLPs with PML-associated mutations bind to glial cells in a neuraminidase-insensitive manner and bind to gangliosides as measured by enzyme-linked immunosorbent assay (ELISA) (50, 52). However, the role of these mutations in JCPyV infection and spread in glial cells has not been addressed.
One hypothesis is that the mutations in VP1 sialic acid-binding sites might render the virus more pathogenic in infected hosts by allowing it to spread more readily to the brain due to reduced nonspecific attachment to sialic acid pseudoreceptors. However, it is also possible that viruses with PML-associated mutations are defective particles produced due to high levels of viral replication or that they are immune escape mutants. Thus, we questioned whether these viruses represent the pathogenic form of the virus that infects glial cells in the CNS. To this end, we utilized a structure-function approach to define the role of these mutations in JCPyV attachment to cellular receptors and infection of glial cells by generating the PML-associated mutations in an infectious viral clone, in pseudoviruses, and in purified VP1 pentamers using the Mad-1 prototype PML strain as the backbone.
RESULTS
JC polyomaviruses with PML-associated mutations are not infectious.
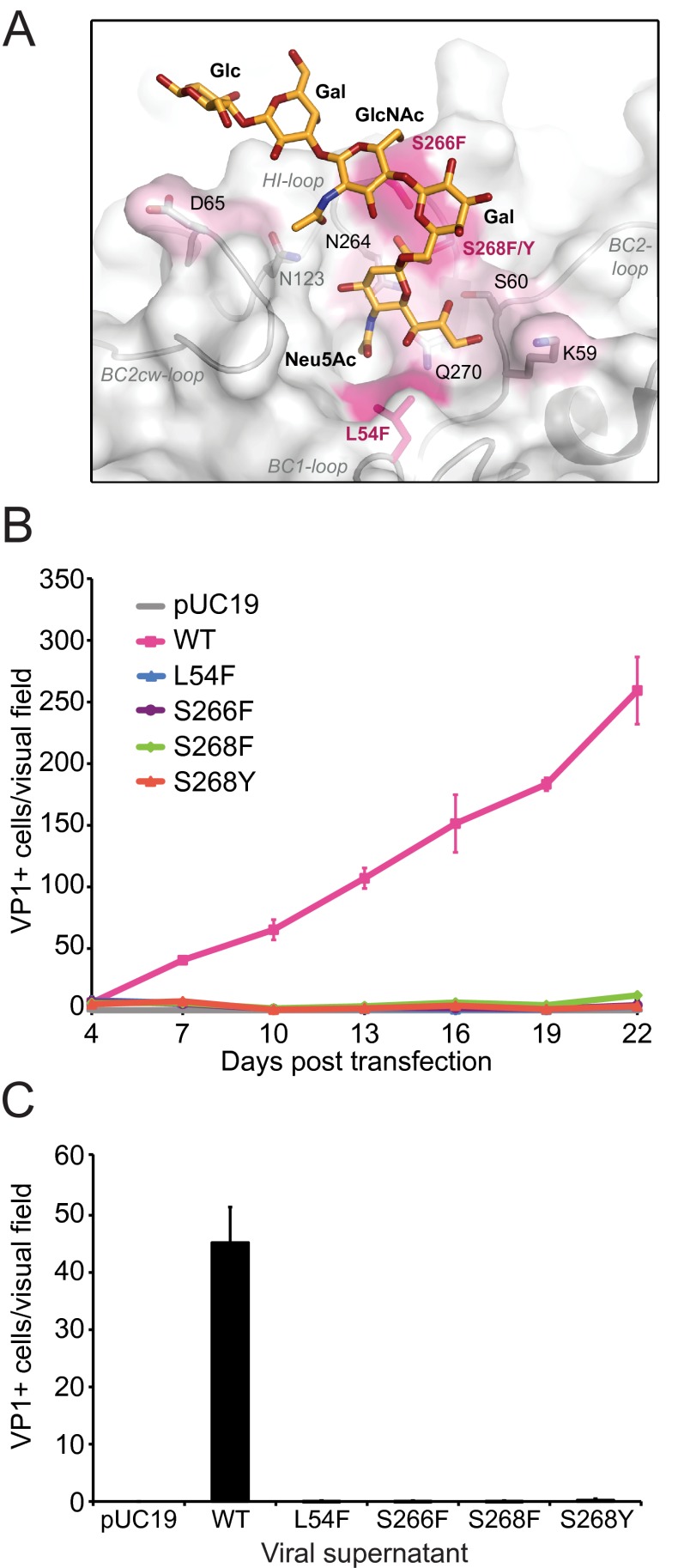
Mutations in JCPyV VP1 arise in individuals with PML at a very high frequency. In fact, 80 to 90% of viral isolates from individuals with PML exhibit mutations in one or more of these sites in VP1, and these mutations are never found in JCPyV isolates from individuals without PML. Furthermore, these mutations are found only in the blood and CSF, but not in the urine, of individuals with PML, indicating that the mutations arise within the host (50, 52–55). The most common PML-associated mutations are L54F and S268F, accounting for approximately 50% of PML-associated mutations, while the mutations S266F and S268Y are less frequent but still common (Table 1) (50, 52). Surprisingly, all four mutations target VP1 residues that form contacts with the terminal sialic acid of the specific receptor motif LSTc (Fig. 1A) (41). To define whether PML-associated VP1 mutations affected JCPyV infectivity, we introduced the most common mutations into the JCPyV infectious genomic clone with a Mad-1 VP1. Viral DNA was transfected into the permissive glial SVG-A cell line and analyzed for growth and spread over 22 days in culture. Infected cells were analyzed by indirect immunofluorescence using an antibody directed against the VP1 capsid protein. Throughout 22 days in culture, only the wild-type JCPyV with a Mad-1 VP1 was capable of growth and spread (Fig. 1B). On day 22, viral supernatants were harvested from cells (in Fig. 1B) and used to infect naive SVG-A cells. Wild-type JCPyV produced infectious viral particles that were released into the supernatant, while viruses with PML-associated mutations did not, indicating that the mutant viruses could not propagate in glial cells (Fig. 1C).
TABLE 1.
Frequency of JCPyV VP1 PML-associated mutations
| Residuesa | % frequency of VP1 mutations in PML patients | Reference |
|---|---|---|
| L54F, K59N, D65H, N264T, S266F/L/T, S268F/Y/C | 81 | 49 |
| L54F, K59M/E/N, N264D/T, S266F/L, S268F/Y/C | 52 | 50 |
| L54F, K59E, D65H, N264D, S266F, S268F/Y, Q270H | 90 | 52 |
| L54F, S60P/T, D65H, N264S, S266F/L, S268F, Q270H | 81 | 53 |
Amino acid numbers are in accordance with the JCPyV Mad-1 strain (1), excluding the methionine at position 1.
FIG 1.
JCPyV with PML-associated mutations are not infectious. (A) PML-associated mutations are highlighted on the wild-type Mad-1 JCPyV VP1 pentamer (surface representation) in complex with LSTc (stick model) (41). Residues in pink indicate mutations used in this study, while those labeled in black represent other PML-associated mutations. (B) Growth of JCPyV VP1 wild-type (WT) and PML-associated mutant viruses. SVG-A cells were transfected with linearized DNA from wild-type and mutant JCPyV. Transfected cells were fixed and stained at day 4 posttransfection and then at 3-day intervals for 22 days by indirect immunofluorescence. Transfected or infected cells were quantified based on nuclear VP1 staining. Each data point represents the average number of infected cells per visual field for 6 fields of view for 3 independent experiments. Error bars indicate standard deviations. (C) Infectivity of supernatants from JCPyV VP1 wild-type and mutant viruses. SVG-A cells were inoculated with supernatants harvested from infected cells at day 22 from panel B. Cells were fixed and stained by indirect immunofluorescence at 72 h postinfection and quantified based on nuclear VP1 staining. The results are presented as the average number of infected cells per visual field for 6 visual fields from 3 individual samples performed in triplicate. Error bars indicate standard deviations.
VP1 pentamers with PML-associated mutations exhibit abolished binding to glial cells.
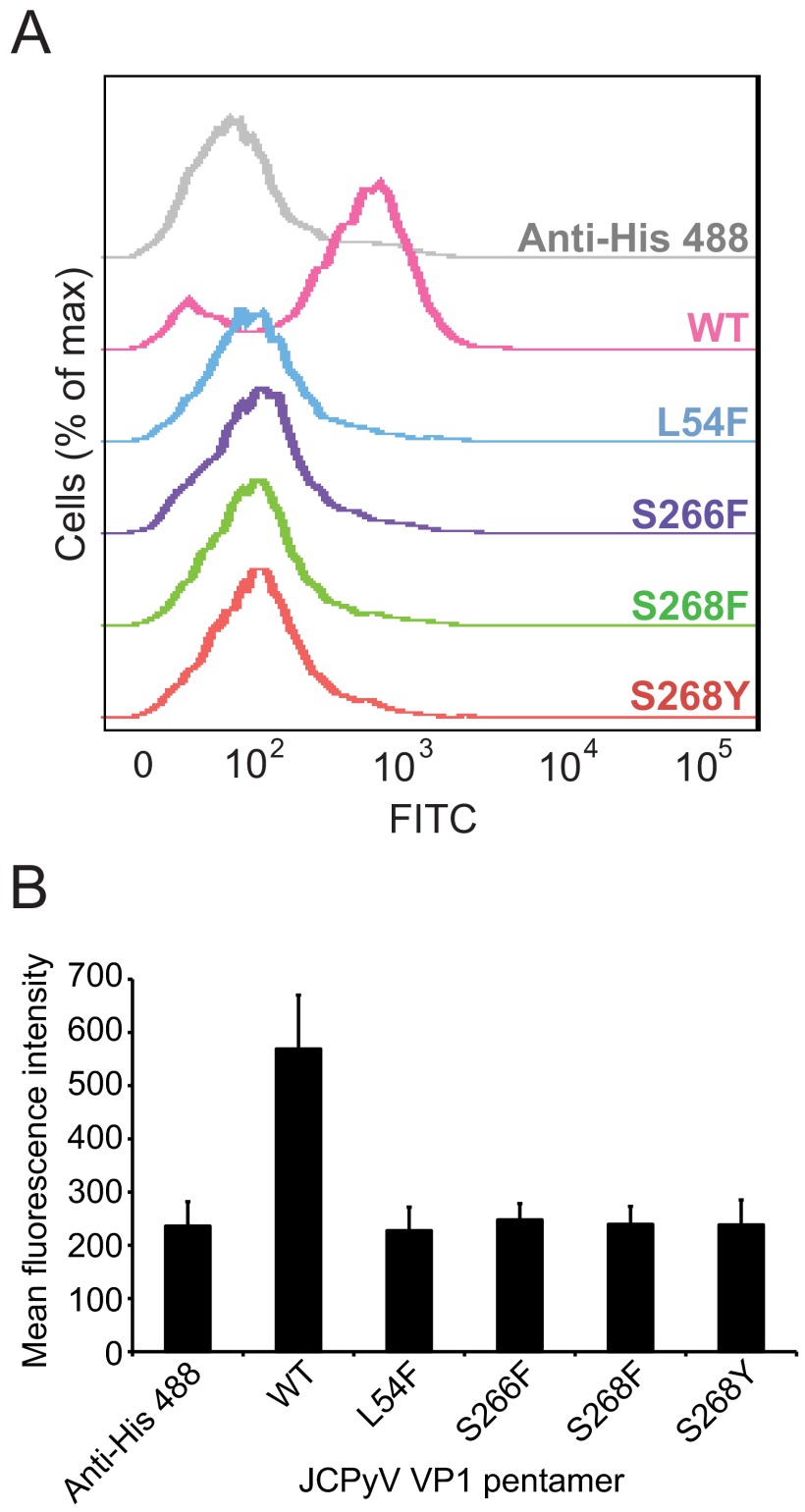
To determine whether the decreased levels of virus growth and infection in the PML-associated mutants correlated with binding to cells, the mutations were introduced into VP1 pentamers. The pentamers serve as a useful tool for studies of JCPyV attachment, entry, and trafficking, as they can bind to cells and traffic through the endosomal compartment to the endoplasmic reticulum with kinetics similar to that of virions (41, 56). Mutated VP1 pentamers were then purified and used to assess binding to SVG-A cells by flow cytometry (Fig. 2). JCPyV wild-type pentamers bind to SVG-A cells, while the VP1 pentamers with PML-associated mutations did not bind to cells, indicating that residues in VP1 that mediate sialic acid binding to the receptor motif LSTc are essential for VP1 engagement of SVG-A cells.
FIG 2.
VP1 pentamers of JCPyV Mad-1 with PML-associated mutations exhibit reduced binding to cells. (A) SVG-A cells were incubated with 100 µg/ml of His-tagged wild-type (WT) or mutant pentamers in PBS, washed, and then incubated with a penta-His Alexa Fluor 488 antibody. Pentamer binding was analyzed by flow cytometry. Histograms represent the fluorescence intensity of Alexa 488 for antibody alone (gray) and pentamer samples for 10,000 gated events. (B) Quantitation of binding of VP1 pentamers with PML-associated mutations. Bar graph represents the mean fluorescence intensity of VP1 pentamers binding to SVG-A cells for 3 independent experiments. Error bars indicate standard deviations.
VP1 pentamers with PML-associated mutations are not capable of binding to LSTc.
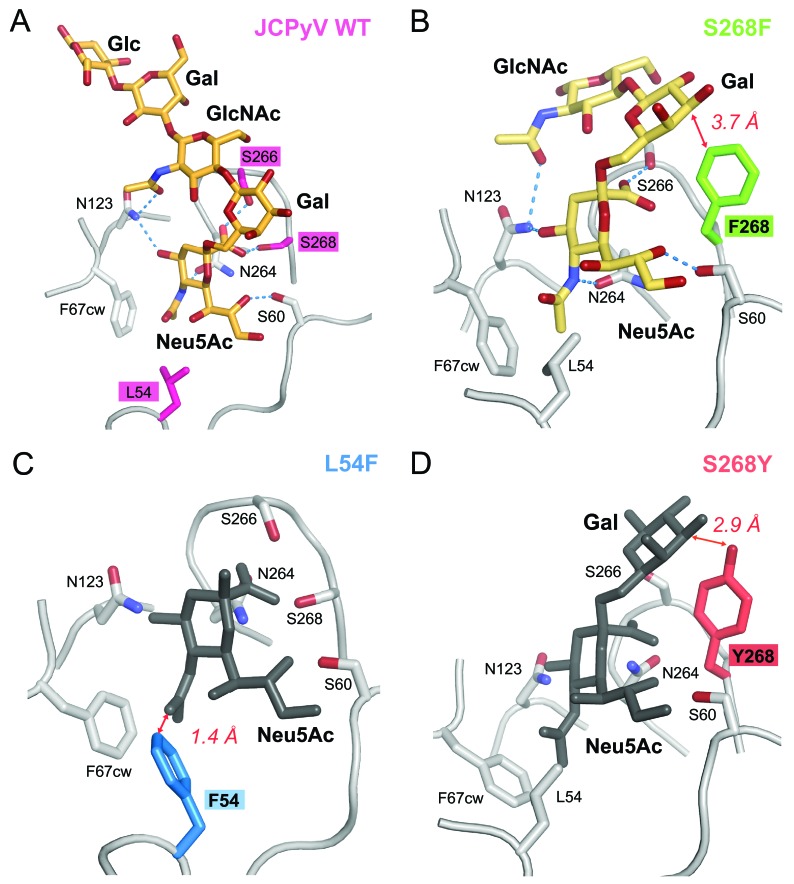
To assess the structural effects of these mutations, we solved crystal structures of three mutant VP1 pentamers: L54F, S268F, and S268Y. In all cases, the crystal lattices are identical to those of the wild-type JCPyV pentamers, with accessible binding sites for LSTc in two of the five VP1 monomers in a given pentamer. In order to analyze whether the mutants can still engage LSTc, crystals of wild-type Mad-1 VP1 pentamers and the L54F, S268F, and S268Y mutants were soaked in LSTc oligosaccharide solution according to the exact procedure used for the complex formation of wild-type VP1 (41), and structures were determined to high resolution (see Table S1 in the supplemental material). Crystal soaking experiments using 5 mM LSTc yielded the JCPyV Mad-1 VP1-LSTc complex structure with two occupied LSTc binding sites per VP1 pentamer (see Fig. S1B) but did not result in additional electron density for carbohydrate moieties in the case of any of the mutated VP1 pentamers. This demonstrates that engagement of LSTc is severely compromised in all three mutants.
We next investigated if any of the VP1 mutants are able to bind LSTc at an increased concentration of 20 mM ligand and an extended soaking time of 2 h. While the L54F and S268Y mutants still do not show any evidence for binding, weak interactions with LSTc can be observed in the S268F mutant. The simulated annealed omit electron density map for S268F clearly shows features of the Neu5Ac-α2,6-Gal-β1,4-GlcNAc portion of LSTc in two LSTc binding sites (see Fig. S1A in the supplemental material). One of these trisaccharides could be built. The structural rearrangements in VP1 upon recognition of LSTc are similar to those seen in the JCPyV Mad-1 VP1-LSTc complex, providing additional evidence for the presence of LSTc (41). N123 interacts via hydrogen bonding with the terminal Neu5Ac and GlcNAc N-acetyl group and is critical for the recognition of LSTc in the L-shaped conformation (Fig. 3A). The N123 side chain is rearranged in the S268F structure in order to accommodate the terminal Neu5Ac (Fig. 3B). Consequently, residues 64 to 68 of the clockwise BC2 loop move to prevent clashes with N123 in the new position. Density for these induced-fit movements as well as partially for the native conformation can be observed in the S268F VP1-LSTc complex structures. These findings are consistent with the occupancy of <1.0 of the carbohydrate ligand within the final complex structure. We conclude that the S268F mutant retains some affinity for LSTc, whereas the other two mutants do not.
FIG 3.
JCPyV VP1 proteins with PML-associated mutations have altered oligosaccharide-binding sites. (A) JCPyV wild-type (WT) VP1 pentamer-LSTc complex. (B) S268F VP1 pentamer-LSTc complex. (C) Unliganded L54F VP1 pentamer. (D) Unliganded S268Y VP1 pentamer. The JCPyV VP1 proteins are shown in cartoon representation. Important residues contributing to ligand binding and specificity are shown in stick representation. Side chains of residues at positions 54, 266, and 268 are highlighted in color. Key hydrogen bonds are shown as blue dashes, and red arrows indicate important distances. Carbohydrate residues are shown in stick representation and colored in orange or light orange (light orange indicates binding with reduced affinity) when present in the complex structures. Carbohydrate moieties depicted in gray are shown only for reference purposes and are obtained by superposing with the JCPyV WT VP1 pentamer-LSTc complex structure.
VP1 pentamers with PML-associated mutations clash with LSTc.
We next examined the effects of the three mutations L54F, S268F, and S268Y on the VP1 structure. Residues L54, S266, and S268 mediate direct contacts or are in close proximity to the terminal Neu5Ac within the LSTc binding site of the wild-type JCPyV VP1-LSTc complex (Protein Data Bank [PDB] accession code 3NXD) (41) (Fig. 3). In order to compare the effects of the mutations on the VP1 structure and its ligand binding properties, the L54F, S268F, and S268Y VP1 structures were superposed onto the JCPyV wild-type VP1-LSTc complex structure. In all three VP1 mutant structures, well-defined electron density could be observed for the respective mutated residue, confirming the presence of the mutation.
Analysis of the S268F VP1 structure shows that the Neu5Ac carboxyl group could still be recognized by S266 via a hydrogen bond, but the hydrogen bond with the side chain of residue 268 within the HI loop would be lost. The side chain of F268 and the α2,6-linked Gal are 3.7 Å apart. Although this is a close contact, it would still allow for engagement of LSTc without leading to severe clashes. Consistent with this, interaction with LSTc was observed with reduced affinity for S268F VP1 in the soaking experiment with 20 mM LSTc (Fig. 3B). Based on B-factor analysis of the bound LSTc and surrounding residues, we expect that the occupancy of LSTc is about 0.6 to 0.8 in one accessible binding site (chain C). The second binding site (chain B) has weaker electron density for LSTc, indicating lower occupancy.
In the case of the L54F VP1 mutant structure, the phenyl side chain of F54 points into the Neu5Ac binding pocket. With a distance of only 1.4 Å between the Neu5Ac moiety in the respective location and the F54 side chain, LSTc binding would lead to severe clashes (Fig. 3C). Finally, the S268Y VP1 structure shows that introduction of the larger tyrosine side chain at position 268 would lead to a close contact of 2.9 Å between the Y268 hydroxyl group and the α2,6-linked Gal. This steric interference likely blocks recognition of the LSTc motif (Fig. 3D).
The S266F mutation also occurs frequently in PML patients. We did not succeed in crystallizing the S266F VP1 pentamer, but modeling suggests that replacement of S266 with a bulky phenylalanine would abolish hydrogen bond formation with the Neu5Ac carboxyl group. In addition, all allowed phenylalanine side chain rotamer conformations would clash with the GlcNAc moiety of the long leg of the L-shaped LSTc motif. Therefore, binding of LSTc is likely also abolished for S266F VP1. We therefore conclude that the most frequently observed VP1 mutations in PML patients directly alter contacts of VP1 with LSTc, with various effects on binding ranging from substantially reduced interactions for the S268F mutant to clashes for the L54F and S268Y (and likely also the S266F) mutants.
JCPyV pseudoviruses with PML-associated mutations are not infectious.
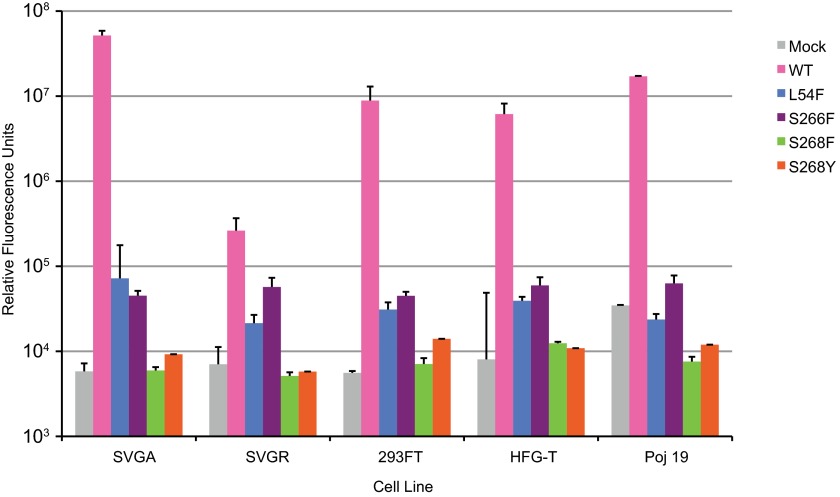
The structural analyses demonstrate that JCPyV pentamers expressing PML mutations are not capable of binding to the functional receptor motif α2,6-linked LSTc (Fig. 3), and the functional data suggest that JCPyV PML mutants cannot bind to or grow in SVG-A cells (Fig. 1 and 2). This led us to hypothesize that JCPyV PML mutants may have low levels of binding and growth that are not easily detected in our established culture model of viral infection. While it is not possible to propagate viruses with growth defects by standard methods, we have established a JCPyV pseudovirus system to help determine the infectivity of mutant viruses. The pseudoviruses are generated by transfecting the viral capsid proteins VP1, VP2, and VP3 into HEK293FT cells together with a reporter plasmid that expresses both green fluorescent protein (GFP) and a secreted form of luciferase (Gaussia luciferase). This assay is robust and provides a sensitive method of detecting the infectivity of mutant viruses that do not propagate under traditional culture methods. The PML-associated mutations were introduced into the Mad-1 VP1 and expressed in the pseudovirus system. Wild-type and mutant purified pseudoviruses were tested for infectivity in the human brain cell types SVG-A (glial), SVG-R (glial variant resistant to JCPyV infection), Poj19II (glial), and HFG-T (human fetal glial cells) or the kidney cell line HEK293FT. Infectivity was measured at 72 h postinfection by luciferase (Fig. 4). The JCPyV pseudoviruses with PML-associated mutations were not infectious in any of the cell types tested, including SVG-A cells, compared to the mock control and wild type (Mad-1) (Fig. 4). SVG-R cells are a variant of SVG cells that are resistant to JCPyV infection despite levels of virus binding equivalent to those of SVG-A cells (57). Thus, the SVG-R cells serve as a useful control to demonstrate the level of background luciferase readings in this assay. Furthermore, the cell lines used had variable levels of 5-HT2AR expression (data not shown), indicating that the presence of 5-HT2AR does not influence infection of viruses with PML-associated mutations. These data confirm that LSTc is a critical receptor motif for JCPyV infection and suggest that 5-HT2AR is mediating a postattachment step in the virus life cycle. However, the expression of T antigen does influence infection by pseudoviruses, as the expression plasmid contains a simian virus 40 (SV40) origin of replication. Therefore, HEK293FT cells which have high levels of T antigen expression are readily infected by the pseudovirus, although kidney cells are generally less susceptible to JCPyV infection in the absence of T antigen in traditional culture models of JCPyV infection (48).
FIG 4.
JCPyV pseudoviruses with PML-associated mutations are not infectious. The cell types shown were plated in a 96-well plate O/N. Medium was aspirated, cells were infected with 1 × 107 particles/ml of wild-type (Mad-1) or mutant pseudovirus in incomplete medium without phenol red at 37°C for 1 h, then complete phenol red-free medium was added, and cells were incubated at 37°C for 72 h. Supernatant from infected cells was collected and analyzed for secreted luciferase using a luminometer. The average relative luciferase units for quadruplicate samples are shown in log scale. Error bars represent standard deviations. These data are representative of 3 experiments performed in triplicate.
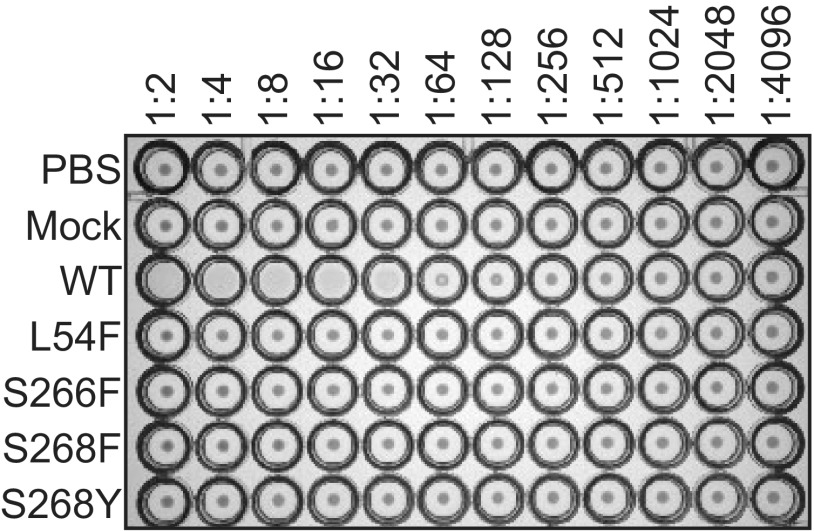
JCPyV pseudoviruses with PML-associated mutations do not hemagglutinate human RBCs.
The viruses and pseudoviruses with PML-associated mutations were not infectious in a number of cell lines tested. Further, purified pentamers do not bind to SVG-A cells and have significantly reduced binding to LSTc as analyzed by X-ray crystallography. In order to determine if the PML-mutant pseudoviruses were capable of binding to other sialic acid receptors, a hemagglutination assay was performed. Wild-type JCPyV hemagglutinates human type O red blood cells (RBCs) (42). Wild-type and PML-associated mutant JCPyV pseudoviruses of equal pseudovirus particle concentrations were tested for the ability to agglutinate RBCs (Fig. 5). The PML mutant pseudoviruses were not capable of hemagglutination of red blood cells, while the wild-type JCPyV pseudovirus resulted in agglutination at a titer of 3.1 × 106 pseudovirus particles. These data suggest that the viruses isolated from individuals with PML which have mutations in VP1 that are critical for binding to LSTc no longer bind to sialic acid.
FIG 5.
JCPyV pseudoviruses with PML-associated mutations do not bind to sialic acid. Wild-type (WT) and mutant pseudoviruses (1 × 107 particles) were added to U-bottomed 96-well plates containing equal volumes of PBS, and 2-fold serial dilutions were made across the plate. Human type O RBCs were added to each well and incubated at 4°C for 4 h. Data are representative of 2 independent experiments.
DISCUSSION
Mutations in the JCPyV capsid protein VP1 frequently arise in patients with PML. In this study, we examined the effect of three frequently occurring mutations on key properties of the virus, including growth, infectivity, capsid protein structure, and receptor binding. We find that all mutations severely compromise the interaction of the virus with its cognate receptor motif, the sialylated LSTc glycan, due to steric interference. As a result, the mutant viruses no longer hemagglutinate, and they no longer grow or infect glial cells. Taken together, these data suggest that although these viruses with PML-associated mutations are frequently found in individuals with PML (52, 53), they are likely not infectious in glial cells in the CNS due to reduced sialic acid binding.
Although viral isolates with VP1 mutations arise in individuals with PML, it has not been demonstrated whether viruses with PML-associated mutations are pathogenic in cells of the CNS. Individuals with PML who develop viruses with PML-associated mutations have higher levels of viral DNA in the CSF (52), yet it is unclear if these viruses are actually capable of increased spread to the brain or increased replication once they have reached the brain. Furthermore, these viruses have not been isolated from JCPyV-infected cells of the brains of individuals with PML but rather from the CSF. Given the avid interaction between JCPyV and cell surface sialic acid, it is possible that the JCPyV isolates with PML-associated mutations are found at a high frequency in the CSF due to their loss of sialic acid binding. The wild-type JCPyV, which retains the ability to bind to sialic acid, might remain cell associated, leading to lower levels of the wild-type virus in the CSF (50, 52). Also, it is possible that PML-associated mutations arise in the viral capsid as a mechanism of immune escape. JCPyV establishes a persistent infection in the kidney of healthy individuals with no significant complications, suggesting that the virus is under immune surveillance. To spread hematogenously to the CNS, the virus may undergo mutations in the viral capsid, making these mutant viruses antigenically distinct and capable of evading the host immune responses, resulting in increased spread. VP1 is the most abundant capsid protein of the virus, and increased antigenicity in the receptor-binding region may allow the pathogenic forms of the virus to spread to the CNS, while the immune system attacks the mutant viruses. Alternatively, mutations in VP1 may arise in the CNS of infected individuals as a mechanism of immune escape, resulting in increased spread and infection of glial cells. Thus, it remains to be demonstrated if the viruses with PML-associated mutations are pathogenic in the brain. Our data demonstrate that JCPyV with PML-associated mutations are not infectious in a panel of glial cell lines.
However, we also cannot exclude the hypothesis that JCPyVs with PML-associated mutations are infectious in the brains of individuals with PML. It is plausible that the loss of sialic acid binding through mutation of specific residues in the viral capsid may be necessary for increased spread to the brain and subsequent infection of glial cells in a sialic acid-independent manner (58, 59). It is reasonable to speculate that reduced binding to sialic acid receptors in the periphery leads to increased spread of JCPyV to the brain, as JCPyV binding to both red blood cells and B lymphocytes is mediated via interactions with sialic acid (Fig. 5) (47). This hypothesis is similar to the scenario demonstrated for mouse polyomavirus (MPyV) in which a mutation in the VP1 sialic acid-binding pocket in a position orthologous to position S268 of JCPyV leads to decreased MPyV binding to RBCs and increases viral dissemination and pathogenicity of the virus, resulting in a lethal outcome (58–61). JCPyV with PML-associated mutations might then engage a non-sialic acid receptor on glial cells in the CNS. It is also possible that the host cell factor necessary for infection by JCPyV with PML-associated mutations is not expressed abundantly on the cell lines that we tested. The crystal structure of JCPyV VP1 revealed a groove on the surface of the pentameric capsid protein that is unique among all polyomaviruses crystallized to date (41, 62, 63). This groove could well be involved in the engagement of JCPyV with other receptor structures on the surface of host cells, and such an interaction might be favored if binding to LSTc is significantly reduced or blocked. There are many differences between the in vitro infection model and a natural in vivo infection that we cannot fully account for in our current tissue culture model of JCPyV infection. JCPyV infects approximately 50% of the population but rarely causes PML and does so only in those who are immunocompromised by HIV infection, AIDS, or immunomodulatory therapies. Therefore, the development of PML must be multifactorial and involve host-specific factors that are influenced in the disease states of HIV infection or immune-mediated diseases such as MS. For instance, the breakdown of the blood-brain barrier and/or use of antiretroviral therapies or natalizumab therapy may cause the upregulation of alternate receptors or host cell factors that allow JCPyV to efficiently attach and enter into glial cells in the CNS. Thus, the tissue culture model of infection may not accurately demonstrate the disease pathogenesis in vivo.
Gorelik and colleagues previously demonstrated that VLPs with PML-associated mutations in the genetic background of the JCPyV genotype 3 have reduced binding to kidney cells, red blood cells, and lymphocytic cells but retain the ability to bind to the glial cells SVG-A cells and astrocytes. Additionally, they demonstrated that introduction of these mutations into a VLP leads to altered receptor usage, consistent with an enhanced affinity for gangliosides, including GM1, GM2, and GD3. However, the JCPyV type 3 VLP binds to asialo-GM1, GD1a, GD1b, GD2, and GT1b (52) and has been reported to bind to GM1 and GM2 (Leonid Gorelik, Consortium for Functional Glycomics [CFG], available online at http://www.functionalglycomics.org, according to CFG policy). This affinity of JCPyV type 3 for gangliosides may have influenced the results of the PML-associated mutations in the type 3 VLPs. Additionally, VLPs of JCPyV have been demonstrated to bind to a panel of gangliosides in viral overlay assays, suggesting that VLPs may nonspecifically engage sialic acid-containing gangliosides, particularly those with α2,6-linkages (64). There are 7 known serotypes of JCPyV and 13 distinct geographic subtypes that have been determined (65). JCPyV isolates from genotypes 1 and 2 are more frequently isolated from individuals than are other genotypes, including type 3 (53, 54). In fact, type 3 strains are not commonly isolated from the CSF of PML patients (50). We analyzed the effect of PML-associated mutations in the Mad-1 prototype strain, the prototype PML-type strain for genotype 1, for which the receptors have been identified (41, 44). Additionally, the amino acid sequences of VP1 of genotypes 1 and 3 differ in up to eight residues (depending on the strain), and some of the mutations are located close to the LSTc binding site. We previously demonstrated that Mad-1 VP1 pentamers do not bind to any ganglioside structures in a glycan array screen (41), and we have not been able to demonstrate that expression of gangliosides influences infection of JCPyV Mad-1 in glial cells (Atwood laboratory, unpublished results). Thus, strain-specific differences in receptor engagement could have influenced the results of the study published by Gorelik et al. (52) when the JCPyV type 3 strain was utilized to study the PML-associated mutations.
Structural analysis of Mad-1 and mutant VP1 pentamers reveals that steric hindrance introduced by PML-associated mutations such as L54F or S268Y abolishes engagement of LSTc, and these mutations would also block binding to other sialylated glycan motifs, independent of their location on gangliosides or glycoproteins or their linkage. Although some mutations, such as S268F, lead to only minor clashes, these mutations would nevertheless drastically reduce the affinity of VP1 for LSTc due to reduced contacts and some steric interference. Consequently, such interactions would likely not be functional for LSTc or any other sialylated glycan whether they are in the genotype 1 or genotype 3 background.
Genotype 3 strains differ from Mad-1 in up to eight amino acid positions. Five of these positions are included in our VP1 pentamer construct (R74, S116, G133, L157, and K163). Residues S116 and G133 are located near the LSTc binding pocket but do not make direct contacts with LSTc, while R74, L157, and K163 are found outside the binding pocket. Additionally, V320, E331, and K344 are found at the C terminus of the full-length VP1 protein and far from the glycan receptor-binding site. Nevertheless, amino acid differences in Mad-1 and genotype 3 that are near the LSTc binding pocket would not change the drastic impact of the mutation L54F or S268F/Y and likely S266F on the interaction with sialylated glycan motifs, as these mutations are located directly in the binding pocket.
Interestingly, engagement of LSTc is likely preserved, perhaps with only moderate decreases in affinity, for less frequently occurring mutations. Reported PML-associated mutations K59M/E/N, S60T, and D65H, which were not addressed in our study, are unlikely to lead to structural changes and would still allow similar contacts with LSTc. In the case of Q270H and N264D/S/T, the hydrogen-bonding network in the Neu5Ac binding pocket would be altered and, thus, these mutations would likely result in an overall reduced affinity for sialylated motifs.
These findings highlight the importance of LSTc as a functional receptor motif. Additionally, we demonstrated that viral isolates with PML-associated mutations have reduced binding to LSTc and other sialic acid motifs, and this reduced binding renders these viruses noninfectious in both kidney and glial cells. Taken together, this work illustrates that engagement of cell surface receptors is an important determinant of tissue tropism and viral pathogenesis for JCPyV infection.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
SVG-A cells are a subclone of the human glial cell line SVG transformed with an origin-defective SV40 mutant (66) and were grown in minimum essential medium (MEM) supplemented to contain 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S) (Mediatech, Inc.) in a humidified incubator at 37°C. SVG-R cells are resistant to JCPyV infection (57) and were grown in MEM supplemented to contain 10% FBS. HEK293FT cells are derived from human embryonal kidney cells transformed with the SV40 large T antigen (Invitrogen Life Technologies) and were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented to contain 10% FBS, 0.1 mM nonessential amino acids (NEAA), 6 mM l-glutamine, 1 mM sodium pyruvate, and 500 mg/ml Geneticin. HFG-T cells are human fetal glial cells transformed with an origin-defective SV40 T antigen (67) grown in modified Eagle’s medium supplemented to contain 10% FBS. Poj19II cells are human fetal glial cells transformed with a replication-defective JCPyV (68) grown in DMEM supplemented to contain 10% FBS. Generation and propagation of the virus strain Mad-1/SVEΔ were previously described (69). JCPyV infection in SVG-A cells was assessed using PAB597, a hybridoma supernatant that produces a monoclonal antibody against JCPyV VP1 (70) and was generously provided by Ed Harlow. Penta-His Alexa Fluor 488 was used at 20 µg/ml (Qiagen).
Viral growth assay.
VP1 mutations were generated in the genomic JCPyV DNA of strain JC12 with a Mad-1 VP1 (68) and subcloned into pUC19 (43). Mutations were introduced by site-directed mutagenesis using an Agilent QuikChange II site-directed mutagenesis kit (Qiagen) according to the manufacturer’s instructions. Sequencing was performed at Genewiz Inc. Primers used for mutagenesis were as follows (with mismatched nucleotides shown in boldface), 5′→3′: L54F, GGTGACCCAGATGAGCATTTTAGGGGTTTTAGTAAGT and ACTTACTAAAACCCCTAAAATGCTCATCTGGGTCACC; S266F, TGTGGCATGTTTACAAACAGGTTTGGTTCCCAGCAG and CTGCTGGGAACCAAACCTGTTTGTAAACATGCCACA; S268F, TTTACAAACAGGTCTGGTTTCCAGCAGTGGAGAGG and CCTCTCCACTGCTGGAAACCAGACCTGTTTGTAAA; S268Y, GTTTACAAACAGGTCTGGTTATCAGCAGTGGAGAGGACTC and GAGTCCTCTCCACTGCTGATAACCAGACCTGTTTGTAAAC.
Ten micrograms of purified plasmid DNA was digested with BamHI (Promega) at 37°C for 2 h to separate the JCPyV genomic DNA from the pUC19 backbone plasmid. Digests were performed in triplicate for each sample and verified by agarose gel electrophoresis. SVG-A cells were plated to 40% confluence in 24-well plates (Corning). Cells in medium without antibiotics were transfected with 1 µg of digested DNA using Fugene (Promega) at a 3:2 ratio (Fugene to DNA). Transfected cells were incubated at 37°C overnight (O/N), and medium containing 5% FBS and 2% P/S was added to cells the next day. Cells were incubated at 37°C and fed with 500 ml of medium containing 5% FBS, 1% P/S, and 1% amphotericin B (Mediatech) or fixed for immunofluorescence staining at day 4 and at 3-day intervals thereafter for 22 days. For the infectivity assay, supernatants were collected from the samples of the growth assay at 22 days posttransfection. SVG-A cells at 70% confluence in 24-well plates (Corning) were infected with 150 µl of virus supernatant at 37°C for 1 h; then 1 ml of medium containing 5% FBS, 1% P/S, and 1% amphotericin B was added to cells; and cells were incubated at 37°C for 72 h. Cells were fixed and stained by indirect immunofluorescence.
Indirect immunofluorescence.
Cells were washed in phosphate-buffered saline (PBS), fixed in cold methanol (MeOH), and incubated at −20°C. Fixed cells were washed in PBS, permeabilized with 0.5% Triton X-100 (TX-100; USB Corporation) at room temperature (RT) for 5 min, incubated with VP1-specific antibody PAB597 (1:10) in PBS at 37°C for 1 h, washed with PBS, incubated with a goat anti-mouse Alexa Fluor 488 (1:1,000)-conjugated antibody in PBS at 37°C for 1 h, and then washed with PBS. Cells were analyzed for nuclear VP1 staining under a 20× objective using an Eclipse TE2000-U microscope (Nikon).
JCPyV pseudovirus production.
Codon optimization of the JCPyV VP1, VP2, and VP3 genes was performed according to the National Cancer Institute Center for Cancer Research Lab of Cellular Oncology Technical Files (http://home.ccr.cancer.gov/LCO/production.asp) to achieve optimal expression in the human-derived cell line 293FT. Genes were synthesized by Blue Heron Biotech, LLC. The VP1 gene was subcloned into the pwP vector in place of the murine polyomavirus (MPyV) VP1 gene. The JCPyV VP2 and VP3 genes were subcloned into the ph2p vectors in place of the MPyV genes (71). The luciferase reporter vector phGluc (72) expresses a secreted form of Gaussia luciferase under the control of the EF1α promoter. All plasmids were obtained from AddGene. Site-specific mutations were made using the Agilent QuikChange II site-directed mutagenesis kit prior to subcloning. Primers for generating PML-associated mutations in VP1 (5′→3′) were as follows, with mismatched nucleotides shown in boldface: L54F, TGGGCGACCCCGATGAACATTTTCGCGGATTC and GAATCCGCGAAAATGTTCATCGGGGTCGCCCA; S266F, GGCATGTTCACAAATCGCTTTGGCTCACAGCAGTGGAGA and CTCCACTGCTGTGAGCCAAAGCGATTTGTGAACATGCC; S268F, CACAAATCGCAGTGGCTTTCAGCAGTGGAGGGGATT and AATCCCCTCCACTGCTGAAAGCCACTGCGATTTGTG; S268Y, CACAAATCGCAGTGGCTACCAGCAGTGGAGGGGATT and AATCCCCTCCACTGCTGGTAGCCACTGCGATTTGTG.
Pseudoviruses were produced by transfection of the VP1, VP2, VP3, and phGluc plasmids into 293FT cells using Fugene 6 transfection reagent (Promega) in a 5:1:1:1 ratio. Mock pseudovirus controls were generated by transfecting HEK293FT cells with control plasmid and the phGluc reporter plasmid in a 7:1 ratio. Cells were harvested 48 h posttransfection by scraping and then pelleted and resuspended in buffer A (10 mM Tris, pH 8, 50 mM NaCl, 0.1 mM CaCl2, 0.01% TX-100) with EDTA-free protease inhibitors (Roche Applied Science). Cells were lysed by three rounds of freezing and thawing, sonicated, and treated with 0.25% deoxycholic acid at 37°C for 30 min. The pH was lowered to 6.0, and the lysates were treated with type V neuraminidase (Sigma) at 37°C for 1 h. The pH was then raised to 7.5, CaCl2 was added, and the lysate was treated with DNase I (New England Biolabs). Pseudoviruses were then purified through an iodixanol gradient by centrifugation at 234,000 × g in an SW55 Ti rotor (Beckman) at 16°C for 3.5 h. The band containing pseudovirus or corresponding control was extracted by syringe.
To determine the titers of pseudoviruses for properly encapsidated genomes, they were again treated with DNase I and protected phGluc was extracted using the DNeasy blood and tissue kit (Qiagen). The titer of the packaged genome was determined using absolute quantification and TaqMan quantitative PCR (qPCR; Applied Biosystems) to create a standard curve using serial dilutions of phGluc. The number of copies for the known plasmid was plotted in a scatter plot against the threshold cycle (CT) value determined for each dilution. A best-fit line was generated, and the trend line equation from regression analysis was used to calculate the relationship between the CT value of the unknown input template and copies of the packaged pseudovirus plasmid. We determined the volume of pseudovirus particles based on the encapsidated genomes to equal 107 viral particles/ml and diluted each sample to use equivalent amounts of pseudovirus to perform experiments.
Pseudovirus luciferase infectivity assay.
Cells were plated in a 96-well plate to 70% confluency and infected with equal particle equivalents (1 × 107 particles/ml) of wild-type and mutant pseudovirus in incomplete medium without phenol red, adjusted for equal volume equivalents with the purification reagent Optiprep (33%). Infected cells were incubated at 37°C for 1 h and washed with PBS, complete medium without phenol red was added, and cells were incubated at 37°C for 72 h. Secreted luciferase was quantitated in 20 to 50 µl of cellular supernatants using the BioLux Gaussia luciferase assay (New England Biolabs) according to the manufacturer’s instructions using an opaque 96-well microplate in a GloMax Multi-Detection System luminometer (Promega) equipped with an autoinjector. Numbers of infected cells were also measured by quantifying GFP-positive cells by fluorescence microscopy using an Eclipse TE2000-U microscope (Nikon).
Generation of pentamers.
cDNA coding for amino acids 22 to 289 of the Mad-1 strain of JCPyV VP1 (UniProtKB entry code P03089) was cloned into the pET15b expression vector (EMD Millipore) in frame with an N-terminal hexahistidine tag (His tag) and a thrombin cleavage site as described previously (41). Mutations were introduced by site-directed mutagenesis with the Agilent QuikChange mutagenesis kit according to the manufacturer’s instructions using primers listed for generation of mutant viruses for viral growth assay. Sequences were verified by Genewiz. VP1 pentamers were purified as described previously (56). Protein concentrations were determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific), and pentamers were concentrated to ~1 mg/ml.
Crystallization.
JCPyV VP1 pentamers carrying mutation L54F, S268F, or S268Y were concentrated to 4.5 mg/ml in 20 mM HEPES (pH 7.5), 150 mM NaCl and crystallized at 20°C using the sitting drop vapor diffusion technique and 100 mM HEPES (pH 7.5), 200 mM KSCN, 12% (wt/vol) polyethylene glycol 3350 (PEG 3350) as the reservoir solution. Drops were set up by mixing the protein solution 1:1 with the reservoir solution and cross-seeded by adding 0.2 µl of a microseeding solution obtained from previous JCPyV VP1 crystals. In order to test for interaction with LSTc oligosaccharide, crystals were soaked in the reservoir solution complemented with 5 mM LSTc (Dextra, United Kingdom) for 3 min or with 20 mM LSTc for 2 h. Subsequently, crystals were transferred for 2 s into a harvesting solution supplemented with 30% (vol/vol) glycerol and 5 mM or 20 mM LSTc, respectively, and then flash-frozen in liquid nitrogen.
Data collection and structure determination.
Data sets were collected at beamline X06DA at SLS (Villigen, Switzerland). Diffraction data were processed with XDS (73), and structures were solved by molecular replacement with Phaser in CCP4 (74, 75). The native JCPyV Mad-1 VP1 structure (PDB accession number 3NXG) lacking solvent molecules was used as the search model. Rigid body and simulated annealing refinement was carried out with Phenix (76) in order to remove model bias. Crystals of L54F, S268F, and S268Y mutant pentamers have the same space group, and similar unit cell parameters, as do wild-type JCPyV Mad-1 VP1 pentamers, with a single VP1 pentamer forming the asymmetric unit in each case. Mutations were introduced into the respective models, and water molecules were added (excluding the LSTc binding site) using Coot (77). Alternating rounds of model building in Coot and restrained refinement, including 5-fold NCS restraints and the translation-libration-screw (TLS) method (78), were performed with Refmac5 (79). In order to test for the presence of LSTc, simulated annealing Fobs-Fcalc electron density maps in the putative LSTc binding site of the mutated VP1 pentamers soaked with LSTc were compared to the respective omit map of the Mad-1 VP1-LSTc complex. In the case of S268F VP1, which showed some electron density for LSTc, the ligand was built and refined using the CCP4 suite library and user-defined restraints for the α2,6-glycosidic bond. Structure figures were prepared with PyMOL (PyMOL Molecular Graphics System, version 1.3; Schrödinger, LLC).
Flow cytometry.
Purified wild-type and mutant JCPyV VP1 pentamers (100 µg/ml) in a 100-µl total volume of PBS were incubated with SVG-A cells in suspension on ice for 2 h with occasional agitation. Cells were washed, pelleted by centrifugation, and suspended in 75 µl of penta-His Alexa Fluor 488 antibody (Qiagen) (20 µg/ml) in PBS on ice for 1 h. Cells were washed, pelleted, resuspended in PBS, and analyzed for pentamer binding using a BD FACSCalibur (Becton, Dickinson and Company) flow cytometer equipped with a 488-nm excitation line. Data were analyzed using BD CellQuestPro (Becton, Dickinson and Company) and FlowJo software (Tree Star, Inc.).
Hemagglutination assay with JC pseudoviruses.
Serial 2-fold dilutions of 1 × 107 particles of wild-type or mutant JC pseudoviruses in a volume of 99 µl were prepared in PBS in a U-bottomed 96-well plate (Corning) (dilution range, 1:2 to 1:4,096). A volume equivalent (99 µl) of 1× PBS was used as a negative control. Human type O negative red blood cells (RBCs) that had been washed with Alsever’s buffer (0.1 M d-glucose, 0.027 M sodium citrate, 0.07 M NaCl, pH 6.5) were added to each well at a volume equivalent of 99 µl, and plates were gently agitated. Plates were incubated at 4°C for 4 h and imaged using a ChemiDoc XRS imager (Bio-Rad).
Protein structure accession numbers.
Coordinates and structure factor amplitudes have been deposited with the RCSB Protein Data Bank (http://www.pdb.org) under accession codes 4JCE (L54F), 4JCF (S268F), and 4JCD (S268Y) of the structures obtained after crystal soaking in harvesting solution supplemented with 20 mM LSTc.
SUPPLEMENTAL MATERIAL
X-ray crystallography studies of L54F, S268F, and S268Y JCPyV VP1 pentamers and their engagement of the LSTc receptor motif. Data collection and structural refinement statistics.
Simulated annealing omit electron density maps of LSTc in the S268F VP1 pentamer-LSTc complex structure (A) and the JCPyV Mad-1 VP1 pentamer-LSTc complex structure (B) shown in a close-up view of the binding pocket. The proteins are shown in cartoon representation with one VP1 monomer highlighted in green (A) or pink (B) and neighboring monomers depicted in gray. A simulated annealed Fobs-Fcalc density omit map contoured at 2.5 σ is shown for 2.5 Å around the respective carbohydrate residues. The oligosaccharide is drawn as a stick model and colored according to atom type (nitrogens in blue, oxygens in red, and carbons in light orange). LSTc (20 mM [light orange] [A] or 5 mM [orange] [B]) was needed for complex formation by crystal soaking. Download
ACKNOWLEDGMENTS
We thank members of the Atwood and Stehle laboratories for critical discussion and review of the manuscript.
Work in the Atwood and Stehle laboratories was supported by PPG 5P01NS065719 (W.J.A. and T.S.) and a Ruth L. Kirschstein National Research Service Award F32NS064870 (M.S.M.) from the National Institute of Neurological Disorders and Stroke.
Footnotes
Citation Maginnis MS, Ströh LJ, Gee GV, O’Hara BA, Derdowski A, Stehle T, Atwood WJ. 2013. Progressive multifocal leukoencephalopathy-associated mutations in the JC polyomavirus capsid disrupt lactoseries tetrasaccharide c binding. mBio 4(3):e00247-13. doi:10.1128/mBio.00247-13.
REFERENCES
- 1. Frisque RJ, Bream GL, Cannella MT. 1984. Human polyomavirus JC virus genome. J. Virol. 51:458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog. 5:e1000363 http://dx.doi.org/10.1371/journal.ppat.1000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH. 2009. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J. Infect. Dis. 199:837–846 [DOI] [PubMed] [Google Scholar]
- 4. Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. 1990. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J. Virol. 64:3139–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 75:3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamza IA, Jurzik L, Stang A, Sure K, Uberla K, Wilhelm M. 2009. Detection of human viruses in rivers of a densely-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 43:2657–2668 [DOI] [PubMed] [Google Scholar]
- 7. Ahmed W, Wan C, Goonetilleke A, Gardner T. 2010. Evaluating sewage-associated JCV and BKV polyomaviruses for sourcing human fecal pollution in a coastal river in Southeast Queensland, Australia. J. Environ. Qual. 39:1743–1750 [DOI] [PubMed] [Google Scholar]
- 8. Monaco MC, Jensen PN, Hou J, Durham LC, Major EO. 1998. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J. Virol. 72:9918–9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dörries K. 1998. Molecular biology and pathogenesis of human polyomavirus infections. Dev. Biol. Stand. 94:71–79 [PubMed] [Google Scholar]
- 10. Houff SA, Major EO, Katz DA, Kufta CV, Sever JL, Pittaluga S, Roberts JR, Gitt J, Saini N, Lux W. 1988. Involvement of JC virus-infected mononuclear cells from the bone marrow and spleen in the pathogenesis of progressive multifocal leukoencephalopathy. N. Engl J. Med. 318:301–305 [DOI] [PubMed] [Google Scholar]
- 11. Monaco MC, Atwood WJ, Gravell M, Tornatore CS, Major EO. 1996. JC virus infection of hematopoietic progenitor cells, primary B lymphocytes, and tonsillar stromal cells: implications for viral latency. J. Virol. 70:7004–7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tan CS, Dezube BJ, Bhargava P, Autissier P, Wüthrich C, Miller J, Koralnik IJ. 2009. Detection of JC virus DNA and proteins in the bone marrow of HIV-positive and HIV-negative patients: implications for viral latency and neurotropic transformation. J. Infect. Dis. 199:881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dörries K, Vogel E, Günther S, Czub S. 1994. Infection of human polyomaviruses JC and BK in peripheral blood leukocytes from immunocompetent individuals. Virology 198:59–70 [DOI] [PubMed] [Google Scholar]
- 14. Dubois V, Lafon ME, Ragnaud JM, Pellegrin JL, Damasio F, Baudouin C, Michaud V, Fleury HJ. 1996. Detection of JC virus DNA in the peripheral blood leukocytes of HIV-infected patients. AIDS 10:353–358 [DOI] [PubMed] [Google Scholar]
- 15. Dubois V, Dutronc H, Lafon ME, Poinsot V, Pellegrin JL, Ragnaud JM, Ferrer AM, Fleury HJ. 1997. Latency and reactivation of JC virus in peripheral blood of human immunodeficiency virus type 1-infected patients. J. Clin. Microbiol. 35:2288–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silverman L, Rubinstein LJ. 1965. Electron microscopic observations on a case of progressive multifocal leukoencephalopathy. Acta Neuropathol. 5:215–224 [DOI] [PubMed] [Google Scholar]
- 17. Zurhein G, Chou SM. 1965. Particles resembling Papova viruses in human cerebral demyelinating disease. Science 148:1477–1479 [DOI] [PubMed] [Google Scholar]
- 18. Spiegel I, Peles E. 2006. A new player in CNS myelination. Neuron 49:777–778 [DOI] [PubMed] [Google Scholar]
- 19. Sorensen A, Moffat K, Thomson C, Barnett SC. 2008. Astrocytes, but not olfactory ensheathing cells or Schwann cells, promote myelination of CNS axons in vitro. Glia 56:750–763 [DOI] [PubMed] [Google Scholar]
- 20. Bradl M, Lassmann H. 2010. Oligodendrocytes: biology and pathology. Acta Neuropathol. 119:37–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Astrom KE, Mancall EL, Richardson EP., Jr. 1958. Progressive multifocal leuko-encephalopathy; a hitherto unrecognized complication of chronic lymphatic leukaemia and Hodgkin’s disease. Brain 81:93–111 [DOI] [PubMed] [Google Scholar]
- 22. Ferenczy MW, Marshall LJ, Nelson CD, Atwood WJ, Nath A, Khalili K, Major EO. 2012. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin. Microbiol. Rev. 25:471–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brew BJ, Davies NW, Cinque P, Clifford DB, Nath A. 2010. Progressive multifocal leukoencephalopathy and other forms of JC virus disease. Nat. Rev. Neurol. 6:667–679 [DOI] [PubMed] [Google Scholar]
- 24. Carson KR, Focosi D, Major EO, Petrini M, Richey EA, West DP, Bennett CL. 2009. Monoclonal antibody-associated progressive multifocal leucoencephalopathy in patients treated with rituximab, natalizumab, and efalizumab: a review from the Research on Adverse Drug Events and Reports (RADAR) Project. Lancet Oncol. 10:816–824 [DOI] [PubMed] [Google Scholar]
- 25. Bloomgren G, Richman S, Hotermans C, Subramanyam M, Goelz S, Natarajan A, Lee S, Plavina T, Scanlon JV, Sandrock A, Bozic C. 2012. Risk of natalizumab-associated progressive multifocal leukoencephalopathy. N. Engl. J. Med. 366:1870–1880 [DOI] [PubMed] [Google Scholar]
- 26. Hellwig K, Gold R. 2011. Progressive multifocal leukoencephalopathy and natalizumab. J. Neurol. 258:1920–1928 [DOI] [PubMed] [Google Scholar]
- 27. Kawamoto E, Nakahashi S, Okamoto T, Imai H, Shimaoka M. 2012. Anti-integrin therapy for multiple sclerosis. J. Autoimmune Dis. 2012:357101 http://dx.doi.org/10.1155/2012/357101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tornatore C, Berger JR, Houff SA, Curfman B, Meyers K, Winfield D, Major EO. 1992. Detection of JC virus DNA in peripheral lymphocytes from patients with and without progressive multifocal leukoencephalopathy. Ann. Neurol. 31:454–462 [DOI] [PubMed] [Google Scholar]
- 29. Atwood WJ, Amemiya K, Traub R, Harms J, Major EO. 1992. Interaction of the human polyomavirus, JCV, with human B-lymphocytes. Virology 190:716–723 [DOI] [PubMed] [Google Scholar]
- 30. Chapagain ML, Nerurkar VR. 2010. Human polyomavirus JC (JCV) infection of human B lymphocytes: a possible mechanism for JCV transmigration across the blood-brain barrier. J. Infect. Dis. 202:184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Daniel AM, Swenson JJ, Mayreddy RP, Khalili K, Frisque RJ. 1996. Sequences within the early and late promoters of archetype JC virus restrict viral DNA replication and infectivity. Virology 216:90–101 [DOI] [PubMed] [Google Scholar]
- 32. Agostini HT, Ryschkewitsch CF, Stoner GL. 1996. Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. J. Clin. Microbiol. 34:159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yogo Y, Zhong S, Shibuya A, Kitamura T, Homma Y. 2008. Transcriptional control region rearrangements associated with the evolution of JC polyomavirus. Virology 380:118–123 [DOI] [PubMed] [Google Scholar]
- 34. Sock E, Renner K, Feist D, Leger H, Wegner M. 1996. Functional comparison of PML-type and archetype strains of JC virus. J. Virol. 70:1512–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frisque RJ. 1983. Nucleotide sequence of the region encompassing the JC virus origin of DNA replication. J. Virol. 46:170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin JD, King DM, Slauch JM, Frisque RJ. 1985. Differences in regulatory sequences of naturally occurring JC virus variants. J. Virol. 53:306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kenney S, Natarajan V, Salzman NP. 1986. Mapping 5′ termini of JC virus late RNA. J. Virol. 58:216–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kenney S, Natarajan V, Selzer G, Salzman NP. 1986. Mapping 5′ termini of JC virus early RNAs. J. Virol. 58:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krebs CJ, McAvoy MT, Kumar G. 1995. The JC virus minimal core promoter is glial cell specific in vivo. J. Virol. 69:2434–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frisque RJ. 1983. Regulatory sequences and virus-cell interactions of JC virus. Prog. Clin. Biol. Res. 105:41–59 [PubMed] [Google Scholar]
- 41. Neu U, Maginnis MS, Palma AS, Ströh LJ, Nelson CD, Feizi T, Atwood WJ, Stehle T. 2010. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe 8:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu CK, Wei G, Atwood WJ. 1998. Infection of glial cells by the human polyomavirus JC is mediated by an N-linked glycoprotein containing terminal alpha(2–6)-linked sialic acids. J. Virol. 72:4643–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gee GV, Tsomaia N, Mierke DF, Atwood WJ. 2004. Modeling a sialic acid binding pocket in the external loops of JC virus VP1. J. Biol. Chem. 279:49172–49176 [DOI] [PubMed] [Google Scholar]
- 44. Elphick GF, Querbes W, Jordan JA, Gee GV, Eash S, Manley K, Dugan A, Stanifer M, Bhatnagar A, Kroeze WK, Roth BL, Atwood WJ. 2004. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science 306:1380–1383 [DOI] [PubMed] [Google Scholar]
- 45. Dugan AS, Gasparovic ML, Atwood WJ. 2008. Direct correlation between sialic acid binding and infection of cells by two human polyomaviruses (JC virus and BK virus). J. Virol. 82:2560–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eash S, Tavares R, Stopa EG, Robbins SH, Brossay L, Atwood WJ. 2004. Differential distribution of the JC virus receptor-type sialic acid in normal human tissues. Am. J. Pathol. 164:419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wei G, Liu CK, Atwood WJ. 2000. JC virus binds to primary human glial cells, tonsillar stromal cells, and B-lymphocytes, but not to T lymphocytes. J. Neurovirol. 6:127–136 [DOI] [PubMed] [Google Scholar]
- 48. Maginnis MS, Haley SA, Gee GV, Atwood WJ. 2010. Role of N-linked glycosylation of the 5-HT2A receptor in JC virus infection. J. Virol. 84:9677–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng HY, Takasaka T, Noda K, Kanazawa A, Mori H, Kabuki T, Joh K, Oh-ishi T, Ikegaya H, Nagashima K, Hall WW, Kitamura T, Yogo Y. 2005. New sequence polymorphisms in the outer loops of the JC polyomavirus major capsid protein (VP1) possibly associated with progressive multifocal leukoencephalopathy. J. Gen. Virol. 86:2035–2045 [DOI] [PubMed] [Google Scholar]
- 50. Sunyaev SR, Lugovskoy A, Simon K, Gorelik L. 2009. Adaptive mutations in the JC virus protein capsid are associated with progressive multifocal leukoencephalopathy (PML). PLoS Genet. 5:e1000368 http://dx.doi.org/10.1371/journal.pgen.1000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Delbue S, Branchetti E, Bertolacci S, Tavazzi E, Marchioni E, Maserati R, Minnucci G, Tremolada S, Vago G, Ferrante P. 2009. JC virus VP1 loop-specific polymorphisms are associated with favorable prognosis for progressive multifocal leukoencephalopathy. J. Neurovirol. 15:51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gorelik L, Reid C, Testa M, Brickelmaier M, Bossolasco S, Pazzi A, Bestetti A, Carmillo P, Wilson E, McAuliffe M, Tonkin C, Carulli JP, Lugovskoy A, Lazzarin A, Sunyaev S, Simon K, Cinque P. 2011. Progressive multifocal leukoencephalopathy (PML) development is associated with mutations in JC virus capsid protein VP1 that change its receptor specificity. J. Infect. Dis. 204:103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reid CE, Li H, Sur G, Carmillo P, Bushnell S, Tizard R, McAuliffe M, Tonkin C, Simon K, Goelz S, Cinque P, Gorelik L, Carulli JP. 2011. Sequencing and analysis of JC virus DNA from natalizumab-treated PML patients. J. Infect. Dis. 204:237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sala M, Vartanian JP, Kousignian P, Delfraissy JF, Taoufik Y, Wain-Hobson S, Gasnault J. 2001. Progressive multifocal leukoencephalopathy in human immunodeficiency virus type 1-infected patients: absence of correlation between JC virus neurovirulence and polymorphisms in the transcriptional control region and the major capsid protein loci. J. Gen. Virol. 82:899–907 [DOI] [PubMed] [Google Scholar]
- 55. Zheng HY, Ikegaya H, Takasaka T, Matsushima-Ohno T, Sakurai M, Kanazawa I, Kishida S, Nagashima K, Kitamura T, Yogo Y. 2005. Characterization of the VP1 loop mutations widespread among JC polyomavirus isolates associated with progressive multifocal leukoencephalopathy. Biochem. Biophys. Res. Commun. 333:996–1002 [DOI] [PubMed] [Google Scholar]
- 56. Nelson CD, Derdowski A, Maginnis MS, O’Hara BA, Atwood WJ. 2012. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology 428:30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gee GV, Manley K, Atwood WJ. 2003. Derivation of a JC virus-resistant human glial cell line: implications for the identification of host cell factors that determine viral tropism. Virology 314:101–109 [DOI] [PubMed] [Google Scholar]
- 58. Bauer PH, Bronson RT, Fung SC, Freund R, Stehle T, Harrison SC, Benjamin TL. 1995. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J. Virol. 69:7925–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bauer PH, Cui C, Liu WR, Stehle T, Harrison SC, DeCaprio JA, Benjamin TL. 1999. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J. Virol. 73:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Freund R, Garcea RL, Sahli R, Benjamin TL. 1991. A single-amino-acid substitution in polyomavirus VP1 correlates with plaque size and hemagglutination behavior. J. Virol. 65:350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dubensky TW, Freund R, Dawe CJ, Benjamin TL. 1991. Polyomavirus replication in mice: influences of VP1 type and route of inoculation. J. Virol. 65:342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neu U, Woellner K, Gauglitz G, Stehle T. 2008. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl. Acad. Sci. U. S. A. 105:5219–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Neu U, Hengel H, Blaum BS, Schowalter RM, Macejak D, Gilbert M, Wakarchuk WW, Imamura A, Ando H, Kiso M, Arnberg N, Garcea RL, Peters T, Buck CB, Stehle T. 2012. Structures of Merkel cell polyomavirus VP1 complexes define a sialic acid binding site required for infection. PLoS Pathog. 8:e1002738 http://dx.doi.org/10.1371/journal.ppat.1002738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Komagome R, Sawa H, Suzuki T, Suzuki Y, Tanaka S, Atwood WJ, Nagashima K. 2002. Oligosaccharides as receptors for JC virus. J. Virol. 76:12992–13000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cubitt CL, Cui X, Agostini HT, Nerurkar VR, Scheirich I, Yanagihara R, Ryschkewitsch CF, Stoner GL. 2001. Predicted amino acid sequences for 100 JCV strains. J. Neurovirol. 7:339–344 [DOI] [PubMed] [Google Scholar]
- 66. Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. 1985. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc. Natl. Acad. Sci. U. S. A. 82:1257–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schweighardt B, Atwood WJ. 2001. HIV type 1 infection of human astrocytes is restricted by inefficient viral entry. AIDS Res. Hum. Retroviruses 17:1133–1142 [DOI] [PubMed] [Google Scholar]
- 68. Chen BJ, Atwood WJ. 2002. Construction of a novel JCV/SV40 hybrid virus (JCSV) reveals a role for the JCV capsid in viral tropism. Virology 300:282–290 [DOI] [PubMed] [Google Scholar]
- 69. Vacante DA, Traub R, Major EO. 1989. Extension of JC virus host range to monkey cells by insertion of a simian virus 40 enhancer into the JC virus regulatory region. Virology 170:353–361 [DOI] [PubMed] [Google Scholar]
- 70. Atwood WJ, Wang L, Durham LC, Amemiya K, Traub RG, Major EO. 1995. Evaluation of the role of cytokine activation in the multiplication of JC virus (JCV) in human fetal glial cells. J. Neurovirol. 1:40–49 [DOI] [PubMed] [Google Scholar]
- 71. Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. 2009. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int. J. Cancer 125:1250–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. 2009. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 5:e1000578 http://dx.doi.org/10.1371/journal.ppat.1000578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450 [DOI] [PubMed] [Google Scholar]
- 79. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
X-ray crystallography studies of L54F, S268F, and S268Y JCPyV VP1 pentamers and their engagement of the LSTc receptor motif. Data collection and structural refinement statistics.
Simulated annealing omit electron density maps of LSTc in the S268F VP1 pentamer-LSTc complex structure (A) and the JCPyV Mad-1 VP1 pentamer-LSTc complex structure (B) shown in a close-up view of the binding pocket. The proteins are shown in cartoon representation with one VP1 monomer highlighted in green (A) or pink (B) and neighboring monomers depicted in gray. A simulated annealed Fobs-Fcalc density omit map contoured at 2.5 σ is shown for 2.5 Å around the respective carbohydrate residues. The oligosaccharide is drawn as a stick model and colored according to atom type (nitrogens in blue, oxygens in red, and carbons in light orange). LSTc (20 mM [light orange] [A] or 5 mM [orange] [B]) was needed for complex formation by crystal soaking. Download