Abstract
The genetic identity and cofactor composition of the bacterial tellurate reductase are currently unknown. In this study, we examined the requirement of molybdopterin biosynthesis and molybdate transporter genes for tellurate reduction in Escherichia coli K-12. The results show that mutants deleted of the moaA, moaB, moaE, or mog gene in the molybdopterin biosynthesis pathway lost the ability to reduce tellurate. Deletion of the modB or modC gene in the molybdate transport pathway also resulted in complete loss of tellurate reduction activity. Genetic complementation by the wild-type sequences restored tellurate reduction activity in the mutant strains. These findings provide genetic evidence that tellurate reduction in E. coli involves a molybdoenzyme.
INTRODUCTION
Tellurium (Te) is a metalloid element used for a variety of industrial applications, including metallurgy, chemical manufacturing, electronics, and nanotechnology (1–4). The disposal of mine tailings and tellurium-containing waste has led to an increase in environmental contamination (5). When released into the environment, tellurium undergoes redox transformations and can be mobilized as the dissolved Te oxyanions tellurate [Te(VI); TeO42−] and tellurite [Te(IV); TeO32−]. These oxyanions are highly toxic to microbiota (6) and cause inhibitory effects to most microorganisms at concentrations as low as 1 μg/ml (1, 4). While naturally occurring Te-resistant bacteria have been isolated that are able to grow in the presence of elevated Te concentrations (7, 8) and several genetic elements have been linked to Te resistance (9), the molecular mechanisms of bacterium-tellurium interactions remain poorly understood.
Microorganisms are known to catalyze the reduction of toxic tellurite into sparingly soluble and less toxic elemental tellurium [Te(0)] (10–15). In comparison, little is known about the microbial reduction of tellurate, even though Te(VI) is the dominant form of Te in the hydrosphere (16, 17). Recently, Shewanella species isolated from deep-ocean hydrothermal vent worms were discovered to respire Te(VI) as a terminal electron acceptor (18), and the anaerobic bacteria Sulfurospirillum barnesii and Bacillus selenitireducens were found to generate energy for growth on lactate by the reduction of Te(VI) to Te(0) (7). A Gram-positive bacterium, Bacillus beveridgei, was recently isolated that can also grow by reducing Te(VI) to Te(0) (8). The genetic identity and cofactor composition of enzymes that catalyze tellurate reduction in these bacteria, as well as other Te(VI) reducers, are currently unknown.
The molybdenum cofactor forms the active site of several important bacterial redox proteins, including the enzymes that catalyze the reduction of selenium and arsenic (15, 19–23), two metalloid elements that share similar chemical characteristics with tellurium. In selenate [Se(VI)]- and arsenate [As(V)]-reducing bacteria, the assembly of the molybdoenzymes is dependent on molybdenum import into the cell and biosynthesis of the molybdopterin cofactor. Previous studies have demonstrated that molybdate uptake in Escherichia coli is facilitated by an ABC-type transporter, and the molybdopterin cofactor is constructed via a biosynthetic pathway encoded by the moa-mog gene system (24, 25). To date, the roles of molybdate transporter and molybdopterin biosynthesis genes in tellurate reduction have not been studied.
In this study, experiments were carried out to investigate the reduction of Te(VI) to Te(0) by E. coli K-12. E. coli mutants carrying single mutations in the molybdopterin biosynthesis (moa-mog) and molybdate transporter (modABC) gene systems were tested for tellurate and tellurite reduction activity. Genetic complementation by the wild-type sequences was also performed to restore activity in the mutant strains that lost the ability to reduce Te(VI). The results provide genetic evidence that the tellurate reductase in E. coli is a molybdopterin-containing enzyme.
MATERIALS AND METHODS
Tellurate reduction experiments.
All strains were grown aerobically and maintained on LB agar (Difco) (see Table S1 in the supplemental material). Tellurate reductase activity for wild-type E. coli strain K-12 was determined by first growing cells in LB broth overnight at 37°C, reaching an optical density at 600 nm (OD600) of 1.6 ± 0.2. Cultures were centrifuged, and a condensed culture was dispensed into flasks of LB containing 50 μM Na2TeO4. Samples were taken at periodic intervals, and the black Te(0) precipitate and cells were removed by filtration (0.45-μm pore size). The total dissolved tellurium remaining in the media was analyzed using inductively coupled plasma optical emission spectrometry (Varian Inc., Palo Alto, CA) at a wavelength of 214.282 nm. Te standards for the calibration curve were prepared with the same LB broth as that used in the experiments. Control experiments were conducted with cells that were heat killed in a water bath at 80°C for 30 min. All experiments were carried out with triplicate cultures.
Tellurate reduction activity was determined for the following E. coli mutant strains: ΔmoaA (EcoGene accession number EG11595), ΔmoaB (EG11596), ΔmoaC (EG11666), ΔmoaD (EG11597), ΔmoaE (EG11598), Δmog (EG11511), ΔmodA (EG12427), ΔmodB (EG10002), ΔmodC (EG10152), Δfnr (EG10325), ΔmenA (EG11880), ΔmenC (EG11532), ΔmenD (EG10579), ΔmenE (EG12437), ΔtatB (EG14322), and ΔtatC (EG11479) (see Table S1 in the supplemental material) (26). Tellurate reduction activity was also determined for moaA, modB, and mog mutants carrying plasmids described in Table S1. Tellurate reductase activity in the mutant strains was determined by growing strains in LB broth overnight at 37°C, reaching an OD600 of 0.85 ± 0.05 before spiking with 50 μM tellurate, and taking a sample after 24 h. Te(0) precipitates were removed by filtration (0.45 μm), and the loss of dissolved tellurium from the media was quantified using the inductively coupled plasma optical emission spectrometry method described above. The mutant strains were also screened for their ability to reduce tellurite and selenate. To test for tellurite reduction activity, overnight LB cultures were spiked with 50 μM Na2TeO3 and visually examined for the formation of black Te(0) after 1 day. To test for selenate reduction activity, the mutant strains were grown in LB broth amended with 1 mM sodium selenate and visually examined for the formation of red elemental selenium [Se(0)].
Tellurate reduction activity was also examined in E. coli mutant strains carrying single mutations of the following genes encoding molybdenum-containing oxidoreductases: dmsA, ynfE, ynfF, bisC, torA, torZ, fdhF, fdoG, fdnG, napA, narG, narZ, ydeP, and yfgD (26). Tellurate reduction in these mutants was determined in overnight LB cultures spiked with 50 μM tellurate and visually examined for the formation of black Te(0) after 1 day.
Genetic complementation.
In order to establish the role of the modA, moaA, and mog genes in tellurate reduction activity, genetic complementations were performed with the E. coli mutants. To clone the moaABCDE operon for complementation of the moaA mutant, a primer set was constructed that consisted of forward primer moa-F1 (5′-GCGAAATAGCACGATCATGACGC-3′), positioned 218 bp upstream of the start codon for the moaA gene, and reverse primer moa-R1 (5′-GCGTAAACGTATGTACTGAGCGG-3′), positioned 44 bp downstream of the stop codon for the moaE gene. The Phusion High-Fidelity PCR kit was used by following the protocol described by the manufacturer (New England BioLabs Inc., Ipswich, MA). The 2,991-bp amplified product was purified using an UltraClean 15 DNA purification kit (Mo Bio, Carlsbad, CA), and then A tails were added to the blunt-ended proofreading enzyme DNA fragments through a second PCR cycle of 72°C for 20 min. The refined product was cloned into the pCR2.1-TOPO vector using the One Shot TOP10 protocol as described by the manufacturer (Life Technologies, Grand Island, NY). The clone was designated pECA27. To clone the modABC operon for complementation of the modB mutant, a primer set was constructed that consisted of forward primer mod-F1 (5′-CAACTTCCTGCTTTTCCTGCCG-3′), positioned 127 bp upstream of the start codon for the moaA gene, and reverse primer mod-R1 (5′-GCCCAGTTCATTTATAGCCACC-3′), positioned 8 bp downstream of the stop codon for the modC gene. The 2,682-bp amplified product was cloned into the pCR2.1-TOPO vector, resulting in pECD25. Finally, to clone the mog mutant, a primer set was constructed that consisted of forward primer mog-F1 (5′-GGTCACGCTACCTCTTCTGAAGC-3′), positioned 89 bp upstream of the start codon for the mog gene, and reverse primer mog-R1 (5′-TTATTCGCTAACGTCGCGTCTTGC-3′), consisting of the last 24 bp of the mog sequence. The 677-bp amplified product was cloned into pCR2.1-TOPO vector, resulting in pECG07. The purified plasmids pECA27, pECD25, and pECG07 were subsequently transformed into JW0764-2, JW0747-1, and JW0008-5, forming ECMOAC, ECMODC, and ECMOGC (see Table S1 in the supplemental material), respectively, by following established procedures (Invitrogen by Life Technologies, Grand Island, NY).
RESULTS
E. coli K-12 cells incubated with Te(VI) removed more than 70% of the dissolved Te from the culture medium within 48 h (Fig. 1). The loss of Te(VI) was concurrent with the formation of black Te(0) precipitates, which was visible after 3 h of incubation. No loss of dissolved Te(VI) was observed in control experiments conducted with heat-killed cells.
Fig 1.
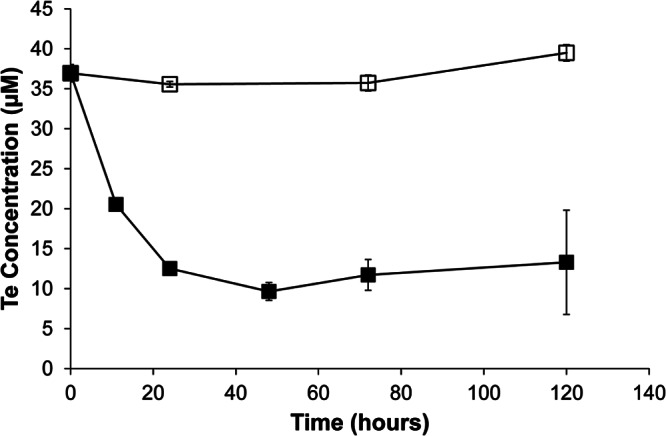
Tellurate reduction by wild-type Escherichia coli K-12. ■, live cells; □, heat-killed cells. Symbols and error bars represent the averages and standard deviations from triplicate experiments.
To determine if molybdopterin biosynthesis genes were required for tellurate reduction, Te(VI) reduction experiments were conducted using E. coli K-12 mutant strains containing single-gene mutations in the moaABCDE operon or the mog gene. Deletion of the moaA gene resulted in the complete loss of tellurate reduction activity (Fig. 2). Tellurate-containing media inoculated with the ΔmoaA mutant strain showed no tellurate loss and did not form black Te(0) precipitates. E. coli mutants deleted of either the moaB or moaE gene also were unable to reduce Te(VI) (Fig. 2). Transformation of pECA27 into the mutant strain JW0747-1, with the moaA deletion, restored the mutant's ability to reduce Te(VI), although the rates of tellurate reduction were slightly lower. The mutant strain JW0008-5, deleted of the mog gene, was also defective in Te(VI) reduction activity (Fig. 2). Complementation of the E. coli mog mutant with pECG07 fully restored the abolished phenotype.
Fig 2.
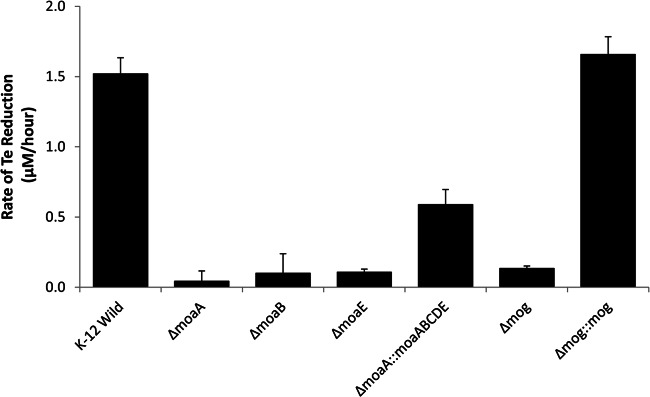
Tellurate reduction in E. coli mutants carrying deletions of molybdopterin biosynthesis genes. The symbol Δ represents the deletion of a gene, and :: represents the genetic complementation of a gene by insertion of a plasmid. Rates of Te reduction by wild-type and mutant strains are shown, and error bars represent standard deviations for parallel triplicate experiments. Shown are values for the E. coli K-12 wild type (K-12 Wild) and mutants JW0764-2 (ΔmoaA), JW0765-1 (ΔmoaB), JW0768-1 (ΔmoaE), and JW0764-2 complemented with pECA27 (ΔmoaA::moaABCDE), mutant JW0008-5 (Δmog), and JW0008-5 complemented with pECG07 (Δmog::mog).
To determine if molybdate transporter genes were required for tellurate reduction, experiments were conducted using E. coli mutant strains containing single gene mutations in the modABC operon. Deletion of either the modB or modC gene resulted in the complete loss of tellurate reduction activity (Fig. 3). Complementation of the E. coli modB mutant with pECD25 fully restored the tellurate reduction activity.
Fig 3.
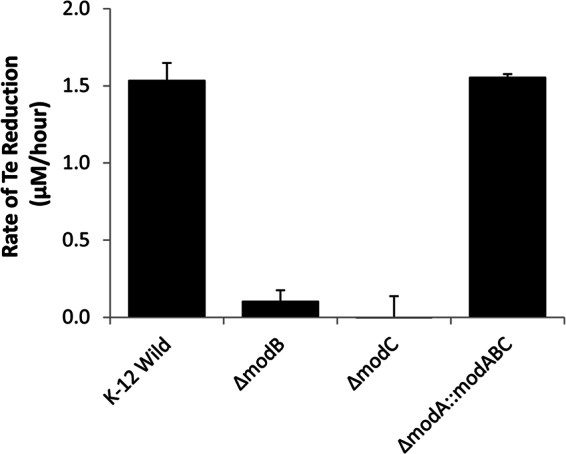
Tellurate reduction in E. coli mutants carrying deletions of molybdate transporter genes. The symbol Δ represents the deletion of a gene, and :: represents the genetic complementation of a gene by insertion of a plasmid. Rates of Te reduction by wild-type and mutant strains are shown, and error bars represent standard deviations for parallel triplicate experiments. Shown are values for the E. coli K-12 wild type (K-12 Wild) and mutants JW0747-1 (ΔmodB), JW0748-2 (ΔmodC), and JW0747-1 complemented with pECD25 (ΔmodA::modABC).
Mutation of molybdopterin biosynthesis genes (moaABCD and mog) and molybdate transporter genes (modABC) had no effect on tellurite reduction activity (Table 1). All mutant strains tested in this study were able to reduce Te(IV) to Te(0), indicating that tellurite reduction is catalyzed by a mechanism that is different from that of the tellurate reduction protein. In contrast, mutation of molybdate uptake and molybdopterin biosynthesis genes abolished Se(VI) reduction activity in E. coli (Table 1). The mutation of the fumarate nitrate reduction regulator gene (fnr) and genes in the twin arginine translocation pathway (tatBC) also resulted in the complete loss of Se(VI) reduction activity but had no effect on the reduction of tellurate or the reduction of tellurite to elemental tellurium (Table 1).
Table 1.
Te and Se reduction activity in E. coli mutants
| Strain | Te(VI) | Te(IV) | Se(IV) |
|---|---|---|---|
| Wild type | + | + | + |
| Δfnr | + | + | − |
| ΔmoaA | − | + | − |
| ΔmoaB | − | + | − |
| ΔmoaC | + | + | − |
| ΔmoaD | + | + | − |
| ΔmoaE | − | + | − |
| ΔtatB | + | + | − |
| ΔtatC | + | + | − |
| ΔmodA | + | + | − |
| ΔmodB | − | + | − |
| ΔmodC | − | + | − |
| Δmog | − | + | − |
| ΔubiH | + | + | − |
Finally, the tellurate reduction activity of E. coli mutants carrying single mutations of genes encoding molybdenum-containing oxidoreductases were tested. Table 2 shows the activity of E. coli mutants with single mutations of the genes dmsA, ynfE, ynfF, bisC, torA, torZ, fdhF, fdoG, fdnG, napA, narG, narZ, ydeP, yheS, and yfgD. All of these mutant strains tested positive for tellurate reduction.
Table 2.
Tellurate reduction by molybdoenzyme mutants
| Gene | Function or gene product | Te(VI) reductiona |
|---|---|---|
| dmsA | Dimethyl sulfoxide reductase | + |
| ynfE | Selenate reductase | + |
| ynfF | Selenate reductase | + |
| bisC | Biotin-d-sulfoxide to biotin | + |
| torA | Trimethylamine N-oxide reductase | + |
| torZ | Homolog of torA | + |
| fdhF | Formate dehydrogenase-H | + |
| fdoG | Formate dehydrogenase-O | + |
| fdnG | Formate dehydrogenase-N | + |
| napA | Periplasmic nitrate reductase | + |
| narG | Membrane-bound nitrate reductase | + |
| narZ | Membrane-bound nitrate reductase | + |
| ydeP | Putative oxidoreductase | + |
| yfgD | Putative oxidoreductase | + |
+, positive activity.
DISCUSSION
Mutants carrying deletions in the molybdopterin biosynthesis pathway lost the ability to reduce tellurate, and complementation by the wild-type sequence restored tellurate reduction activity in the ΔmoaA and Δmog mutant strains (Fig. 2). In E. coli, the protein product of moaA is responsible for converting GTP to cyclic pyranopterin monophosphate (cPMP) (25). Because cPMP is a critical intermediate in the biosynthesis of molybdopterin, mutation of the moaA gene prohibits the formation of the molybdenum cofactor. Similarly, the product of moaE is an essential component of the MPT synthase protein that inserts two sulfur atoms into cPMP. Mutation of the moaE gene prevents insertion of the dithiolene functional groups, which are required for Mo ligation by cPMP. In the final step of molybdenum cofactor synthesis, molybdenum is chelated and incorporated into molybdopterin by the action of the mog protein. Deletion of the mog gene precludes the incorporation of molybdenum into molybdopterin; thus, it inhibits the formation of an active tellurate reductase.
Mutants carrying deletions of modB and modC, genes that are required for molybdate transport, also lost the ability to reduce tellurate (Fig. 3). The protein product of modB provides the transmembrane channel necessary to transfer molybdate across the cytoplasmic membrane (24). Mutants carrying modB gene deletions thus are unable to transport molybdate ions from the periplasm into the cell. ModC is the ATPase subunit that serves as an energizing protein of the molybdate transport system. Mutation of the modC gene hampers ATP binding and energization of molybdate transporter. The loss of the molybdate transporter system into the cell impedes the delivery of molybdenum to molybdopterin, which further supports the hypothesis that the active site of the tellurate reductase contains the molybdenum cofactor.
In addition to the loss of Te(VI) reduction activity, mutation of the molybdate uptake and molybdopterin biosynthesis genes in E. coli also eliminated the ability to reduce Se(VI) (Table 1). Selenate reduction in E. coli is mediated by the molybdopterin-containing nitrate reductase, as well as the protein products of the genes ynfE and ynfF (15, 27). The ynfE and ynfF proteins are also predicted to bind a molybdopterin cofactor; thus, they require molybdenum for their catalytic activity. The molybdopterin-containing selenate reductases in E. coli exhibit functions similar to those of the membrane-bound respiratory dimethylsulfoxide (DMSO) reductase. First, the selenate reductase carries an N-terminal twin-arginine translocation (TAT) signal sequence. The experimental data indicate that mutation of the genes tatB and tatC in E. coli, which obstructs protein export via the TAT system, causes complete loss of Se(VI) reduction activity (Table 1). This is similar to the TAT-dependent selenate reductase in Enterobacter cloacae SLD1a-1 (28). Second, the E. coli selenate reductase is connected to the electron transport chain using electrons supplied by menaquinol to reduce selenate to selenite. Previously, we demonstrated that E. coli mutants carrying a deletion of either the menD, menC, or menE gene in the menaquinone biosynthesis pathway was unable to reduce selenate (29). Finally, upstream of the ynfE gene is a conserved sequence for the FNR (fumarate nitrate reduction regulator) protein binding site, and deletion of the fnr gene from E. coli abolishes selenate reduction activity (Table 1). The expression of the ynfE and ynfF genes are similar to that of the selenate reductase gene in E. cloacae SLD1a-1, which is regulated by the global anaerobic regulator FNR (30).
A suite of E. coli mutants were examined for Te(VI) and Te(IV) reductase activity, and comparison to selenate reduction provides insights into the characteristics of the tellurate reductase. The deletion of the tat genes does not eliminate tellurate reductase activity (Table 1), suggesting that E. coli harbors a tat-independent tellurate reductase. One possibility is that the tellurate reductase is similar to the tat-independent nitrate reductase (narG), which is a molybdopterin-containing enzyme that catalyzes oxyanion reduction inside the cytoplasm. Furthermore, deletion of the fnr gene does not inhibit tellurate reduction (Table 1), indicating that unlike the selenate reductase, E. coli does not regulate the expression of the tellurate reductase under anaerobic conditions. We were unable to grow E. coli using tellurate as the sole terminal electron acceptor (data not shown), and it does not appear that tellurate reduction is an anaerobic respiratory process. Therefore, it is doubtful that the tellurate reductase in E. coli is associated with the electron transport chain.
Analysis of the E. coli genome indicates that there are 15 genes that encode putative molybdoenzymes (Table 2). In an attempt to identify the tellurate reductase, we tested if single deletions of these molybdoenzyme genes resulted in loss of Te reduction activity. None of the mutants that were examined lost the ability to reduce tellurate. This suggests that multiple different molybdoenzymes are able to catalyze tellurate reduction; therefore, deletion of a single molybdoenzyme gene does not abolish Te(VI)-reducing activity. If this is the case, then tellurate reduction might be a secondary activity of a molybdoenzyme for which the primary function is the reduction of another substrate.
An interesting question is whether or not other tellurate-reducing bacteria, including Te(VI)-respiring microorganisms, also employ a molybdopterin-containing enzyme for tellurate reduction. Intriguingly, genome sequences for Te(VI)-respiring bacteria Bacillus selenitireducens and Sulfurospirillum barnesii contain the modABC transporter genes for molybdate uptake and the moaE gene for molybdopterin biosynthesis. Pending the establishment of genetic systems in these microorganisms, our results suggest that mutation of molybdate transporter and molybdopterin biosynthesis genes is an effective method for determining if the respiratory tellurate reductase is a molybdopterin-containing enzyme.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation (EAR 0843295).
We thank Hung-Kuang Chang and Madhavi Parikh for their laboratory assistance.
Footnotes
Published ahead of print 8 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03996-12.
REFERENCES
- 1.Taylor DE. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111–115 [DOI] [PubMed] [Google Scholar]
- 2.Chivers T. 1996. Tellurium compounds of the main-group elements: progress and prospects. J. Chem. Soc. Dalton Trans. 1996:1185–1194 [Google Scholar]
- 3.Tang Z, Kotov NA, Giersig M. 2002. Spontaneous organization of single CdTe nanoparticles into luminescent nanowires. Science 297:237–240 [DOI] [PubMed] [Google Scholar]
- 4.Turner RJ, Borghese R, Zannoni D. 2012. Microbial processing of tellurium as a tool in biotechnology. Biotechnol. Adv. 30:954–963 [DOI] [PubMed] [Google Scholar]
- 5.Wray DS. 1998. The impact of unconfined mine tailings and anthropogenic pollution on a semi-arid environment–an initial study of the Rodalquilar mining district, south east Spain. Environ. Geochem. Health 20:29–38 [Google Scholar]
- 6.Turner RJ, Weiner JH, Taylor DE. 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 145:2549–2557 [DOI] [PubMed] [Google Scholar]
- 7.Baesman SM, Bullen TD, Dewald J, Zhang D, Curran S, Islam FS, Beveridge TJ, Oremland RS. 2007. Formation of tellurium nanocrystals during anaerobic growth of bacteria that use Te oxyanions as respiratory electron acceptors. Appl. Environ. Microbiol. 73:2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baesman SM, Stolz JF, Kulp TR, Oremland RS. 2009. Enrichment and isolation of Bacillus beveridgei sp. nov., a facultative anaerobic haloalkaliphile from Mono Lake, California, that respires oxyanions of tellurium, selenium, and arsenic. Extremophiles 13:695–705 [DOI] [PubMed] [Google Scholar]
- 9.Chasteen TG, Fuentes DE, Tantaleán JC, Vásquez CC. 2009. Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33:820–832 [DOI] [PubMed] [Google Scholar]
- 10.Castro ME, Molina RC, Díaz WA, Pradenas GA, Vásquez CC. 2009. Expression of Aeromonas caviae ST pyruvate dehydrogenase complex components mediate tellurite resistance in Escherichia coli. Biochem. Biophys. Res. Commun. 380:148–152 [DOI] [PubMed] [Google Scholar]
- 11.Chiong M, Barra R, González E, Vásquez C. 1988. Resistance of Thermus spp. to potassium tellurite. Appl. Environ. Microbiol. 54:610–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison JJ, Ceri H, Stremick C, Turner RJ. 2004. Differences in biofilm and planktonic cell mediated reduction of metalloid oxyanions. FEMS Microbiol. Lett. 235:357–362 [DOI] [PubMed] [Google Scholar]
- 13.Moore MD, Kaplan S. 1992. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 174:1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Summers AO, Jacoby GA. 1977. Plasmid-determined resistance to tellurium compounds. J. Bacteriol. 129:276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avazéri C, Turner RJ, Pommier J, Weiner JH, Giordano G, Verméglio A. 1997. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 143:1181–1189 [DOI] [PubMed] [Google Scholar]
- 16.Lee DS, Edmond JM. 1985. Tellurium species in seawater. Nature 313:782–785 [Google Scholar]
- 17.Huang C, Hu B. 2008. Speciation of inorganic tellurium from seawater by ICP-MS following magnetic SPE separation and preconcentration. J. Separation Sci. 31:760–767 [DOI] [PubMed] [Google Scholar]
- 18.Csotonyi JT, Stackebrandt E, Yurkov V. 2006. Anaerobic respiration on tellurate and other metalloids in bacteria from hydrothermal vent fields in the eastern pacific ocean. Appl. Environ. Microbiol. 72:4950–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bébien M, Kirsch J, Méjean V, Verméglio A. 2002. Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 148:3365–3372 [DOI] [PubMed] [Google Scholar]
- 20.Malasarn D, Keeffe JR, Newman DK. 2008. Characterization of the arsenate respiratory reductase from Shewanella sp. strain ANA-3. J. Bacteriol. 190:135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 100:10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroda M, Yamashita M, Miwa E, Imao K, Fujimoto N, Ono H, Nagano K, Sei K, Ike M. 2011. Molecular cloning and characterization of the srdBCA operon, encoding the respiratory selenate reductase complex, from the selenate-reducing bacterium Bacillus selenatarsenatis SF-1. J. Bacteriol. 193:2141–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krafft T, Bowen A, Theis F, Macy JM. 2000. Cloning and sequencing of the genes encoding the periplasmic-cytochrome B-containing selenate reductase of Thauera selenatis. DNA Sequence 10:365–377 [DOI] [PubMed] [Google Scholar]
- 24.Self WT, Grunden AM, Hasona A, Shanmugam KT. 2001. Molybdate transport. Res. Microbiol. 152:311–321 [DOI] [PubMed] [Google Scholar]
- 25.Leimkühler S, Wuebbens MM, Rajagopalan KV. 2011. The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coordination Chem. Rev. 255:1129–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko K, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guymer D, Maillard J, Sargent F. 2009. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 191:519–528 [DOI] [PubMed] [Google Scholar]
- 28.Ma J, Kobayashi DY, Yee N. 2007. Chemical kinetic and molecular genetic study of selenium oxyanion reduction by Enterobacter cloacae SLD1a-1. Environ. Sci. Technol. 41:7795–7801 [DOI] [PubMed] [Google Scholar]
- 29.Ma J, Kobayashi DY, Yee N. 2009. Role of menaquinone biosynthesis genes in selenate reduction by Enterobacter cloacae SLD1a-1 and Escherichia coli K12. Environ. Microbiol. 11:149–158 [DOI] [PubMed] [Google Scholar]
- 30.Yee N, Ma J, Dalia A, Boonfueng T, Kobayashi DY. 2007. Se(VI) reduction and the precipitation of Se(0) by the facultative bacterium Enterobacter cloacae SLD1a-1 are regulated by FNR. Appl. Environ. Microbiol. 73:1914–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


