Abstract
“Candidatus Midichloria mitochondrii” is an intramitochondrial bacterium of the order Rickettsiales associated with the sheep tick Ixodes ricinus. Bacteria phylogenetically related to “Ca. Midichloria mitochondrii” (midichloria and like organisms [MALOs]) have been shown to be associated with a wide range of hosts, from amoebae to a variety of animals, including humans. Despite numerous studies focused on specific members of the MALO group, no comprehensive phylogenetic and statistical analyses have so far been performed on the group as a whole. Here, we present a multidisciplinary investigation based on 16S rRNA gene sequences using both phylogenetic and statistical methods, thereby analyzing MALOs in the overall framework of the Rickettsiales. This study revealed that (i) MALOs form a monophyletic group; (ii) the MALO group is structured into distinct subgroups, verifying current genera as significant evolutionary units and identifying several subclades that could represent novel genera; (iii) the MALO group ranks at the level of described Rickettsiales families, leading to the proposal of the novel family “Candidatus Midichloriaceae.” In addition, based on the phylogenetic trees generated, we present an evolutionary scenario to interpret the distribution and life history transitions of these microorganisms associated with highly divergent eukaryotic hosts: we suggest that aquatic/environmental protista have acted as evolutionary reservoirs for members of this novel family, from which one or more lineages with the capacity of infecting metazoa have evolved.
INTRODUCTION
“Candidatus Midichloria mitochondrii” is an intracellular bacterium associated with the sheep tick Ixodes ricinus (1), the main vector of Lyme borreliosis and other diseases in Europe (2). “Ca. Midichloria mitochondrii” presents an unusual lifestyle, surviving and multiplying inside the tick mitochondria (3). Analysis of 16S rRNA gene sequences revealed that the bacterium constitutes a deep branch of the order Rickettsiales (1). In the second volume of Bergey's Manual of Systematic Bacteriology, the order Rickettsiales encompasses three families: Rickettsiaceae, Anaplasmataceae, and Holosporaceae (4). Recently, oceanic environmental bacteria known as the SAR11 group have been proposed to form a fourth family of Rickettsiales, the Pelagibacteraceae (5; see also references 6 and 7). Based on phylogenetic results, “Ca. Midichloria mitochondrii” is not attributable to any of the families/main lineages of this order (8). In the last decade, projects aiming at detecting and cataloguing the bacterial diversity in environmental and biological samples have revealed that “Ca. Midichloria mitochondrii” is likely just “the tip of the iceberg” of an emerging novel group of intracellular bacteria. These bacterial lineages phylogenetically allied to “Ca. Midichloria mitochondrii” are associated with a wide range of hosts scattered throughout the eukaryotic tree of life, from arthropods, such as ticks, fleas, and stink bugs, to ciliates, amoebae, cnidarians, sponges, fish, and humans (3, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19). Following Mariconti et al. (15), we refer to the bacteria phylogenetically related to “Ca. Midichloria mitochondrii” as MALOs (midichloria and like organisms). Besides the ecological and evolutionary interest of MALOs, which derives from their widespread distribution and peculiar intramitochondrial lifestyle (at least in ticks), we emphasize that there is growing evidence for the infectivity of these bacteria to vertebrates, including humans, and for their immunological and potentially pathogenic roles (11, 14, 15). As for the intramitochondrial niche of “Ca. Midichloria mitochondrii,” analysis of both the symbiont and the tick mitochondrial genomes has not yet revealed any clues leading to an understanding of the biology of this peculiar type of symbiosis (20, 21, 22).
Despite numerous investigations focused on specific members of the MALO group associated with novel hosts and the description of novel genera and species allied to the group, no comprehensive phylogenetic, statistical, and ecological studies have so far been performed on the MALOs as a whole in the overall framework of the Rickettsiales. Here, we present a study aiming at defining whether the MALO group represents a coherent and monophyletic clade, whether it is structured into subgroups that could represent genera, and whether the overall clade could represent a novel family of the order Rickettsiales.
MATERIALS AND METHODS
Overview of the analytical approach.
Our study is based on analysis of all of the 16S rRNA gene sequences available for taxonomically described Rickettsiales (as of August 2012) plus 16S rRNA gene sequences available for MALOs, selecting only complete or almost complete gene sequences. We performed (i) phylogenetic analyses aimed at verifying the monophyly of the MALO group; (ii) analyses aimed at detecting evolutionarily significant units within the Rickettsiales and the MALO clades (through the generalized mixed Yule coalescent [GMYC] method [23, 24]); (iii) a principal-coordinate analysis (PCoA) on the 16S rRNA gene sequences; and (iv) an analysis of molecular variance (AMOVA) between the four Rickettsiales families (4, 5), with MALOs examined as a separate family or as members of existing families, with tests of the significance of the groups defined.
Taxon selection, alignment strategies, and phylogenetic analyses.
A data set of complete/almost complete 16S rRNA gene sequences for taxonomically well-identified taxa belonging to Rickettsiales and from MALOs were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/; August 2012). The 16S rRNA starting data set was composed of 103 species/operational taxonomic units (OTUs) consisting of 7 non-Rickettsiales alphaproteobacteria, 8 OTUs from Pelagibacteraceae, 3 OTUs from Holosporaceae, 32 species/OTUs of Rickettsiaceae (genera Rickettsia, Orientia, and Cryptoprodotis), and 23 species of Anaplasmataceae (genera Neorickettsia, Erhlichia, Neoerhlichia, Anaplasma, and Wolbachia), plus 1 undescribed OTU and 29 species/OTUs from MALOs. The accession numbers and taxonomic status of all of the species/OTUs used in the analysis are reported in Table S1 in the supplemental material. The retrieved data set was subsequently subjected to different alignment methods in order to explore variations of the informative sites in the alignments. The data set was aligned using two algorithms for multiple-sequence alignment, MUSCLE (25) with default settings and MAFFT version 6 (http://mafft.cbrc.jp/alignment/server/ [26]) using a G-INS-i search strategy (27), a 200PAM scoring matrix (considering the divergence between the analyzed organisms), and default parameters for the remaining settings. The data sets obtained, identified with “A” for the one obtained using MUSCLE and “B” for the one obtained with MAFFT, were trimmed using Gblocks (28) with different parameters: “conservative” (c) mode, which did not allow gap positions within the final block, and “liberal” (l) mode, which allowed gap positions within the final block and allowed less strict flanking positions. These approaches resulted in four alignments, on which the phylogenetic analyses were performed. The four alignments were designated “Ac” (aligned by MUSCLE plus Gblocks conservative mode), “Al” (aligned by MUSCLE plus Gblocks liberal mode), “Bc” (aligned by MAFFT plus Gblocks conservative mode), and “Bl” (aligned by MAFFT plus Gblocks liberal mode). Phylogenetic analyses were performed on each of the data sets using the following methods: the distance matrix-based method neighbor joining (NJ) and the character state-based approaches maximum likelihood (ML) and Bayesian inference (BI). NJ trees were inferred using MEGA 5 (29) implementing the Tamura-Nei (30) model of nucleotide substitutions, including transitions and transversions, rate variation among sites modeled with a gamma distribution (shape parameter = 1), and gaps treated with partial deletion. Node support was estimated using 1,000 bootstrap replicates. For ML and BI, the evolutionary models best fitting the analyzed data sets were selected with jModeltest 0.1.1 (31) using the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Both criteria led to the selection of the General Time Reversible (GTR) (32) with proportions of invariable sites (I) and gamma distribution (Γ) as the best model of nucleotide substitutions. ML analyses were performed using PhyML version 3.0 (33) with the following options: GTR (32) as the nucleotide substitution model, optimized proportions of invariable sites, estimated nucleotide frequency, optimized rate variation across sites into six substitution rate categories, estimated gamma shape parameter, the best of nearest neighbor interchange (NNI) and subtree pruning and regrafting (SPR) tree-searching operations, and nonparametric bootstrap analysis (1,000 replicates). Bayesian inference was performed with MrBayes 3.2 (34) on the Web-based Bioportal (35). In MrBayes, all the Markov chain Monte Carlo analyses were implemented in two runs of 10 million generations with four chains each, sample frequency settled every 1,000 generations, and the GTR substitution model (32), estimated proportions of invariable sites, and gamma distribution with 6 categories as parameters of the likelihood model. The convergence of each run was verified with Tracer 1.4 (36), and 0.25% of the sampled trees were discarded as burn-in.
Detection of ESUs.
The GMYC method (23, 24) was applied to the 16S rRNA gene tree obtained with ML on alignment Al in order to identify evolutionary significant units (ESUs) (37). The null hypothesis assumes that all samples belong to a single entity, while the alternative to be tested assumes that the samples are divided into n independently evolving units; the log-likelihood ratio test is performed to compare the likelihoods of the two models. The method is implemented in the R package GMYC in “splits” (SPecies LImits by Threshold Statistics, available at http://r-forge.r-project.org/projects/splits/). The ML tree inferred on the alignment Al was processed in order to drop all branches with 0 length. This operation resulted in a tree with 71 terminal nodes and 69 internal nodes. The tree was converted to ultrametric using penalized likelihood with a smoothing parameter of 1 (selected after cross-validation of values between 0.1 and 100) as implemented in r8s 1.7 (38).
PCoA.
We used PCoA (39), a commonly used ordination method for data organized into distance matrices (40), to place the analyzed species/OTUs in a new coordinate system. The pairwise nucleotide distance matrix was calculated on alignment Al, which displayed the highest number of likelihood-informative sites, using MEGA 5 (29) following the removal of the outgroup taxa (i.e., on the remaining 96 species/OTUs). The implemented model of nucleotide substitutions was the Tamura-Nei model (30), including transitions and transversions, rate variation among sites modeled with a gamma distribution (shape parameter = 1), and gaps treated with partial deletion. The PCoA was performed with the software MVSP 3.1 (41) using a Euclidean metric and no data transformation.
AMOVA.
AMOVA was performed to quantify the genetic variation between and within the families of Rickettsiales plus MALOs. AMOVA (42, 43, 44) was performed on the nucleotide distance matrix used as input for PCoA analysis (which used as input alignment Al). AMOVA was performed considering three different possible scenarios, based on phylogenetic results, in order to explore the variability of the molecular variances between/within the families of Rickettsiales plus MALOs in the three different situations. The three scenarios were defined as follows: (i) MALOs assumed to be part of Rickettsiaceae, with the data set divided into four groups corresponding to the four described families; (ii) MALOs considered part of Anaplasmataceae, again with four groups corresponding to the families; and (iii) MALOs considered separately, with the data set divided into five groups corresponding to the four described families plus MALOs.
Analysis of similarity between the described families plus MALOs.
In order to assess whether the five groups identified by phylogenetic analysis, GMYC, and PCoA were significantly different, and to validate our decision to elevate MALOs to the family rank, the genetic pairwise distance matrix used for PCoA and AMOVA was subjected to a nonparametric one-way analysis of similarity (ANOSIM) (45) with permutations (999). The R value, obtained by the ANOSIM analysis, is a measure of similarity between groups; an R value of 1 indicates that members of the selected group are more similar to each other than to members of other groups. ANOSIM analysis was performed using the R package ANOSIM in “vegan” (46).
Between- and within-group nucleotide distances.
For the five groups examined, i.e., MALOs and the four families of the Rickettsiales, the intragroup mean nucleotide distances and the between-group mean distances were calculated on the p distance matrix with complete deletion of gaps, using MEGA 5 (29).
RESULTS AND DISCUSSION
Phylogenetic analyses.
The different alignment algorithms and masking strategies resulted in the following four aligned data sets: two alignments without gaps (Ac and Bc, 868 and 867 bp in size, respectively) and two alignments with gaps (Al and Bl, 1,396 and 1,361 bp in size, respectively). Features of the four data sets obtained with different alignment algorithms and masking procedures (i.e., the size, the likelihood-informative sites, the parsimony-informative sites, and sites without polymorphism) are reported in Table 1. Phylogenetic analyses based on the four data sets consistently indicated that MALOs are monophyletic, as are the four described families of the Rickettsiales (Rickettsiaceae, Anaplasmataceae, Holosporaceae, and Pelagibacteraceae) (Fig. 1 and 2; see Fig. S1 to S12 in the supplemental material). All but one of the phylogenetic analyses based on the four data sets using different inference methods (BI, ML, and NJ) agreed with each other in terms of the relationships between the Rickettsiales families. The consensus cladogram of 11 of the 12 phylogenetic relationships inferred from the four data sets and analyzed by the three methods is shown in Fig. 1 (one topology was slightly different in that MALOs formed a sister group of Rickettsiaceae). Differences in the values of branch support were found among the trees inferred from the four data sets, as shown in Fig. 1. The alignments obtained by the conservative masking strategy (Ac and Bc) contained fewer informative sites (403 likelihood-informative sites) than the alignments obtained using a liberal masking strategy (Al and Bl, with 813 and 787 likelihood-informative sites, respectively), resulting in lower phylogenetic resolution of the former strategy at the family level and below. The results obtained (excluding the positions of MALOs) were in agreement with previously published topologies inferred from concatenated gene and protein alignments (5, 20, 47). Figure 2 shows the unrooted phylogeny inferred from Al (the data set with the highest number of informative sites, 813) using the Bayesian method. In our analyses based on 16S rRNA gene sequences, MALOs were placed as the sister group of Anaplasmataceae, which is concordant with some reports based on 16S rRNA gene sequences (1, 13) and multiple-protein-sequence alignments (8) but differs from other reports that found MALOs to be a sister group of Rickettsiaceae (8, 16, 20). As shown in Fig. 2, the phylogeny of the Rickettsiales appears as a typical evolutionary star radiation with short internal branches supporting long clade branches. Reconstruction of the relationships between main clades evolved by star radiation is generally difficult to establish (48). Phylogenetic relationships within the groups Holosporaceae, Anaplasmataceae, and MALOs were well resolved, while the exact positioning of some lineages in Rickettsiaceae was elusive. The results inferred from all four data sets by both the Bayesian and maximum-likelihood methods exhibited consistent relationships between the taxa/OTUs within the MALO group (Fig. 2 and 3).
Table 1.
Features of the16S rRNA gene sequence alignment data sets
| Data set | Alignment algorithm | Gblocks strategy | Size (nt) | No. (%) of sites |
||
|---|---|---|---|---|---|---|
| Likelihood informative | Parsimony informative | Invariant | ||||
| Ac | MUSCLE | Conservative | 868 | 403 (46.4) | 360 (41.5) | 448 (51.6) |
| Al | MUSCLE | Liberal | 1,396 | 813 (58.2) | 820 (58.7) | 635 (45.5) |
| Bc | MAFFT | Conservative | 867 | 403 (46.5) | 362 (41.8) | 447 (51.6) |
| Bl | MAFFT | Liberal | 1,361 | 787 (57.8) | 787 (57.8) | 574 (42.2) |
Fig 1.
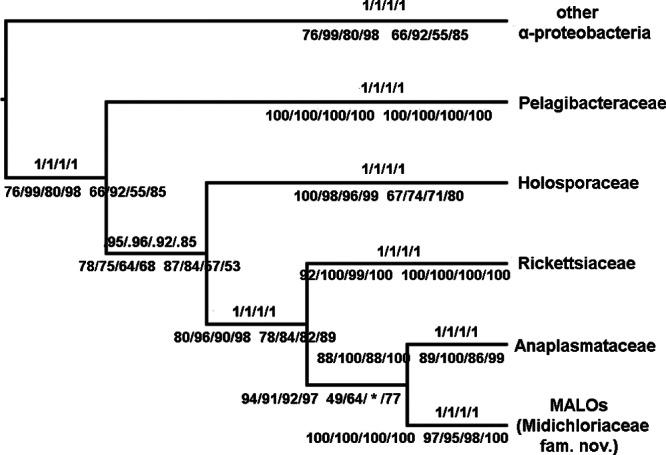
Majority rule consensus cladogram of the four Rickettsiales families plus MALOs based on the four 16S rRNA gene sequence data sets (Ac, Al, Bc, and Bl) implementing BI, ML, and NJ inference methods. Support values inferred from the four aligned data sets are reported on the branches, with the Bayesian posterior probability above and the maximum-likelihood (left) and NJ (right) bootstrap values below. The asterisk represents the sole branch that does not agree with the majority rule consensus cladogram (i.e., MALOs were found to be the sister clade of Rickettsiaceae in the tree inferred with NJ on data set Bc).
Fig 2.
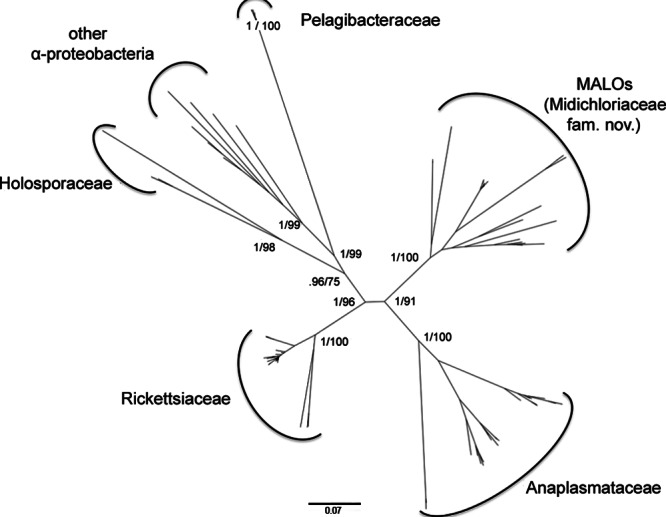
Unrooted Bayesian phylogram of the Rickettsiales based on the 16S rRNA gene sequence Al alignment. Bayesian posterior probability and ML bootstrap values are reported for the main lineages. (Phylograms obtained with BI, ML, and NJ on the four alignment data sets are shown in Fig. S1 to S12 in the supplemental material.) The scale bar indicates the distance in substitutions per site.
Fig 3.
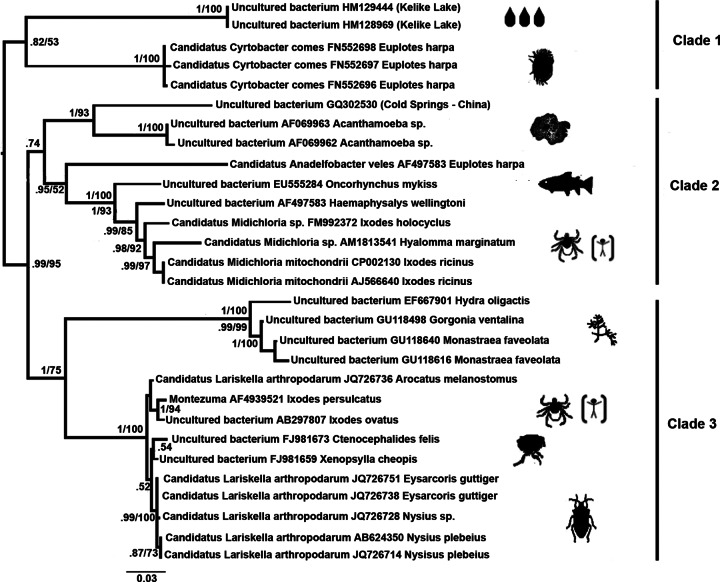
Bayesian phylogram of MALOs/“Ca. Midichloriaceae” fam. nov., obtained from the 16S rRNA gene sequence Al data set. The values on the nodes are the Bayesian posterior probabilities and maximum-likelihood bootstrap percentages (values below 50% are not reported). The names after the accession numbers are those of the hosts of the included MALO bacteria. Where 16S rRNA gene sequences were obtained from environmental samples, the source is indicated in parentheses. Visual representations of hosts (or sources) of the bacteria are mapped beside the terminal tips. The scale bar indicates the distance in substitutions per site.
An evolutionary scenario for MALOs, from aquatic protists to metazoa.
Figure 3 shows the phylogenetic relationship of MALOs only, along with their source hosts and environmental origins. The phylogenetic relationships among the MALO taxa/OTUs were generally well resolved and supported (Fig. 2 and 3). The basal branch within MALOs is the lineage leading to bacteria harbored by brackish water ciliates and bacteria from water samples from Kelike Lake in Tibet (it is possible that these are symbionts of some aquatic eukaryote). It is notable that host taxonomic groups were generally coherent; for example, we identified MALO clades exclusively associated with cnidarians, arthropods, amoebae, and ciliates. However, there are some exceptions, e.g., “Candidatus Anadelfobacter veles” was not grouped with the other symbionts of ciliates (i.e., “Candidatus Cyrtobacter comes”) but clustered with the group of MALOs associated with ticks. The taxonomic distribution of MALO hosts was quite diverse, ranging from amoebae to vertebrates. We speculate that aquatic eukaryotic microorganisms like ciliates and amoebae, which feed on bacteria and are likely prone to establish stable interactions with phagocytosed bacteria (49, 50, 51), might represent a sort of evolutionary reservoir of MALOs from which the lineages infecting animals could have evolved, plausibly more than once. The deepest branch in the MALO tree leads to symbionts associated with aquatic protists (Fig. 3, clade 1). Furthermore, a sister group relationship is also observed between symbionts of other protists and those of metazoans, i.e., vertebrates and ticks (Fig. 3, clade 2). Finally, there is also a group of MALOs associated with filter-feeding aquatic invertebrates, that is, a sister group to arthropod-associated MALOs (Fig. 3, clade 3). These relationships indicate the potential for the origin of metazoan-associated MALOs from water-dwelling organisms. Regarding clade 2 in Fig. 3, one hypothesis to explain the trajectory of MALOs from aquatic/environmental protista/amoebae to ticks could be the infection (even transient) of a vertebrate host, from which ticks obtain their blood meals. Amoebae demonstrated to harbor MALOs have been isolated from humans (9) and belong to the genus Acanthamoeba, a well-known group of environmental protists capable of infecting vertebrates. The possibility that bacteria from the MALO group are infectious to vertebrates is supported by several independent studies, from the well-established case of the rainbow trout (11) to the results indicating the circulation of “Ca. Midichloria mitochondrii” in humans (15) (see the introduction). Therefore, although it is not certain at this stage that MALOs are infectious and pathogenic to vertebrates, it is reasonable to conclude that vertebrates are at least transient hosts for these bacteria (thus, in Fig. 3, we have indicated a vertebrate, i.e., Homo sapiens, as a possible host for MALOs from clades 2 and 3). In summary, the scenario that we propose is that aquatic/environmental protista have acted as evolutionary reservoirs of MALOs, from which one or more lineages have evolved with the capacity to infect metazoa.
Detection of evolutionary significant units within the order Rickettsiales.
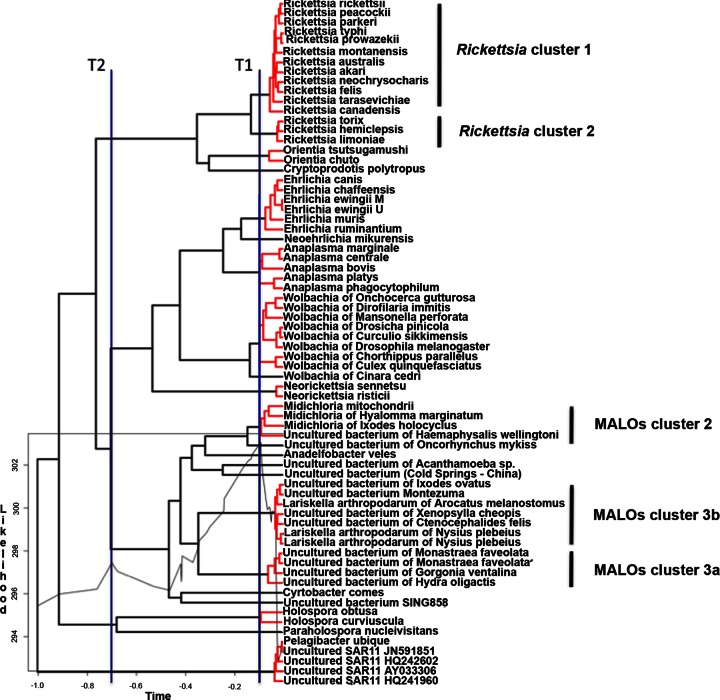
In addition to the above-mentioned phylogenetic analyses, we also applied the GMYC method (23), a tool developed for the detection of ESUs within a given data set based on a maximum-likelihood approach that has recently also been applied to a 16S rRNA gene sequence data set from prokaryotes (37). The main results of GMYC analysis are shown in Fig. 4. The GMYC model exhibited a significantly better likelihood (L) than the null model (log LGMYC = 303; log LNULL = 295.4; 2ΔL = 15.2; χ2 test, P = 0.0016) and identified a transition (T1) in the branching rate at the threshold time −0.094 from the present (see the lineage through time plot in Fig. S13 in the supplemental material), which detected 12 ML ESU clusters for a total of 22 ML entities (cluster range, 11 to 14; entity range, 18 to 26). Analyzing the peaks on the likelihood plot, we derived a second threshold line (T2; threshold time, −0.7 from the present; log L = 297.5). The two threshold lines, albeit generated independently of any taxonomic/nomenclature information, correspond to the genus level (T1) and to the family rank (Fig. 4). Considering T1 in detail, there is an overall correspondence throughout the order Rickettsiales between the ESUs identified by this analysis and described genera. Minor differences are observed in the genus Rickettsia, where GMYC recognizes two ESUs, the first encompassing most of the species so far described (Rickettsia cluster 1, which includes human pathogens), and a second (Rickettsia cluster 2), which encompasses Rickettsia endosymbionts of leaches (Torix tukubanatorix and Hemiclepsis marginata) and insects (Rickettsia limoniae). Rickettsia cluster 2 could perhaps be recognized as a novel genus, in agreement with the results of previous studies (52, 53). With regard to MALOs, GMYC recognizes three groups of sequences that are defined at the T1 threshold as ESUs, i.e., MALO cluster 2 (from I. ricinus and other ticks), MALO cluster 3a (from cnidarians and sponges), and MALO cluster 3b (from Hemiptera bugs, fleas, and ticks). In addition, GMYC identified six single-lineage ESUs at the T1 level, corresponding to two described genera (i.e., “Candidatus Cyrtobacter” and “Candidatus Anadelfobacter” from ciliates) and four undescribed lineages (from two types of environmental samples, Acanthamoeba sp., and rainbow trout). In summary, threshold level T1, which shows good correspondence with the genus level for all of the other Rickettsiales, indicates the existence of nine lineages that might deserve to be ranked at the genus level within the MALO group, supporting the four published genera within the group (i.e., “Candidatus Midichloria,” “Candidatus Lariskella,” “Ca. Anadelfobacter,” and “Ca. Cyrtobacter”) and indicating that five further genera could be proposed. Interestingly, T2, the second identified likelihood peak, identifies five evolutionary lineages that correspond to the four described families of the Rickettsiales plus MALOs (Fig. 4). Thus, at the family level, the GMYC method also generates results that match the current classification of Rickettsiales and indicates that the MALO group should to be elevated to the family rank. We therefore performed a series of analyses to obtain further support for this indication.
Fig 4.
Maximum-likelihood ultrametric tree, obtained from the Al data set, depicting the identified ESUs within Rickettsiales plus MALOs/“Ca. Midichloriaceae” fam. nov. (the tree was processed in order to drop all branches with 0 length; see Materials and Methods). Clusters of ESUs are highlighted in red; black terminal tips indicate the identified single entities. The likelihood-through-time plot is mapped on the tree. The vertical blue line T1 shows the maximum-likelihood transition point of the switch in branching rates. T2 shows a second likelihood peak corresponding to the family rank on the tree. The scale bar on the x axis indicates the time unit from the root of the ultrametric tree (time −1.0) to the tips (time 0).
Statistical analyses of the main clades of the order Rickettsiales.
The results of the PCoA performed on the distance matrix generated on Al are presented graphically in Fig. 5 (PCoA case score plot). After the analysis of the eigenvalues and of the scree plot, the first three principal components were considered. These components explain a total of 89.1% of the variation (1st component, 52.6%; 2nd component, 24.2%; and 3rd component, 12.3%). Figure 5 shows the case score plot for the first three principal components, in which five groups of OTUs that correspond to the five clades of Rickettsiales (i.e., the four described families plus MALOs) are displayed with different shapes. We note that all five clades of Rickettsiales are well isolated. Interestingly all of the members of MALOs are grouped together, while there are members of Rickettsiaceae and Anaplasmataceae that are located rather far from the centroid of the corresponding family. Within Rickettsiaceae, the deviating taxa are the two species of Orientia and “Ca. Cryptoprodotis polytropus.” In agreement with this result, in the phylogenetic tree, these species stem as long basal branches and are quite divergent from the members of the genus Rickettsia (Fig. 2). A similar situation arises in both Anaplasmataceae and Holosporaceae, where the two species of Neorickettsia and “Ca. Paraholospora nucleivisitans,” which constitute basal and long phylogenetic branches within the families (Fig. 2), appear quite divergent in the PCoA case score plot.
Fig 5.
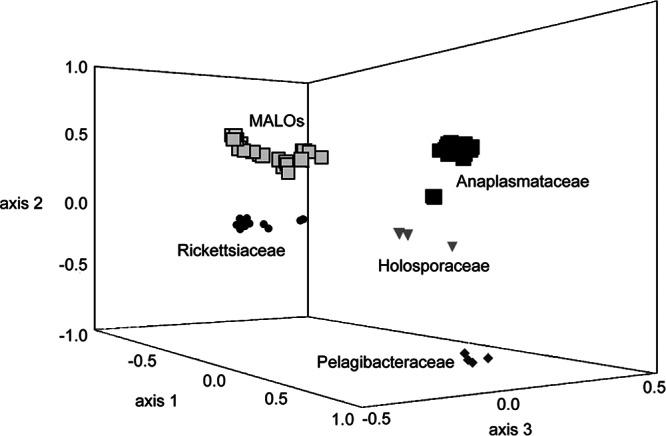
Principal-coordinate analysis based on Euclidean distances of the pairwise nucleotide distance matrix obtained from the 16S rRNA gene sequence Al data set. The explained variances are as follows: 1st axis, 52.6%; 2nd axis, 24.2%; 3rd axis, 12.3%. The four Rickettsiales families plus MALOs are labeled.
AMOVAs were performed on the nucleotide pairwise genetic distance matrix (implementing the Tamura-Nei [30] model of nucleotide substitutions; see Materials and Methods) estimated on Al (after removal of the outgroups) simulating three possible scenarios for the ranking and positioning of MALOs: (i) assumed to be part of the Rickettsiaceae, (ii) assumed to be part of the Anaplasmataceae, or (iii) assumed to be a separate family (see Materials and Methods for details). The results of the AMOVAs (shown in Table 2) indicate that when MALOs are considered a separate family, the explained percentage of variance among families is 63.1%, i.e., well above that explained when MALOs were grouped into the Anaplasmataceae (55.5%) or the Rickettsiaceae (49.6%). In parallel, the explained percentage of variation within families is lower when MALOs are considered a separate group (Table 2). In other words, the assumption of MALOs as a separate group at the family rank maximizes the intergroup variance and minimizes that within the groups.
Table 2.
AMOVA
| Tested hypothesis | Variation among groups |
Variation within groups |
||
|---|---|---|---|---|
| Variance component | Variation explained (%) | Variance component | Variation explained (%) | |
| MALOs assumed as part of Anaplasmataceae | 0.60 | 55.5 | 0.48 | 44.5 |
| MALOs assumed as part of Rickettsiaceae | 0.54 | 49.6 | 0.55 | 50.4 |
| MALOs assumed as separate group | 0.63 | 63.1 | 0.37 | 36.9 |
The ANOSIM performed on the estimated genetic pairwise distance matrix on alignment Al confirms significant diversity between the five groups examined here, i.e., the four families of Rickettsiales plus MALOs (R = 0.976; significance, P = 0.001). Finally, Table 3 presents a summary of the distances within and between the Rickettsiales families and the MALO group.
Table 3.
Nucleotide p distances within and between the four families of Rickettsiales plus MALOs
| Family |
p distancea |
||||
|---|---|---|---|---|---|
| MALOs | Anaplasmataceae | Rickettsiaceae | Pelagibacteraceae | Holosporaceae | |
| MALOs | 8.7 (0.5) [0,14.7] | ||||
| Anaplasmataceae | 14.6 (0.9) | 9 (0.5) [0.1, 15.6] | |||
| Rickettsiaceae | 13.6 (0.9) | 14.8 (0.9) | 2.7 (0.2) [0, 10.2] | ||
| Pelagibacteraceae | 19.4 (1.1) | 20.2 (1.1) | 19.8 (1.2) | 0.8 (0.2) [0.1, 1.3] | |
| Holosporaceae | 17.7 (0.9) | 17.6 (0.9) | 16.5 (1) | 21.1 (1.2) | 12.7 (0.7) [3.2, 17.7] |
Distances are expressed as percentages and were calculated on alignment Al (1,396 nucleotides); below the diagonal, mean p distances between the five clades; on the diagonal, within-group mean p distances. Standard deviations are shown in parentheses and minimum and maximum values in square brackets.
In summary, the results obtained by different approaches (i.e., phylogeny, GMYC, PCoA, and AMOVA) indicate that bacteria belonging to the MALO group deserve to be ranked at the family level. We thus propose to create a novel family in the order Rickettsiales, i.e., “Candidatus Midichloriaceae.”
Taxonomy.
“Candidatus Midichloriaceae” fam. nov. (Mi.di.chlo.ri.a.ce′a.e. N.L. n. “Candidatus Midichloria,” type genus of the family; suffix -aceae, ending to denote a family; N.L. fem. pl. n. “Candidatus Midichloriaceae,” the family of the genus “Candidatus Midichloria”). The new family encompasses bacteria associated with a wide range of hosts, from protists to vertebrates, including humans; all of the members of this family that have so far been investigated by transmission electron microscopy have been shown to be intracellular, with a typical Gram-negative cell wall.
Concluding remarks.
The novel family described here, “Candidatus Midichloriaceae,” is to be regarded as a group of organisms of potential medical and veterinary relevance, considering their capacity to induce an immune response in tick-exposed human subjects and the detection of their DNA in tick salivary glands and in tissue/blood samples from different vertebrates (11, 14, 15). The results obtained by phylogenetic analyses for members of “Candidatus Midichloriaceae” and their hosts suggest a possible scenario of life history transitions from MALOs infecting water-dwelling protists to those infecting different types of metazoa (as discussed above). The idea that arthropod- and vertebrate-associated intracellular bacteria arose from aquatic/environmental protists is not new (9). In addition to the scenario discussed here for the “Candidatus Midichloriaceae,” there is also evidence for the deep branching of a lineage of the Rickettsiaceae that infects aquatic protista (53). Considering that current evidence places the family of ciliate-infecting bacteria Holosporaceae as the sister group of the lineage leading to Rickettsiaceae, Anaplasmataceae, and “Candidatus Midichloriaceae,” there is overall evidence indicating that intracellular Rickettsiales were originally associated with aquatic/environmental protista that served (and potentially still serve) as an ecological and evolutionary reservoir for the Rickettsiales infecting animals.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge D. Fontaneto for his critical input and discussion and J. P. Euzéby for taxonomical advice.
This work was partially supported by MIUR PRIN 2008 to C. Bazzocchi (national coordinator, F. Verni).
Footnotes
Published ahead of print 15 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03971-12.
ADDENDUM IN PROOF
On the same day this study was accepted for publication, an advanced online publication presented data on a novel bacterium phylogenetically related to “Ca. M. mitochondrii” (T. Driscoll et al., Genome Biol. Evol. doi:10.1093/gbe/evt036, 2013) and referred to this species and related organisms as “Midichloriaceae,” following a previous informal proposal to rank this bacterial group at the family level, discussed in a recent book chapter (J. J. Gillespie, E. Nordberg, A. F. Azad, and B. W. Sobral, p. 84–141, in A. F. Azad and G. H. Palmer, ed., Intracellular pathogens II: Rickettsiales, 2012).
REFERENCES
- 1.Sassera D, Beninati T, Bandi C, Bouman EA, Sacchi L, Fabbi M, Lo N. 2006. ‘Candidatus Midichloria mitochondrii', an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int. J. Syst. Evol. Microbiol. 56:2535–2540 [DOI] [PubMed] [Google Scholar]
- 2.Rizzoli A, Hauffe H, Carpi G, Vourc HG, Neteler M, Rosa R. 2011. Lyme borreliosis in Europe. Euro Surveill. 16:19906. [PubMed] [Google Scholar]
- 3.Beninati T, Lo N, Sacchi L, Genchi C, Noda H, Bandi C. 2004. A novel alphaproteobacterium resides in the mitochondria of ovarian cells of the tick Ixodes ricinus. Appl. Environ. Microbiol. 70:2596–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumler J, Walker D. 2005. Order II. Rickettsiales Gieszczykiewicz 1939, 25AL emend. Dumler, Barbet, Bekker, Dasch, Palmer, Ray, Rikihisa and Rurangirwa 2001, 2156, p 96–160. In Garrity GM, Brenner D, Krieg N, Staley J. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer-Verlag, New York, NY [Google Scholar]
- 5.Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappe MS, Giovannoni SJ. 2011. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM. 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgiades K, Madoui MA, Le P, Robert C, Raoult D. 2011. Phylogenomic analysis of Odyssella thessalonicensis fortifies the common origin of Rickettsiales, Pelagibacter ubique and Reclimonas americana mitochondrion. PLoS One 6:e24857 doi:10.1371/journal.pone.0024857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Ezpeleta N, Embley TM. 2012. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS One 7:e30520 doi:10.1371/journal.pone.0030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritsche TR, Horn M, Seyedirashti S, Gautom RK, Schleifer KH, Wagner M. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraune S, Bosch TC. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U. S. A. 104:13146–13151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd SJ, LaPatra SE, Snekvik KR, St-Hilaire S, Cain KD, Call DR. 2008. Strawberry disease lesions in rainbow trout from southern Idaho are associated with DNA from a Rickettsia-like organism. Dis. Aquat. Organ. 82:111–118 [DOI] [PubMed] [Google Scholar]
- 12.Erickson DL, Anderson NE, Cromar LM, Jolley A. 2009. Bacterial communities associated with flea vectors of plague. J. Med. Entomol. 46:1532–1536 [DOI] [PubMed] [Google Scholar]
- 13.Vannini C, Ferrantini F, Schleifer KH, Ludwig W, Verni F, Petroni G. 2010. “Candidatus anadelfobacter veles” and “Candidatus Cyrtobacter comes,” two new rickettsiales species hosted by the protist ciliate Euplotes harpa (Ciliophora, Spirotrichea). Appl. Environ. Microbiol. 76:4047–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mediannikov O, Ivanov LI, Nishikawa M, Saito R, Sidel'nikov IN, Zdanovskaia NI, Mokretsova EV, Tarasevich IV, Suzuki H. 2004. Microorganism “Montezuma” of the order Rickettsiales: the potential causative agent of tick-borne disease in the Far East of Russia. Zh. Mikrobiol. Epidemiol. Immunobiol. l:7–13 (In Russian.) [PubMed] [Google Scholar]
- 15.Mariconti M, Epis S, Gaibani P, Dalla Valle C, Sassera D, Tomao P, Fabbi M, Castelli F, Marone P, Sambri V, Bazzocchi C, Bandi C. 2012. Humans parasitized by the hard tick Ixodes ricinus are seropositive to Midichloria mitochondrii: is Midichloria a novel pathogen, or just a marker of tick bite? Pathog. Glob. Health 106:391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuura Y, Kikuchi Y, Meng XY, Koga R, Fukatsu T. 2012. Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl. Environ. Microbiol. 78:4149–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunagawa S, Woodley C, Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554 doi:10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epis S, Sassera D, Beninati T, Lo N, Beati L, Piesman J, Rinaldi L, McCoy KD, Torina A, Sacchi L, Clementi E, Genchi M, Magnino S, Bandi C. 2008. Midichloria mitochondrii is widespread in hard ticks (Ixodidae) and resides in the mitochondria of phylogenetically diverse species. Parasitology 135:485–494 [DOI] [PubMed] [Google Scholar]
- 19.Boscaro V, Petroni G, Ristori A, Verni F, Vannini C. 2013. “Candidatus Defluviella procrastinata” and “Candidatus Cyrtobacter zanobii”, two novel ciliate endosymbionts belonging to the “Midichloria clade”. Microb. Ecol. 65:302–310 [DOI] [PubMed] [Google Scholar]
- 20.Sassera D, Lo N, Epis S, D'Auria G, Montagna M, Comandatore F, Horner D, Pereto J, Luciano AM, Franciosi F, Ferri E, Crotti E, Bazzocchi C, Daffonchio D, Sacchi L, Moya A, Latorre A, Bandi C. 2011. Phylogenomic evidence for the presence of a flagellum and cbb(3) oxidase in the free-living mitochondrial ancestor. Mol. Biol. Evol. 28:3285–3296 [DOI] [PubMed] [Google Scholar]
- 21.Montagna M, Sassera D, Griggio F, Epis S, Bandi C, Gissi C. 2012. Tick-box for 3′-end formation of mitochondrial transcripts in ixodida, basal chelicerates and Drosophila. PLoS One 7:e47538 doi:10.1371/journal.pone.0047538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariconti M, Epis S, Sacchi L, Biggiogera M, Sassera D, Genchi M, Alberti E, Montagna M, Bandi C, Bazzocchi C. 2012. A study on the presence of flagella in the order Rickettsiales: the case of ‘Candidatus Midichloria mitochondrii.' Microbiology 158:1677–1683 [DOI] [PubMed] [Google Scholar]
- 23.Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP. 2006. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55:595–609 [DOI] [PubMed] [Google Scholar]
- 24.Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG. 2007. Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 5:e87 doi:10.1371/journal.pbio.0050087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K, Toh H. 2008. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics 9:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526 [DOI] [PubMed] [Google Scholar]
- 31.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 32.Lanave C, Preparata G, Saccone C, Serio G. 1984. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20:86–93 [DOI] [PubMed] [Google Scholar]
- 33.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 34.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Skjaeveland A, Orr RJ, Enger P, Ruden T, Mevik BH, Burki F, Botnen A, Shalchian-Tabrizi K. 2009. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics 10:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barraclough TG, Hughes M, Ashford-Hodges N, Fujisawa T. 2009. Inferring evolutionarily significant units of bacterial diversity from broad environmental surveys of single-locus data. Biol. Lett. 5:425–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution, divergence times in the absence of a molecular clock. Bioinformatics 19:301–302 [DOI] [PubMed] [Google Scholar]
- 39.Gower J. 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338 [Google Scholar]
- 40.Sneath PHA, Sokal RR. 1973. Numerical taxonomy, the principles and practice of numerical classification. W. H. Freeman and Company, San Francisco, CA [Google Scholar]
- 41.Kovach WL. 1999. M.V.S.P. A multi-variate statistical package for Windows, version 3.1. Services KC, Pentraeth, Wales [Google Scholar]
- 42.Weir BS. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370 [DOI] [PubMed] [Google Scholar]
- 43.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wier BS. 1996. Genetic data analysis II: methods for discrete population Genetic Data. Sinauer Associates, Sunderland, MA [Google Scholar]
- 45.Clarke K. 1993. Non-parametric multivariate analysis of changes in community structure. Aust. J. Ecol. 18:117–143 [Google Scholar]
- 46.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 6:927–930 [Google Scholar]
- 47.Williams KP, Sobral BW, Dickerman AW. 2007. A robust species tree for the alphaproteobacteria. J. Bacteriol. 189:4578–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bordenstein SR, Paraskevopoulos C, Dunning Hotopp JC, Sapountzis P, Lo N, Bandi C, Tettelin H, Werren JH, Bourtzis K. 2009. Parasitism and mutualism in Wolbachia: what the phylogenomic trees can and cannot say. Mol. Biol. Evol. 26:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barker J, Brown MR. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253–1259 [DOI] [PubMed] [Google Scholar]
- 50.Brown M, Barker J. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7:46–50 [DOI] [PubMed] [Google Scholar]
- 51.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kikuchi Y, Sameshima S, Kitade O, Kojima J, Fukatsu T. 2002. Novel clade of Rickettsia spp. from leeches. Appl. Environ. Microbiol. 68:999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vannini C, Petroni G, Verni F, Rosati G. 2005. A bacterium belonging to the Rickettsiaceae family inhabits the cytoplasm of the marine ciliate Diophrys appendiculata (Ciliophora, Hypotrichia). Microb. Ecol. 49:434–442 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.