Abstract
In this study, we achieved the efficient synthesis of 2-O-d-glucopyranosyl-l-ascorbic acid (AA-2G) from soluble starch by fusing a carbohydrate-binding module (CBM) from Alkalimonas amylolytica α-amylase (CBMAmy) to cyclodextrin glycosyltransferase (CGTase) from Paenibacillus macerans. One fusion enzyme, CGT-CBMAmy, was constructed by fusing the CBMAmy to the C-terminal region of CGTase, and the other fusion enzyme, CGTΔE-CBMAmy, was obtained by replacing the E domain of CGTase with CBMAmy. The two fusion enzymes were then used to synthesize AA-2G from soluble starch as a cheap and easily soluble glycosyl donor. Under the optimal conditions, the AA-2G yields produced using CGTΔE-CBMAmy and CGT-CBMAmy were 2.01 g/liter and 3.03 g/liter, respectively, which were 3.94- and 5.94-fold of the yield from the wild-type CGTase (0.51 g/liter). The reaction kinetics of the two fusion enzymes were analyzed and modeled to confirm the enhanced specificity toward soluble starch. It was also found that, compared to the wild-type CGTase, the two fusion enzymes had relatively high hydrolysis and disproportionation activities, factors that favor AA-2G synthesis. Finally, it was speculated that the enhancement of soluble starch specificity may be related to the changes of substrate binding ability and the substrate binding sites between the CBM and the starch granule.
INTRODUCTION
l-Ascorbic acid (l-AA), which is also called vitamin C, plays an important role in many biological processes, such as collagen formation, carnitine synthesis, iron absorption, and chronic disease prevention (1–4). However, its applications are extremely limited because of its instability in aqueous solution, especially in the presence of heat, light, Cu2+, and ascorbate oxidase (5–8). Therefore, many l-AA derivatives have been manufactured to improve the stability of l-AA by chemical synthesis or biotransformation (9). Among all of the l-AA derivatives, 2-O-d-glucopyranosyl-l-ascorbic acid (AA-2G) is considered to be a superior substitute for l-AA because of its excellent properties, such as high nonreducibility, excellent antioxidant ability, and effortless release of l-AA and glucose (7). In addition to being stable in vitro and highly active in vivo, AA-2G exhibits lipophilicity and may be used as a component in cosmetics (9).
AA-2G is mainly produced by enzymatic transformation, and the commonly used enzyme is cyclodextrin glycosyltransferase (CGTase) (EC 2.4.1.19), which can transfer the glycosyl residue from a glycosyl donor to the C-2 position of l-AA with an α-1,2-linkage (9). Many saccharides, such as α- and β-cyclodextrins, maltodextrin, and starch, can be used as glycosyl donors (10). It has been demonstrated that α- and β-cyclodextrins are the best glycosyl donors for AA-2G production (10–12). However, because of the high cost of α-cyclodextrin and the low solubility of β-cyclodextrin in aqueous solution, neither is suitable for the large-scale production of AA-2G (9). Therefore, the selection of a new, inexpensive, and highly soluble glycosyl donor for AA-2G enzymatic production is attracting increasing interest. In our previous work, the maltodextrin specificity of CGTase from Paenibacillus macerans was enhanced via site saturation mutations of lysine 47, tyrosine 195, tyrosine 260, and glutamine 265; maltodextrin is a cheap and highly soluble glycosyl donor, and the mutations led to improved AA-2G productivity (13, 14). Though extensive work has been conducted, the improvement in AA-2G productivity by this approach is limited, and it is necessary to find other engineering strategies. In this study, we attempted to improve the specificity of CGTase toward soluble starch by fusion protein engineering, using soluble starch as the glycosyl donor for AA-2G synthesis.
The microbial hydrolytic enzymes that catalyze the degradation of starch granules typically possess a modular architecture and often contain noncatalytic ancillary domains referred to as carbohydrate-binding modules (CBMs). The CBMs target specific polysaccharide structures (15). A CBM is defined as a contiguous amino acid sequence from a carbohydrate-active enzyme that folds as a separate domain and shows carbohydrate-binding ability (15). CBMs have been categorized into 54 different families based on amino acid sequence similarities (http://www.cazy.org/fam/accCBM.html), and the members of each family share a structural fold (16). CGTases and various starch hydrolases belong to the CBM20 family, whose common feature is that they are joined to catalytic modules associated with starch or glycogen metabolism. CBM20 members with affinity for starch are commonly referred to as starch-binding domains (SBDs). Interestingly, there is strong evidence to suggest that the SBD of an amylolytic enzyme is an independent domain and maintains its original conformation even when it is separated from the catalytic center (17). Based on the three-dimensional structures (18–20), the basic framework of the amylolytic enzymes consists of three common domains: A, B, and C. Domain A possesses a catalytic core (β/α)8 structure, followed by a domain consisting of β-strands folded in a Greek-key motif (domain C). Domain B is inserted between the third β-strand and the third α-helix of a (β/α)8-barrel. Two additional domains, D and E, subsequent to the C-terminal region of domain C, are present in CGTases, and the E domain is involved in the binding of granular starch (19). Domain E of CGTase is an independent domain and retains starch-binding activity when separated from the other four domains (A to D) in CGTase (21).
CBMs usually have independent structures and specific functions. Therefore, many CBMs are used with other enzymes as fusion proteins to improve properties, such as catalytic efficiency and substrate specificity. For instance, Mamo et al. fused the CBM from Thermotoga neapolitana with a family 10 xylanase from Bacillus halodurans S7 to enhance the hydrolytic efficiency on insoluble xylan (22). Kittur et al. created a chimeric xylanase by fusing a family 2b CBM from Streptomyces thermoviolaceus STX-II to the carboxyl terminus of XynB, a thermostable and single-domain family 10 xylanase from Thermotoga maritima, to increase its catalytic activity (23). Zhang et al. fused the CBMs from Thermobifida fusca cellulose Cel6A and Cellulomonas fimi cellulose CenA to the carboxyl terminus of T. fusca cutinase to improve scouring efficiency (24).
A recent study indicates that the C-terminal region located downstream from the topological domain C of the α-amylase exhibits high similarity to the E domain of the bacterial CGTases involved in binding starch (25). Therefore, in this work, aimed at improving the binding capacity of CGTase to soluble starch, two fusion enzymes were constructed by fusing the CBM from alkaliphilic Alkalimonas amylolytica α-amylase (CBMAmy) to the carboxyl terminus (CGT-CBMAmy) or replacing the E domain of P. macerans CGTase with CBMAmy (CGTΔE-CBMAmy). In addition, the reaction kinetics of the two fusion enzymes were investigated to confirm the improvement in soluble starch specificity, and the influence of fusion on the cyclization, disproportionation, and hydrolysis activities of CGTase was explored. Finally, we speculated on the possible mechanism responsible for the enhanced soluble starch specificity of the fusion enzymes. This is the first report on enhancing the substrate specificity toward soluble starch by fusing CBMAmy with CGTase. The fused CGTases have great potential for the industrial production of AA-2G with soluble starch as a cheap and highly soluble glycosyl donor.
MATERIALS AND METHODS
Bacterial strains, plasmids, and materials.
Plasmids α-amylase/pET-22b(+) and cgt/pET-20b(+) were constructed in our previous work (13, 26). Plasmid α-amylase/pET-22b(+) was used as the gene source of CBM from alkaliphilic A. amylolytica N10 strain α-amylase (CBMAmy), and plasmid cgt/pET-20b(+) was used as the gene source of CGTase from the P. macerans strain. Escherichia coli JM109 was used as the host for plasmid construction, E. coli BL 21(DE3) was used as the expression host, and pET-20b(+) was used as the cloning and expression vector.
PrimeSTAR HS DNA polymerase, restriction endonucleases, and PCR reagents were purchased from TakaRa (Dalian, China). The competent cells of E. coli JM109 and E. coli BL21(DE3) were purchased from TakaRa (Dalian, China). DNA sequencing was performed by Sangon (Shanghai, China). AA-2G was purchased from Wako Pure Chemical Industries, Ltd. (Wako, Japan), and l-AA was purchased from Jiangshan Pharmaceutical (Jiangsu, China).
Construction of vectors for expression of the CGTase-CBM fusion protein.
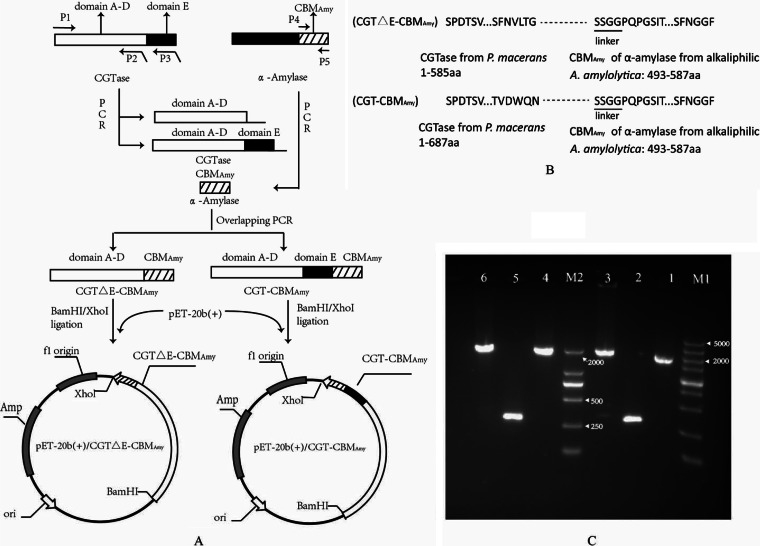
The genes encoding CGTase (NCBI accession no. AF047363) and CBMAmy (NCBI accession no. AY268953) were amplified using plasmids cgt/pET-20b(+) and α-amylase/pET-22b(+) as the templates, respectively. Overlapping PCR was used to fuse CBMAmy to the C terminus of CGTase and to replace domain E of CGTase with CBMAmy. The sequences of the primers are listed in Table 1. The fusion procedure is shown in Fig. 1A, and the amino acid composition of the fusion proteins is shown in Fig. 1B. A BamHI restriction site was introduced at the 5′ end of primer P1, and an XhoI restriction site was introduced at the 3′ end of primer P5. The primers were synthesized by Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China).
Table 1.
Oligonucleotide primers used for the construction of the fusion enzymes
| Primer | Sequence (5′→3′)a |
|---|---|
| P1 | CGGGATCCGTCACCCGATACGAGCGTGGACA |
| P2 | CGGACCACCACTGGACCCCGTCAGTACATTGAAGCTTTTG |
| P3 | CGGACCACCACTGGAATTTTGCCAGTCCACCGTCACC |
| P4 | TCCAGTGGTGGTCCGCAGC |
| P5 | CCGCTCGAGGCCACCATTAAAACTGGGATGC |
The underlined letters denote the coding sequences of the restriction endonuclease sites; 15-bp segments of the overlapping regions are in boldface and underlined.
Fig 1.
Construction of the fusion enzymes CGTΔE-CBMAmy and CGT-CBMAmy. (A) The carbohydrate-binding module (CBM) from alkaliphilic A. amylolytica α-amylase (CBMAmy) was fused to the carboxyl terminus of P. macerans cyclodextrin glycosyltransferase (CGTase) or was used to replace the E domain of P. macerans CGTase using overlapping extension PCR. (B) Amino acid composition of the fusion proteins. (C) Agarose gel electrophoresis of the overlapping extension PCR. M1, DL5000 DNA marker; M2, DL2000 DNA marker; lane 1, the gene fragment of domains A to D of CGTase; lanes 2 and 5, the gene fragment of CBMAmy; lane 3, the gene fragment of the fusion gene coding for CGTΔE-CBMAmy; lane 4, the gene fragment of CGTase; lane 6, the gene fragment of the fusion gene coding for CGT-CBMAmy.
The gel-purified PCR product was digested with BamHI and XhoI and then ligated into the similarly restriction-digested expression vector pET-20b(+). The ligation mixture was used to transform chemically competent E. coli JM109. The plasmid isolated from these transformants was confirmed by DNA sequencing. The plasmids with the correct sequences for CGTΔE-CBMAmy and CGT-CBMAmy were named pET-20b(+)/CGTΔE-CBMAmy and pET-20b(+)/CGT-CBMAmy, respectively. The plasmids were then transformed into E. coli BL21(DE3) for expression.
Preparation and purification of CGTase and the fusion enzymes.
The wild-type CGTase and the fusion enzymes CGTΔE-CBMAmy and CGT-CBMAmy were prepared as previously reported (27). The recombinant E. coli BL21(DE3) was inoculated into 20 ml Luria-Bertani (LB) medium containing 100 μg/ml ampicillin and grown at 37°C overnight. Then the seed culture was inoculated into the flask at a ratio of 4% (vol/vol) for fermentation. The fermentation medium had a pH of 7.0 and contained the following (g/liter): glucose, 8; lactose, 0.5; peptone, 12; yeast extract, 24; K2HPO4, 16.43; KH2PO4, 2.31; CaCl2, 0.28; glycerol, 4; and ampicillin, 0.1. The flask culture was incubated on a rotary shaker (200 rpm) at 25°C for 90 h. The expression of CGTases was induced with 0.01 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) when the optical density at 600 nm (OD600) reached 0.6. The broth was centrifuged at 10, 000 × g and 4°C for 5 min, and the supernatant was purified and used for the subsequent transformation. The purification of the crude enzyme solution was carried out using a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen, Chatsworth, CA) as described in reference 28.
Biosynthesis and analysis of AA-2G.
The purified wild-type and fusion CGTases were diluted with 50 mM phosphate buffer (pH 6.5) to a protein concentration of 1 mg/ml and were then mixed with the substrates (l-AA and soluble starch), the final concentration of which was 0.5% (wt/vol). The mixture was incubated at 37°C for 24 h under dark and oxygen-free conditions. The glucoamylase (10 U/ml) was finally added to the reaction mixture and incubated at 65°C and pH 5.5 for 24 h to hydrolyze the reaction intermediate AA-2Gn (where “n” represents the number of glycosyls attached to l-AA) to AA-2G. AA-2G was analyzed using the method described previously (10). On the basis of the initial transformation conditions (temperature, 37°C; pH 5.5), the effects of reaction temperature (20, 28, 36, 44, and 52°C) and pH (acetic acid-sodium acetate buffer, pH 4.0, 4.5, 5.0, 5.5, and 6.0; phosphate buffer, pH 6.0, 6.5, 7.0, and 8.0) on the biosynthesis of AA-2G by the wild-type CGTase and fusion enzymes were also investigated.
The kinetic analysis of the wild-type and mutant CGTases for AA-2G biosynthesis (using l-AA and soluble starch as the substrates) was performed by measuring the amount of AA-2G produced using different concentrations of soluble starch (0.1, 0.5, 1, 2, and 4 g/liter) while fixing the concentration of l-AA (0.1, 0.5, 1, 2, and 4 g/liter), and the obtained results were subjected to kinetic analysis using SigmaPlot (Jandel Scientific). The equations presented below were used to fit the experimental data to determine which kinetic mechanism applies to the transglycosylation reactions catalyzed by CGTase.
The “ping-pong” mechanism is represented by equation 1
| (1) |
and the substrate inhibition mechanism is represented by equation 2
| (2) |
where v is the reaction rate [the amount of AA-2G formed by 1 mg of enzyme per h, (g/liter AA-2G)/(h · mg enzyme)], Vmax is the maximal reaction rate [(g/liter AA-2G)/(h · mg enzyme)], a and b are the donor (soluble starch) and acceptor (l-AA) concentrations (g/liter), respectively, KmA and KmB are the affinity constants for the substrates l-AA and soluble starch, respectively, and KiB is the inhibitor constant for the substrate soluble starch.
Analysis of CGTase activity.
The α-cyclodextrin-forming activity was determined using the methyl orange method described by Li et al. (27). Namely, 0.1 ml of the purified wild-type or mutant CGTase diluted with 50 mM phosphate buffer to 1 mg/ml was added into 0.9 ml of 3% (wt/vol) soluble starch in 50 mM phosphate buffer (pH 6.0). After incubation at 40°C for 10 min, the reaction was terminated by the addition of 1.0 M HCl (1.0 ml). Finally, 1.0 ml of 0.1 mM methyl orange in 50 mM phosphate buffer (pH 6.0) was added, and the absorbance at 505 nm was measured after incubation at 16°C for 20 min. One unit of α-cyclodextrin-forming activity was defined as the amount of enzyme that was able to produce 1 μmol of α-cyclodextrin per minute. The hydrolyzing activity was analyzed by the starch-degrading method (29). The purified CGTase (1 mg/ml) was incubated with 1% (wt/vol) soluble starch solution in 50 mM phosphate buffer at pH 6.5 and 50°C for 10 min. One unit of hydrolyzing activity was defined as the amount of enzyme producing 1 μmol of reducing sugar (determined by the 3,5-dinitrosalicylic acid method) per minute. The disproportionation activity was determined as described previously (29), and the reaction mixture contained 6 mM 4-nitrophenyl-α-d-maltoheptaoside-4-6-O-ethylidene (EPS) (Megazyme, County Wicklow, Ireland) as the donor substrate, 10 mM maltose as the acceptor substrate, and 1 mg/ml purified CGTase in 10 mM citrate buffer (pH 6.0). The mixtures were incubated at 50°C for 10 min. One unit of activity was defined as the amount of enzyme converting 1 μmol of EPS per minute. Protein concentrations were determined using the Bradford method with a Bradford protein assay kit and bovine serum albumin as a standard (Beyotime, Jiangsu, China).
Structure modeling of the wild-type CGTase and fusion enzymes and molecular docking with malt disaccharide.
The homology models of the wild-type and mutant CGTases from P. macerans were constructed using the crystallographic structure of B. circulans strain 251 CGTase (Protein Data Bank [PDB] accession code 1CDG) (19) as the template by the program suite MODELLER 9v2 (30). The models had 67.9% identity to the template. All graphical molecular representations were generated using Accelrys Discovery Studio Client 2.5. Structural alignment was done according to the combinatorial extension method by using the server http://cl.sdsc.edu/ (31). The stereochemical quality of the model was examined with PROCHECK (12), Verify3D (3), and ProQ (30), having 93% of the residues in the most favored regions and 0.5% in disallowed regions. The root mean square deviation (RMSD) between the template and model alpha carbon backbones was calculated using the combinatorial extension method (31). The overall structures of the model and template were very similar, with an RMSD of 0.7 Å. Molecular docking was performed between the obtained three-dimensional structures of the CBMs and the malt disaccharide molecular structure (http://davapc1.bioch.dundee.ac.uk/prodrg/) using the Autodock 4.2 software with a genetic algorithm.
RESULTS AND DISCUSSION
Construction and purification of the fusion enzymes.
Two fusion enzymes, CGT-CBMAmy and CGTΔE-CBMAmy, were constructed by either introducing the CBM of alkaliphilic A. amylolytica α-amylase into the C-terminal region of P. macerans CGTase or replacing its E domain through overlapping PCR amplification. As shown in Fig. 1C, the genes of CGT-CBMAmy and CGTΔE-CBMAmy were 2,346 and 2,040 bp, respectively. These fused genes were subsequently inserted into the expression vector pET-20b(+), which encodes a C-terminal His6 tag and an N-terminal signal peptide PelB to allow the section of expressed proteins. The resulting constructs, pET-20b(+)/CGT-CBMAmy and pET-20b(+)/CGTΔE-CBMAmy, were used for protein expression in E. coil BL21(DE3). Both fusion enzymes were purified by Ni-Sepharose affinity chromatography. SDS-PAGE results demonstrated that their molecular masses were about 84 and 72 kDa for CGT-CBMAmy and CGTΔE-CBMAmy, respectively (data not shown).
Synthesis of AA-2G by the wild-type CGTase and fusion enzymes.
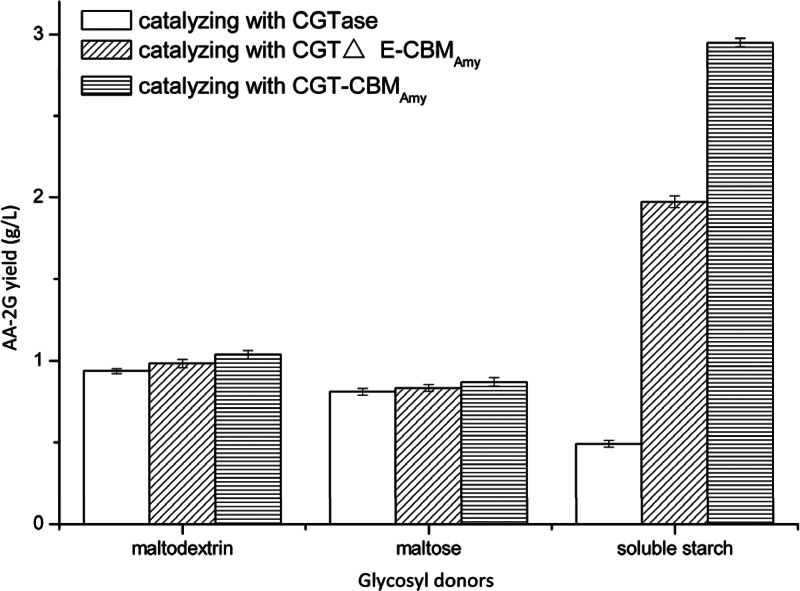
Maltodextrin, maltose, and soluble starch were used as the glycosyl donors for the production of AA-2G by the wild-type CGTase and the fusion enzymes CGT-CBMAmy and CGTΔE-CBMAmy. As shown in Fig. 2, when using maltodextrin, maltose, and soluble starch as the glycosyl donors, the AA-2G yields produced by the wild-type CGTase were 0.94, 0.81, and 0.49 g/liter, respectively. The AA-2G yield produced by both of the fusion enzymes was almost the same as that produced by the wild-type CGTase when using maltodextrin and maltose as the glycosyl donors. However, when using soluble starch as the substrate, the AA-2G yields produced by the fusion enzymes CGT-CBMAmy and CGTΔE-CBMAmy were 5.94- and 3.94-fold of that produced by the wild-type CGTase, respectively.
Fig 2.
Comparison of the AA-2G yields of the wild-type CGTase and the fusion enzymes with maltodextrin, maltose, and soluble starch as the glycosyl donors. The reaction temperature and pH were 37°C and 5.5, respectively.
Influence of reaction temperature and pH on the enzymatic synthesis of AA-2G.
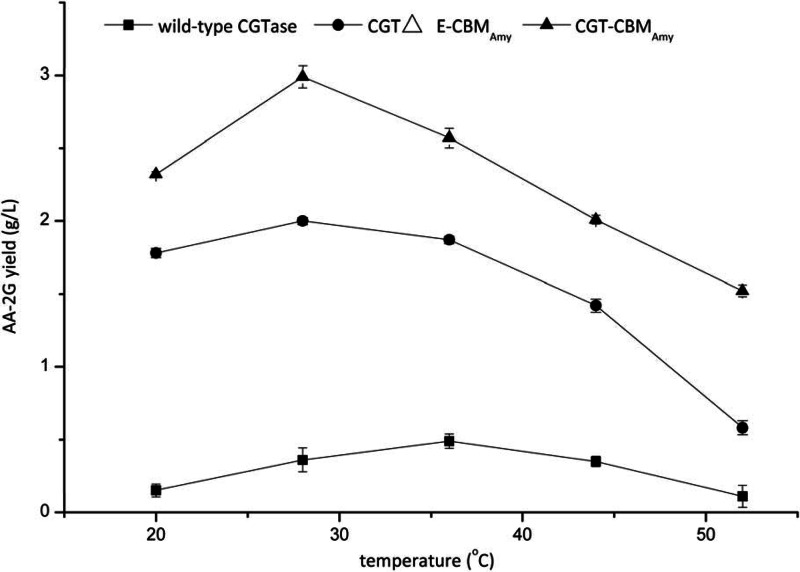
The influence of temperature on AA-2G synthesis by the wild-type and fusion enzymes was investigated. As shown in Fig. 3, when using soluble starch as the substrate, the AA-2G yield produced by the wild-type CGTase was greatest at 36°C, which was the same when using maltodextrin (13, 14) and β-cyclodextrin as the glycosyl donors (10). However, for the fusion enzymes, CGT-CBMAmy and CGTΔE-CBMAmy, the optimal temperature for AA-2G synthesis was 28°C.
Fig 3.
Influence of reaction temperature on AA-2G synthesis by the wild-type CGTase and fusion enzymes with soluble starch as the glycosyl donor.
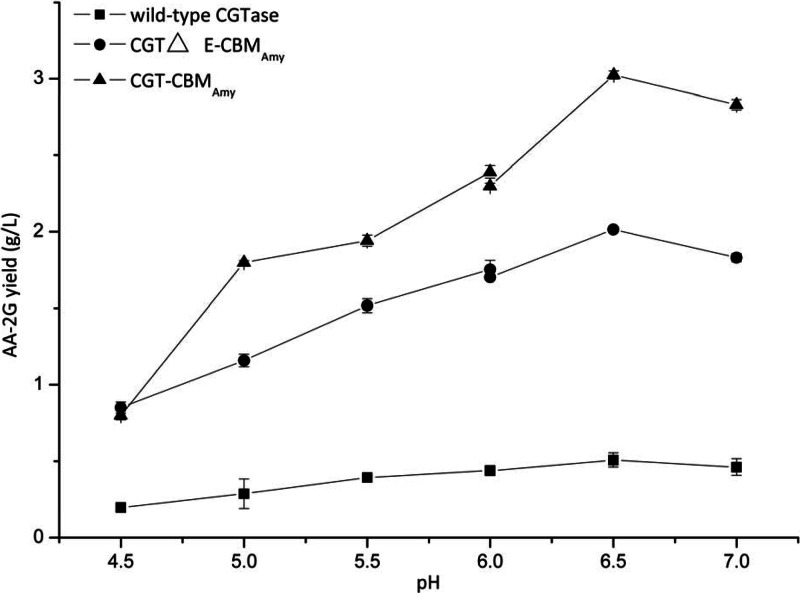
Figure 4 shows the influence of pH on AA-2G synthesis with soluble starch as the glycosyl donor by the wild-type and fusion enzymes. It can be seen that the optimal pH of the wild-type CGTase and fusion enzymes CGT-CBMAmy and CGTΔE-CBMAmy was 6.5, which was also the optimal pH for AA-2G synthesis with maltodextrin as the glycosyl donor (13, 14). However, this differed from the optimal pH of AA-2G biosynthesis using β-cyclodextrin as the glycosyl donor, which was 5.5 (10).
Fig 4.
Influence of reaction pH on AA-2G synthesis by the wild-type CGTase and fusion enzymes with soluble starch as the glycosyl donor (acetic acid-sodium acetate buffer, pH 4.0, 4.5, 5.0, 5.5, and 6.0; phosphate buffer, pH 6.0, 6.5, 7.0, and 8.0).
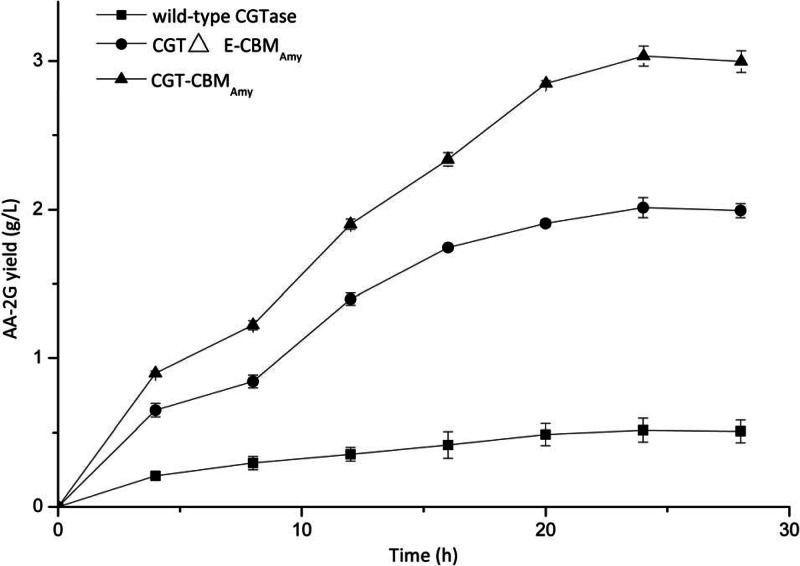
Figure 5 shows the time profiles of AA-2G synthesis using the wild-type and fusion enzymes under the optimal conditions. At the initial stage of the reaction, the amount of AA-2G increased dramatically. At 24 h, the AA-2G yields produced by the wild-type CGTase, CGT-CBMAmy, and CGTΔE-CBMAmy reached their greatest values: 0.51, 2.01, and 3.03 g/liter, respectively.
Fig 5.
Time profiles of AA-2G synthesis by the wild-type CGTase and fusion enzymes with l-AA and soluble starch as the substrates. The reaction conditions were optimal for each enzyme: wild-type CGTase (temperature, 37°C; pH 6.5); CGTΔE-CBMAmy and CGT-CBMAmy (temperature, 28°C; pH 6.5).
Analysis and modeling of reaction kinetics of wild-type CGTase and fusion CGTases.
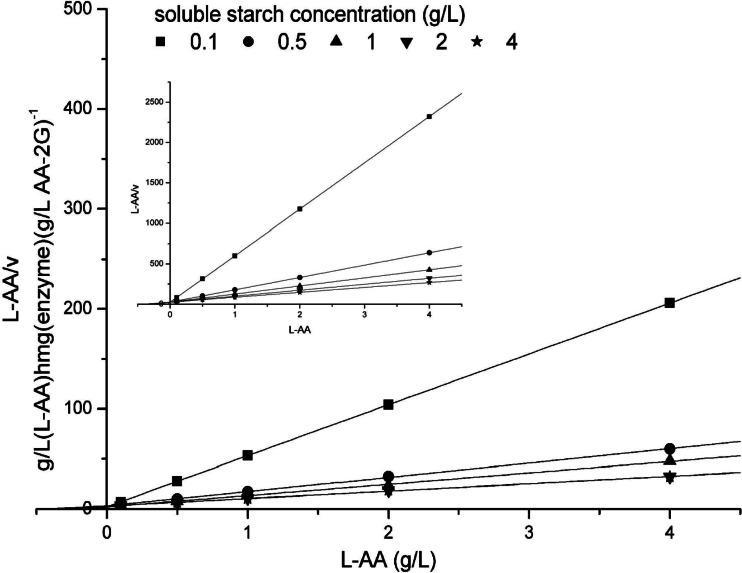
The Hanes-Woolf plot was used to model the reaction kinetics of the fusion enzyme CGT-CBMAmy (Fig. 6). The experimental data corresponded well to the values calculated using equation 2, which represented a substrate inhibition mechanism, indicating that a too-high concentration of soluble starch inhibited the fusion enzyme CGT-CBMAmy activity for AA-2G synthesis. For the wild-type CGTase, the experimental data (Fig. 6, inset) corresponded well to the calculated values by equation 1, which indicated a normal ping-pong type of kinetics. This was the same with the reaction kinetics of the wild-type CGTase with maltodextrin as the glycosyl donor (13). In addition, the experimental data for the fusion enzyme CGTΔE-CBMAmy fitted best to the values calculated using equation 2 (data not shown), and the detailed kinetic parameters are listed in Table 2. As can be seen, the maximal reaction rates (Vmax) of the fusion enzymes were higher than the rate of the wild-type CGTase. Compared to the wild-type CGTase, the Km of the fusion enzymes CGTΔE-CBMAmy and CGT-CBMAmy with soluble starch as the substrate decreased by 12.9 and 30.3%, respectively, while the kcat/Km increased by 3.87- and 8.23-fold, respectively. The Km for the substrate l-AA of the fusion enzymes decreased by 8 and 22.5%, while their catalytic efficiencies increased by 3.62- and 7.33-fold, respectively, compared to that of the wild-type CGTase. The kinetic results indicated that the affinities and catalytic efficiencies of both fusion enzymes toward soluble starch and l-AA increased compared to those of the wild-type CGTase. The inhibitor constant Ki (soluble starch) values indicated that the substrate inhibition by soluble starch was most significant for the fusion enzyme CGTΔE-CBMAmy.
Fig 6.
Hanes-Woolf plots of the AA-2G biosynthesis by the wild-type CGTase (insert) and fusion enzyme CGT-CBMAmy.
Table 2.
Kinetic parameters of the wild-type CGTase and two fusion enzymesa
| Enzyme | Vmax (g · liter−1 AA-2G/h · mg enzyme) | kcat (h−1) |
l-AA |
Soluble starch |
|||
|---|---|---|---|---|---|---|---|
| Km (g/liter) | kcat/Km (liters/g · h) | Km (g/liter) | kcat/Km (liters/g · h) | Ki (g/liter) | |||
| Wild type | 0.021 ± 0.016 | 1.575 | 0.545 ± 0.022 | 2.88 | 1.107 ± 0.029 | 1.422 | NDb |
| CGTΔE-CBMAmy | 0.089 ± 0.009 | 6.675 | 0.501 ± 0.015 | 13.32 | 0.964 ± 0.022 | 6.924 | 18.3 |
| CGT-CBMAmy | 0.135 ± 0.010 | 10.125 | 0.422 ± 0.018 | 23.99 | 0.771 ± 0.021 | 13.132 | 23.1 |
Each value represents the mean of three independent measurements, and the deviations from the mean were below 5%.
ND, not detectable.
The cyclization, hydrolysis, and disproportionation activities of the fusion enzymes were investigated. As shown in Table 3, both of the fusion enzymes nearly lost the ability of cyclization (α-cyclodextrin-forming activity). Compared with the wild-type CGTase, the fusion enzymes showed hydrolysis (starch-degrading) activity increases of 0.86- and 1.36-fold, respectively. Also, the disproportionation activities of the fusion enzymes increased by nearly 2- and 3-fold, respectively.
Table 3.
Comparison of the reaction activities, AA-2G yields, and stabilities of the wild-type and fusion enzymes
| Enzyme | Relative activity (%)a |
||
|---|---|---|---|
| Cyclization (α-cyclodextrin-forming activity) | Hydrolysis (starch-degrading activity) | Disproportionation | |
| Wild type | 100 | 100 | 100 |
| CGTΔE-CBM>Amy | NDb | 186 ± 10 | 198.0 ± 1.0 |
| CGT-CBMAmy | ND | 236 ± 6 | 306.7 ± 0.8 |
The cyclization, hydrolysis, and disproportionation reaction activities for the wild-type CGTase were 165 ± 5, 8.3 ± 0.1, and 806 ± 8 U/mg, respectively, and were defined as 100% of the relative activity. Each value represents the mean of triple independent measurements, and the deviations from the mean were below 5%.
ND, not detectable.
The enzymatic synthesis of AA-2G by CGTase with soluble starch as the glycosyl donor may comprise two reactions: the long-chain starch is hydrolyzed to oligosaccharides, and the glycosyl from the oligosaccharides is transferred to the acceptor l-AA by disproportionation. The change trends for the hydrolysis and disproportionation activity of the fusion enzymes were in accordance with the increase in AA-2G yield, indicating that the hydrolysis and disproportionation of fusion enzymes might be the main reaction for AA-2G biosynthesis with soluble starch as the glycosyl donor. However, for the oligosaccharides such as maltose and maltodextrin, the disproportionation might be the main reaction. The low binding ability between the oligosaccharides and the CBM region of enzymes may lead to the low AA-2G titers with maltose and maltodextrin as glycosyl donors (Fig. 2).
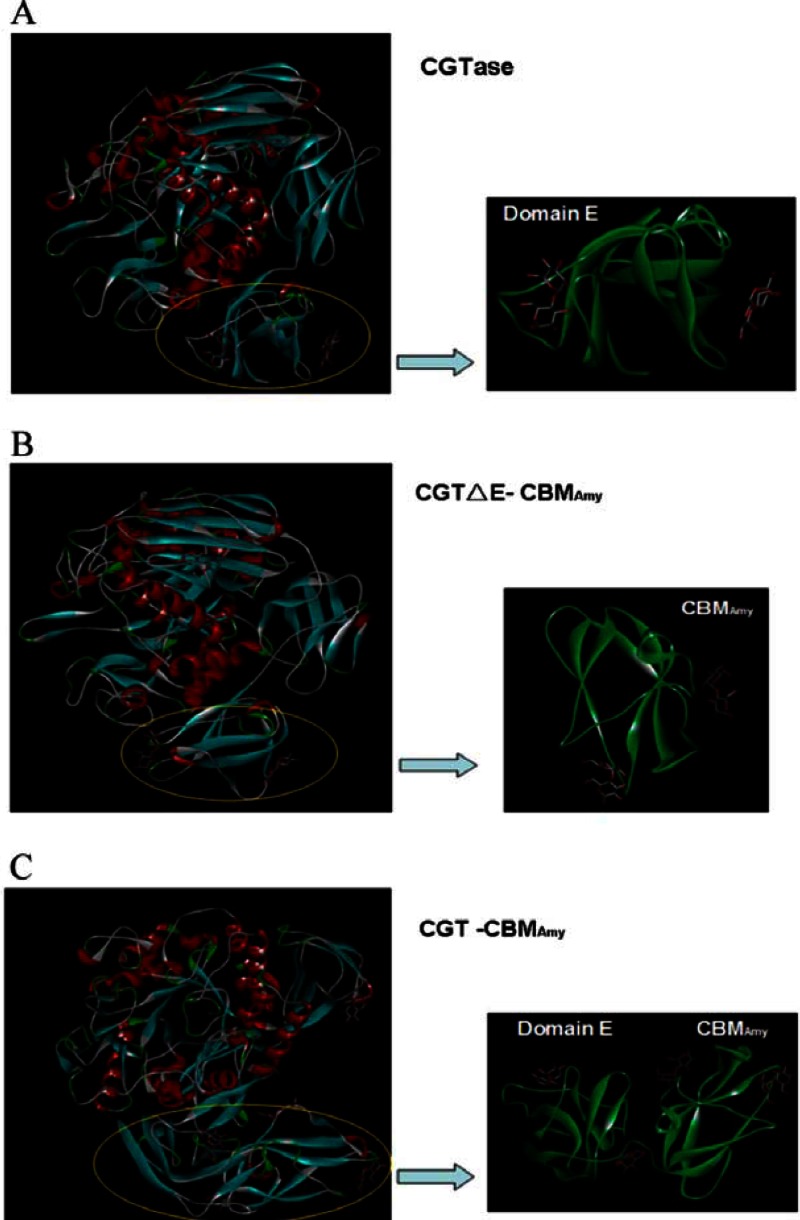
In this work, we found that when the single E domain of P. macerans CGTase was replaced by the CBM of alkaliphilic A. amylolytica α-amylase (CGTΔE-CBMAmy), the AA-2G yield with soluble starch as the glycosyl donor increased dramatically. In contrast, the AA-2G yields with maltodextrin and maltose as glycosyl donors did not change significantly (Fig. 2). This might reveal that the binding ability between the CBM of alkaliphilic A. amylolytica α-amylase and starch granules was stronger than with the native domain E of CGTase. As shown in Fig. 7A and B, the binding free energy of molecular docking of the wild-type CGTase with the malt disaccharide (−1.93 kJ/mol) was higher than that of the fusion enzyme CGTΔE-CBMAmy with the malt disaccharide (−3.99 kJ/mol), which may be additional evidence for the explanation above. Interestingly, it seems that the fusion of CBM to the C-terminal region of the P. macerans CGTase (CGT-CBMAmy) was more efficient for AA-2G synthesis than the replacement of the E domain with CBM when soluble starch was the glycosyl donor. The most likely explanation is that the starch granule binding ability was enhanced by the increase of carbohydrate binding sites for the fusion enzyme CGT-CBMAmy, which had four carbohydrate binding sites (Fig. 7C). The decrease in Km and increase in kcat/Km (Table 2) verifies this explanation to some extent.
Fig 7.
Molecular docking of the wild-type CGTase and fusion enzymes with malt disaccharide.
In summary, two fusion enzymes, CGTΔE-CBMAmy and CGT-CBMAmy, were constructed to enhance soluble starch substrate specificity for AA-2G biosynthesis. The enhanced specificity toward soluble starch was confirmed by reaction kinetics. Compared to the commonly used glycosyl donors α- and β-cyclodextrin, soluble starch is preferable as a substrate for AA-2G synthesis because it is inexpensive and water soluble. Therefore, work including system engineering, enzyme immobilization, and transformation optimization would be useful to further improve AA-2G yield with soluble starch as the substrate.
ACKNOWLEDGMENTS
This work was financially supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (111-2-06), the National High Technology Research and Development Program of China (863 Program, 2011AA100905; and 973 Program, 2012CB720802 and 2012CB720806).
Footnotes
Published ahead of print 15 March 2013
REFERENCES
- 1.Englard S, Seifter S. 1986. The biochemical functions of ascorbic acid. Annu. Rev. Nutr. 6:365–406 [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara S, Kakihara H, Sakaguchi K, Imanaka T. 1992. Analysis of mutations in cyclodextrin glucanotransferase from Bacillus stearothermophilus which affect cyclization characteristics and thermostability. J. Bacteriol. 174:7478–7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacob RA, Sotoudeh G. 2002. Vitamin C function and status in chronic disease. Nutr. Clin. Care 5:66–74 [DOI] [PubMed] [Google Scholar]
- 4.Naidu KA. 2003. Vitamin C in human health and disease is still a mystery? An overview. Nutr. J. 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li ZF, Zhang JY, Wang M, Gu ZB, Du GC, Li JK, Wu J, Chen J. 2009. Mutations at subsite-3 in cyclodextrin glycosyltransferase from Paenibacillus macerans enhancing alpha-cyclodextrin specificity. Appl. Microbiol. Biotechnol. 83:483–490 [DOI] [PubMed] [Google Scholar]
- 6.Penninga D, Strokopytov B, Rozeboom HJ, Lawson CL, Dijkstra BW, Bergsma J, Dijkhuizen L. 1995. Site-directed mutations in tyrosine 195 of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 affect activity and product specificity. Biochemistry 34:3368–3376 [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto I, Muto N, Murakami K, Suga S, Yamaguchi H. 1990. l-Ascorbic acid alpha-glucoside formed by regioselective transglucosylation with rat intestinal and rice seed alpha-glucosidases: its improved stability and structure determination. Chem. Pharm. Bull. (Tokyo) 38:3020–3023 [DOI] [PubMed] [Google Scholar]
- 8.Zhang ZC, Li JH, Liu L, Sun J, Hua ZZ, Du GC, Chen JA. 2011. Enzymatic transformation of 2-O-alpha-d-glucopyranosyl-l-ascorbic acid (AA-2G) by immobilized alpha-cyclodextrin glucanotransferase from recombinant Escherichia coli. J. Mol. Catal. B. Enzym. 68:223–229 [Google Scholar]
- 9.Han R, Liu L, Li J, Du G, Chen J. 2012. Functions, applications and production of 2-O-d-glucopyranosyl-l-ascorbic acid. Appl. Microbiol. Biotechnol. 95:313–320 [DOI] [PubMed] [Google Scholar]
- 10.Zhang ZC, Li JH, Liu L, Sun J, Hua ZZ, Du GC, Chen JA. 2011. Enzymatic transformation of 2-O-alpha-d-glucopyranosyl-l-ascorbic acid by alpha-cyclodextrin glucanotransferase from recombinant Escherichia coli. Biotechnol. Bioprocess Eng. 16:107–113 [Google Scholar]
- 11.Jun HK, Bae KM, Kim SK. 2001. Production of 2-O-alpha-d-glucopyranosyl l-ascorbic acid using cyclodextrin glucanotransferase from Paenibacillus sp. Biotechnol. Lett. 23:1793–1797 [Google Scholar]
- 12.Tanaka M, Muto N, Yamamoto I. 1991. Characterization of Bacillus stearothermophilus cyclodextrin glucanotransferase in ascorbic acid 2-O-alpha-glucoside formation. Biochim. Biophys. Acta 1078:127–132 [DOI] [PubMed] [Google Scholar]
- 13.Han R, Liu L, Shin HD, Chen RR, Du G, Chen J. 6 November 2012. Site-saturation engineering of lysine 47 in cyclodextrin glycosyltransferase from Paenibacillus macerans to enhance substrate specificity towards maltodextrin for enzymatic synthesis of 2-O-d-glucopyranosyl-l-ascorbic acid (AA-2G). Appl. Microbiol. Biotechnol. [Epub ahead of print.] doi:10.1007/s00253-012-4514-1. [DOI] [PubMed]
- 14.Han R, Liu L, Shin HD, Chen RR, Du G, Chen J. 2013. Systems engineering of tyrosine 195, tyrosine 260, and glutamine 265 in cyclodextrin glycosyltransferase from Paenibacillus macerans to enhance maltodextrin specificity for 2-O-d-glucopyranosyl-l-ascorbic acid synthesis. Appl. Environ. Microbiol. 79:672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christiansen C, Abou Hachem M, Janeček Š Viksø-Nielsen A, Blennow A, Svensson B. 2009. The carbohydrate-binding module family 20-diversity, structure, and function. FEBS J. 276:5006–5029 [DOI] [PubMed] [Google Scholar]
- 16.Coutinho PM. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p 15–23 In Ohmiya K, Hayashi K, Sakka K, Kobayashi C, Karita S, Kimura T. (ed), Genetics, biochemistry, and ecology of cellulose degradation. Uni Publishers, Tokyo, Japan [Google Scholar]
- 17.Lin LL, Lo HF, Chi MC, Ku KL. 2003. Functional expression of the raw starch-binding domain of Bacillus sp. strain TS-23 α-amylase in recombinant Escherichia coli. Starch 55:197–202 [Google Scholar]
- 18.Matsuura Y, Kusunoki M, Harada W, Kakudo M. 1984. Structure and possible catalytic residues of Taka-amylase A. J. Biochem. 95:697–702 [DOI] [PubMed] [Google Scholar]
- 19.Lawson CL, Van Montfort R, Strokopytov B, Rozeboom HJ, Kalk KH, de Vries GE, Penninga D, Dijkhuizen L, Dijkstra BW. 1994. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J. Mol. Biol. 236:590–600 [DOI] [PubMed] [Google Scholar]
- 20.Kim JS, Cha SS, Kim HJ, Kim TJ, Ha NC, Oh ST, Cho HS, Cho MJ, Kim MJ, Lee HS. 1999. Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. J. Biol. Chem. 274:26279–26286 [DOI] [PubMed] [Google Scholar]
- 21.Dalmia BK, Schütte K, Nikolov ZL. 1995. Domain E of Bacillus macerans cyclodextrin glucanotransferase: an independent starch-binding domain. Biotechnol. Bioeng. 47:575–584 [DOI] [PubMed] [Google Scholar]
- 22.Mamo GR, Hatti-Kaul Mattiasson B. 2007. Fusion of carbohydrate binding modules from Thermotoga neapolitana with a family 10 xylanase from Bacillus halodurans S7. Extremophiles 11:169–177 [DOI] [PubMed] [Google Scholar]
- 23.Kittur FS, Mangala SL, Rus'd AA, Kitaoka M, Tsujibo H, Hayashi K. 2003. Fusion of family 2b carbohydrate-binding module increases the catalytic activity of a xylanase from Thermotoga maritima to soluble xylan. FEBS Lett. 549:147–151 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Chen S, Xu M, Cavaco-Paulo A, Wu J, Chen J. 2010. Characterization of Thermobifida fusca cutinase-carbohydrate-binding module fusion proteins and their potential application in bioscouring. Appl. Environ. Microbiol. 76:6870–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo HF, Lin LL, Chiang WY, Chie MC, Hsu WH, Chang CT. 2002. Deletion analysis of the C-terminal region of the a-amylase of Bacillus sp. strain TS-23. Arch. Microbiol. 178:115–123 [DOI] [PubMed] [Google Scholar]
- 26.Yang H, Liu L, Shin H, Chen RR, Li J, Du G, Chen J. 4 December 2012. Comparative analysis of heterologous expression, biochemical characterization optimal production of an alkaline α-amylase from alkaliphilic Alkalimonas amylolytica in Escherichia coli and Pichia pastoris. Biotechnol. Prog. [Epub ahead of print.] doi:10.1002/btpr.1657. [DOI] [PubMed]
- 27.Li Z, Li B, Gu Z, Du G, Wu J, Chen J. 2010. Extracellular expression and biochemical characterization of alpha-cyclodextrin glycosyltransferase from Paenibacillus macerans. Carbohydr. Res. 345:886–892 [DOI] [PubMed] [Google Scholar]
- 28.Li ZF, Zhang JY, Sun Q, Wang M, Gu ZB, Du GC, Wu J, Chen J. 2009. Mutations of lysine 47 in cyclodextrin glycosyltransferase from Paenibacillus macerans enhance beta-cyclodextrin specificity. J. Agric. Food. Chem. 57:8386–8391 [DOI] [PubMed] [Google Scholar]
- 29.van der Veen BA, Uitdehaag JC, Dijkstra BW, Dijkhuizen L. 2000. The role of arginine 47 in the cyclization and coupling reactions of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 implications for product inhibition and product specificity. Eur. J. Biochem. 267:3432–3441 [DOI] [PubMed] [Google Scholar]
- 30.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 15:5.6.1–5.6.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shindyalov IN, Bourne PE. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11:739–747 [DOI] [PubMed] [Google Scholar]