Abstract
Accumulation of xylitol in xylose fermentation with engineered Saccharomyces cerevisiae presents a major problem that hampers economically feasible production of biofuels from cellulosic plant biomass. In particular, substantial production of xylitol due to unbalanced redox cofactor usage by xylose reductase (XR) and xylitol dehydrogenase (XDH) leads to low yields of ethanol. While previous research focused on manipulating intracellular enzymatic reactions to improve xylose metabolism, this study demonstrated a new strategy to reduce xylitol formation and increase carbon flux toward target products by controlling the process of xylitol secretion. Using xylitol-producing S. cerevisiae strains expressing XR only, we determined the role of aquaglyceroporin Fps1p in xylitol export by characterizing extracellular and intracellular xylitol. In addition, when FPS1 was deleted in a poorly xylose-fermenting strain with unbalanced XR and XDH activities, the xylitol yield was decreased by 71% and the ethanol yield was substantially increased by nearly four times. Experiments with our optimized xylose-fermenting strain also showed that FPS1 deletion reduced xylitol production by 21% to 30% and increased ethanol yields by 3% to 10% under various fermentation conditions. Deletion of FPS1 decreased the xylose consumption rate under anaerobic conditions, but the effect was not significant in fermentation at high cell density. Deletion of FPS1 resulted in higher intracellular xylitol concentrations but did not significantly change the intracellular NAD+/NADH ratio in xylose-fermenting strains. The results demonstrate that Fps1p is involved in xylitol export in S. cerevisiae and present a new gene deletion target, FPS1, and a mechanism different from those previously reported to engineer yeast for improved xylose fermentation.
INTRODUCTION
The need to replace conventional fossil fuels with alternative renewable fuels is increasing in the face of growing demand for energy and rising concerns about greenhouse gas emissions (1). Lignocellulosic biomass from nonfood stocks, such as agricultural and forestry residues, has been recognized as a promising and sustainable source for producing liquid biofuels (2–5). Xylose is the second most abundant sugar in lignocellulosic biomass, comprising up to 35% of the total carbohydrates (6). Therefore, efficient utilization of xylose has to be ensured for economically feasible production of lignocellulosic biofuels.
Saccharomyces cerevisiae is a widely used microorganism for biofuel production (7). However, wild-type S. cerevisiae cannot metabolize xylose because of the lack of a pathway to efficiently convert xylose to d-xylulose, which the organism can metabolize via the pentose phosphate pathway after phosphorylation (6). Two types of xylose-assimilating pathways have been identified and used to engineer xylose-utilizing S. cerevisiae strains. One is the redox cofactor-dependent xylose reductase (XR)/xylitol dehydrogenase (XDH) pathway (8–15), and the other is the redox-neutral xylose isomerase (XI) pathway (16–23). Introduction of the XR/XDH pathway into S. cerevisiae by metabolic engineering approaches has been widely studied, but redox imbalance is a key problem, because XR can use both NADPH and NADH, while XDH uses NAD+ exclusively (6, 24). The cofactor imbalance may lead to substantial xylitol accumulation and low ethanol yields (8, 9, 25, 26). Expressing the XI pathway can avoid the cofactor imbalance problem under anaerobic conditions, but xylitol accumulation has also been observed in strains expressing XI (17, 18, 20), because the nonspecific aldose reductase encoded by the GRE3 gene can produce xylitol from xylose (27). Various rational approaches have been used to reduce xylitol accumulation and improve xylose utilization, such as optimizing the expression levels of xylose-assimilating reactions (26), engineering the cofactor preference of XR/XDH enzymes (28–33), perturbing the pentose phosphate pathway by gene knockout or overexpression (34–39), or deleting GRE3 in strains expressing the XI pathway (21, 40, 41). While extensive previous efforts focused on manipulating intracellular metabolic reactions to improve xylose utilization and reduce by-product (e.g., xylitol) accumulation, controlling the xylitol export process might also be a meaningful strategy for reducing its formation and increasing carbon flux toward target products. However, a xylose transporter in S. cerevisiae has not been reported.
The major intrinsic protein (MIP) family is a group of integral membrane channel proteins found in a wide range of organisms from bacteria to humans (42). Fps1p has been identified in S. cerevisiae as a yeast member of the MIP family and is known as an aquaglyceroporin involved in glycerol transport by facilitated diffusion (43). It has also been found that Fps1p is involved in the uptake of acetic acid (44, 45) and arsenite and antimonite (45) and the regulation of osmotolerance (46). Since xylitol and glycerol share similar structures with linear polyols and Fps1p has shown relatively broad transport capacity, we hypothesized that Fps1p might be involved in xylitol transport. The Escherichia coli aquaglyceroporin GlpF has been reported to mediate the transport of a range of linear polyalcohols, including glycerol and xylitol (47). Also, a study has shown that expression of hyperactive Fps1-Δ1 in a gpd1Δ gpd2Δ S. cerevisiae mutant strain could mediate the uptake of xylitol at a much lower rate than glycerol uptake, but the wild-type Fps1 did not mediate detectable uptake of xylitol in the same assay (48). A transport study using secretory vesicles prepared from cells overexpressing Fps1p showed its capability for xylitol transport at a rate 1 order of magnitude lower than that for glycerol transport (48). However, the actual role of Fps1p in xylitol transport in living yeast cells is unclear. This study aimed to determine the role of Fps1p in xylitol transport in engineered S. cerevisiae strains during xylose metabolism and the effect of deleting the FPS1 gene on xylose fermentation for biofuel production.
MATERIALS AND METHODS
Plasmid and strain construction.
The plasmids and strains used in this study are summarized in Table 1. The S. cerevisiae FPS1 gene, PCR amplified from the genomic DNA of the D452-2 strain using primer pairs FPS1-f and FPS1-r, was cloned into the yeast integrative plasmid pRS403 under the control of the TDH3 promoter and CYC1 terminator using SpeI and SalI restriction enzyme sites, yielding the plasmid pRS403-FPS1. The TOP10 E. coli strain for cloning was grown in Luria-Bertani medium at 37°C, and 50 μg/ml of ampicillin was added to the medium when required. Transformation of the plasmid constructs into S. cerevisiae strain D10 overexpressing XYL1 was performed using the yeast EZ-Transformation kit (BIO 101, Vista, CA). Positive transformants were selected on synthetic complete medium containing 20 g/liter glucose (SCD) with amino acids or nucleotides added as necessary. A yeast strain overexpressing the FPS1 gene (D10F) and a control strain (D10c) were obtained, and introduction of the overexpression cassette was confirmed by diagnostic PCR with primer pairs targeting the TDH3 promoter (TDH3-f) and FPS1 (Table 2). The loxP-KanMX-loxP cassette for FPS1 gene deletion was PCR amplified with primers that target the KanMX marker gene on the plasmid pUG6 (49), with about 50-bp sequences homologous to sequence upstream (FK-f) and downstream (FK-r) of the FPS1 gene (Table 2). Transformation of PCR products into S. cerevisiae was performed using lithium acetate (LiAc)-polyethylene glycol (PEG) methods (50). Positive transformants were selected on YP medium (10 g/liter of yeast extract and 20 g/liter of peptone) containing 20 g/liter of d-glucose (YPD) with 200 μg/ml of Geneticin G418. Diagnostic PCR with primer targeting ∼750 bp upstream of the FPS1 gene (Fd-f) and the KanMX-specific primer (KanMX-r) was performed to confirm successful deletion. All other strains used in the study are listed in Table 1. Specifically, the recombinant xylose-fermenting S. cerevisiae strain SR8 was constructed previously in our laboratory (51) through (i) heterologous expression of XYL1 (coding for XR), XYL2 (coding for XDH), and XYL3 (coding for XK) from Scheffersomyces stipitis in S. cerevisiae D452-2 and optimization of the expression levels of XR, XDH, and XK; (ii) laboratory evolution on xylose; and (iii) deletion of ALD6, coding for acetaldehyde dehydrogenase.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| D452-2 | MATa leu2 his3 ura3 can1 | 70 |
| D10 | D452-2 leu2::LEU2 pYS10 | 53 |
| D10-fps1Δ | D10 fps1Δ::KanMX | This study |
| SR6 | D10 ura3::URA3 pSR6-X123 | 51 |
| SR6-fps1Δ | SR6 fps1Δ::KanMX | |
| SR8 | SR6 his1::HIS1 pSR3-X23, evolved, and ALD6 deletion by ald6::AUR1-C pAUR_d_ALD6 | 51 |
| SR8-fps1Δ | SR8 fps1Δ::KanMX | |
| D10F | D10 his3::HIS3 p403-FPS1 | This study |
| D10c | D10 his3::HIS3 pRS403 | This study |
| Plasmids | ||
| pYS10 | pRS305 TDH3P-XYL1-TDH3T | 11 |
| pSR6-X123 | pRS306 TDH3P-XYL1-TDH3T PGK1P-XYL2-PGK1T TDH3P-XYL3-TDH3T | 51 |
| pSR3-X23 | pRS403 PGK1P-XYL2-PGK1T TDH3P-XYL3-TDH3T | 51 |
| pAUR_d_ALD6 | pAUR101 containing the truncated ALD6 gene | 54 |
| pRS403 | Yeast integrative vector with HIS3 marker | 70 |
| p403-FPS1 | pRS403 TDH3P-FPS1-CYC1T | This study |
Table 2.
Primers for PCR amplification
| Primer | Sequencea | Comment |
|---|---|---|
| FK-f | TATTTTACCAAGTACGCTCGAGGGTACATTCTAATGCATTAAAAGACagctgaagcttcgtacgc | FPS1 deletion cassette using a KanMX marker |
| FK-r | GCAGTATTTTTTTCTATCAGTCTATATTATTTGTTTCTTTTTCTTGTCTGTTTTCgcataggccactagtggatc | |
| Fd-f | GTACATAACCGTAGGAAGGTACG | FPS1 deletion confirmation |
| KanMX-r | CTTTTCCTTACCCATGGTTGT | |
| FPS1-f | GCCACTAGTAAAAATGAGTAATCCTCAAAAAGCTC | FPS1 overexpression |
| FPS1-r | GCCCTCGAGTTATTTATTGCTGCCATTATATGAT | |
| TDH3-f | AGTTTATCATTATCAATACTCGCCATT |
Nucleotide sequences in lowercase represent the sequence targeting the loxP-KanMX-loxP cassette used for FPS1 deletion. Underlined sequences represent restriction enzyme sites.
Culture conditions and fermentation experiments.
Yeast strains were routinely cultivated at 30°C in YPD. Fermentation experiments under oxygen-limited conditions were performed in 50 ml of fermentation medium in a 250-ml Erlenmeyer flask at 30°C and 100 rpm. Anaerobic batch fermentation experiments were performed at 30°C in serum bottles sealed with butyl rubber stoppers. The media were prepared by flushing with nitrogen that had passed through a heated, reduced copper column to remove oxygen. Various fermentation media were used, depending on the purpose of the experiments, including YP medium containing glucose (20 g/liter) (YPD20), YP medium containing xylose (40 g/liter) (YPX40), and YP medium containing glucose (20 g/liter) and xylose (20 g/liter or 40 g/liter) (YPD20X20 or YPD20X40, respectively). For experiment setup, precultured cells in YPD medium were centrifuged and washed twice with sterilized water. The harvested cells were inoculated into fermentation medium, and initial cell densities were adjusted to around an optical density at 600 nm (OD600) of 1 normally or an OD600 of 10 for high-cell-density fermentations. All fermentations were performed in duplicate at 30°C and 100 rpm. Culture samples were taken from fermentation experiments to measure the OD600 and metabolite concentrations. For anaerobic-fermentation experiments, samples were taken by sterile syringe and with 26-gauge (26G) needles (BD, Franklin Lakes, NJ).
Intracellular xylitol and glycerol measurement.
Extracts of intracellular glycerol and xylitol were prepared in a way slightly modified from a previous method (52). The SR8-fps1Δ and SR8 strains were grown in YPX40 medium under anaerobic conditions, and the D10-fps1Δ and D10 strains were grown in YPD20X20 under oxygen-limited conditions, all in duplicate. For each culture, four 10-ml samples were withdrawn during the xylose consumption stage, as for the D10 and D10-fps1Δ strains (at 24 h), or at mid-exponential phase in culture incubated with xylose, as for the SR8 and SR8-fps1Δ strains. The cells were pelleted and quickly washed twice with 40 ml cold YP medium and centrifuged again at 4°C. The cells were resuspended in 0.5 ml 50 mM Tris-HCl, pH 7.5. The cell pellets were boiled for 15 min to extract intracellular glycerol and xylitol. As a control, samples without boiling were also analyzed to consider potential contamination of extracellular glycerol and xylitol. Samples were then centrifuged to remove cellular debris, and the supernatant was used for measuring glycerol and xylitol concentrations by high-performance liquid chromatography (HPLC). The total protein in each sample was measured with a bicinchoninic acid (BCA) protein assay kit (Pierce), and the value was used to normalize the measured metabolite concentrations.
Intracellular NAD(H) and NADP(H) analysis.
Intracellular cofactor analysis was performed to compare strain SR8-fps1Δ and strain SR8 in anaerobic fermentation in YPX40. Yeast culture samples were taken during the mid-exponential phase of cell growth. Intracellular NAD+, NADH, NADP+, or NADPH was extracted and measured using an Enzychrom NAD+/NADH assay or NADP+/NADPH assay kit (Bioassay Systems, Hayward, CA) according to the manufacturer's instructions.
Analytical methods.
Xylose, glucose, glycerol, xylitol, acetate, and ethanol concentrations were quantified by high-performance liquid chromatography (HPLC) (Agilent Technologies 1200 Series equipped with a refractive-index detector). The Rezex ROA-Organic Acid H+ (8%) column (Phenomenex Inc., Torrance, CA) was used. The mobile phase (0.005 N H2SO4) was eluted at a flow rate of 0.6 ml/min at 50°C. Cell growth was monitored by the optical density at 600 nm using a UV-visible spectrophotometer (Biomate 5; Thermo, NY).
RESULTS
Effect of FPS1 deletion on xylitol secretion.
An S. cerevisiae strain for producing xylitol was constructed by expressing only XR encoded by the XYL1 gene from S. stipitis and was given the strain name D10 (Table 1). Lacking the enzyme to further metabolize xylitol (i.e., XDH), the D10 strain accumulated xylitol as the end product from xylose metabolism, and a 1:1 molar ratio of production of xylitol over xylose consumption was observed (53). Thus, we deleted the FPS1 gene in the D10 strain to evaluate how xylitol excretion could be influenced. The D10-fps1Δ strain and the wild-type D10 strain were examined for differential xylitol excretion in YP medium with xylose and glucose, where glucose was necessary to serve as the carbon and energy source and to provide cofactors for the XR reaction, as xylose could not be metabolized beyond xylitol. Both the D10 and D10-fps1Δ strains fermented glucose to ethanol and accumulated biomass first, followed by the conversion of xylose to xylitol, where a considerable difference between the two strains was shown (Fig. 1A and B). The D10 strain converted all consumed xylose into xylitol within 80 h (Fig. 1A), but in sharp contrast, the D10-fps1Δ strain consumed xylose at an extremely low rate, and only 4 g/liter of extracellular xylitol was produced at the end of the experiment (Fig. 1B). Glycerol concentrations in both cultures were not significant.
Fig 1.
Fermentation profiles of the engineered S. cerevisiae D10 strain and D10-fps1Δ strain in YP medium containing xylose (20 g/liter) and glucose (20 g/liter) (A and B) or YP medium containing glucose (20 g/liter) (C and D) under oxygen-limited conditions. The results are the means of duplicate experiments; the error bars indicate standard deviations and are not visible when smaller than the symbol size. ■, glucose; ◆, xylose; ▲, xylitol; ▼, glycerol; ●, ethanol; ○, OD600.
Notably, there was a significant delay in ethanol consumption by the D10-fps1Δ strain, while the D10 strain metabolized ethanol immediately after glucose depletion and concurrently converted xylose to xylitol. This observation gave rise to the possibility that deficiency of xylitol production by the D10-fps1Δ strain might be due to FPS1 deletion having negatively affected ethanol uptake and thus resulted in a lack of carbon/energy sources and cofactors for cell metabolism rather than direct involvement of Fps1p in xylitol excretion. In order to exclude this possibility, we tested the ethanol reassimilation capability of the D10-fps1Δ and D10 strains in medium containing only glucose. When incubated with glucose under oxygen-limited conditions, S. cerevisiae first consumes glucose and then metabolizes ethanol produced from glucose fermentation. If FPS1 deletion hampered ethanol uptake, ethanol consumption would be slowed, as observed in Fig. 1A and B. However, the experimental results showed that the D10-fps1Δ strain had an ethanol concentration profile and cell growth similar to those of the D10 strain (Fig. 1C and D), suggesting that the impaired xylitol production by the D10-fps1Δ strain was not due to the limitation in ethanol consumption, i.e., availability of a carbon source and cofactors.
Intracellular xylitol concentrations were determined from culture samples taken at 24 h. Because the D10 strain and the D10-fps1Δ strain consumed different amounts of xylose, the intracellular xylitol concentration was normalized by xylose consumption to illustrate the relative xylitol accumulation inside the cell. The D10-fps1Δ strain accumulated twice as much xylitol inside the cell as the D10 strain (Fig. 2), indicating that FPS1 deletion could block xylitol export. Therefore, the most likely explanation for the deficiency of xylitol excretion by the D10-fps1Δ strain is that lack of Fps1p hindered xylitol export and accumulation of intracellular xylitol impeded the XR reaction.
Fig 2.
Intracellular xylitol concentration (g/g dry cell mass) normalized by xylose consumption (g/g dry cell mass) in samples of strains D10 and D10-fps1Δ growing in YP medium containing xylose (20 g/liter) and glucose (20 g/liter) under oxygen-limited conditions (final unit after normalization, g/g xylose). The results are the means of duplicate experiments; the error bars indicate standard deviations.
Effect of FPS1 overexpression on xylitol secretion.
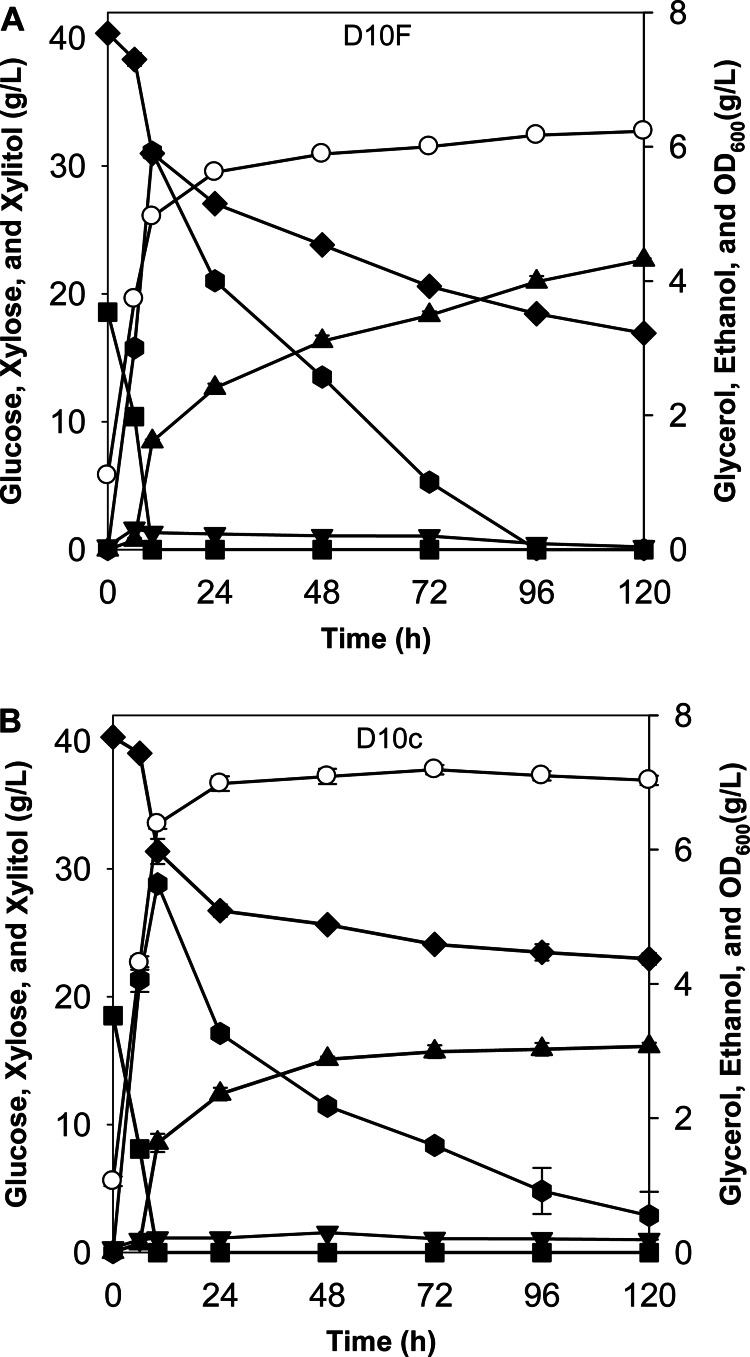
In order to cross-validate the role of Fps1p in xylitol excretion in S. cerevisiae, we also investigated the effects of FPS1 overexpression in the D10 strain. An FPS1 overexpression cassette was integrated into the D10 strain, resulting in the D10F strain, and the control strain D10c was also constructed by transformation with an empty integrative plasmid (Table 1). As expected, the FPS1-overexpressing strain (D10F) converted xylose into xylitol faster than the control strain (D10c), as shown in Fig. 3. The specific xylitol productivity by the D10F strain (0.152 ± 0.002 h−1) was significantly higher than that by D10c (0.091 ± 0.001 h−1), suggesting that overexpression of FPS1 enhanced the flux of xylitol secretion.
Fig 3.
Fermentation profiles for the engineered S. cerevisiae strain D10F (A) and the control strain D10c (B) in YP medium containing xylose (40 g/liter) and glucose (20 g/liter) under oxygen-limited conditions. The results are the means of duplicate experiments; the error bars indicate standard deviations and are not visible when smaller than the symbol size. ■, glucose; ◆, xylose; ▲, xylitol; ▼, glycerol; ●, ethanol; ○, OD600.
Effects of FPS1 deletion on xylose fermentation under oxygen-limited conditions.
Based on the above observations, we hypothesized that FPS1 deletion might increase the yield of target products (e.g., ethanol) from xylose fermentation by confining xylitol inside the cell and might push the carbon flux in the desired direction. To test this hypothesis, we used an engineered xylose-fermenting S. cerevisiae strain, SR6, that accumulated substantial amounts of xylitol during xylose fermentation because of unbalanced expression of S. stipitis XR and XDH. As shown in Fig. 4A, the SR6 strain accumulated nearly 16 g/liter xylitol during fermentation of 40 g/liter xylose, while ethanol production was extremely low (ethanol/xylose yield [Yethanol/xylose] = 0.555 ± 0.001 g ethanol/g xylose). When FPS1 was deleted in the SR6 strain, xylitol production by the SR6-fps1Δ strain was drastically reduced to less than 5 g/liter when 40 g/liter xylose was depleted, and the ethanol yield (Yethanol/xylose = 0.206 ± 0.002) was 4-fold higher than that of the SR6 strain (Table 3). It is evident that elimination of Fps1p can lead to more efficient operation of xylose assimilation through XR and XDH reactions, despite the redox cofactor imbalance. In addition, the glycerol yield decreased from 0.079 ± 0.001 g glycerol/g xylose in fermentation by the SR6 strain to 0.014 ± 0.001 g glycerol/g xylose for the SR6-fps1Δ strain. This result was consistent with the known function of Fps1p in facilitating glycerol transport. Interestingly, the SR6-fps1Δ strain grew faster (Table 3) and reached a higher OD600 at the end of fermentation (Fig. 4) than the SR6 strain, most likely because the consumed xylose was used more efficiently with less xylitol secretion.
Fig 4.
Fermentation profiles of engineered S. cerevisiae strain SR6 (A) and strain SR6-fps1Δ (B) in YP medium containing xylose (40 g/liter) under oxygen-limited conditions. The results are the means of duplicate experiments; the error bars indicate standard deviations and are not visible when smaller than the symbol size. ■, xylose; ▲, xylitol; ▼, glycerol; ●, ethanol; ×, acetate; ○, OD600.
Table 3.
Fermentation performances of the fps1Δ strains and the reference strains under different conditions
| Parametera | Value (mean ± SD) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Oxygen-limited conditions (initial OD = 1) | Anaerobic conditions |
|||||||
| Initial OD = 1 |
Initial OD = 10 |
|||||||
| SR6 | SR6-fps1Δ | SR8 | SR8-fps1Δ | SR8 | SR8-fps1Δ | SR8 | SR8-fps1Δ | |
| Yxylitol(g/g) | 0.378 ± 0.003 | 0.108 ± 0.001 | 0.070 ± 0.001 | 0.055 ± 0.0008 | 0.12 ± 0.002 | 0.084 ± 0.002 | 0.111 ± 0.001 | 0.089 ± 0.0006 |
| Yglycerol(g/g) | 0.079 ± 0.001 | 0.014 ± 0.001 | 0.056 ± 0.0005 | 0.047 ± 0.001 | 0.131 ± 0.002 | 0.112 ± 0.002 | 0.647 ± 0.001 | 0.611 ± 0.001 |
| Yethanol(g/g) | 0.055 ± 0.001 | 0.206 ± 0.002 | 0.332 ± 0.002 | 0.341 ± 0.002 | 0.320 ± 0.001 | 0.351 ± 0.001 | 0.348 ± 0.002 | 0.365 ± 0.002 |
| Vethanol (g/liter · h−1) | 0.054 ± 0.001 | 0.182 ± 0.002 | 0.480 ± 0.005 | 0.474 ± 0.003 | 0.172 ± 0.001 | 0.064 ± 0.001 | 1.10 ± 0.005 | 1.13 ± 0.02 |
| Pethanol* (h−1) | 0.012 ± 0.0006 | 0.027 ± 0.001 | 0.216 ± 0.002 | 0.215 ± 0.004 | 0.130 ± 0.002 | 0.073 ± 0.001 | 0.269 ± 0.002 | 0.279 ± 0.009 |
| μmax (h−1) | 0.022 ± 0.0005 | 0.025 ± 0.001 | 0.125 ± 0.001 | 0.118 ± 0.001 | 0.040 ± 0.0006 | 0.023 ± 0.0005 | 0.042 ± 0.0005 | 0.034 ± 0.001 |
| rxylose* (h−1) | 0.217 ± 0.003 | 0.204 ± 0.001 | 0.647 ± 0.008 | 0.633 ± 0.009 | 0.50 ± 0.005 | 0.31 ± 0.02 | 0.781 ± 0.001 | 0.769 ± 0.001 |
Yxylitol, xylitol yield (g xylitol/g xylose); Yglycrol, glycerol yield (g glycerol/g xylose); Yethanol, ethanol yield (g ethanol/g xylose); Vethanol, volumetric ethanol productivity (g · liter−1 · h−1); Pethanol*, specific ethanol productivity (g ethanol g cell−1 · h−1); μmax, maximum specific growth rate (h−1); rXylose*, specific xylose consumption rate (g xylose g cell−1 · h−1).
The effect of FPS1 deletion for decreasing xylitol production and increasing the ethanol yield was also demonstrated using a different engineered S. cerevisiae strain (SR8). The SR8 strain was rationally engineered and evolved to ferment xylose efficiently and rapidly (51). The SR8 strain can finish complete fermentation of 40 g/liter xylose within 30 h with an initial inoculation at an OD of 1 under oxygen-limited conditions. An FPS1 deletion mutant of the SR8 strain consumed xylose at a rate similar to that of the parental SR8 strain under oxygen-limited conditions but exhibited a significantly higher ethanol yield and lower yields of xylitol and glycerol (Table 3) (P < 0.005), which was consistent with the results from the fermentation experiments with the SR6 strain described in Fig. 4.
Effects of FPS1 deletion on xylose fermentation under anaerobic conditions.
Xylitol accumulation during xylose fermentation through the XR/XDH pathway is more problematic under anaerobic conditions than under oxygen-limited conditions due to the lack of oxygen to regenerate NAD+. Despite the challenge, anaerobic fermentation is preferred in industrial applications because of the prohibitively high cost of aeration. The SR8 strain is capable of fermentation and growth on xylose alone under strict anaerobic conditions, providing a platform to test the potential effects of FPS1 deletion on improving anaerobic xylose fermentation. The SR8 strain was able to ferment xylose as the sole substrate to ethanol under anaerobic conditions at a respectable rate and with a yield of 0.320 ± 0.001 g ethanol/g xylose (Table 3). When FPS1 was deleted in the SR8 strain, the SR8-fps1Δ strain showed both a significant increase of the ethanol yield to 0.351 ± 0.001 g ethanol/g xylose and lower by-product accumulation (Table 3). In particular, the xylitol yield in fermentation by the SR8 strain was 23% higher than that by the SR8-fps1Δ strain. While anaerobic xylose fermentation by SR8-fps1Δ took longer than that by the SR8 strain with a starting cell mass at an OD600 of 1; both the SR8 and SR8-fps1Δ strains were able to consume 40 g/liter of xylose within 20 h and had similar specific xylose consumption rates (Table 3) when a higher starting cell mass (OD600 = 10) was used.
The intracellular concentrations of both xylitol and glycerol in the SR8-fps1Δ strain were considerably higher than those in the SR8 strain (Fig. 5A), demonstrating again that FPS1 deletion could block xylitol export, as well as glycerol transport. NAD+, NADH, NADP+, and NADPH from the culture samples of the SR8 and SR8-fps1Δ strains were also analyzed. While the NADP+/NADPH ratio was slightly lower in the SR8-fps1Δ strain than in the SR8 strain, there was no significant difference in the NAD+/NADH ratio (Fig. 5B).
Fig 5.
Intracellular xylitol and glycerol concentration (g/g dry cell mass) (A) and redox cofactor NAD(P)+/NAD(P)H ratio (B) in samples of strain SR8 and SR8-fps1Δ growing in YP medium containing xylose (40 g/liter) under anaerobic conditions. The results are the means of duplicate experiments; the error bars indicate standard deviations.
DISCUSSION
Accumulation of xylitol during xylose fermentation by engineered S. cerevisiae with the XR/XDH pathway has been a problem for a long time (6), especially when oxygen availability is limited for NADH reoxidation. Thus, many studies attempted to optimize the expression levels of the genes coding for the XR/XDH pathway or to engineer XR and XDH proteins with balanced cofactor preferences to reduce xylitol accumulation and increase the ethanol yield (19, 28, 54–60). While previous efforts to improve xylose fermentation mostly focused on manipulation of enzymatic reactions related to xylose metabolism, this study demonstrated an innovative approach that controls the carbon flux by accumulating the intermediate product (i.e., xylitol) inside the cell to facilitate the reaction toward the target direction. It was suggested previously that increasing the intracellular xylitol concentration could help conversion of xylitol to xylulose, and an unpublished investigation regarding the possible role of Fps1p was mentioned (61). The comprehensive experimental study reported here shows the involvement of Fps1p in xylitol transport in S. cerevisiae, demonstrating a new function of this MIP family protein. The results revealed that deletion of FPS1 can substantially reduce xylitol formation and increase ethanol production from xylose fermentation by engineered S. cerevisiae. The strategy demonstrated here will contribute to current efforts to develop efficient xylose-utilizing yeast strains for economically feasible production of cellulosic biofuels.
The fps1Δ strains in this study consistently exhibited lower xylitol excretion than parental strains harboring the wild-type FPS1, suggesting that Fps1p is involved in xylitol accumulation in the medium regardless of strain background. Similar results were also observed for glycerol production. Still, we found that both xylitol and glycerol production by fps1Δ strains was not eliminated. A prior study showed that glycerol uptake by S. cerevisiae could be attributed to two components: one is facilitated transport mediated by Fps1p, and the other is free diffusion through the phospholipid bilayer of the cell membrane (43). Similarly, free diffusion may be a reason for xylitol excretion, as well as for glycerol excretion, in the fps1Δ strains here. Notably, the deletion of FPS1 showed a more drastic effect in reducing xylitol production when the redox imbalance was more severe. For example, the SR6 strain with unbalanced XR and XDH enzyme activities generated xylitol as the major product and produced very little ethanol, but the corresponding fps1Δ mutant produced nearly 70% less xylitol than a parental strain (SR6). In comparison, xylitol reduction by FPS1 deletion in the SR8 strain, which has optimized XR/XDH expression levels and evolved to ferment xylose with a low xylitol yield, was not as significant as that in the SR6 strain under the same fermentation conditions. These results indicate that FPS1p-dependent facilitated diffusion may play a major role in xylitol excretion when the intracellular xylitol concentration is relatively high.
The effect of FPS1 deletion on xylose fermentation in recombinant S. cerevisiae with the XR/XDH pathway was evaluated under both oxygen-limited conditions and strictly anaerobic conditions. The fps1Δ strains (e.g., SR6-fps1Δ and SR8-fps1Δ) did not show a growth defect under oxygen-limited conditions compared to the parental strains. In contrast, anaerobic experiments with the SR8-fps1Δ strain showed significantly reduced cell growth and xylose consumption rates, even though increasing biomass inoculation could overcome this problem (Table 3). It is likely that deletion of FPS1 in engineered yeast strains here hampered the glycerol production pathway, and the capability to deal with the redox imbalance problem in xylose fermentation under anaerobic conditions became less efficient. However, there was no significant difference in the NAD+/NADH ratio between the two strains SR8-fps1Δ and SR8, indicating that FPS1 deletion might not drastically influence the cofactor balance or that some alternative pathway other than glycerol formation could contribute to redox balancing (62). The reason for slightly higher NADP+/NADPH in the SR8-fps1Δ strain is unclear. One possible explanation might be that increased carbon flux to the pentose phosphate pathway due to less xylitol excretion produced more NADPH.
It is worth noting that the natural xylose-utilizing yeast S. stipitis can ferment xylose to ethanol at near the theoretical yields and produces negligible amounts of xylitol during xylose fermentation (63–65). Unlike S. stipitis, most natural xylose-fermenting yeasts produce considerable amounts of extracellular xylitol (64), and two main reasons were reported in previous studies. For one thing, XR in S. stipitis, as well as another xylose-fermenting yeast, Pachysolen tannophilus, can use both NADH and NADPH as cofactors, which alleviates the redox imbalance compared to yeast strains whose XR is strictly NADPH specific (66). For another, S. stipitis still produces less xylitol than P. tannophilus, although they both have dual-cofactor-specific XR, which has been explained by the existence of a complex oxidative respiratory system unique to S. stipitis. There is a noncytochrome electron transport chain in S. stipitis serving as a redox sink for reoxidizing surplus NADH and reducing xylitol production (67). This may also be a reason for the observation that recombinant S. cerevisiae expressing the same level of XR and XDH enzyme activities as in S. stipitis produces xylitol at a much higher level than S. stipitis (68). However, besides these two well-known mechanisms, another possibility to account for less xylitol production in S. stipitis is that the cell membrane of the microorganism may not normally, or only very inefficiently, export xylitol. A previous study investigated the distribution of intermediates and products during d-{1-13C} xylose metabolism in S. stipitis and revealed that xylitol was the major intermediate accumulating inside the cell but that no xylitol was produced extracellularly (69). In addition, a BLAST search of FPS1 against the genome sequence of S. stipitis did not a show significant match. Taken together, these findings suggest the need for future research to investigate the xylitol excretion capability of S. stipitis compared with other xylitol-excreting strains and to gain further insight into how the natural xylose-fermenting yeast may possibly use the strategy of limiting xylitol export to efficiently metabolize xylose.
ACKNOWLEDGMENT
This work was supported by funding from the Energy Biosciences Institute to Y.-S.J.
Footnotes
Published ahead of print 8 March 2013
REFERENCES
- 1.U. S. Department of Energy 2011. Report on the first quadrennial technology review. U.S. Department of Energy, Washington, DC [Google Scholar]
- 2.Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM. 2006. Ethanol can contribute to energy and environmental goals. Science 311:506–508 [DOI] [PubMed] [Google Scholar]
- 3.Gray KA, Zhao L, Emptage M. 2006. Bioethanol. Curr. Opin. Chem. Biol. 10:141–146 [DOI] [PubMed] [Google Scholar]
- 4.Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund MF, Liden G, Zacchi G. 2006. Bio-ethanol: the fuel of tomorrow from the residues of today. Trends Biotechnol. 24:549–556 [DOI] [PubMed] [Google Scholar]
- 5.Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T. 2006. The path forward for biofuels and biomaterials. Science 311:484–489 [DOI] [PubMed] [Google Scholar]
- 6.Jeffries TW, Jin YS. 2004. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 63:495–509 [DOI] [PubMed] [Google Scholar]
- 7.Olsson L, Nielsen J. 2000. The role of metabolic engineering in the improvement of Saccharomyces cerevisiae: utilization of industrial media. Enzyme Microb. Technol. 26:785–792 [DOI] [PubMed] [Google Scholar]
- 8.Kotter P, Ciriacy M. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 38:776–783 [Google Scholar]
- 9.Tantirungkij M, Nakashima N, Seki T, Yoshida T. 1993. Construction of xylose-assimilating Saccharomyces cerevisiae. J. Ferment. Bioeng. 75:83–88 [Google Scholar]
- 10.Eliasson A, Hofmeyr JHS, Pedler S, Hahn-Hagerdal B. 2001. The xylose reductase/xylitol dehydrogenase/xylulokinase ratio affects product formation in recombinant xylose-utilising Saccharomyces cerevisiae. Enzyme Microb. Technol. 29:288–297 [Google Scholar]
- 11.Jin YS, Jeffries TW. 2003. Changing flux of xylose metabolites by altering expression of xylose reductase and xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 106:277–286 [DOI] [PubMed] [Google Scholar]
- 12.Karhumaa K, Fromanger R, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2007. High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 73:1039–1046 [DOI] [PubMed] [Google Scholar]
- 13.Jeppsson M, Träff-Bjerre KL, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund MF. 2003. Effect of enhanced xylose reductase activity on xylose consumption and product distribution in xylose-fermenting recombinant Saccharomyces cerevisiae. FEMS Yeast Res. 3:167–175 [DOI] [PubMed] [Google Scholar]
- 14.Matsushika A, Sawayama S. 2008. Efficient bioethanol production from xylose by recombinant Saccharomyces cerevisiae requires high activity of xylose reductase and moderate xylulokinase activity. J. Biosci. Bioeng. 106:306–309 [DOI] [PubMed] [Google Scholar]
- 15.Toivari MH, Salusjarvi L, Ruohonen L, Penttila M. 2004. Endogenous xylose pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 70:3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H, Cheng J-S, Wang BL, Fink GR, Stephanopoulos G. 2012. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng. 14:611–622 [DOI] [PubMed] [Google Scholar]
- 17.Ha SJ, Kim SR, Choi JH, Park MS, Jin YS. 2011. Xylitol does not inhibit xylose fermentation by engineered Saccharomyces cerevisiae expressing xylA as severely as it inhibits xylose isomerase reaction in vitro. Appl. Microbiol. Biotechnol. 92:77–84 [DOI] [PubMed] [Google Scholar]
- 18.Brat D, Boles E, Wiedemann B. 2009. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75:2304–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karhumaa K, Sanchez RG, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2007. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell. Fact. 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MSM, De Laat WTAM, Den Ridder JJJ, Op Den Camp HJM, Van Dijken JP, Pronk JT. 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae. FEMS Yeast Res. 4:69–78 [DOI] [PubMed] [Google Scholar]
- 21.Kuyper M, Hartog MMP, Toirkens MJ, Almering MJH, Winkler AA, Van Dijken JP, Pronk JT. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5:399–409 [DOI] [PubMed] [Google Scholar]
- 22.Lönn A, Träff-Bjerre KL, Cordero Otero RR, Van Zyl WH, Hahn-Hägerdal B. 2003. Xylose isomerase activity influences xylose fermentation with recombinant Saccharomyces cerevisiae strains expressing mutated xylA from Thermus thermophilus. Enzyme Microb. Technol. 32:567–573 [Google Scholar]
- 23.Walfridsson M, Bao X, Anderlund M, Lilius G, Bülow L, Hahn-Hagerdal B. 1996. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl. Environ. Microbiol. 62:4648–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:147–177 [DOI] [PubMed] [Google Scholar]
- 25.Ho NWY, Chen Z, Brainard AP. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walfridsson M, Anderlund M, Bao X, Hahn-Hagerdal B. 1997. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl. Microbiol. Biotechnol. 48:218–224 [DOI] [PubMed] [Google Scholar]
- 27.Kuhn A, Vanzyl C, Vantonder A, Prior BA. 1995. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl. Environ. Microbiol. 61:1580–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengtsson O, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2009. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krahulec S, Petschacher B, Wallner M, Longus K, Klimacek M, Nidetzky B. 2010. Fermentation of mixed glucose-xylose substrates by engineered strains of Saccharomyces cerevisiae: role of the coenzyme specificity of xylose reductase, and effect of glucose on xylose utilization. Microb. Cell. Fact. 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsushika A, Watanabe S, Kodaki T, Makino K, Sawayama S. 2008. Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP(+)-dependent xylitol dehydrogenase, and xylulokinase. J. Biosci. Bioeng. 105:296–299 [DOI] [PubMed] [Google Scholar]
- 31.Petschacher B, Nidetzky B. 2008. Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb. Cell. Fact. 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe S, Abu Saleh A, Pack SP, Annaluru N, Kodaki T, Makino K. 2007. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 153:3044–3054 [DOI] [PubMed] [Google Scholar]
- 33.Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hagerdal R, Gorwa-Grauslund MF. 2006. The expression of a Pichia stipitis xylose reductase mutant with higher Km for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 93:665–673 [DOI] [PubMed] [Google Scholar]
- 34.Jin YS, Alper H, Yang YT, Stephanopoulos G. 2005. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl. Environ. Microbiol. 71:8249–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2005. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22:359–368 [DOI] [PubMed] [Google Scholar]
- 36.Lu C, Jeffries T. 2007. Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 73:6072–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonderegger M, Schümperli M, Sauer U. 2004. Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 70:2892–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson B, Hahn-Hagerdal B. 2002. Overproduction of pentose phosphate pathway enzymes using a new CRE-loxP expression vector for repeated genomic integration in Saccharomyces cerevisiae. Yeast 19:225–231 [DOI] [PubMed] [Google Scholar]
- 39.Walfridsson M, Hallborn J, Penttila M, Keranen S, Hahn-Hagerdal B. 1995. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl. Environ. Microbiol. 61:4184–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traff KL, Cordero RRO, van Zyl WH, Hahn-Hagerdal B. 2001. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl. Environ. Microbiol. 67:5668–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanino T, Hotta A, Ito T, Ishii J, Yamada R, Hasunuma T, Ogino C, Ohmura N, Ohshima T, Kondo A. 2010. Construction of a xylose-metabolizing yeast by genome integration of xylose isomerase gene and investigation of the effect of xylitol on fermentation. Appl. Microbiol. Biotechnol. 88:1215–1221 [DOI] [PubMed] [Google Scholar]
- 42.Reizer J, Reizer A, Saier MH. 1993. The MIP family of integral membrane channel proteins: sequence comparisons, evolutionary relationships, reconstructed pathway of evolution, and proposed functional differentiation of the two repeated halves of the proteins. Crit. Rev. Biochem. Mol. Biol. 28:235–257 [DOI] [PubMed] [Google Scholar]
- 43.Luyten K, Albertyn J, Skibbe WF, Prior BA, Ramos J, Thevelein JM, Hohmann S. 1995. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 14:1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mollapour M, Piper PW. 2007. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol. Cell. Biol. 27:6446–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wysocki R, Chery CC, Wawrzycka D, Van Hulle M, Cornelis R, Thevelein JM, Tamas MJ. 2001. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol. Microbiol. 40:1391–1401 [DOI] [PubMed] [Google Scholar]
- 46.Tamas MJ, Luyten K, Sutherland FCW, Hernandez A, Albertyn J, Valadi H, Li H, Prior BA, Killan SG, Ramos J, Gustafsson L, Thevelein JM, Hohmann S. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31:1087–1104 [DOI] [PubMed] [Google Scholar]
- 47.Fu DX, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM. 2000. Structure of a glycerol-conducting channel and the basis for its selectivity. Science 290:481–486 [DOI] [PubMed] [Google Scholar]
- 48.Karlgren S, Pettersson N, Nordlander B, Mathai JC, Brodsky JL, Zeidel ML, Bill RM, Hohmann S. 2005. Conditional osmotic stress in yeast: a system to study transport through aquaglyceroporins and osmostress signaling. J. Biol. Chem. 280:7186–7193 [DOI] [PubMed] [Google Scholar]
- 49.Guldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gietz RD, Schiestl RH. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2:31–34 [DOI] [PubMed] [Google Scholar]
- 51.Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin YS. 2013. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8:e57048 doi:10.1371/journal.pone.0057048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao W, Deschenes RJ, Fassler JS. 1999. Intracellular glycerol levels modulate the activity of Sln1p, a Saccharomyces cerevisiae two-component regulator. J. Biol. Chem. 274:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh EJ, Ha S-J, Rin Kim S, Lee W-H, Galazka JM, Cate JHD, Jin Y- S. 2013. Enhanced xylitol production through simultaneous co-utilization of cellobiose and xylose by engineered Saccharomyces cerevisiae. Metab. Eng. 15:226–234 [DOI] [PubMed] [Google Scholar]
- 54.Lee SH, Kodaki T, Park YC, Seo JH. 2012. Effects of NADH-preferring xylose reductase expression on ethanol production from xylose in xylose-metabolizing recombinant Saccharomyces cerevisiae. J. Biotechnol. 158:184–191 [DOI] [PubMed] [Google Scholar]
- 55.Matsushika A, Watanabe S, Kodaki T, Makino K, Inoue H, Murakami K, Takimura O, Sawayama S. 2008. Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81:243–255 [DOI] [PubMed] [Google Scholar]
- 56.Krahulec S, Klimacek M, Nidetzky B. 2012. Analysis and prediction of the physiological effects of altered coenzyme specificity in xylose reductase and xylitol dehydrogenase during xylose fermentation by Saccharomyces cerevisiae. J. Biotechnol. 158:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khoury GA, Fazelinia H, Chin JW, Pantazes RJ, Cirino PC, Maranas CD. 2009. Computational design of Candida boidinii xylose reductase for altered cofactor specificity. Protein Sci. 18:2125–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim SR, Ha S-J, Kong II, Jin Y-S. 2012. High expression of XYL2 coding for xylitol dehydrogenase is necessary for efficient xylose fermentation by engineered Saccharomyces cerevisiae. Metab. Eng. 14:336–343 [DOI] [PubMed] [Google Scholar]
- 59.Matsushika A, Sawayama S. 2011. Comparative study on a series of recombinant flocculent Saccharomyces cerevisiae strains with different expression levels of xylose reductase and xylulokinase. Enzyme Microb. Technol. 48:466–471 [DOI] [PubMed] [Google Scholar]
- 60.Parachin NS, Bergdahl B, van Niel EWJ, Gorwa-Grauslund MF. 2011. Kinetic modelling reveals current limitations in the production of ethanol from xylose by recombinant Saccharomyces cerevisiae. Metab. Eng. 13:508–517 [DOI] [PubMed] [Google Scholar]
- 61.Hahn-Hagerdal B, Wahlbom CF, Gárdonyi M, van Zyl WH, Cordero Otero RR, Jönsson LJ. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73:53–84 [DOI] [PubMed] [Google Scholar]
- 62.Nissen TL, Kielland-Brandt MC, Nielsen J, Villadsen J. 2000. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab. Eng. 2:69–77 [DOI] [PubMed] [Google Scholar]
- 63.Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz J, Lindquist E, Dehal P, Shapiro H, Jin YS, Passoth V, Richardson PM. 2007. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 25:319–326 [DOI] [PubMed] [Google Scholar]
- 64.Agbogbo FK, Coward-Kelly G. 2008. Cellulosic ethanol production using the naturally occurring xylose-fermenting yeast, Pichia stipitis. Biotechnol. Lett. 30:1515–1524 [DOI] [PubMed] [Google Scholar]
- 65.Jin YS, Cruz J, Jeffries TW. 2005. Xylitol production by a Pichia stipitis D-xylulokinase mutant. Appl. Microbiol. Biotechnol. 68:42–45 [DOI] [PubMed] [Google Scholar]
- 66.Verduyn C, Vankleef R, Frank J, Schreuder H, Vandijken JP, Scheffers WA. 1985. Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem. J. 226:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeppsson H, Alexander NJ, Hahnhagerdal B. 1995. Existence of cyanide-insensitive respiration in the yeast Pichia stipitis and its possible influence on product formation during xylose utilization. Appl. Environ. Microbiol. 61:2596–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hahn-Hagerdal B, Jeppsson H, Olsson L, Mohagheghi A. 1994. An interlaboratory comparison of the performance of ethanol-producing micro-organisms in a xylose-rich acid hydrolysate. Appl. Microbiol. Biotechnol. 41:62–72 [Google Scholar]
- 69.Ligthelm ME, Prior BA, Dupreez JC, Brandt V. 1988. An investigation of D-(1-C-13) xylose metabolism in Pichia stipitis under aerobic and anaerobic conditions. Appl. Microbiol. Biotechnol. 28:293–296 [Google Scholar]
- 70.Mumberg D, Muller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122 [DOI] [PubMed] [Google Scholar]