Abstract
HIV-1 dual infection (DI) and CXCR4 (X4) coreceptor usage are associated with accelerated disease progression but frequency and dynamics of coreceptor usage during DI is unknown. Ultradeep sequencing was used to interrogate for DI and infer coreceptor usage in longitudinal blood samples of 102 subjects. At baseline, X4 usage was high (23 subjects harbored X4 variants) and was not associated with infection duration or DI. Coreceptor usage changed over time in 12 of 47 participants, and X4 usage emerged in 4 of 41 monoinfections vs 2 of 5 superinfections (P = .12), suggesting a weak statistical trend toward occurrence of superinfection and acquiring X4 usage.
Keywords: HIV-1 dual infection, HIV-1 coinfection, HIV-1 superinfection, coreceptor tropism, coreceptor usage, ultradeep pyrosequencing, next-generation sequencing, genotypic tropism prediction, genotypic coreceptor usage prediction
Human immunodeficiency virus type 1 (HIV-1) dual infection (DI) is manifested as ≥2 distinct viral subpopulations within the same host. DI cases can be divided into coinfection, that is, acquisition of at least 2 strains from different source partners simultaneously or within a brief period of time, and superinfection, that is, acquisition of a second strain after an immune response to the first infection has been established [1]. DI has been associated with increased viral load, faster decline in CD4+ T cells, and shorter time to AIDS diagnosis [2]. Similarly, infection with CXCR4 (X4) coreceptor-using virus has been linked to decreased CD4+ T-cell counts [3] and accelerated disease progression [4]. Although a strong preference for viral variants using the CCR5 (R5) coreceptor is displayed during HIV-1 transmission, the use of the alternative X4 coreceptor emerges in approximately half of individuals over the course of disease [5].
Ultradeep pyrosequencing (UDS) is a useful technique for increasing the sensitivity of HIV-1 coreceptor usage prediction [6] and for uncovering cases of DI [2]. This study exploits UDS and computational analyses to quantify the impact of DI on coreceptor usage in a large primary infection cohort of men who have sex with men (MSM).
METHODS
Study Participants and Clinical Data
This study included MSM enrolled in the San Diego Primary Infection Cohort between January 1998 and January 2007, who deferred antiretroviral therapy (ART) for at least 6 months after their estimated date of infection (EDI) and had at least 2 blood plasma samples available for UDS, one of which was collected within 3 years of EDI. At all time points, CD4 cell counts (LabCorp), and blood plasma HIV-1 RNA levels (Amplicor HIV-1 Monitor Test, Roche Molecular Systems, Inc) were quantified. EDI was calculated using baseline measurements following an established protocol [7].
Sequencing and Dual Infection Screening
HIV-1 RNA was isolated from blood plasma (QIAamp Viral RNA Mini Kit, Qiagen, Hilden, Germany), complementary DNA (cDNA) generated (RETROscript Kit, Applied Biosystems/Ambion, Austin, TX), and UDS (454 GS FLX Titanium Roche, Branford, CT) of env, gag, and pol was performed as described elsewhere [2]. For each sample, the cDNA template input was calculated assuming 43% reverse transcription efficiency and was expressed as the number of templates (log10) for the first round of nested polymerase chain reaction (PCR). Average efficiency was validated by quantifying the input cDNA for 19 random samples from the total sample pool with real-time quantitative PCR (qPCR) and comparing these results with the HIV RNA viral load measurements (data not shown) [8]. UDS data were interrogated for DI with a previously published bioinformatics pipeline [2]. Notably, UDS can detect circulating minority variants as low as 0.25% of the population, and recent studies have found even lower limits of detection [9].
Genotypic Coreceptor Usage Prediction
Coreceptor usage was predicted using an online prediction tool, geno2pheno 454 [6], for all samples containing the V3 env region. Based on the sensitivity of UDS, samples were conservatively classified as bearing X4-capable virus when geno2pheno predicted >1% of the viral population as X4 variants. Based on optimization studies by other groups [10, 11], a geno2pheno false positive rate (FPR) level of 5% was selected. Prediction agreement with phenotypic coreceptor usage was in line with previous studies [12].
Statistical Analyses
Fisher exact test was applied to detect associations between categorical data. Spearman rank correlation analysis was used to test for association between continuous variables. The differences in continuous variables (age, blood HIV RNA level, and CD4 cell count) between categories were evaluated with the Mann-Whitney test. For all statistical tests, a 2-tailed P value ≤ .05 was considered statistically significant.
RESULTS
Study Cohort
HIV-infected participants (n = 102) were predominantly white (82%) men with a median age of 31 years (interquartile range [IQR], 25.3–37 years) who reported sex with other men as their main risk factor for HIV transmission. At the first sampled time point, the median CD4+ T-cell count was 509 cells/mL (IQR, 412–708 cells/mL), the median time of baseline sample from EDI was 85 days (IQR, 45–85 days), and the median blood plasma level of HIV-1 was 4.76 HIV-1 RNA log10 copies/mL (IQR, 4.27–5.24 log10 copies/mL). The median calculated cDNA input in the first-round nested PCR was 3.42 log10 copies/10 µL (IQR: 3.13–3.90 log10 copies), and the median number of UDS reads passing quality filters was 4450 (IQR, 1911–8722). Based on analysis of UDS data, 16 cases of DI (11 coinfections and 5 superinfections) were identified and investigated further.
Coreceptor Usage Prediction and DI
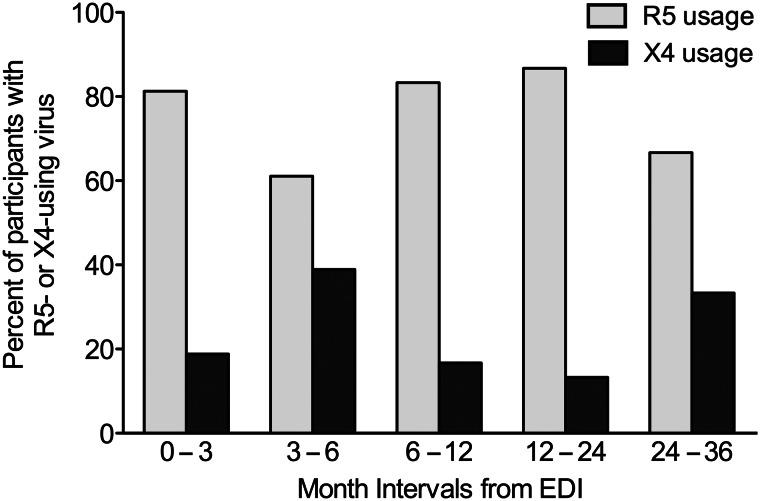
At the first sampled time point, 23 of 102 (22.5%) participants harbored detectable X4-using variants in their viral population, and within the first 3 months from EDI (N = 48), the prevalence of X4 usage was 18.8% (Figure 1). The median intrasample proportion of predicted X4 variants in participants harboring X4 usage was 11.9% (IQR, 1.8%–89.0%) observed at a median time since EDI of 3.1 months (IQR, 2.8–9.3 months). No association between detection of X4 variants and coinfection was found at baseline (Fisher exact test, P > .29). The intrasample proportion of predicted X4-using variants was not significantly associated with time since EDI (Spearman's rho, P > .29). No difference was found in age, viral load, or CD4+ T-cell count between coinfection and monoinfection groups (Mann-Whitney Test, P > .29). No differences were found in age, viral load, or CD4+ T-cell count between samples with predicted R5 and X4 viral populations (Mann-Whitney Test, P > .29).
Figure 1.
Prevalence of predicted R5 and X4 coreceptor usage. Participants first-sampled within 3 years after EDI harbored mostly R5 usage (light gray bars), but 23 of 102 (22.5%) had detectable X4 variants (dark gray bars). A significant prevalence of X4 usage was detected at all time intervals, even in the first 3 months after infection (18.8%). Abbreviations: EDI, estimated date of infection; R5 usage, viral population demonstrated R5 coreceptor usage; X4 usage, viral population with >1% X4 variants detected at a false positive rate of 5%.
In total, 41 monoinfected, one coinfected, and 5 superinfected individuals were followed longitudinally. Superinfection occurred at a median of 20.1 months from EDI (IQR, 7.0–27.7 months), with a median follow-up of 24.3 months (IQR, 7.0–32.5 months). Median follow-up for the monoinfection group was 12.4 months (IQR, 4.6–29.3 months). During follow-up, the viral populations in 12 of 47 subjects changed coreceptor usage (6 toward X4, and 6 toward R5). Viral coreceptor usage in 5 superinfected (and 1 coinfected) subjects did not have statistically significantly different rates of change (toward R5 or X4) compared to monoinfected individuals. For the 41 monoinfections, inferred coreceptor usage changed from R5 to X4 in 4 participants. For the 5 superinfections, predictions changed from R5 to X4 in 2 participants (Fisher exact test, P = .12). Viral coreceptor usage did not change in the single coinfected participant.
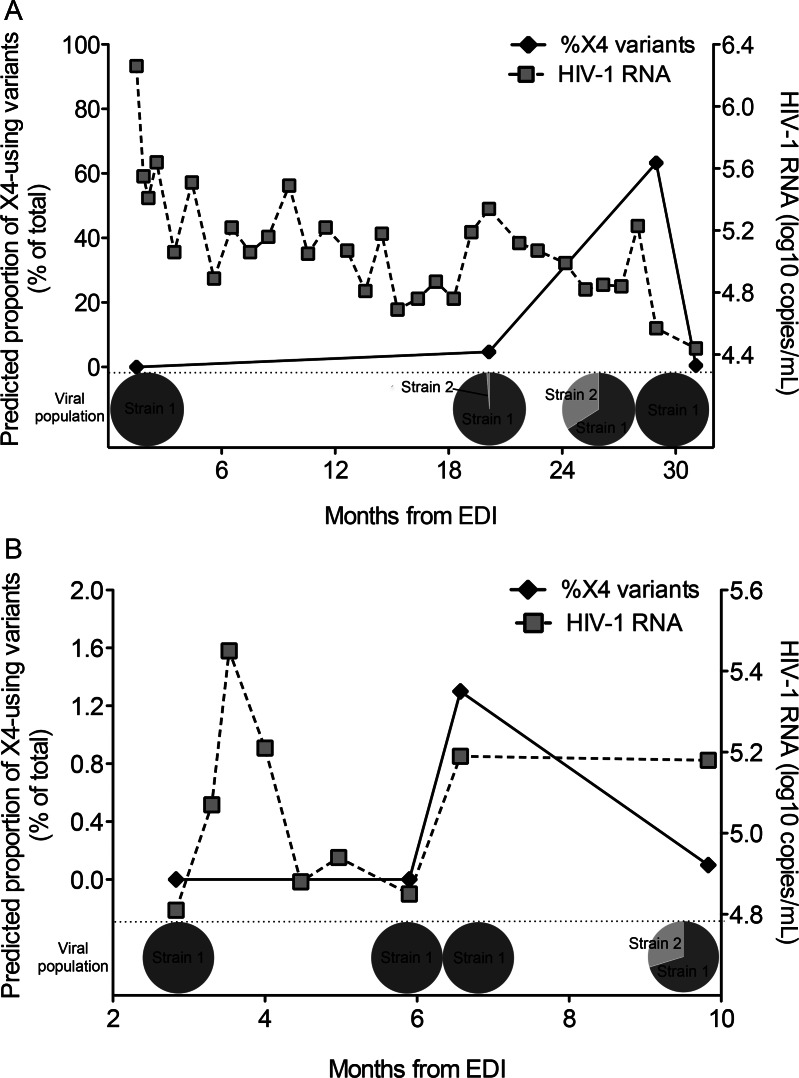
Interrogation of the first superinfection case where coreceptor usage changed (G59) revealed an R5-using viral population at 1.5 months after the EDI; however, X4-using variants were detected 18 months later (Figure 2A). Simultaneously, DI was confirmed. Geno2pheno predicted X4-using variants in the second but not the initial strain. Phylogenetic analysis demonstrated both strains present from 20 to 29 months after the EDI; however, the second strain disappeared at 31 months after the EDI, and concurrently the viral population returned to R5. In the second case (L78), the viral population was R5 at baseline, then low-level X4 variants were detected at 6.6 months after the EDI, but the superinfecting strain was not identified until 3 months later. At this time point, X4 variants decreased below 1% (Figure 2B).
Figure 2.
HIV-1 superinfection and inferred X4 coreceptor usage over time. In case G59 A, an increase in the proportion of X4 usage (black solid diamonds) coincided with the detection of a superinfecting strain (pie charts), and X4 variants disappeared when this strain was no longer detected. In case L78 B, low-level detection of X4 usage occurred right before detection of the superinfecting strain, and decreased below 1% when the second strain was detected. HIV-1 RNA viral load (gray squares) over time for each participant is superimposed. Abbreviations: EDI, estimated date of infection; HIV-1, human immunodeficiency virus type 1; X4 usage, viral population with >1% X4 variants detected at a false positive rate of 5%.
DISCUSSION
The use of next-generation sequencing in this study provided the opportunity to identify with more sensitivity both HIV-1 DI and minority variants with different coreceptor usage in a cohort of patients followed longitudinally after primary infection. The cohort had a high prevalence of inferred X4 usage at the first sampled time point (22.5%), similar to previous reports using UDS [12]. Baseline X4 usage in the cohort was not associated with DI or time after EDI. The analysis of subjects followed over time found that viral populations in 26% of subjects changed coreceptor usage during follow-up (13% toward X4, and 13% toward R5). When compared to a similar group of 41 monoinfected individuals followed longitudinally, the viral coreceptor usage in 5 superinfected (and 1 coinfected) subjects did not have statistically significantly different rates of change toward either R5 or X4. However, coreceptor usage did change toward X4 (>1% intrasample X4 variants) in 4 of 41 (9.8%) monoinfections vs 2 of 5 (40%) superinfections (P = .12), suggesting a possible association between X4 usage gain and superinfection.
In the superinfection cases where inferred coreceptor usage changed, the second strain harbored X4 variants, and in the case of transient superinfection, X4 variants became undetectable with the disappearance of the second strain. Since superinfection can occur at any time during primary infection [2], high-risk (ie, having multiple sex partners) R5-using infected individuals could develop X4-capable superinfection, even in the earliest stages of infection. If superinfection is transient, coreceptor usage can also revert to the prior phenotype as observed here. This rapid change in viral genetic diversity associated with DI has implications for coreceptor tropism testing in the clinical setting, which assumes that the viral population at the time of testing is an accurate representation of the moment when CCR5 antagonists are actually introduced into an individual's antiretroviral regimen.
A limitation of this study is the lack of standardized cutoffs, both for the FPR level and for the X4 variant proportion above which an individual sample can be called X4 with confidence. Previous work in the MOTIVATE studies determined that for their UDS informatics pipelines, the FPR was 3.5% when a viral population had >2% predicted X4 variants [12]. These results set a good benchmark that should be independently replicated as the field matures. Another limitation is that while UDS is very sensitive at detecting DI, it is subject to experimental biases when preceded by PCR amplification [13]. A separate analysis of 29 cohort participants showed a correlation between env maximum diversity and infection duration, whereas no association was observed between viral diversity and HIV RNA viral load or template input (data not shown). These observations suggest that the generated UDS reads are representative of the actual plasma virus populations in vivo. Other limitations include the limited number of participants with identifiable DI and the follow-up difference compared to monoinfected subjects, both of which can confound the detection of statistically significant differences in viral coreceptor usage; as well as the inevitable sampling bias that occurs in clinically collected specimens at discrete time points.
In summary, this study generated one of the largest sets of UDS data on a well-characterized cohort of HIV-1 infected MSM and analyzed the frequency and associations between DI and inferred X4-using virus. These observations raise the possibility of superinfection as a risk factor for more rapid development of X4-tropic infection. The study's findings can guide future lines of research, such as the actual frequency with which superinfection can change viral coreceptor usage, and the identification of genetic determinants of coreceptor usage outside the V3 loop.
Notes
Acknowledgments. We are grateful to Mehdi Bouhaddou for his assistance with data organization and analysis, Alexander Thielen for his technical support and helpful discussion, Caroline Ignacio for her wonderful technical support, and Demetrius Dela Cruz for his administrative assistance, as well as all the participants in the San Diego Primary Infection Cohort. Phenotypic HIV coreceptor tropism assays in this study were performed at Monogram Biosciences Clinical Reference Laboratory.
Financial support. This work was supported by the Department of Veterans Affairs and grants from: the National Institutes of Health (AI090970, AI100665, AI080353, MH097520, DA034978, MH83552, AI36214, MH62512, AI43638, AI74621, TW008908, AI69432, AI096113, AI47745, GM093939), the International AIDS Vaccine Initiative (IAVI); and the James B. Pendleton Charitable Trust. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential Conflicts of interest. D. D. R. has served as a consultant for Biota, Bristol-Myers Squibb, Chimerix, Gen-Probe, Gilead Sciences, Merck & Co, Monogram Biosciences, and Tobira Therapeutics. D. M. S. has received research support from ViiV Pharmaceuticals and has served as a consultant to Gen-Probe. S. K. P. has served as a consultant to Monogram Biosciences and Gen-Probe. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Smith DM, Richman DD, Little SJ. HIV superinfection. J Infect Dis. 2005;192:438–44. doi: 10.1086/431682. [DOI] [PubMed] [Google Scholar]
- 2.Pacold ME, Pond SL, Wagner GA, et al. Clinical, virologic, and immunologic correlates of HIV-1 intraclade B dual infection among men who have sex with men. AIDS. 2012;26:157–65. doi: 10.1097/QAD.0b013e32834dcd26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalmet K, Dauwe K, Foquet L, et al. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J Infect Dis. 2012;205:174–84. doi: 10.1093/infdis/jir714. [DOI] [PubMed] [Google Scholar]
- 4.Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–74. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 5.Bozzette SA, McCutchan JA, Spector SA, Wright B, Richman DD. A cross-sectional comparison of persons with syncytium- and non-syncytium-inducing human immunodeficiency virus. J Infect Dis. 1993;168:1374–9. doi: 10.1093/infdis/168.6.1374. [DOI] [PubMed] [Google Scholar]
- 6.Thielen A, Lengauer T. Geno2pheno[454]: a Web server for the prediction of HIV-1 coreceptor usage from next-generation sequencing data. Intervirology. 2012;55:113–7. doi: 10.1159/000332002. [DOI] [PubMed] [Google Scholar]
- 7.Little SJ, Frost SD, Wong JK, et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol. 2008;82:5510–8. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gianella S, Delport W, Pacold ME, et al. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol. 2011;85:8359–67. doi: 10.1128/JVI.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mild M, Hedskog C, Jernberg J, Albert J. Performance of ultra-deep pyrosequencing in analysis of HIV-1 pol gene variation. PLoS One. 2011;6:e22741. doi: 10.1371/journal.pone.0022741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swenson LC, Moores A, Low AJ, et al. Improved detection of CXCR4-using HIV by V3 genotyping: application of population-based and “deep” sequencing to plasma RNA and proviral DNA. J Acquir Immune Defic Syndr. 2010;54:506–10. doi: 10.1097/QAI.0b013e3181d0558f. [DOI] [PubMed] [Google Scholar]
- 11.Daumer M, Kaiser R, Klein R, Lengauer T, Thiele B, Thielen A. Genotypic tropism testing by massively parallel sequencing: qualitative and quantitative analysis. BMC Med Inform Decis Mak. 2011;11:30. doi: 10.1186/1472-6947-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swenson LC, Mo T, Dong WW, et al. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis. 2011;203:237–45. doi: 10.1093/infdis/jiq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes JP, Totten P. Estimating the accuracy of polymerase chain reaction-based tests using endpoint dilution. Biometrics. 2003;59:505–11. doi: 10.1111/1541-0420.00060. [DOI] [PubMed] [Google Scholar]