Abstract
We determined the chemical structure of lipoteichoic acid (LTA) from Lactobacillus gasseri JCM 1131T. The repeating unit was comprised of glycerolphosphate and 2-alanylglycerolphosphate. The glycolipid anchor was tetrahexosylglycerol with two or three acyl groups. To our knowledge, this is the first demonstration of a tetrahexose structure in an LTA glycolipid anchor.
TEXT
Clarification of the cell envelope structure of probiotic lactic acid bacteria is important for understanding their host-microbe interactions. In a comparison of the genomes from 12 species of lactobacilli, large proportions (19.9 to 29.3%) of the putative total proteins were estimated to be cell surface proteins, including membrane proteins (1). In particular, the genomes of Lactobacillus acidophilus group members (e.g., L. acidophilus, Lactobacillus johnsonii, and Lactobacillus gasseri), which are often used as probiotics, encode a large number of LPXTG-motif proteins, approximately half of which are estimated to have mucus-binding properties (1). Peptidoglycan and teichoic acids are also important molecules, as they are recognized by host immune cells.
Teichoic acids are classified as wall teichoic acid (WTA) and lipoteichoic acid (LTA). WTA links covalently to the N-acetylmuramic acid residues in cell wall peptidoglycan, while LTA is anchored to the cell membrane through its glycolipid moiety. In particular, LTA has been shown to act as a ligand for Toll-like receptor 2 on host immune cells (2) and to induce production of proinflammatory cytokines (3). LTA is also suggested to work as an adhesion molecule with human intestinal epithelial cells (4). Fibronectin may be a host receptor for LTA (5). However, the structural information regarding LTA is insufficient, given the complex nature of this molecule.
LTA is an anionic polymer comprised of a repeating glycerophosphate (GroP) backbone that is connected to a glycolipid (in many cases, dihexosyldiacylglycerol) through a phosphodiester linkage. The C-2 hydroxyl group of the GroP residue is often substituted by d-alanine and/or hexoses. Strain-level variation has been detected in both the degree of polymerization and the substitution ratio of the repeating GroP unit (6, 7).
Within the abovementioned L. acidophilus group members, structural information on LTA is limited and information is completely lacking for L. gasseri, although this species is often used as a probiotic. L. gasseri has been isolated not only from the intestine but also from the oral cavity, vagina, urine, and blood, suggesting a strong association between this bacterium and the human body. Thus, the structure of LTA from L. gasseri needs to be clarified to promote our understanding of the interaction of this bacterium with the host. Therefore, in this study, we determined the chemical structure of LTA from L. gasseri JCM 1131T.
L. gasseri JCM 1131T was grown to log phase in 0.5× Difco Lactobacilli MRS broth (Becton, Dickinson and Co., Franklin Lakes, NJ). The cells were collected and disrupted, and the LTA was purified by n-butanol extraction followed by hydrophobic interaction chromatography (HIC) as described previously with some modifications (8). As a result, 38 mg of LTA was obtained from 25 g of wet cells. The repeating unit of LTA was analyzed with one- and two-dimensional nuclear magnetic resonance (NMR) spectroscopy. The glycolipid anchor fraction of LTA was prepared by treatment with 98% (vol/vol) acetic acid at 100°C for 3 h. The carbohydrate portion of the glycolipid anchor was obtained via deacylation by treatment with 20% (wt/vol) ammonia at room temperature for 12 h. The glycolipid anchor and its carbohydrate portion were analyzed with matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), which was performed as described previously, with some modifications (9, 10). The chemical composition of the glycolipid anchor, including anchor sugars and fatty acids, was analyzed with gas chromatography (GC) as described previously (11) (see the supplemental material for detailed descriptions of all these experimental conditions).
Structure of the repeating unit from L. gasseri JCM 1131T LTA.
The structure of the repeating unit was determined by one- and two-dimensional NMR analyses. The peaks detected by 1H NMR and 13C distortionless enhancement by polarization transfer with an angle parameter of 135° (DEPT-135) NMR (see Fig. S1A, B, and C in the supplemental material) were attributable to the GroP residue and the 2-alanyl-GroP (2-AlaGroP) residue with a substitution at the C-2 hydroxyl group of glycerol to d-alanine (Table 1), as deduced from previous reports (9, 10, 12). No hexoses were detected as substituents. The assignments were supported by correlations with the results of correlation spectroscopy (COSY) (see Fig. S1A). The ratio of d-alanine substitution on the GroP residue was estimated to be 31% based on the intensity of the peaks in the 1H NMR spectrum (Table 1). The average number of repeating units was estimated to be 20 to 30 according to the ratio of the peak area of protons in the GroP residue and protons bound to the carbons next to the double-bonded carbons in the unsaturated fatty acid residues (δ, 1.7 to 2.2 ppm) in the 1H NMR spectrum.
Table 1.
Assignments for the 1H and 13C DEPT-135 NMR spectra of the repeating-unit region of L. gasseri JCM 1131T LTA
| Residue | Proton (1H) | δ (ppm) | Intensitya | Carbon (13C) | δ (ppm) |
|---|---|---|---|---|---|
| Unsubstituted glycerol | H-1, 3 | 3.90 | 1.38H (2H × 0.69) | C-1, 3 | 68.98 |
| 3.96 | 1.38H (2H × 0.69) | ||||
| H-2 | 4.05 | 0.69H (1H × 0.69) | C-2 | 72.32 | |
| Alanine-substituted glycerol | H-1, 3 | 4.11 | 1.24H (4H × 0.31) | C-1, 3 | 66.43 |
| H-2 | 5.40 | 0.31H (1H × 0.31) | C-2 | 77.01 | |
| Substituted alanine | C-1 | 172.84b | |||
| H-2 | 4.29 | 0.31H (1H × 0.31) | C-2 | 51.70 | |
| H-3 | 1.63 | 0.93H (3H × 0.31) | C-3 | 18.08 |
The intensity was adjusted by taking a proton of one repeating unit as 1.
This peak was detected by heteronuclear multiple-bond correlation.
The typical LTA of Gram-positive bacteria has been reported to contain a GroP backbone with frequent substitution of the C-2 hydroxyl groups by d-alanine and/or hexoses such as glucose, galactose, and N-acetylglucosamine (6). L. gasseri JCM 1131T LTA also possesses a poly-GroP backbone with d-alanine substitution. Thus, L. gasseri JCM 1131T showed a typical repeating-unit structure. The GroP repeating-unit structures in other lactobacilli have also been reported. In Lactobacillus delbrueckii subsp. lactis strains ATCC 15808, Ads-5, and LL78 (7); Lactobacillus rhamnosus GG (13); Lactobacillus reuteri 100-23 (14); and Lactobacillus plantarum NCIMB 8826 (15), 27 to 79% of the GroP residues are substituted with d-alanine. Among these, a hexose substitution (in all cases, glucose) was detected in L. delbrueckii subsp. lactis strains ATCC 15808 and LL78 and in L. reuteri 100-23 at 3 to 27%.
Structure of the glycolipid anchor from L. gasseri JCM 1131T LTA.
MALDI-TOF MS of the carbohydrate portion of the glycolipid anchor gave a peak at m/z 763.41, which was attributable to the (M + Na)+ molecular ion of tetrahexosylglycerol (Fig. 1A). The tetrahexose was found to be composed of galactose and glucose at a molar ratio of 3:1 by GC. The fatty acid composition of LTA was determined by GC. Oleic acid [C18:1(n-9)] was a major constituent, representing 70.0% of the total fatty acids. In addition, palmitic acid (C16:0, 18.5%) and stearic acid (C18:0, 7.3%) were detected. MALDI-TOF MS of the glycolipid anchor gave peaks which were divided into two groups with low (group 1) and high (group 2) molecular masses (Fig. 1B). These lines of evidence strongly suggest that the peaks in groups 1 and 2 were attributable to tetrahexosyldiacylglycerol (Hex4DAG) and acyltetrahexosyldiacylglycerol (acylHex4DAG), respectively, although the presence of acyltetrahexosylmonoacylglycerol cannot be excluded (16). For example, the peaks at m/z 1,292.01 and m/z 1,265.97 were attributable to Hex4DAG containing C18:1(n-9)/C18:1(n-9) and C18:1(n-9)/C16:0, respectively. The peaks at m/z 1,557.40 and m/z 1,530.40 were attributable to acylHex4DAG containing C18:1(n-9)/C18:1(n-9)/C18:1(n-9) and C18:1(n-9)/C18:1(n-9)/C16:0, respectively. Similar peak groups corresponding to trihexosyldiacylglycerol (Hex3DAG) and acyltrihexosyldiacylglycerol (acylHex3DAG) have been reported in L. plantarum LTA (9). The peak assignments for L. gasseri JCM 1131T LTA were supported by a difference of 162 in molecular mass, corresponding to one hexose residue, compared to the mass spectrum of L. plantarum LTA (9). No equivalent peaks corresponding to Hex3 structures were observed in L. gasseri JCM 1131T (Fig. 1B).
Fig 1.
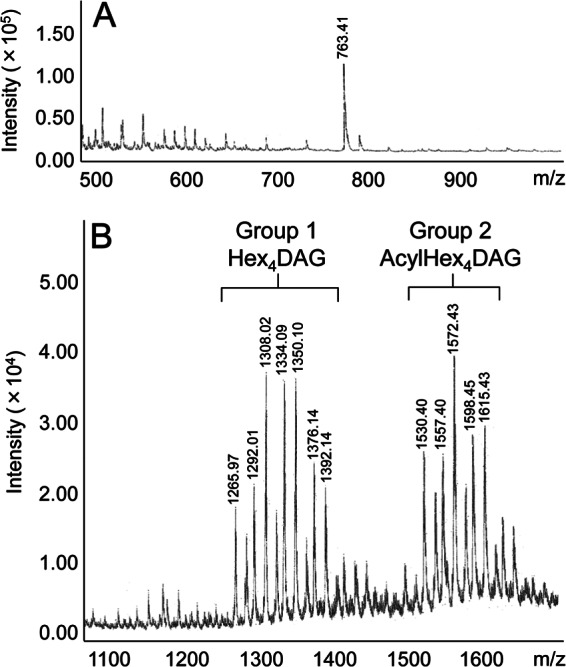
MALDI-TOF mass spectra of the glycolipid anchor fraction in L. gasseri JCM 1131T LTA. (A) Carbohydrate portion. (B) Glycolipid anchor.
The glycolipid anchor structures found in L. gasseri JCM 1131T LTA, Hex4DAG and acylHex4DAG, are unique. To our knowledge, a Hex4 structure has not been reported in a glycolipid anchor of LTA. Hex2 structures are the most common in Gram-positive bacteria, including many staphylococci, bacilli, and streptococci. The LTA glycolipid anchor structures reported to date in four species of lactobacilli, L. plantarum KCTC 10887BP (9), Lactobacillus helveticus DSM 20075T (17), L. rhamnosus DSM 20021T (formerly Lactobacillus casei) (18), and Lactobacillus pentosus DSM 20314T (formerly L. plantarum) (19), have been identified as Hex3. The uniqueness of the Hex4 structure found in L. gasseri JCM 1131T might have some influence on the nature of the cell surface, leading to specific biological activities compared to those of other lactobacilli. The sugar composition was found to be glucose and galactose, which is common to the Hex3 structure of LTA in the abovementioned four species of lactobacilli. On the other hand, the Hex2 structure reported in many Gram-positive bacteria is composed exclusively of glucose.
Furthermore, the structure of acylHex4DAG is unique in that it has three acyl groups. In most cases, the number of acyl groups has been reported to be two in Gram-positive bacteria. The distribution of glycolipid anchors having three acyl groups is relatively limited; it includes the abovementioned four species of lactobacilli and some lactococci, including Lactococcus lactis subsp. lactis NCDO 712 (formerly Streptococcus lactis) (20) and Lactococcus garvieae Kiel 42172 (formerly S. lactis) (21). Acyl groups are an important determinant of host receptor recognition, namely, Toll-like receptors (22). Thus, the number of acyl groups has a marked influence on the host-microbe interaction. For example, a loss of one acyl group from a two-acyl-group-type glycolipid anchor of LTA dramatically reduced cytokine production in in vitro cell culture experiments (23). Thus, the biological activity of acylHex4DAG in the host-microbe interaction seems interesting.
The fatty acid composition of the glycolipid anchor in L. gasseri JCM 1131T LTA was found to comprise 70% C18:1(n-9) and 19% C16:0, similar to the values reported for L. rhamnosus DSM 20021T LTA [51% C18:1(n-9) and 27% C16:0] (18). Lactobacilli often require C18:1(n-9) as a growth factor; this is true for L. gasseri JCM 1131T (24). The Tween 80 in MRS broth serves as a source of C18:1(n-9). Thus, L. gasseri JCM 1131T is enriched with C18:1(n-9) in its glycolipid anchor to meet the requirements of cell physiology. The incorporation of Tween 80-derived C18:1(n-9) in the cell membrane of lactobacilli has been reported previously (25), suggesting that the fatty acid composition of the glycolipid anchor in LTA is influenced by the fatty acid composition of the culture medium.
Conclusion.
The overall chemical structure of LTA from L. gasseri JCM 1131T is illustrated in Fig. 2. The LTA is composed of a poly-GroP backbone. Approximately 30% of the C-2 hydroxyl groups in the GroP residues are substituted by d-alanine without substitution by a hexose. The glycolipid anchor is identified as Hex4DAG and acylHex4DAG, having galactose and glucose as the anchor sugars at an approximate molar ratio of 3:1. The anchor lipid contains predominantly C18:1(n-9) in addition to C16:0 and C18:0 as minor components. The novel Hex4DAG and acylHex4DAG structures found in the glycolipid anchor of LTA suggest that L. gasseri JCM 1131T engages in a different type of host-microbe interaction from those of other gut microbes, especially in terms of modulation of the host immune system; however, the exact mechanism remains to be elucidated.
Fig 2.
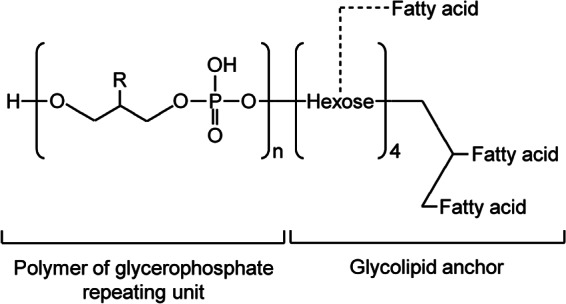
Putative chemical structure of L. gasseri JCM 1131T LTA. For R, hydroxyl and d-alanyl groups were found at a ratio of 69:31. The hexoses identified were galactose and glucose at a ratio of 3:1. The fatty acids were oleic acid (70%), palmitic acid (19%), stearic acid (7%), and others (4%). n = 20 to 30.
Supplementary Material
ACKNOWLEDGMENTS
We thank Akari Kikuchi (Department of Applied Biology and Chemistry, Faculty of Applied Bio-Science, Tokyo University of Agriculture, Tokyo, Japan) for making the LTA preparation, Eri Fukushi (GC-MS and NMR Laboratory, School of Agriculture, Hokkaido University, Hokkaido, Japan) for conducting the NMR analysis, and Izuru Nagashima and Hiroki Shimizu (Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology [AIST], Hokkaido, Japan) for conducting MALDI-TOF MS.
This work was supported in part by a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science (no. 21380053 to A.Y.).
Footnotes
Published ahead of print 15 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00243-13.
REFERENCES
- 1. Barinov A, Bolotin A, Langella P, Maguin E, van de Guchte M. 2011. Genomics of the genus Lactobacillus, p 3–32 In Sonomoto K, Yokota A. (ed), Lactic acid bacteria and bifidobacteria: current progress in advanced research. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 2. Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274: 17406–17409 [DOI] [PubMed] [Google Scholar]
- 3. Mohamadzadeh M, Pfeiler EA, Brown JB, Zadeh M, Gramarossa M, Managlia E, Bere P, Sarraj B, Khan MW, Pakanati KC, Ansari MJ, O'Flaherty S, Barrett T, Klaenhammer TR. 2011. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. U. S. A. 108: 4623–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Granato D, Perotti F, Masserey I, Rouvet M, Golliard M, Servin A, Brassart D. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 65: 1071–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simpson WA, Courtney HS, Ofek I. 1987. Interactions of fibronectin with streptococci: the role of fibronectin as a receptor for Streptococcus pyogenes. Rev. Infect. Dis. 9(Suppl 4): S351–S359 [DOI] [PubMed] [Google Scholar]
- 6. Fischer W. 1988. Physiology of lipoteichoic acids in bacteria. Adv. Microb. Physiol. 29: 233–302 [DOI] [PubMed] [Google Scholar]
- 7. Räisänen L, Draing C, Pfitzenmaier M, Schubert K, Jaakonsaari T, von Aulock S, Hartung T, Alatossava T. 2007. Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on d-alanyl and α-glucose substitution of poly(glycerophosphate) backbones. J. Bacteriol. 189: 4135–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morath S, Geyer A, Hartung T. 2001. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193: 393–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jang KS, Baik JE, Han SH, Chung DK, Kim BG. 2011. Multi-spectrometric analyses of lipoteichoic acids isolated from Lactobacillus plantarum. Biochem. Biophys. Res. Commun. 407: 823–830 [DOI] [PubMed] [Google Scholar]
- 10. Cot M, Ray A, Gilleron M, Vercellone A, Larrouy-Maumus G, Armau E, Gauthier S, Tiraby G, Puzo G, Nigou J. 2011. Lipoteichoic acid in Streptomyces hygroscopicus: structural model and immunomodulatory activities. PLoS One 6: e26316 doi:10.1371/journal.pone.0026316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morita N, Nishida T, Tanaka M, Yano Y, Okuyama H. 2005. Enhancement of polyunsaturated fatty acid production by cerulenin treatment in polyunsaturated fatty acid-producing bacteria. Biotechnol. Lett. 27: 389–393 [DOI] [PubMed] [Google Scholar]
- 12. Batley M, Redmond JW, Wicken AJ. 1987. Nuclear magnetic resonance spectra of lipoteichoic acid. Biochim. Biophys. Acta 901: 127–137 [DOI] [PubMed] [Google Scholar]
- 13. Vélez MP, Verhoeven TLA, Draing C, von Aulock S, Pfitzenmaier M, Geyer A, Lambrichts I, Grangette C, Pot B, Vanderleyden J, de Keersmaecker SCJ. 2007. Functional analysis of d-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73: 3595–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walter J, Loach DM, Alqumber M, Rockel C, Hermann C, Pfitzenmaier M, Tannock GW. 2007. d-Alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 9: 1750–1760 [DOI] [PubMed] [Google Scholar]
- 15. Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. 2005. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl. Acad. Sci. U. S. A. 102: 10321–10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwasaki H, Shimada A, Ito E. 1986. Comparative studies of lipoteichoic acids from several Bacillus strains. J. Bacteriol. 167: 508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer W, Koch HU, Rösel P, Fiedler F. 1980. Alanine ester-containing native lipoteichoic acids do not act as lipoteichoic acid carrier. J. Biol. Chem. 255: 4557–4562 [PubMed] [Google Scholar]
- 18. Nakano M, Fischer W. 1978. Trihexosyldiacylglycerol and acyltrihexosyldiacylglycerol as lipid anchors of the lipoteichoic acid of Lactobacillus casei DSM 20021. Hoppe-Seyler's Z. Physiol. Chem. 359: S1–S11 [DOI] [PubMed] [Google Scholar]
- 19. Fischer W, Mannsfeld T, Hagen G. 1990. On the basic structure of poly(glycerophosphate) lipoteichoic acids. Biochem. Cell Biol. 68: 33–43 [DOI] [PubMed] [Google Scholar]
- 20. Laine RA, Fischer W. 1978. On the relationship between glycerophosphoglycolipids and lipoteichoic acids of Gram-positive bacteria. III. Di(glycerophospho)-acylkojibiosyldiacylglycerol and related compounds from Streptococcus lactis NCDO 712. Biochim. Biophys. Acta 529: 250–262 [DOI] [PubMed] [Google Scholar]
- 21. Koch HU, Fischer W. 1978. Acyldiglucosyldiacylglycerol-containing lipoteichoic acid with a poly(3-O-galabiosyl-2-O-galactosyl-sn-glycero-1-phosphate) chain from Streptococcus lactis Kiel 42172. Biochemistry 17: 5275–5281 [DOI] [PubMed] [Google Scholar]
- 22. Kang JY, Nan X, Jin MS, Youn SJ, Ryu YH, Mah S, Han SH, Lee H, Paik SG, Lee JO. 2009. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31: 873–884 [DOI] [PubMed] [Google Scholar]
- 23. Morath S, Stadelmaier A, Geyer A, Schmidt RR, Hartung T. 2002. Synthetic lipoteichoic acid from Staphylococcus aureus is a potent stimulus of cytokine release. J. Exp. Med. 195: 1635–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sawatari Y, Hirano T, Yokota A. 2006. Development of food grade media for the preparation of Lactobacillus plantarum starter culture. J. Gen. Appl. Microbiol. 52: 349–356 [DOI] [PubMed] [Google Scholar]
- 25. Veerkamp JH. 1971. Fatty acid composition of Bifidobacterium and Lactobacillus strains. J. Bacteriol. 108: 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


