Abstract
Erwinia amylovora bacteriophages (phages) belonging to the Myoviridae and Podoviridae families demonstrated a preference for either high-exopolysaccharide-producing (HEP) or low-exopolysaccharide-producing (LEP) bacterial hosts when grown on artificial medium without or with sugar supplementation. Myoviridae phages produced clear plaques on LEP hosts and turbid plaques on HEP hosts. The reverse preference was demonstrated by most Podoviridae phages, where clear plaques were seen on HEP hosts. Efficiency of plating (EOP) was determined by comparing phage growth on the original isolation host to the that on the LEP or HEP host. Nine of 10 Myoviridae phages showed highest EOPs on LEP hosts, and 8 of 11 Podoviridae phages had highest EOPs on HEP hosts. Increasing the production of EPS on sugar-supplemented medium or decreasing production by knocking out the synthesis of amylovoran or levan, the two EPSs produced by E. amylovora, indicated that these components play crucial roles in phage infection. Amylovoran was virtually essential for proliferation of most Podoviridae phages when phage population growth was compared to the wild type. Decreased levan production resulted in a significant reduction of progeny from phages in the Myoviridae family. Thus, Podoviridae phages are adapted to hosts that produce high levels of exopolysaccharides and are dependent on host-produced amylovoran for pathogenesis. Myoviridae phages are adapted to hosts that produce lower levels of exopolysaccharides and host-produced levan.
INTRODUCTION
The common characteristic of infection of rosaceous plants by Erwinia amylovora, the fire blight pathogen, is the appearance of wilt, necrosis, and the production of copious amount of exudates, or “ooze.” In the 1930s, experiments established that the ooze was bacterial in origin and responsible for the induction of wilt symptoms in pear shoot cuttings (1). A majority of the field samples of E. amylovora examined by Billing (2) were encapsulated by the ooze, with less than 1% having no capsule. Goodman et al. (3) named the ooze polysaccharide amylovorin, to reflect its toxic effect on plant tissues. Amylovorin was subsequently renamed amylovoran to be consistent with polysaccharide nomenclature (4). In due course, the linkages between E. amylovora's capsule, slime, polysaccharides, ooze, and pathogenicity were made (5, 6, 7, 8, 9, 10). Growth of the bacterium on sucrose-, glucose-, or sorbitol-enriched media results in the production of excess quantities of two exopolysaccharides (EPSs), one acidic and one neutral (2, 6, 7). The acidic EPS, amylovoran, may form a capsule, slime, and/or float free in the liquid medium (5, 11). Amylovoran is a heterogeneous polymer consisting of repeating units of one glucuronic acid and four galactose residues (12). The ams region of the genome controls the production of the EPS (13), with rcsA and rcsB genes being required for synthesis (13, 14). Mutation in rcsA or rcsB results in reduced amylovoran production (15). The neutral EPS, levan, is synthesized in the presence of sucrose by the enzyme levansucrase, which cleaves the sugar and polymerizes fructose into a polyfructan (β-2,6-d-fructofuranan) (10, 11, 16). Levan synthesis takes place extracellularly through the action of the enzyme encoded by the lsc gene (10, 17).
EPSs play multifaceted and complex roles in the interactions between bacteria and their environments (18, 19). Functions include key roles in bacterial virulence and pathogenesis (20, 21, 22, 23, 24), surface adhesion (18), as a major constituent of the biofilm glycocalyx (18), and as a component that renders the cell susceptible (25) or resistant (26, 27, 28) to bacteriophage (phage) attack. EPSs prevent cellular desiccation (3, 25, 29, 30) by keeping nutrients and water in close proximity to the bacterial cell (29, 30). The protective nature of the EPSs of E. amylovora is exhibited in the ability of the pathogen to “hide” from the plant host defenses (22). Amylovoran mutants are avirulent, producing no disease symptoms (4, 11), and thus EPS is an essential contributor to E. amylovora pathogenicity (4, 23) and virulence (2, 6, 7, 15, 20, 21). Mutants deficient for levansucrase produce disease symptoms on immature pears and therefore remain pathogenic (10, 17). EPSs also play an important role in bacterial biofilms, and Koczan et al. (19) showed that, for E. amylovora, both amylovoran and levan are required for biofilm formation. In vitro, cells lacking amylovoran were unable to attach to a growth substrate to initiate biofilm formation, and levan-deficient mutants showed reduced biofilm formation.
EPSs have been reported to delay or prevent phage adsorption for several bacterial species, presumably by providing a physical barrier between the phage and outer membrane receptors (26, 27, 28). Certain phages, however, have evolved to recognize specifically and bind bacterial extracellular polymers (31, 32, 33, 34). Erwinia spp. phages normally produce clear translucent plaques indicative of a strong infection in vitro (6, 7, 20, 35, 36, 37, 38). Many phage infections, however, result in the production of turbid or hazy plaques (2, 36, 37, 38) that contain a thin layer of bacteria on the plaque surface. Historically, hazy plaques were attributed to the presence of lysogenic bacteria resulting from infections with temperate phages (2, 37, 38). The main purpose of this study was to determine the roles of amylovoran and levan in plaque morphology, preference of phages for specific hosts, and phage productivity. Lytic phages are currently being evaluated as biological control agents for the control of E. amylovora in the orchard (35, 36, 39, 40, 41, 42). The flower stigma is the primary entry site for E. amylovora during fire blight infection and provides the nutrient-rich environment in which the pathogen and biological control agent interact (43, 44, 45, 46). This sugar-rich microenvironment stimulates EPS production by the bacterium and could potentially alter the efficacy of a phage-based biopesticide. The present work provides information that is critical to the selection of phages developed as biocontrol mixtures or cocktails. The choice of phages will directly influence field efficacy and the prevention of bacterial resistance.
MATERIALS AND METHODS
Bacterial isolates, bacteriophages, and plasmids.
Bacterial isolates, phages, and plasmids used in this study are listed in Table 1. Phage isolates were stored in 0.8% nutrient broth (Difco) at 4°C. The phages, part of the AAFC Vineland Master Collection, were characterized previously by restriction fragment length polymorphism analysis and transmission electron microscope observations into groups and families (36). New additions were placed into families based on information from real-time PCR probe-primer sets designed to detect Erwinia spp. phages belonging to the Myoviridae and Podoviridae (39, 47). E. amylovora was grown in nutrient broth (NB; Difco Laboratories, Sparks, MD), nutrient agar (NA; Laboratories, Sparks, MD), NA supplemented with 0.5% sucrose (NAS), or Luria-Bertani medium (LB; Difco Laboratories, Sparks, MD) at 27°C. To increase EPS production, NB was supplemented with 0.5% sucrose and 1% sorbitol (NBSS). For transformation selection, antibiotics were added to the culture medium at the following concentrations: kanamycin (Km), 20 μg/ml; chloramphenicol (Cm), 20 μg/ml; ampicillin (Ap), 100 μg/ml. Datsenko and Wanner (48) previously detailed the properties of the plasmids used in the present study for the production of deletion mutants.
Table 1.
Erwinia amylovora isolates, bacteriophages, and plasmids used in the study
| Isolate or plasmid | Descriptiona | Reference(s) or source |
|---|---|---|
| Erwinia amylovora | ||
| Ea110Rb | Wild type | 37 |
| Ea110RΔrcsB | Kmr, ΔrcsB | 47 |
| Ea110RΔlsc | Cmr, Δlsc | 47 |
| Ea29-7 | Wild type | 36 |
| Ea29-7ΔrcsB | Kmr, ΔrcsB | 47 |
| Ea29-7Δlsc | Cmr, Δlsc | 47 |
| EaD-7 | Wild type | 51 |
| EaD-7ΔrcsB | Kmr, ΔrcsB | This study |
| EaD-7Δlsc | Cmr, Δlsc | This study |
| Ea6-4 | Wild type | 51 |
| Ea6-4ΔrcsB | Kmr, ΔrcsB | 47 |
| Ea6-4Δlsc | Cmr, Δlsc | 47 |
| Ea6-4ΔrcsB Δlsc | Kmr Cmr, ΔrcsB Δlsc | 47 |
| Ea17-1-1 | Wild type | 51 |
| Ea17-1-1ΔrcsB | Kmr, ΔrcsB | 47 |
| Ea17-1-1Δlsc | Cmr, Δlsc | This study |
| Ea17-1-1ΔrcsB Δlsc | Kmr Cmr, ΔrcsB, Δlsc | This study |
| EaG-5 | Wild type | 51 |
| Ea G-5ΔrcsB | Kmr, ΔrcsB | This study |
| EaG-5Δlsc | Cmr, Δlsc | 47 |
| Ea G-5ΔrcsB Δlsc | Kmr Cmr, Δrcs, Δlsc | This study |
| Bacteriophages | ||
| ϕEa21-4 | Myoviridae | 40 |
| ϕEa1(h) | Podoviridae | 37 |
| ϕEa35-7 | Siphoviridae | 36 |
| 46 phage isolates | All three families | 36, 47 |
| Plasmids | ||
| pKD46 | Apr, PBAD γ β exo pSC101 oriTS | 48 |
| pKD13 | Kmr, FRT cat FRT PS1 PS2 oriR6K rgbN | 48 |
| pKD32 | Cmr, FRT cat FRT PS1 PS2 oriR6K rgbN | 48 |
ΔrcsB, amylovoran deficient; Δlsc, levan deficient.
Rifampin-resistant isolate of E. amylovora 110.
Phage manipulations.
The host ranges of Erwinia spp. phages were determined by the ability to form plaques on E. amylovora isolates and by plaque morphology. Clear plaques indicated high host sensitivity, turbid plaques indicated partial lysis, and no plaques indicated a nonhost. Efficiency of plating (EOP), the ratio of PFU/ml obtained with an assay host to the PFU/ml obtained with the isolation host, was calculated using the double layer plaque method (49). Assay host refers to the tested host, and isolation host refers to the E. amylovora isolate with which Gill et al. (36) initially isolated the phage. Phage population growth was monitored by inoculating liquid medium with phage and bacteria at a multiplicity of infection of 1. The cultures were incubated for 24 h at 27°C, lysates were prepared, and titers were determined by using the double agar overlay method (49).
Generation of EPS-deficient mutants.
A modified PCR deletion method (24, 48) was used to generate E. amylovora mutants with deletions of the rscB gene (ΔrscB), the regulator of capsular synthesis, the lsc gene (Δlsc), which encodes levansucrase, or double deletions of both genes (ΔrscB Δlsc). For mutant production, linear recombination constructs consisting of either a kanamycin (Kmr) or chloramphenicol (Cmr) resistance gene flanked by 50-nucleotide homology arms targeting either lsc or rcsB were created by PCR. The Kmr gene was amplified from pKD13 with the rcsB-Km primers, and the Cmr gene was amplified from pKD32 (48) with the lsc-Cm primers. Plasmids (obtained from The E. coli Genetics Stock Center, Yale University) and primers used to generate recombination constructs are listed in Table 1 and Table S1 in the supplemental material, respectively. Each PCR mixture contained final concentrations of 200 nM each primer, 1× polymerase buffer (NEB, Ipswich, MA), 2.5 U Taq polymerase (NEB), 300 μM each deoxynucleoside triphosphate (NEB), 3 mM MgCl2, and 2 μl of DNA template. Reactions were run in a GeneAmp 9700 thermal cycler (Applied Biosystems, Foster City, CA) under the following conditions: 95°C for 5 min, and 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s. PCR products were purified using a gel purification kit (Norgen Biotek, St. Catharines, ON, Canada).
Prior to introduction of a recombination construct, E. amylovora cells were made electrocompetent by washing 3 times in 35 ml ice-cold sterile distilled H2O and transformed with plasmid pKD46 by electroporation. Resulting transformants were screened by selecting for ampicillin resistance on LB agar. The pKD46 transformants were grown overnight at 27°C, reinoculated in LB broth containing 0.1% arabinose to induce the red recombinase (48), grown to exponential phase (optical density at 600 nm [OD600], 0.8), and made electrocompetent again by washing as described above. Cells were transformed with a recombination construct using electroporation and recovered in 1 ml of SOC medium (50) for 1 to 4 h at 27°C before plating on LB agar with the appropriate selective antibiotic. Electroporations were performed at 2.5 kV and 25 μF with the pulse controller set at 200 Ω. Recombinants were selected on LB containing kanamycin or chloramphenicol. In the resulting mutants (Table 1), the Cmr or Kmr insert replaced part of the coding region of the target gene by site-specific recombination in the flanking regions. To evict the temperature-sensitive pKD46 from mutants, cells were briefly heated to 37°C for 10 min, plated on LB lacking ampicillin, and grown overnight at 27°C. Replica plating on LB and LB-ampicillin agar was used to differentiate the colonies that lost the ampicillin resistance carried on pKD46.
EPS measurement.
Levan and amylovoran production levels were measured in 1-ml supernatant aliquots obtained from liquid cultures grown in nutrient broth (Difco) at 27°C with shaking. Levan production was not measured directly. Instead, levansucrase in supernatants was detected by the addition of 1 ml of 50 mM sodium phosphate, 2 M sucrose, and 0.05% sodium azide and incubation for 24 h at 27°C (15). The amylovoran concentration was determined by precipitating the substance from the culture filtrates with 50 mg/ml of cetylpyridinium chloride and measuring turbidity (the OD600) (5).
RESULTS
EPS production in wild-type isolates.
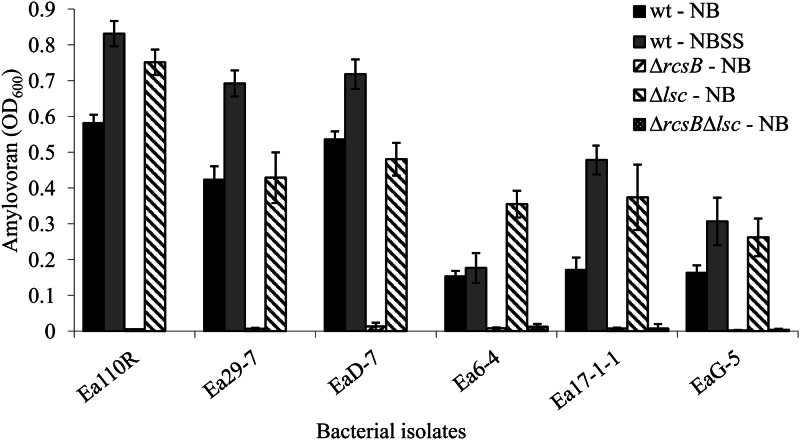
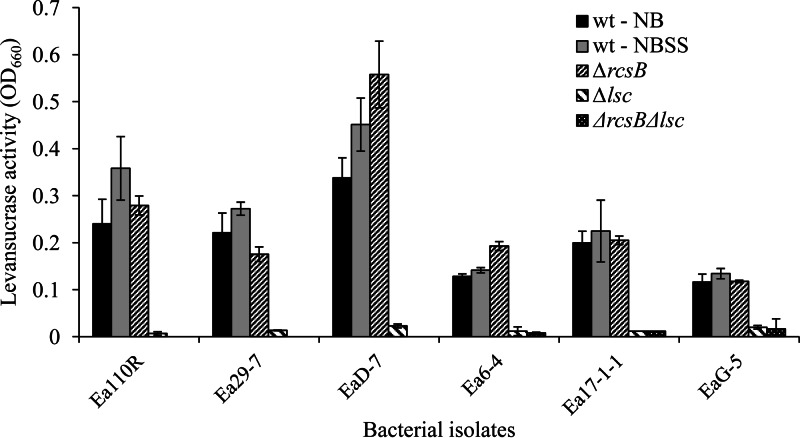
Wild-type E. amylovora isolates Ea110R, Ea29-7, and EaD-7 produced higher levels of amylovoran in NB than isolates Ea6-4, Ea17-1-1, and EaG-5 (Fig. 1). Amendment of the growth medium with 0.5% sucrose and 1% sorbitol resulted in increased amylovoran in all isolates except for Ea6-4. Levan production, measured by levansucrase activity, was higher for Ea110R, Ea29-7, and EaD-7 and lower for Ea6-4, Ea17-1-1, and EaG-5 (Fig. 2). Addition of sorbitol and sucrose to the medium resulted in no statistical difference for Ea110R, Ea29-7, and EaD-7 and modest increases in levansucrase activity for Ea6-4, Ea17-1-1, and EaG-5. Based on these results, Ea110R, Ea29-7, and EaD-7 were grouped as high EPS producers (HEP), and Ea6-4, Ea17-1-1, and EaG-5 were considered low EPS producers (LEP).
Fig 1.
Production of amylovoran by the wild type and ΔrscB, Δlsc, and ΔrscB Δlsc mutants of Erwinia amylovora. Amylovoran was precipitated from culture filtrates with 50 mg/ml of cetylpyridinium chloride, and turbidity (the OD600) was measured (5). Each bar represents the mean ± standard deviation of three independent measurements.
Fig 2.
Production of levansucrase by the wild type and ΔrscB, Δlsc, and ΔrscB Δlsc mutants of Erwinia amylovora. Activities were measured in the culture medium after removal of the bacterial cells by centrifugation (15). Each bar represents the mean ± standard deviation of three independent measurements.
EPS production and plaque morphology.
Myoviridae phages, classified as group 1 (36), produced clear plaques on the LEP hosts (Ea6-4, Ea17-1-1, and EaG-5) (Table 2). On HEP hosts (Ea110R, Ea29-7, and EaD-7), these phages either produced turbid plaques or completely failed to infect. The opposite pattern was found for most (8 of 11) of the Podoviridae phages. These phages produced large clear plaques on HEP isolates and turbid or no plaques on LEP hosts. The analysis included only two Siphoviridae phages, and no consistent pattern for host preference was evident, as ϕEa10-5 produced clear plaques on all hosts and ϕEa35-7 produced clear plaques only on HEP hosts. To determine if plaque morphology would be changed by host modification via passing the phage through a nonpreferred host, four phages were grown in Ea110R (HEP group) and Ea6-4 (LEP group). Plaque appearance was unchanged on the same six hosts listed in Table 2 for any phage grown in either host. Thus, plaque morphologies on either host type were not altered by the passage through a nonpreferred host for any of the four phages (data not shown).
Table 2.
Effects of E. amylovora HEP and LEP cells on bacteriophage plaque appearance and efficiency of plating
| Family and phage isolate | Isolation hosta | Phage groupb | Plaque appearance (EOP)c |
Clear plaque formationsd: | |||||
|---|---|---|---|---|---|---|---|---|---|
| HEP |
LEP |
||||||||
| Ea110R | Ea29-7 | EaD-7 | Ea6-4 | Ea17-1-1 | EaG-5 | ||||
| Myoviridae | |||||||||
| ϕEa9-6 | Ea17-1-1 | 1* | T (0.5) | N (0) | T (0.1) | C (1.0) | C (1.0) | C (1.4) | LEP |
| ϕEa9-7 | Ea6-4 | 1* | T (1.0) | T (1.0) | T (0.2) | C (1.0) | C (1.4) | C (1.0) | LEP |
| ϕEa10-1 | Ea17-1-1 | 1* | T (0.3) | T (—) | T (0.1) | C (0.8) | C (1.0) | C (0.5) | LEP |
| ϕEa10-2 | Ea6-4 | 1 | T (0.5) | N (0) | T (0.4) | C (1.0) | C (1.9) | C (1.0) | LEP |
| ϕEa10-4 | EaG-5 | 1 | T (0.4) | T (0.6) | T (0.3) | C (0.6) | C (1.8) | C (1.0) | LEP |
| ϕEa21-2 | EaG-5 | 1 | T (1.0) | T (0.3) | T (0.8) | C (1.3) | C (1.5) | C (1.0) | LEP |
| ϕEa21-4 | Ea6-4 | 1 | T (1.2) | T (1.3) | T (0.7) | C (1.0) | C (1.2) | C (0.9) | LEP |
| ϕEa35-2 | Ea17-1-1 | 1 | T (0.3) | N (0) | N (0) | C (0.6) | C (1.0) | C (0.9) | LEP |
| Siphoviridae | |||||||||
| ϕEa10-5 | Ea110R | [2] | C (1.0) | C (0.5) | C (0.6) | C (1.6) | C (1.9) | C (2.5) | Both |
| ϕEa35-7 | Ea29-7 | 2 | C (0.8) | C (1.0) | C (0.9) | N (0) | T (—) | T (—) | LEP |
| Podoviridae | |||||||||
| ϕEa10-8 | Ea29-7 | 3A | C (1.5) | C (1.0) | C (1.0) | T (—) | T (—) | T (—) | HEP |
| ϕEa10-11 | Ea17-1-1 | 3A | C (0.5) | C (0.2) | C (0.3) | C (0.5) | C (1.0) | C (0.6) | Both |
| ϕEa31-3 | Ea29-7 | 3A | C (1.0) | C (0.9) | C (0.7) | N (0) | T (0.3) | N (0) | HEP |
| ϕEa46-2 | EaD-7 | 3B | C (1.3) | C (1.1) | C (1.0) | T (—) | T (—) | T (—) | HEP |
| ϕEa1(h)e | Ea110R | 3C | C (1.0) | C (2.3) | C (1.2) | C (2.6) | T (—) | C (3.2) | Both |
| ϕEa9-4 | EaG-5 | 5 | T (0.5) | N (0) | T (0.2) | C (0.3) | C (0.9) | C (1.0) | LEP |
| ϕEa51-4 | Ea29-7 | 6 | C (1.5) | C (1.0) | C (0.9) | T (—) | T (—) | T (—) | HEP |
| ϕEa31-7 | Ea29-7 | * | C (0.9) | C (1.0) | C (1.4) | T (—) | T (—) | T (—) | HEP |
| ϕEa31-8 | EaD-7 | * | C (1.5) | C (0.9) | C (1.0) | T (—) | T (—) | T (—) | HEP |
| ϕEa45-1B | Ea29-7 | * | C (0.8) | C (1.0) | C (1.0) | N (0) | T (—) | T (—) | HEP |
| ϕEa46-1A2 | EaD-7 | * | C (3.0) | C (2.5) | C (1.0) | T (—) | T (0.7) | T (—) | HEP |
Original bacterial isolation host, as published in reference 36.
Bacteriophages were placed into families and groups based on RFLP patterns and transmission electron micrographs (36). * denotes the sole use of real-time PCR to place the phage into the Myoviridae or Podoviridae (47); [2] indicates a presumptive Siphoviridae phage based on nondetection by real-time PCR (47).
Plaque appearance categories: C, clear; T, turbid/hazy; N, no plaques. EOP is the phage titer on the bacterial isolate divided by the phage titer on the isolation host.
General category of bacterial host, based on HEP or LEP status. —, plaques were present but could not be counted and distinguished due to their hazy appearance.
Ea1(h) was obtained from A. L. Jones (38).
Plaque morphology and efficiency of plating.
To correlate plaque morphology with sensitivity or partial resistance to a phage, EOP was determined for several phage-host combinations (Table 2; EOP values are indicated in parentheses after the plaque morphology descriptor). The average EOP (n = 24; combinations where the plaques were too faint to be counted accurately were excluded from this total) of Myoviridae phages on LEP hosts Ea6-4, Ea17-1-1, and EaG-5 was 1.08, and on HEP hosts Ea110R, Ea29-7, and Ea D-7 the average EOP (n = 23) was 0.48. This difference was significant (P < 0.001; Mann-Whitney U test). Podoviridae phages had an average EOP of 0.85 on LEP hosts (n = 13) and 1.06 on HEP hosts (n = 33), a significant difference (P < 0.05). For Siphoviridae, ϕEa35-7 had a preference for high-EPS hosts (EOPs of 0.9 for HEP and 0 for LEP), and for ϕEa10-5 the EOP was 2.0 on the LEP hosts and 0.7 on HEP isolates. A strong correlation between plaque morphology and EOP was evident. Of the 71 phage-host combinations that resulted in clear, countable plaques, 60 (85%) had EOP values greater than 0.7. Only 8 of the 23 combinations (35%) that produced turbid yet countable plaques reached this criterion.
Enhanced EPS production and phage population growth.
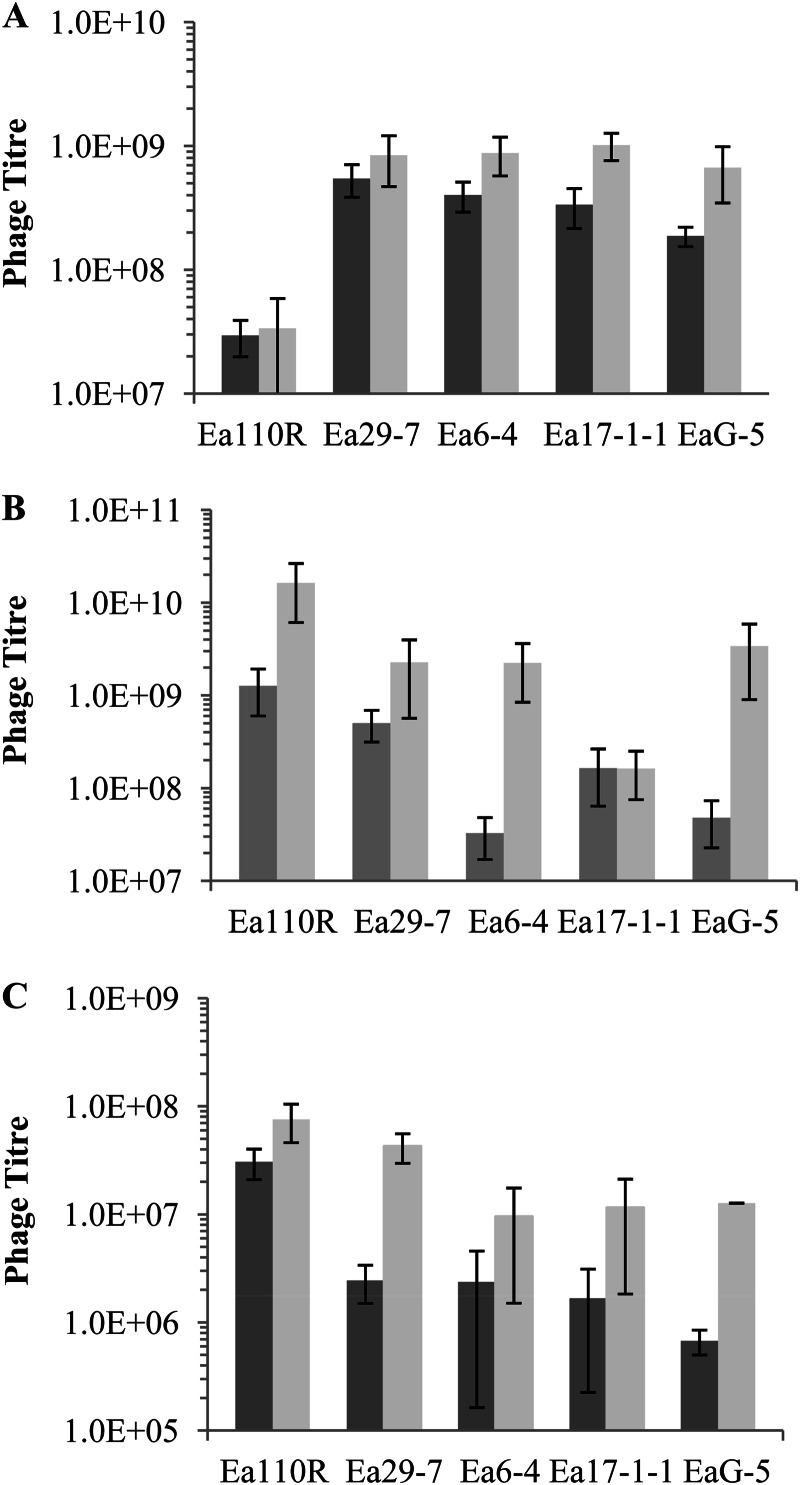
Single representatives from the three phage families were monitored for population propagation on five E. amylovora hosts on NA or NAS to increase EPS production (Fig. 3). The Myoviridae phage ϕEa21-4 multiplied equally well on Ea29-7 (HEP) and Ea6-4, Ea17-1-1, and EaG-5 (all LEP), but substantially less well with the HEP isolate Ea110R (Fig. 3A). With the exception of the growth on Ea110R, these outcomes were consistent with EOP observations, which indicated that ϕEa21-4 was able to infect most hosts equally well. Growth on NAS increased titers 2.3-fold (NA versus NAS, P = 0.02), a result that did not correlate well with plaque morphology, as this phage produced clear plaques on LEP hosts. This increase, however, was far smaller than the responses of the other two phages (see below) and may simply have been due to increased vigor of the hosts on the more-nutritive medium. The Siphoviridae phage ϕEa35-7 (Fig. 3B) propagated best on the HEP hosts Ea110R and Ea29-7 when grown on NA (HEP versus LEP, P < 0.001), a finding consistent with clear plaque morphology and high EOP results (Table 2). Phage production increased dramatically in four of the five cultures grown on supplemented medium. The average population size for all five hosts was approximately 30 times higher on NAS than on nonsupplemented NA (P < 0.001), a result consistent with the observation that this phage preferred hosts that produce large amounts of EPS. The contribution of improved host vigor on NAS to increased phage production cannot be determined presently. However, based on the magnitude of the change compared to ϕEa21-4, it is most probable that the major factor in the increase was enhanced EPS production. The Podoviridae phage ϕEa1(h) (Fig. 3C) produced substantially fewer progeny, compared to the other two phages, on all hosts, but it showed significantly more production on HEP hosts in the absence of sucrose (HEP versus LEP, P < 0.01) or presence of sucrose (HEP versus LEP, P < 0.001). The addition of sucrose to the growth medium had a strong effect and increased phage titers on all hosts, with an average increase of almost 6-fold (NA versus NAS, P < 0.01). As with the Siphoviridae phage, the contribution of increased host vigor cannot be estimated accurately, but it is likely that most of the increase is a result of increased EPS. The similar production on all hosts was consistent with clear plaque formation and strong EOPs for most isolates (Table 2).
Fig 3.
Population titers of Erwinia amylovora bacteriophages from five host isolates grown on NA or NAS. (A) ϕEa21-4 (Myoviridae); (B) ϕEa35-7 (Siphoviridae); (C) ϕEa1(h) (Podoviridae). Results are means and standard deviations from three independent experiments.
EPS-deficient mutants, decreased EPS production, and phage population growth.
Mutants with defects in the synthesis of amylovoran and/or levan were produced in order to understand the roles of each of the two EPSs in phage infection. Amylovoran-deficient mutants with a deletion of the rcsB gene and levan-deficient mutants with a deletion of the lcs gene were generated for all isolates (Table 1). Stable rcsB and lsc double deletion derivatives were recovered from the LEP isolates Ea6-4, Ea17-1-1, and EaG-5. Despite numerous attempts, including several alterations to the mutagenesis procedures, double deletion mutants of the HEP isolates were not obtained, suggesting that this genotype is lethal in these bacteria. All mutational changes were confirmed by PCR.
Amylovoran production was negligible in all ΔrcsB strains (Fig. 1), and levansucrase activity was almost undetectable for all Δlsc strains (Fig. 2). The ΔrcsB Δlsc double mutants produced extremely low amounts of both EPSs. Four of the six levansucrase mutants, Ea110RΔlsc, Ea6-4Δlsc, Ea17-1-1Δlsc, and EaG-5Δlsc, demonstrated increases in amylovoran accumulation compared to their wild-type progenitor. Most rcsB mutants did not show any significant change in levansucrase activity compared to the nonmutant wild type.
Phage populations were substantially decreased for all phages on either ΔrcsB or Δlsc strains (Table 3). Myoviridae and Siphoviridae phages produced significantly fewer progeny on the Δlsc strains than on the ΔrcsB strains (P < 0.001 and P < 0.01, respectively). Decreased amylovoran production in the ΔrcsB strains resulted in significantly reduced progeny for all of the Podoviridae phages (P < 0.001). A few phages in all three families showed apparently reduced progeny production in both mutant backgrounds, although the results were not statistically significant.
Table 3.
Efficiencies of plating of bacteriophages from the Myoviridae, Siphoviridae, and Podoviridae on ΔrcsB, Δlsc, and wild-type isolates of Erwinia amylovora
| Family and phage isolate | Isolation host | Efficiency of platinga |
|
|---|---|---|---|
| ΔrcsBb | Δlscc | ||
| Myoviridae | |||
| ϕEa10-1 | Ea17-1-1 | 0.25 | 0.06 |
| ϕEa21-2 | Ea17-1-1 | 1.11 | 0.12 |
| ϕEa21-4 | Ea 6-4 | 1.38 | 0.02 |
| Siphoviridae | |||
| ϕEa10-19 | Ea17-1-1 | 0.64 | 0.05 |
| ϕEa35-7 | Ea110R | 5.34 | 0.26 |
| Podoviridae | |||
| ϕEa10-6 | Ea17-1-1 | 0 | 0.98 |
| ϕEa31-3 | Ea29-7 | 0 | 0.34 |
| ϕEa35-3 | Ea110R | 0.0005 | 0.23 |
| ϕEa45-1B | Ea29-7 | 0.0007 | 1.84 |
| ϕEa46-1A2 | Ea110R | 0.0003 | 0.34 |
| ϕEa50-3 | Ea29-7 | 0.0012 | 1.35 |
| ϕEa51-7 | Ea29-7 | 0.0026 | 1.44 |
Number of plaques on the mutant host strain divided by the number of plaques on the wild-type bacterial host. Results are from three independent experiments.
Amylovoran-deficient mutant.
Levan-deficient mutant.
DISCUSSION
The production of clear or turbid plaques on a host lawn appeared to be a good indication of the ability of a phage to attack an E. amylovora wild-type isolate. This characteristic correlated well with a high EOP and population growth for most phages. A possible explanation for the production of turbid plaques is that a portion of the population has undergone lysogeny, resulting in resistance to homologous phages (52, 53). However, none of the hosts contained detectable prophage DNA, based on real-time PCR screening and/or production of virions through induction (47). A more reasonable explanation for differences in plaque appearance is host preference. Billing (2) observed in 1960 that some phages produce confluent lysis and plaque halos on encapsulated E. amylovora isolates and turbid lysis on hosts with reduced encapsulation. Other phages showed the opposite pattern. In the present study, we found that most Podoviridae phages appear to be adapted to hosts that produce relatively high amounts of EPS and Myoviridae phages appear to be adapted to hosts that have lower levels of EPS (Table 2). Individual phages in the Siphoviridae may be adapted to one type of host or the other; however, no generalization on host preference can be made for this family due to the small sample size as well as the lack of a clear pattern within our sample.
Alteration of the amount of EPS produced, either by increasing production on supplemented medium or by knocking out synthesis of either type in the deletion mutants, indicated that phage efficacy is dependent on both of these substances. Amylovoran was virtually essential for the proliferation of most phages in the Podoviridae with progeny production dropping to less than 1% in the ΔrcsB mutants compared to wild-type progenitors. For Myoviridae and Siphoviridae phages, however, levan may be the primary site of interaction, as growth on the amylovoran knockout strain showed little to no decrease in progeny production while the levan knockout reduced proliferation significantly (Table 3). The individual roles of amylovoran and levan in E. amylovora phage-host interactions have not been described, and the involvement of EPS in Erwinia spp. phage infection is limited to a few early reports (2, 6, 7, 20). Some phages have evolved to recognize specific polysaccharides of other bacterial species (34, 54, 55). Certain Escherichia coli phages bind only when a capsular EPS with a serotype-specific surface K antigen is present on a potential host (54, 55, 56, 57). Removing the capsular EPS from cells results in a lack of K antigen-specific phage lysis (55). Phages that attack Vibrio cholerae have also been found to specifically lyse EPS-producing strains (58), and the capsular polysaccharide surrounding the cells of Streptococcus thermophilus could also play a role in the cellular adsorption of specific phages (32). For Erwinia spp. phages, it is likely that amylovoran and levan serve as binding sites, similar to these examples.
Many EPS-specific phages carry depolymerases that can recognize and degrade specific polymers as part of the virion tail (56). Podoviridae phages carry depolymerases that degrade amylovoran (59, 60). Depolymerases are thought to allow the virion to gain access to the cell surface, where it most likely binds to an outer membrane receptor. Removal of the EPS barrier should improve phage growth by exposing an outer membrane receptor, but that was not found to be the case in the present study for phages that are known to produce depolymerase. If such a secondary receptor exists in E. amylovora, binding to EPS is a necessary step prior to attachment to the membrane receptor. There is the possibility that the rcsB deletion also altered some other component that acts as the cell surface receptor. There is evidence for similar secondary effects. Mutations in the rcs system of E. amylovora have been found to increase resistance to the antimicrobial peptide polymyxin B, which requires cell membrane binding to be effective (61).
Myoviridae and Siphoviridae phages do not express depolymerase activities in culture and appear to lack the gene (39, 40). It is not known how these phages subsequently bypass the EPS barrier and infect their hosts. Adaptation of these phages to LEP hosts appears to be a reasonable strategy for propagation, simply because the EPS barrier is smaller than that on an HEP bacterium.
For several bacterial species, most of the functions ascribed to EPS are of a protective nature, including inhibition of phage attack (28). EPS is presumed to provide a physical barrier between infecting phages and cell surface receptors (27, 62). A protective function has been reported for Rhizobium meliloti, as EPS nonspecifically prevented phage adsorption (8). The physical removal of the K1 EPS capsule restored phage susceptibility of Escherichia coli (57). EPS was also found to be responsible for the inhibition of phage adsorption to Lactococcus lactis (27). These observations suggest that the production of EPS may be a near-universal antiphage defense mechanism, providing a physical barrier to the cell surface receptors. The EPSs of E. amylovora, however, do not appear to provide such a protective role and may simply provide an attachment site.
Current information suggests that amylovoran and levan production levels are loosely coregulated. Amylovoran biosynthesis is controlled by the ams operon, which includes the rcsA and rcsB genes (61). Overexpression of rcsA or rcsB reduces levansucrase synthesis (10). Mutations that impair rcsA function can have variable effects on levan synthesis, reducing levansucrase activity or not. rcsB mutations generally do not effect levan production (15). The present results with the rcsB deletion strains that showed no change in levansucrase activities support this conclusion (Fig. 2). Mutation of the levansucrase gene, lsc, has been reported to have no effect on amylovoran production (10). Four of the lsc deletion mutants in this study, however, showed increases in amylovoran production, ranging from 29 to 119% (Fig. 1). The difference between the two studies can be explained by either an environmental effect, as Geier and Geider (10) tested production on medium supplemented with sorbitol to stimulate EPS production while the present measurements were on bacteria grown on nonsupplemented nutrient agar, or by unknown genetic differences in the isolates used in the two studies. While it is presently not possible to determine the nature of the cross talk between the two EPS biosynthetic pathways, it is apparent that isolates differ in overall EPS production but still strive to maintain a minimal level of at least one of the two types (15). The difficulty in recovering ΔrcsB Δlsc strains of the HEP isolates supported this conclusion. Furthermore, coregulated synthesis of the EPSs would allow the bacteria to avoid unnecessary energy expenditures associated with EPS overproduction.
This study showed that the pathogenicity and presumably adsorption of phages that attack E. amylovora was mediated by amylovoran or levan. If in fact the phage receptor for certain Erwinia spp. phages is amylovoran, the obvious way for the cell to become phage resistant is to change the polymer's composition or abolish its production. However, amylovoran produced by E. amylovora is correlated with pathogenicity, as deficient mutants are avirulent (4, 5, 9, 10, 17, 21, 63, 64, 65). If amylovoran composition were altered by mutation, it would likely also reduce virulence of the bacterium to the plant host. This in turn would greatly reduce the chance of survival of the bacterium. Thus, the adaptation of phages in the Podoviridae to amylovoran is a very effective strategy for their own survival.
The fire blight pathogen populations increase exponentially in the sugar-rich stigma surface under optimal environmental conditions. The bacterial capsule, specifically the constituent EPS amylovoran, plays an important role in bacterial virulence and pathogenicity. Our research has demonstrated that Erwinia spp. phages are highly adapted to the pathogen and the plant environment. Phage populations increase significantly in the presence of sugars, and amylovoran plays a key role in the ability of the Podoviridae phages to infect the HEP bacterial host. In contrast, the Myoviridae phages preferentially infect the LEPs and respond to the presence of levan on the host. To obtain a phage-mediated biopesticide with high efficacy, these phage-host interactions will need to be taken into consideration.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a research grant from Agriculture and Agri-Food Canada. Financial support for graduate students was provided by Brock University.
We thank Kathy Whybourne for technical assistance and editorial advice.
Footnotes
Published ahead of print 15 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00067-13.
REFERENCES
- 1.Hildebrandt EM. 1939. Studies on fire-blight ooze. Phytopathology 29:142–156 [Google Scholar]
- 2.Billing E. 1960. An association between capsulation and phage sensitivity in Erwinia amylovora. Nature 186:819–820 [DOI] [PubMed] [Google Scholar]
- 3.Goodman RN, Huang JS, Huang PY. 1974. Host-specific phytotoxic polysaccharide from apple tissue infected by Erwinia amylovora. Science 183:1081–1082 [DOI] [PubMed] [Google Scholar]
- 4.Bellemann P, Geider K. 1992. Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biological characterization. J. Gen. Microbiol. 138:931–940 [DOI] [PubMed] [Google Scholar]
- 5.Bellemann P, Bereswill S, Berger S, Geider K. 1994. Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int. J. Biol. Macromol. 16:290–296 [DOI] [PubMed] [Google Scholar]
- 6.Bennett RA, Billing E. 1978. Capsulation and virulence in Erwinia amylovora. Ann. Appl. Biol. 89:41–45 [Google Scholar]
- 7.Bennett RA, Billing E. 1980. Origin of polysaccharide component of ooze from plants infected with Erwinia amylovora. J. Gen. Microbiol. 116:341–349 [Google Scholar]
- 8.Defives C, Werquin MM, Mary P, Hornez JP. 1996. Roles of exopolysaccharides and lipopolysaccharides in the adsorption of the Siphovirus phage NM8 to Rhizobium meliloti M11S cells. Curr. Microbiol. 33:371–376 [DOI] [PubMed] [Google Scholar]
- 9.Geider K, Geier G, Bellemann P, Bernhard F, Bugert P, Metzger M. 1993. Exopolysaccharides in pathogenicity of Erwinia amylovora. Acta Hortic. 338:255–262 [Google Scholar]
- 10.Geier G, Geider K. 1993. Characterization and influence on virulence of the levansucrase gene from the fire blight pathogen Erwinia amylovora. Physiol. Mol. Plant Pathol. 42:387–404 [Google Scholar]
- 11.Denny TP. 1995. Involvement of bacterial polysaccharides in plant pathogenesis. Phytopathology 33:173–197 [DOI] [PubMed] [Google Scholar]
- 12.Nimtz M, Mort A, Domke T, Wray V, Zhang YQF, Coplin D, Geider K. 1996. Structure of amylovoran, the capsular exopolysaccharide from the fire blight pathogen Erwinia amylovora. Carbohydr. Res. 287:59–76 [DOI] [PubMed] [Google Scholar]
- 13.Bugert P, Geider K. 1995. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol. Microbiol. 15:917–933 [DOI] [PubMed] [Google Scholar]
- 14.Bugert P, Bereswill S, Geider K. 1996. Regulation and structure of the ams-region involved in exopolysaccharide synthesis of Erwinia amylovora. Acta Hortic. 411:275–280 [Google Scholar]
- 15.Bereswill S, Geider K. 1997. Characterization of the rcsB gene from Erwinia amylovora and its influence on exopolysaccharide synthesis and virulence of the fire blight pathogen. J. Bacteriol. 179:1354–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geider K, Aldridge P, Bereswill S, Bugert P, Langlotz C. 1996. Characterization of exopolysaccharide synthesis by Erwinia amylovora. Acta Hortic. 411:259–263 [Google Scholar]
- 17.Gross M, Geier G, Rudolp K, Geider K. 1992. Levan and levansucrase synthesized by the fireblight pathogen Erwinia amylovora. Physiol. Mol. Plant Pathol. 40:371–381 [Google Scholar]
- 18.Costerton JW, Cheng K-J, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435–464 [DOI] [PubMed] [Google Scholar]
- 19.Koczan JM, McGrath MJ, Zhao Y, Sundin GW. 2009. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications to pathogenicity. Phytopathology 99:1237–1244 [DOI] [PubMed] [Google Scholar]
- 20.Ayers AR, Ayers SB, Goodman RN. 1979. Extracellular polysaccharide of Erwinia amylovora: a correlation with virulence. Appl. Environ. Microbiol. 38:659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SA, Ngugi HK, Halbrendt NO, O'Keefe G, Lehman B, Travis JW, Sinn JP, McNellis TW. 2010. Virulence characteristics accounting for fire blight disease severity in apple trees and seedlings. Phytopathology 100:539–550 [DOI] [PubMed] [Google Scholar]
- 22.Leigh JA, Coplin DL. 1992. Exopolysaccharides in plant-bacterial interactions. Annu. Rev. Microbiol. 46:307–346 [DOI] [PubMed] [Google Scholar]
- 23.Steinberger EM, Beer SV. 1988. Creation and complementation of pathogenicity mutants of Erwinia amylovora. Mol. Plant Microbe Interact. 1:135–144 [Google Scholar]
- 24.Zhao Y, Sundin GW, Wang D. 2009. Construction and analysis of pathogenicity island deletion mutants of Erwinia amylovora. Can. J. Microbiol. 55:457–464 [DOI] [PubMed] [Google Scholar]
- 25.Hughes KA, Sutherland IW, Jones MV. 1998. Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology 144:3039–3047 [DOI] [PubMed] [Google Scholar]
- 26.Cerning J. 1990. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol. Rev. 87:113–130 [DOI] [PubMed] [Google Scholar]
- 27.Forde A, Fitzgerald GF. 1999. Analysis of exopolysaccharide (EPS) production mediated by the bacteriophage adsorption blocking plasmid, pCI658, isolated from Lactococcus lactis ssp. cremoris HO2. Int. Dairy J. 9:465–472 [Google Scholar]
- 28.Looijesteijn PJ, Trapet L, de Vries E, Abee T, Hugenholtz J. 2001. Physiological function of exopolysaccharides produced by Lactococcus lactis. Int. J. Food Microbiol. 64:71–80 [DOI] [PubMed] [Google Scholar]
- 29.Sutherland IW. 1988. Bacterial surface polysaccharides: structure and function. Int. Rev. Cytol. 113:187–231 [DOI] [PubMed] [Google Scholar]
- 30.Whitfield C. 1988. Bacterial extracellular polysaccharides. Can. J. Microbiol. 34:415–420 [DOI] [PubMed] [Google Scholar]
- 31.Deveau H, Van Calsteren M-R, Moineau SS. 2002. Effect of exopolysaccharides on phage-host interactions in Lactococcus lactis. Appl. Environ. Microbiol. 68:4364–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia P, Martinez B, Obeso JM, Rodriguez A. 2008. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47:479–485 [DOI] [PubMed] [Google Scholar]
- 33.Leiman PG, Kostyuchenko VA, Shneider MM, Kurochkina LP, Mesyanzhinov VV, Rossmann MG. 2000. Structure of bacteriophage T4 gene product 11, the interface between the baseplate and short tail fibers. J. Mol. Biol. 301:975–985 [DOI] [PubMed] [Google Scholar]
- 34.Scholl D, Adhya S, Merril C. 2005. Escherichia coli K1's capsule is a barrier to bacteriophage T7. Appl. Environ. Microbiol. 71:4872–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boulé J, Sholberg PL, Lehman SM, O'Gorman DT, Svircev AM. 2011. Isolation and characterization of eight bacteriophages infecting Erwinia amylovora and their potential as biological control agents in British Columbia, Canada. Can. J. Plant Pathol. 33:308–317 [Google Scholar]
- 36.Gill JJ, Svircev AM, Smith R, Castle AJ. 2003. Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 69:2133–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie DF, Klos EJ. 1977. Isolation of Erwinia amylovora bacteriophage from aerial parts of apple trees. Phytopathology 67:101–104 [Google Scholar]
- 38.Schnabel EL, Jones AL. 2001. Isolation and characterization of five Erwinia amylovora bacteriophages and assessment of phage resistance in strains of Erwinia amylovora. Appl. Environ. Microbiol. 67:59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehman SM. 2007. Development of a bacteriophage-based biopesticide for fire blight. Ph.D. thesis. Brock University, St. Catharines, Ontario, Canada [Google Scholar]
- 40.Lehman SM, Kropinski AM, Castle AJ, Svircev AM. 2009. The complete genome of the broad-host-range Erwinia amylovora phage Ea21-4 and its relationship to Salmonella phage Felix 01. Appl. Environ. Microbiol. 75:2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svircev AM, Castle AJ, Lehman SM. 2010. Bacteriophages for control of phytopathogens in food production systems, p 79–102 In Sabour PM, Griffiths MW. (ed), Bacteriophages in the control of food- and waterborne pathogens. ASM Press, Washington, DC [Google Scholar]
- 42.Svircev AM, Lehman SM, Sholberg P, Roach D, Castle AJ. 2011. Phage biopesticides and soil bacteria: multilayered and complex interactions, p 215–235 In Witzany G. (ed), Biocommunication in soil microorganisms. Springer, Heidelberg, Germany [Google Scholar]
- 43.Stockwell VO, Johnson KB, Johnson VW. 2006. Colonization of flowers by Pseudomonas fluorescens A506 formulated in a biopolymer gel. Acta Hortic. 704:293–299 [Google Scholar]
- 44.Stockwell VO, Johnson KB, Loper JE. 1998. Establishment of bacterial antagonists of Erwinia amylovora on pear and apple blossoms as influenced by inoculum preparation. Phytopathology 88:506–513 [DOI] [PubMed] [Google Scholar]
- 45.Stockwell VO, Johnson KB, Sugar D, Loper JE. 2002. Antibiosis contributes to biological control of fire blight by Pantoea agglomerans strain Eh252 in orchards. Phytopathology 92:1202–1209 [DOI] [PubMed] [Google Scholar]
- 46.Stockwell VO, Johnson KB, Sugar D, Loper JE. 2010. Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology 100:1330–1339 [DOI] [PubMed] [Google Scholar]
- 47.Roach DR. 2011. Erwinia amylovora bacteriophage resistance. Ph.D. thesis. Brock University, St. Catharines, Ontario, Canada [Google Scholar]
- 48.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams MH. 1959. Bacteriophages. Interscience Publishers, New York, NY [Google Scholar]
- 50.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 51.Jeng RS, Svircev AM, Myers AL, Believa L, Hunter DM, Hubbes M. 2001. The use of 16S and 16S-23S rDNA to easily detect and differentiate common Gram-negative orchard epiphytes. J. Microbiol. Methods 44:69–77 [DOI] [PubMed] [Google Scholar]
- 52.Abedon ST. 2008. Bacteriophage ecology: population growth, evolution, and impact of bacterial viruses. Cambridge University Press, Cambridge, England [Google Scholar]
- 53.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70:217–248 [DOI] [PubMed] [Google Scholar]
- 54.Bayer ME, Thurow H, Bayer MH. 1979. Penetration of the polysaccharide capsule of Escherichia coli (Bi 161/42) by bacteriophage K29. Virology 94:95–118 [DOI] [PubMed] [Google Scholar]
- 55.Stirm S. 1968. Escherichia coli K bacteriophages, I. Isolation and introductory characterization of five Escherichia coli K bacteriophages. J. Virol. 2:1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindberg AA. 1977. Bacterial surface carbohydrates and bacteriophage adsorption, p 289–356. In Sutherland I. (ed), Surface carbohydrates of the prokaryotic cell. Academic Press, London, England [Google Scholar]
- 57.Pelkonen S, Aalto J, Finne J. 1992. Differential activities of bacteriophage depolymerase on bacterial polysaccharide: binding is essential but degradation is inhibitory in phage infection of K-1 defective Escherichia coli. J. Bacteriol. 174:7757–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albert MJ, Bhuiyan NA, Rahman A, Ghosh AN, Hultenby K, Weintraub A, Nahar S, Kibriya AKMG, Ansaruzzaman M, Shimada T. 1996. Phage specific for Vibrio cholerae O139 Bengal. J. Clin. Microbiol. 34:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim W-S, Geider K. 2000. Characterization of a viral EPS-depolymerase, a potential tool for control of fire blight. Phytopathology 90:1263–1268 [DOI] [PubMed] [Google Scholar]
- 60.Vandenbergh PA, Wright AM, Vidaver AK. 1985. Partial purification and characterization of a polysaccharide depolymerase associated with phage-infected Erwinia amylovora. Appl. Environ. Microbiol. 49:994–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang D, Korban SS, Zhao Y. 2009. The rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora. Mol. Plant Pathol. 10:277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 63.Aldridge P, Metzger M, Geider K. 1997. Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol. Gen. Genet. 256:611–619 [DOI] [PubMed] [Google Scholar]
- 64.Kim W-S, Hildebrand M, Jock S, Geider K. 2001. Molecular comparison of pathogenic bacteria from pear trees in Japan and the fire blight pathogen Erwinia amylovora. Microbiology 147:2951–2959 [DOI] [PubMed] [Google Scholar]
- 65.Oh C-S, Beer SV. 2005. Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253:185–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.