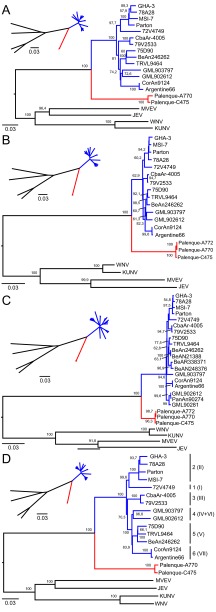
FIG 2.
Genetic and probabilistic distances of SLEV-Palenque from other viruses of the Japanese encephalitis serocomplex. NJ and ML phylogenies were invested for the NS3 (A), NS5 (B), and E (C) genes, as well as for the complete open reading frame (ORF) (D). ML analyses were performed with the GTR substitution model (NS5, E) and the HKY85 substitution model (NS3, ORF) with invariant sites, four gamma categories, and 1,000 bootstrap replicates with PHYML as implemented in Geneious. Bootstrap values of >60% are shown. NJ analyses were done with the p-distance model in Geneious and are shown on a smaller scale. Genetic lineages are named in accordance with the report by Auguste et al. (28), and genotypes as previously published by Kramer and Chandler (26) are shown in parentheses in panel D. Cosmopolitan strains are blue, sylvatic SLEV strains are red, and representative members of the Japanese encephalitis serocomplex are black.