Abstract
We analyzed 636 patients with diverse myeloproliferative neoplasms or myelodysplastic/myeloproliferative neoplasms for mutations of the Casitas B-cell lymphoma gene (CBLmut) in exons 8 and 9 and performed correlations to other genetic alterations. CBLmut were detected in 63 of 636 (9.9%) of these selected patients. CBLmut were more frequent in myelodysplastic/myeloproliferative neoplasms than myeloproliferative neoplasms (51 of 328, 15.5% vs. 12 of 291, 4.1%; P<0.001). Frequency was 48 of 278 (17.3%) in chronic myelomonocytic leukemia and 3 of 33 (9.1%) in unclassifiable myelodysplastic/myeloproliferative neoplasms. CBLmut was not detected in polycythemia vera, primary myelofibrosis, essential thrombocythemia, or refractory anemia with ring sideroblasts and marked thrombocytosis. CBLmut were underrepresented in JAK2V617F mutated as compared to JAK2V617wt cases (P<0.001), and mutually exclusive of JAK2exon12mut and MPLW515mut. CBLmut were associated with monosomy 7 (P=0.008) and TET2mut (P=0.003). In chronic myelomonocytic leukemia, CBLmut had no significant impact on survival outcomes. Therefore, CBLmut are frequent in chronic myelomonocytic leukemia, absent in classical myeloproliferative neoplasms, and are only exceptionally found in coincidence with JAK-STAT pathway activating mutations.
Keywords: CBL mutations, chronic myelomonocytic leukemia, myeloproliferative neoplasms, prognosis, sequencing
Introduction
The Casitas B-cell lymphoma gene (CBL) (on chromosome 11q23.3) contains several functional domains. One of these, the C-terminal domain, gives rise to the ubiquitin activity site of the Cbl protein. By ubiquitination, the Cbl protein is targeting multiple sites of receptor tyrosine kinases, e.g. PDGFR or FLT3, resulting in negative modulation of tyrosine kinase signaling.1 Mutations in CBL (CBLmut) were first identified due to acquired uniparental disomy (UPD) of 11q in myeloid neoplasms.1-3 These mutations lead to dysregulation of receptor tyrosine kinases and have the potential to transform hematopoietic cells by constitutively activating the FLT3 pathway.4 With regards to the myeloid entities which can be affected by these mutations, Dunbar et al. identified CBLmut in 7 of 12 patients with uniparental disomy (UPD) of 11q in a cohort of 301 patients with different myeloid disorders including MDS, the MDS/MPN overlap category, MPNs, and acute myeloid leukemia (AML).2 Grand et al. found CBLmut in 8% of atypical chronic myeloid leukemia (aCML), 6% of myelofibrosis, and 1% of hypereosinophilic syndrome/chronic eosinophilic leukemia (HES/CEL) cases.3 Beer et al. documented a patient in whom a CBLmut was detectable in megakaryocytes two years before transformation from MPN to AML.5 Very heterogeneous frequencies of CBLmut were reported in chronic myelomonocytic leukemia (CMML) ranging from 5%6 to 22%.7 Detailed analysis in other entities has been scarce. To evaluate the role of CBLmut in diverse MPNs and myelodysplastic/myeloproliferative neoplasms (MDS/MPN), we analyzed CBLmut in a large cohort of 636 adult patients and performed correlation studies with other molecular mutations, karyotypes, and clinical outcomes.
Design and Methods
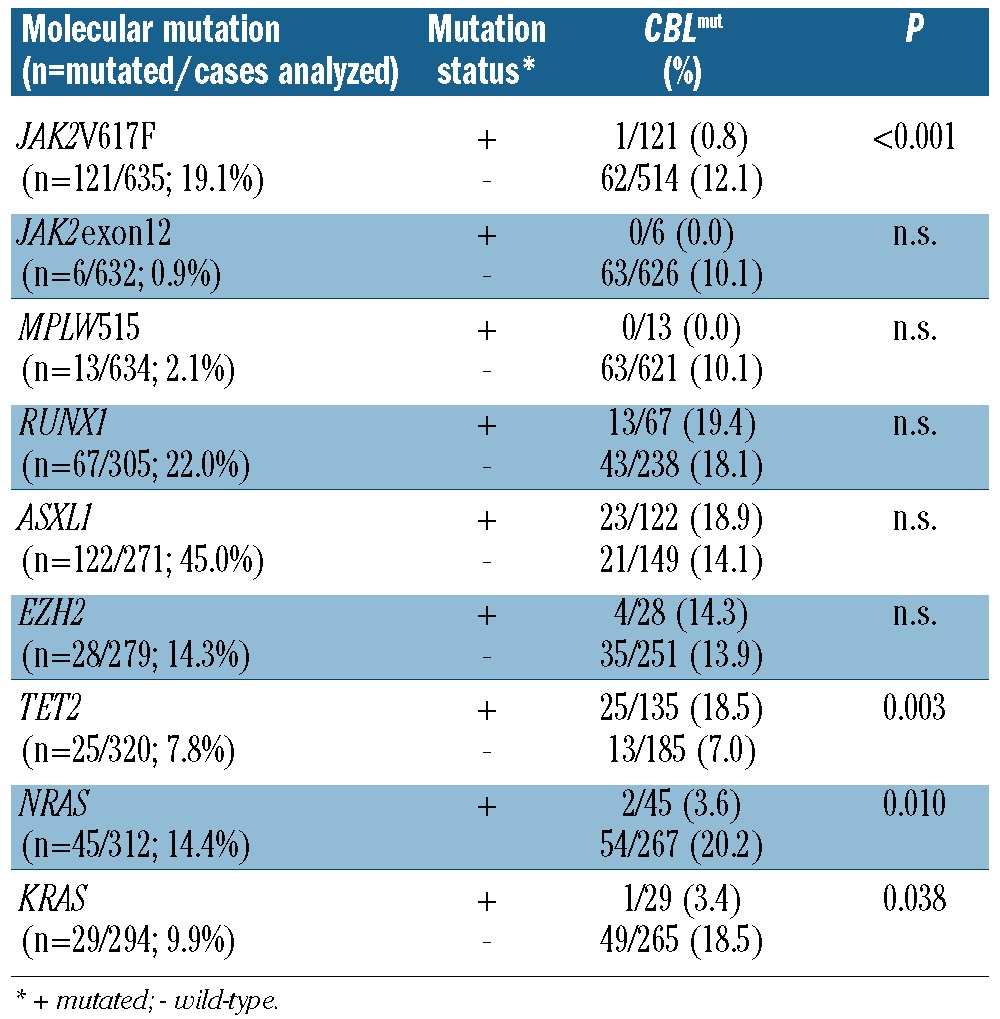
The study cohort was made up of 636 patients: 291 patients had MPNs (polycythemia vera, PV, n=32; essential thrombocythemia, ET, n=48; primary myelofibrosis, PMF, n=19; unclassifiable MPN, n=175; so-called ‘advanced MPN’ (corresponding to an accelerated phase of an MPN or s-AML following a previous MPN n=17). A total of 328 patients had disorders from the WHO overlap category of myelodysplastic/myeloproliferative neoplasms (CMML-1, n=194; CMML-2, n=84; unclassified MDS/MPN, n=33; RARS-T, n=17),8 and 17 patients had HES/CEL. Details of some of the CMML7 (81 of 278) and RARS-T9 patients have been published previously and 4 CMML cases have recently been published elsewhere for clinical and histological analysis.10 Demographic data and blood values are shown in Table 1. Diagnoses were performed according to the WHO.8 There were 389 males and 247 females (male/female ratio 1.6) with a median age of 70.7 years (range 18.4-93.3). Patients were selected according to the availability of cytomorphology, cytogenetics and molecular genetic characterization. Samples were referred to the MLL Munich Leukemia Laboratory in the period from August 2005 to April 2011. Patients gave their written consent. The study was approved by the Internal Review Board of the Munich Leukemia Laboratory in accordance with the Declaration of Helsinki.
Table 1.
CBLmut frequency, demographic data, and peripheral blood values in the different entities.
Bone marrow and/or peripheral blood samples underwent May Grünwald Giemsa staining and cytochemistry with myeloperoxidase (MPO) and non-specific esterase (NSE).11 Chromosome banding analysis was carried out in all 636 cases, combined with fluorescence in situ hybridization (FISH) when necessary.12 Patients were assigned to the following cytogenetic subgroups: normal karyotype, -Y (in male patients), gain of 1q, chromosome 7 abnormalities, trisomy 8 as sole abnormality, 12p deletion, 20q deletion, complex karyotype (defined by ≥3 chromosomal abnormalities), reciprocal translocations, other trisomies, and other alterations (Online Supplementary Table S1).
CBLmut analysis was performed by direct Sanger sequencing covering exons 8-9.3 Mutation loads were estimated visually from electropherograms of forward and reverse reactions as generated by Sanger sequencing and confirmed by pyrosequencing7 in half of the cases with good correlations. In addition, BCR-ABL1 was excluded by multiplex RT (reverse transcription)-PCR in all patients.13 Mutation analysis was carried out in subsets of patients: JAK2V617F (n=635),14JAK2 exon 12 mutations (n=632),15MPL (n=634),16RUNX1 (n=305),17EZH2 (n=279),18TET2 (n=320),7NRAS (n=312),19KRAS (n=294),7 and ASXL1 (n=271; by direct Sanger sequencing of exon 12).20
Overall survival was the interval from the first evaluation of the patient's sample in the Munich Leukemia Laboratory to death. Median overall survival (OS) was calculated according to Kaplan Meier and compared by two-sided log rank test. Dichotomous variables were compared by the χ2 test, continuous variables by Student's t-test. SPSS (version 19.0.0, IBM, Ehningen, Germany) software was used for statistical analysis.
Results and Discussion
In the total cohort, CBLmut were detected in 63 of 636 (9.9%) patients. Localization of the mutations in the LINKER and RING domain (exons 8-9) is shown in Figure 1A. When the different diagnostic entities were compared, CBLmut were more frequent in MDS/MPN than in MPN (51 of 328, 15.5% vs. 12 of 291, 4.1%; P<0.001). In MDS/MPN, the frequency of CBLmut was highest in CMML with 48 of 278 (17.3%) of all cases (CMML-1: 36 of 194, 18.6%; CMML-2: 12 of 84, 14.3%) being followed by MDS/MPNu (3 of 33, 9.1%). No CBLmut was identified in the 17 RARS-T patients. Therefore, the high frequency in MDS/MPN was due to the overrepresentation in CMML. Within the MPN category, the frequency was highest in MPNu (11 of 175, 6.4%) and advanced MPN (1 of 17, 5.9%). No CBLmut was identified in PV (n=32), PMF (n=19), ET (n=48) or HES/CEL (n=17). Taken together, due to their high frequency in CMML, CBLmut showed a higher frequency in the overlap MDS/MPN category as compared to the MPN category, and were not detected within clearly defined entities such as ET, PV, PMF, HES/CEL, or RARS-T.
Figure 1.
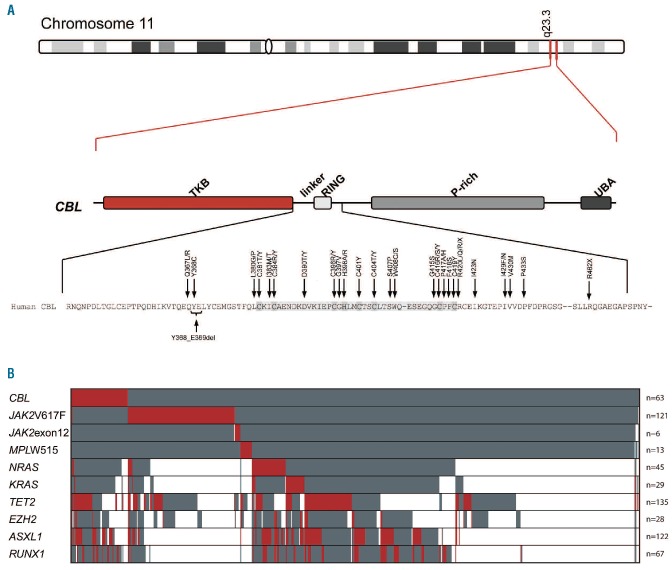
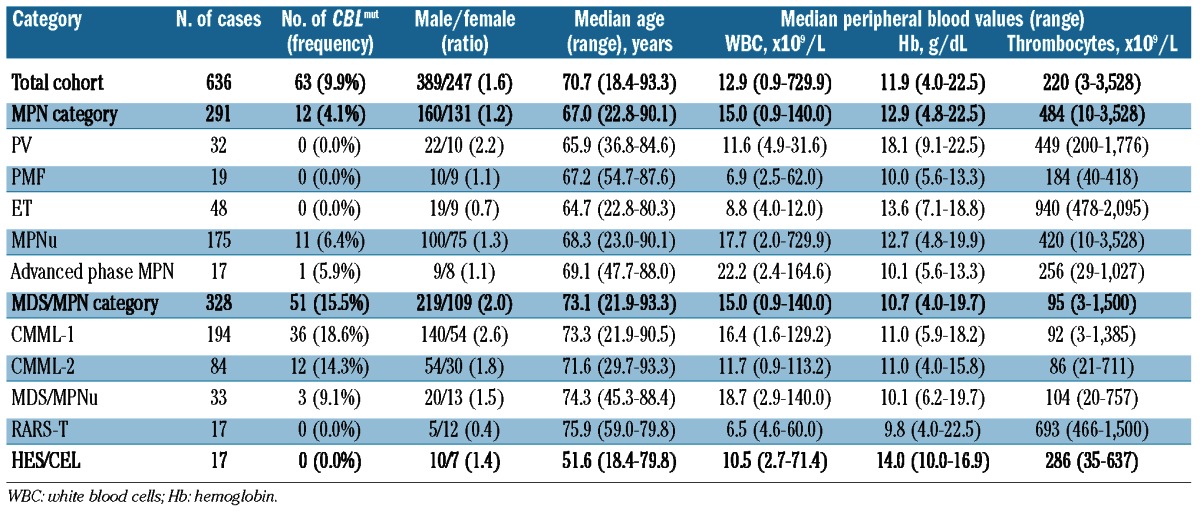
(A) Illustration of the localization of the different mutations within the CBL gene and of the corresponding amino acids detected in the patients of this study. The highest conserved domains are marked in dark gray. Top position of CBL at chromosomal band 11q23.3. Middle panel: structure of the gene according to Grand et al.3 with TKB (tyrosine kinase binding) domain, linker and Ring domain, P-rich (Proline rich) domain and UBA (ubiquitin-associated) domain. Bottom amino acid exchanges detected in our cohort. (B) Distribution and frequency of CBLmut and other molecular mutations in the total cohort of 636 patients. Red indicates a mutation within the respective gene, gray indicates no mutation. White cells indicate that the respective gene mutation was not analyzed for this patient. Patients are presented vertically.
CBLmut were strongly underrepresented in JAK2V617F mutated as compared to JAK2V617wt patients (1 of 121, 0.8% vs. 62 of 514, 12.1%; P<0.001). CBLmut was detected concomitantly with JAK2V617F in only one case. This case showed a high load of CBLmut alleles in contrast to a low mutation JAK2V617F level of 1%. CBLmut were mutually exclusive of JAK2exon12 (n=6) and MPL (n=13) mutations. The frequency of CBLmut was lower in NRASmut as compared to NRASwt cases (2 of 45, 3.6% vs. 54 of 267, 20.2%; P=0.010) and KRASmut as compared to KRASwt cases (1 of 29, 3.4% vs. 49 of 265, 18.5%; P=0.038). In contrast, CBLmut showed a significantly higher frequency in TET2mut cases as compared to TET2wt (25 of 135, 18.5% vs. 13 of 185, 7.0%; P=0.003) (Table 2; Figure 1B). There was no significant difference in CBLmut dependence on the RUNX1, ASXL1, and EZH2 mutation status (Table 2).
Table 2.
Frequency of CBLmut in different molecular subgroups (P values were calculated by χ2 test). CBLmut rates are given within the molecular subgroups as defined by JAK2V617F, JAK2exon12, MPLW515, RUNX1, ASXL1, EZH2, TET2, and NRAS mutation status.
Summarizing these results, presence of CBLmut with the JAK2V617F was extremely rare, and CBLmut seem to show mutual exclusiveness of JAK2exon12 and MPLW515 mutations. This gives rise to the hypothesis that CBLmut do not play a role in the ‘classical’ MPNs, although larger numbers of patients and the whole CBL gene would have to be analyzed for definite conclusions to be drawn. Furthermore, CBLmut were significantly underrepresented in NRASmut (P=0.010) and KRASmut (P=0.038) patients in our study. Also in pediatric JMML, no CBLmut case was detected in 91 patients with RAS pathway activating mutations (P<0.001)21 and a single double mutated case only was identified in CMML.21 Therefore, CBLmut and JAK-STAT activating mutations largely seem to exclude each other, although, here again, larger numbers of patients and the whole CBL gene would have to be analyzed for definite conclusions to be drawn. This is in accordance with the function of CBL, as it is involved in negative modulation of tyrosine kinase signaling, and, therefore, does itself finally end up in the JAK-STAT pathway. CBLmut in addition to another JAK-STAT activating mutation would probably not result in a further growth advantage for the respective cell. In contrast, Aranaz et al. found the same frequencies of CBLmut in patients with JAK2V617F-positive and -negative MPNs; however, these were only a very few cases each.22 This suggests that such a coincidence still might occur very rarely, and it is still debatable as to whether in these rare cases two different subclones coexist. As we found a CBLmut rate of 5.9% in advanced MPNs in our study, it may be speculated that the respective mutations may contribute to disease progression in the MPNs, which is also in accordance with data on blast phase of chronic myeloid leukemia.23,24
Correlation of CBLmut with different cytogenetic subgroups (Online Supplementary Table S1) revealed the highest frequency in patients with monosomy 7. CBLmut were more frequent in patients with monosomy 7 (4 of 9, 44.4%) when compared to all remaining cases (59 of 627, 9.4%; P=0.008). CBLmut showed no significant correlations with other frequent cytogenetic subgroups, i.e. normal karyotypes, trisomy 8, or loss of Y chromosome.
Of all 63 CBLmut patients, 56 (88.9%) had only one CBLmut. Of these 56, 37 had a mutation/wild-type load of 50% or less and 19 had a load of over 50%. Eight (12.7%) cases had two different CBLmut in parallel. These cases were reanalyzed by pyrosequencing for better quantification of the mutation load, which in all cases was more than 50%. Combination of mutations and load were as follows: 1) p.Ile423Asn (38%) + p.Val430Met (40%); 2) p.Cys404Tyr (31%) + p.Arg420Gln (36%); 3) p.Cys384Arg (84%) + p.Met400Arg (7%); 4) p.His398Arg (90%) + p.Ile429_Phe434del (6%); 5) p.Cys416Ser (43%) + p.Arg420Gly (38%); 6) p.Arg420Gln (15%) + p.Arg420X (72%); 7) p.Gly415Ser (38%) + p.Arg462X (42%); 8) p.Asp390Tyr (40%) + splicing of exon 9 (31%). In all 4 cases in whom the mutations were located on the same amplicon, they were shown to appear on different alleles. Based on these data, it was not possible to draw definite conclusions as to whether these mutations were in different clones or whether both alleles of one clone were mutated. The mean mutation load in all patients was 55.0±26.0%. There was no significant difference in mean mutation load between CMML patients and the other CBLmut patients (59.0±29.1% vs. 53.1±24.4%; n.s.).
Most (n=57, 90.5%) alterations were missense mutations. Three cases had small deletions (p.Tyr368_Glu369del; p.Leu370_Tyr371del; and p.Ile429_Phe434del), 2 further cases revealed a stop mutation (p.Arg420X, and p.Arg462X), and one case an exon 9 splice mutation. Some mutations were recurrent in our cohort, such as p.Arg420Gln (n=4), p.Phe418Ser (n=3), p.Arg420Leu (n=2), p.Cys404Tyr (n=2), p.Cys416Arg (n=2), p.Ile383Met (n=2), p.Ile429Asn (n=2), and p.Leu380Pro (n=2), whereas all others were detected in single cases only (Figure 1A).
Biological characteristics and peripheral blood values were compared between CBLmut and CBL wild-type (CBLwt) cases in the CMML cohort (n=278). The male/female ratio was higher in the CBLmut CMML patients than in CBLwt patients (5.0 vs. 2.0; P=0.025). No significant differences were found regarding median age or peripheral blood parameters between CBLmut and CBLwt cases in CMML (Online Supplementary Table S2).
Because of the high prevalence of CBLmut in CMML, outcome analysis was performed only in this subcohort. Clinical follow-up data were available in 176 of 278 CMML patients (36 CBLmut, 140 CBLwt). Median overall survival (OS) of the whole CMML cohort was 29.9 months (CMML-1: median OS not reached; CMML-2: median OS 29.6 months; n.s.). Within the whole CMML cohort, there was no significant difference between OS of patients with CBLmut and that of those with CBLwt (median 32.4 vs. 29.9 months). When the CMML-1 cohort (follow-up data available in 112 patients) was investigated separately, CBLmut patients had shorter OS than CBLwt (median 25.4 months vs. median not reached; P=0.227), but this difference did not reach significance. In the CMML-2 cohort (n=64 patients with survival data), survival outcomes were very similar between CBLmut and CBLwt patients (32.4 vs. 24.8 months; n.s.; Online Supplementary Figure S1). Corresponding to our previous analysis, including some of the patients from this study,7 patients with CBLmut had shorter OS when compared to those with CBLwt in the CMML-1 cohort; but this difference did not reach significance. Therefore, the prognostic value of CBLmut in CMML and in the MPNs, and its contribution to disease progression, deserves further investigation.25
In conclusion, CBLmut are overrepresented in CMML when compared to the MPNs. They rarely occur together with JAK-STAT pathway activating mutations, but are frequently seen with other genetic markers, e.g. mutations of the TET2 gene. Because of the high frequency for CMML and certain exclusion patterns with other mutations, CBLmut analysis is a useful additive tool for differential diagnosis.
Supplementary Material
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures: The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460(7257):904-8 [DOI] [PubMed] [Google Scholar]
- 2.Dunbar AJ, Gondek LP, O'Keefe CL, Makishima H, Rataul MS, Szpurka H, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68 (24):10349-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113(24):6182-92 [DOI] [PubMed] [Google Scholar]
- 4.Reindl C, Quentmeier H, Petropoulos K, Greif PA, Benthaus T, Argiropoulos B, et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res. 2009;15 (7):2238-47 [DOI] [PubMed] [Google Scholar]
- 5.Beer PA, Delhommeau F, LeCouedic JP, Dawson MA, Chen E, Bareford D, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115 (14):2891-900 [DOI] [PubMed] [Google Scholar]
- 6.Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J, et al. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol. 2009;27(36):6109-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, et al. Next-Generation Sequencing Technology Reveals a Characteristic Pattern of Molecular Mutations in 72.8% of Chronic Myelomonocytic Leukemia by Detecting Frequent Alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28(24):3858-65 [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed. Lyon: International Agency for Research on Cancer. (IARC), 2008 [Google Scholar]
- 9.Flach J, Dicker F, Schnittger S, Kohlmann A, Haferlach T, Haferlach C. Mutations of JAK2 and TET2, but not CBL are detectable in a high portion of patients with refractory anemia with ring sideroblasts and thrombocytosis. Haematologica. 2010;95 (3):518-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwaab J, Ernst T, Erben P, Rinke J, Schnittger S, Ströbel P, et al. Activating CBL mutations are associated with a distinct MDS/MPN phenotype. Ann Hematol. 2012;91(11):1713-20 [DOI] [PubMed] [Google Scholar]
- 11.Loeffler H, Rastetter J, Haferlach T. Atlas of Clinical Hematology, 6th ed. Heidelberg: Springer, 2004 [Google Scholar]
- 12.Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U, et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia. 2002;16(1):53-9 [DOI] [PubMed] [Google Scholar]
- 13.Cross NC, Melo JV, Feng L, Goldman JM. An optimized multiplex polymerase chain reaction (PCR) for detection of BCR-ABL fusion mRNAs in haematological disorders. Leukemia. 1994;8(1):186-9 [PubMed] [Google Scholar]
- 14.Schnittger S, Bacher U, Kern W, Schroder M, Haferlach T, Schoch C. Report on two novel nucleotide exchanges in the JAK2 pseudokinase domain: D620E and E627E. Leukemia. 2006;20(12):2195-7 [DOI] [PubMed] [Google Scholar]
- 15.Schnittger S, Bacher U, Haferlach C, Geer T, Muller P, Mittermuller J, et al. Detection of JAK2 exon 12 mutations in 15 patients with JAK2V617F negative polycythemia vera. Haematologica. 2009;94(3):414-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnittger S, Bacher U, Haferlach C, Dengler R, Krober A, Kern W, et al. Detection of an MPLW515 mutation in a case with features of both essential thrombocythemia and refractory anemia with ringed sideroblasts and thrombocytosis. Leukemia. 2008;22(2):453-5 [DOI] [PubMed] [Google Scholar]
- 17.Dicker F, Haferlach C, Kern W, Haferlach T, Schnittger S. Trisomy 13 is strongly associated with AML1/RUNX1 mutations and increased FLT3 expression in acute myeloid leukemia. Blood. 2007;110(4):1308-16 [DOI] [PubMed] [Google Scholar]
- 18.Grossmann V, Kohlmann A, Eder C, Haferlach C, Kern W, Cross NC, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25(5):877-9 [DOI] [PubMed] [Google Scholar]
- 19.Nakao M, Janssen JW, Seriu T, Bartram CR. Rapid and reliable detection of N-ras mutations in acute lymphoblastic leukemia by melting curve analysis using LightCycler technology. Leukemia. 2000;14(2):312-5 [DOI] [PubMed] [Google Scholar]
- 20.Schnittger S, Eder C, Alpermann T, Fasan A, Grossmann V, Kohlmann A, et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an extremely poor outcome. Blood. 2011; 118(21):416a. [DOI] [PubMed] [Google Scholar]
- 21.Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114(9):1859-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aranaz P, Hurtado C, Erquiaga I, Migueliz I, Ormazabal C, Cristobal I, et al. CBL mutations in myeloproliferative neoplasms are also found in its proline-rich domain and in patients with the V617FJAK2. Haematologica. 2012;97(8):1234-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makishima H, Jankowska AM, McDevitt MA, O'Keefe C, Dujardin S, Cazzolli H, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117 (21):e198-e206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossmann V, Kohlmann A, Zenger M, Schindela S, Eder C, Weissmann S, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25(3):557-60 [DOI] [PubMed] [Google Scholar]
- 25.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24(6):1128-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.