Summary
MCL1, which encodes the anti-apoptotic protein MCL1, is among the most frequently amplified genes in human cancer. A chemical genomic screen identified compounds, including anthracyclines, that decreased MCL1 expression. Genomic profiling indicated that those compounds were global transcriptional repressors that preferentially affect MCL1 due to its short mRNA half-life. Transcriptional repressors and MCL1 shRNAs induced apoptosis in the same cancer cell lines and could be rescued by physiological levels of ectopic MCL1 expression. Repression of MCL1 released the pro-apoptotic protein BAK from MCL1, and Bak deficiency conferred resistance to transcriptional repressors. A computational model, validated in vivo, indicated that high BCL-xL expression confers resistance to MCL1 repression, thereby identifying a patient selection strategy for the clinical development of MCL1 inhibitors.
INTRODUCTION
Inhibition of apoptosis is a critical step in the pathogenesis of cancers, and is a major barrier to effective treatment (Adams and Cory, 2007; Danial and Korsmeyer, 2004). It is now thought that one or more components of the apoptosis pathway are dysregulated in all cancers (Hanahan and Weinberg, 2011) either by genetic mutation of the genes encoding these proteins (e.g. point mutations, copy number abnormalities, or chromosomal translocation), or by other mechanisms (e.g. epigenetic mechanisms or upstream oncogenic mutations). Despite this central importance in the development and maintenance of cancer, few apoptosis-targeted therapeutics have reached clinical evaluation.
Of particular importance is the BCL2 family of proteins. Highly conserved from worm to human, these proteins control the activation of downstream caspases, which are the major effectors of apoptosis. The BCL2 family can be divided into three main subclasses, defined in part by the homology shared within four conserved regions termed BCL2 homology (BH) domains (Adams and Cory, 2007; Danial and Korsmeyer, 2004). The “multidomain” pro-apoptotic members BAX and BAK possess BH1-3 domains, and together constitute a requisite gateway to the intrinsic apoptosis pathway (Lindsten et al., 2000; Wei et al., 2001). In contrast, the pro-apoptotic proteins, such as BIM, PUMA and NOXA, share homology only within the BH3 amphipathic α-helical death domain, prompting the title “BH3-only”. Anti-apoptotic family members such as BCL2, BCL-xL and MCL1 show conservation in all four BH domains. The BH1, BH2 and BH3 domains of those proteins are in close proximity and create a hydrophobic pocket that can accommodate the BH3 domain of a pro-apoptotic member (Danial and Korsmeyer, 2004; Petros et al., 2004).
Despite overwhelming genetic and functional evidence implicating the BCL2-family proteins as therapeutic targets, effective therapeutic inhibitors of these proteins have been difficult to develop. Elegant NMR-based structural biology efforts led to development of the small-molecule BCL2/BCL-xL inhibitor ABT-737 (Oltersdorf et al., 2005) and its analog ABT-263, now in early clinical trials (Tse et al., 2008). While it is expected that ABT-263 or related compounds will have clinical activity in BCL2- or BCL-xL-dependent tumors, it is clear that many tumors do not depend on these proteins, but rather rely on other anti-apoptotic factors such as MCL1 (Lin et al., 2006; van Delft et al., 2006).
MCL1 has only recently been recognized as an important therapeutic target in cancer. MCL1 is highly expressed in a variety of human cancers (Krajewska et al., 1996a; Krajewska et al., 1996b). Its expression has been linked to tumor development (Zhou et al., 2001) and resistance to anti-cancer therapies. For example, over-expression of MCL1 is a major resistance mechanism for the experimental BCL2/BCL-xL inhibitor ABT-737 (Chen et al., 2007; Keuling et al., 2009; van Delft et al., 2006), and MCL1 has been similarly implicated in the resistance of non-BCL2-family-targeted therapy (Wei et al., 2006). Importantly, we recently reported that amplification of the MCL1 locus is one of the most frequent somatic genetic events in human cancer, further pointing to its centrality in the pathogenesis of malignancy (Beroukhim et al., 2010). While the development of MCL1 inhibitors has been of considerable interest, no such inhibitors have yet reached the clinic. A particularly promising strategy, however, was recently reported by Walensky and colleagues, whereby ‘stapled’ helical MCL1 BH3 peptides function as effective MCL1 inhibitors in pre-clinical models (Stewart et al., 2010). Whether such stapled peptides will make for effective clinical therapeutics remains to be established. Furthermore, no biomarkers for patient selection have been discovered for MCL1 inhibitors. Therefore, we used a chemical genomic strategy to identify MCL1-downregulating small-molecules and to discover biomarkers of MCL1 dependency.
RESULTS
Gene-expression-based high-throughput screen identifies small-molecules repressing MCL1 expression
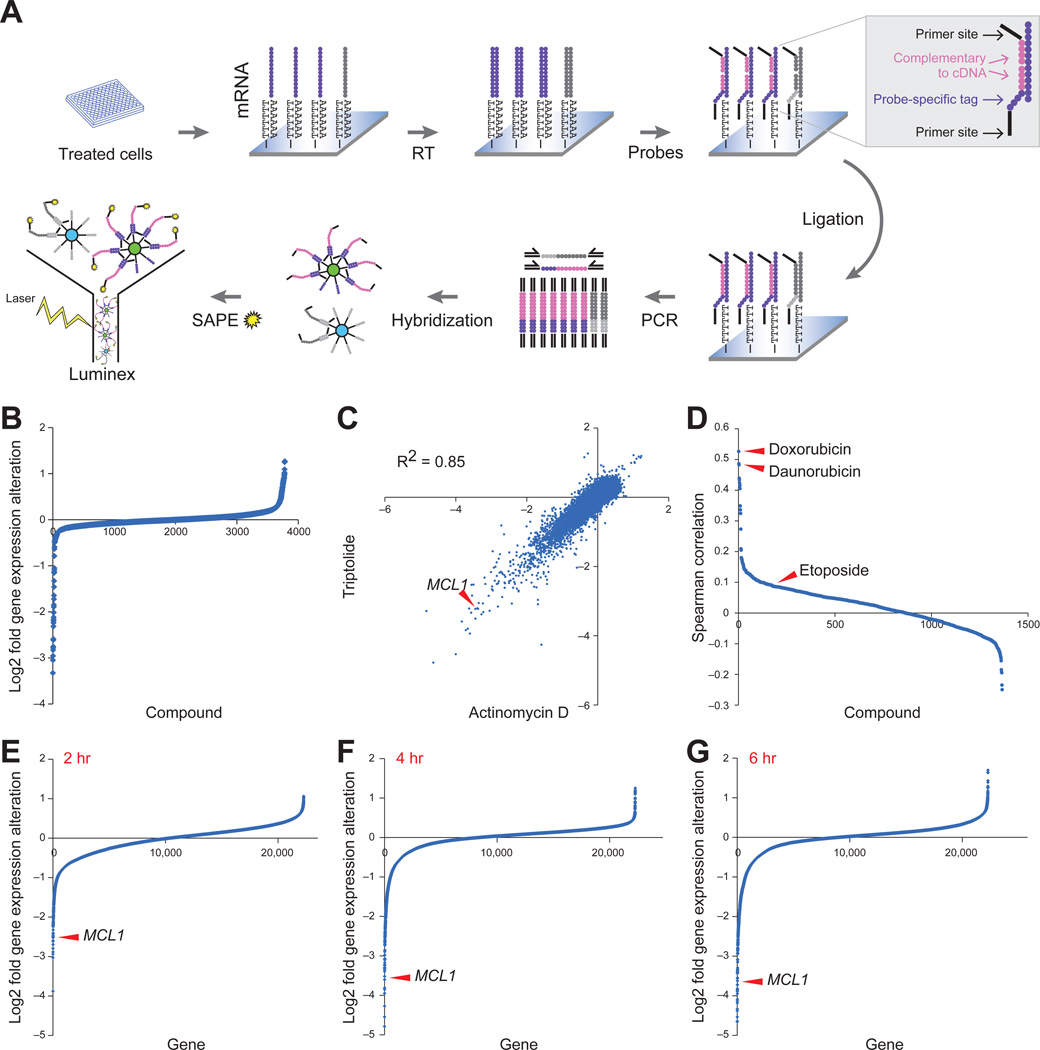
MCL1 is frequently amplified in human cancers (Beroukhim et al., 2010), and is highly expressed across a panel of 729 human cancer cell lines (Figure S1A). We hypothesized that it might be possible to discover small-molecules that decrease MCL1 expression, thereby activating the apoptosis cascade in MCL1-dependent tumors. We therefore developed an assay to profile the mRNA levels of MCL1 and 48 other apoptosis-related genes using the Luminex bead-based method (Hieronymus et al., 2006; Peck et al., 2006) (Figure 1A, Table S1). We profiled many apoptosis-related genes in addition to MCL1 in order to identify compounds that preferentially repress MCL1 while preserving expression of the pro-apoptotic factors.
Figure 1. Bead-based high-throughput gene expression screening identified MCL1 repression by transcriptional inhibitor compounds.
A. Illustration of screening procedure. mRNA levels of MCL1 and 48 other apoptotic genes were measured in MCF7 cells 8 hours after treatment with 2,922 small molecules.
B. MCL1 expression modulation by 2,922 compounds. Compounds and DMSO controls were sorted by MCL1 expression repression. The y-axis displays log2 gene expression fold change.
C. Gene expression profiling by Affymetrix microarrays of MCF7 cells with triptolide (500 nM) and actinomycin D (2.5 µM) for 4 hours. Both the x- and y-axis displays log2 gene expression fold alteration.
D. The 1,317 compounds used to treat MCF7 cells in the Connectivity Map were displayed in descending order of their correlation with triptolide (calculated as the Spearman rank correlation of the differential gene expression across the Affymetrix U133A chip).
E-G. Gene expression repression by triptolide at 2 hours (E), 4 hours (F) and 6 hours (G). The genes were ranked by the extent of repression. The y-axis displays log2 gene expression fold alteration. Arrowheads indicate MCL1.
See also Figure S1 and Tables S1 and S2.
We carried out a pilot screen using MCF7 breast cancer cells treated with 2,922 small-molecule compounds including 530 FDA approved drugs. We used MCF7 cells, which are deficient in caspase-3, to avoid identifying compounds that repress MCL1 expression through feedback apoptosis mechanisms. We also performed the assay at an early time point (8 hours post-treatment) for this reason. We counter-screened against compounds that caused significant cell death at 8 hours using an LDH viability assay, reasoning that such compounds must not be acting by classical apoptosis-inducing mechanisms.
Twenty-four compounds (0.8%) decreased MCL1 expression at least two-fold (Figure 1B). All 24 compounds reduced MCL1 expression more than any of the other 48 apoptosis-related genes assayed, suggesting at least some degree of preferential activity against MCL1. We selected 14 commercially available compounds for further testing. Seven of these exhibited significant dose-related repression of MCL1 expression. The 7 compounds included the natural product triptolide, the transcription inhibitors 5,6-dichlorobenzimidazole riboside (DRB) and actinomycin D, the kinase inhibitor 5-iodotubercidin, and the anthracyclines doxorubicin, daunorubicin and epirubicin. Despite having distinct reported mechanisms of action (Table S2), treatment of these compounds resulted in decreased MCL1 expression in multiple cell lines, suggesting a common mechanism of MCL1 repression across cancer types (Figure S1B).
Small molecules that repress MCL1 share transcriptional profiles
We compared genome-wide expression profiles of cells following treatment with candidate compounds to det1ermine whether they shared a common mechanism of action. We performed genome-wide gene expression profiling in MCF7 cells following treatment with triptolide and actinomycin D. The expression changes induced by triptolide and actinomycin D were highly similar (R2=0.85), suggesting that, like actinomycin D, triptolide likely functions as a transcriptional inhibitor (Figure 1C). Consistent with this observation, triptolide was recently reported to bind to XPB, a subunit of TFIIH (Titov et al., 2011), and inhibit phosphorylation of the C-terminal tail of RNA polymerase II, which results in transcriptional inhibition (Leuenroth and Crews, 2008).
Using the Connectivity Map database containing expression profiles of 1,366 compounds (www.broadinstitute.org/cmap) (Lamb et al., 2006), the triptolide induced profile showed a high degree of similarity to both doxorubicin and daunorubicin (ranked 1 and 2 of 1,366, respectively, using Spearman correlation) (Figure 1D). The anti-cancer effect of anthracyclines has long been attributed to inhibition of DNA topoisomerase II (Desmedt et al., 2011; Moretti et al., 2009). However, the DNA topoisomerase II inhibitor etoposide induced a transcriptional profile distinct from that induced by triptolide (Figure 1D). Taken together, these results strongly suggest that the compounds that emerged from our MCL1-repression screen, including the anthracyclines, function as global transcriptional repressors. We therefore refer to them as Transcriptional Repressor (TR) compounds.
Strikingly, the TR compounds showed dramatic preferential activity against MCL1 compared to the rest of the transcriptome. For example, MCL1 was in the top 0.05 percentile of triptolide-repressed genes (Figure 1E-1G), and the MCL1 transcript was repressed more than 5-fold within 2 hours of treatment (Figure 1E). On the contrary, none of the other BCL2 family genes was repressed more than two fold. Consistent with the reported short half-life of MCL1 protein (30 minutes) (Adams and Cooper, 2007), inhibition of MCL1 mRNA caused a rapid decrease in MCL1 protein levels that occurred prior to PARP cleavage, a marker for caspase activation (Figure S1C).
TR compounds share a pattern of cell killing and can be rescued by physiologically relevant levels of MCL1
Based on the shared mechanisms suggested above, we hypothesized that if MCL1 repression is a biologically-relevant target of TR compounds, then these compounds should induce apoptosis in the same cancer cell lines. We therefore measured caspase activation and cell viability of 74 non-small cell lung cancer (NSCLC) and 33 breast cancer cell lines following treatment with actinomycin D, doxorubicin, triptolide, and flavopiridol. Flavopiridol has previously been reported to repress MCL1 expression via inhibition of CDK9 (Chen et al., 2005).
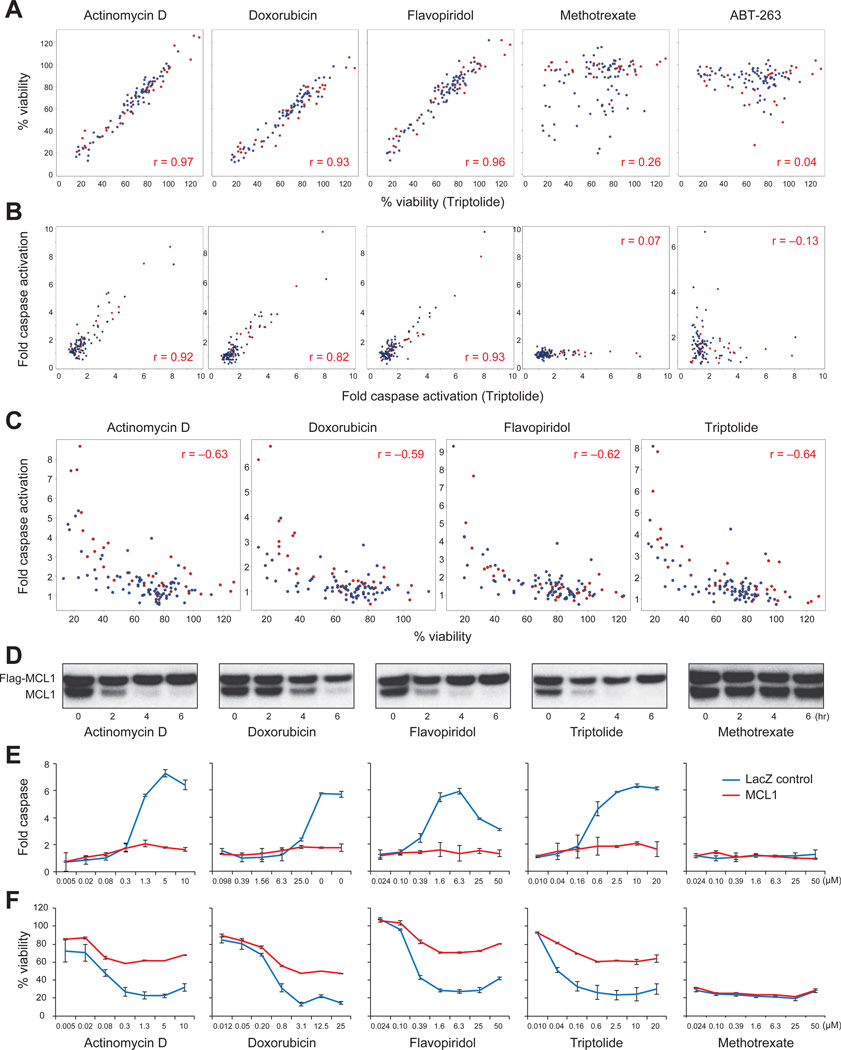
Responses to the TR compounds were highly correlated when measured both by caspase activation and cell viability (Pearson r >0.82 and 0.93, respectively, when compared to triptolide) (Figure 2A-B). As expected, cell viability was highly correlated with caspase activation for each TR compound (Figure 2C, Pearson r >0.59), indicating that the TR compounds impair cell viability via apoptosis. By contrast, compounds that kill cells via different mechanisms, such as methotrexate and etoposide, demonstrated different patterns of cytotoxicity (Figure 2A-B, S2A). Despite the fact that TR compounds repress the expression of many genes, ectopic expression of physiological levels of MCL1 rescued cells from TR compound treatment (Figure 2D-F). In contrast, ectopic expression of MCL1 had no such rescue effect for other classes of compounds, such as methotrexate (Figure 2D-F).
Figure 2. TR compounds shared a pattern of cell killing.
A-B. 74 NSCLC (blue dots) and 33 breast (red dots) cancer cell lines were treated with small molecules. The effect of triptolide (x-axis) is compared with the effect of other compounds (y-axis), measured both by cell viability after 24 hours of treatment (A) and fold-caspase-3 activation after 6 hours of treatment (B). Concentrations of the compounds for displayed data were: actinomycin D (1.25 µM), doxorubicin (12.5 µM), flavopiridol (6.25 µM), methotrexate (6.25 µM), ABT-263 (6.25 µM), triptolide (2.5 µM).
C. Correlation between cell viability and caspase activation.
D. Expression levels of endogenous MCL1 and FLAG-MCL1 in HMC-1-8 cells after treatment with the indicated compounds were evaluated by Western blot. For each panel, cells were treated with the compound for 0, 2, 4 and 6 hours (left to right for each compound).
E-F. Ectopic expression of physiological levels of FLAG-MCL1 rescued HMC-1-8 cells from TR compounds, but not methotrexate, as measured by caspase activation at 6 hours (E) and cell viability at 24 hours (F). Error bars indicate standard deviation of duplicate measurements.
See also Figure S2.
If TRs block global transcription, we hypothesized that combination treatment with TR compounds would counteract the effects of compounds that kill cells by inducing the expression of pro-apoptotic proteins. The proteasome inhibitor bortezomib induces apoptosis through the induction of the pro-apoptotic protein NOXA (Gomez-Bougie et al., 2007; Voortman et al., 2007). As predicted, treatment with the TR compounds doxorubicin, actinomycin D or triptolide rescued cells from the apoptotic effects of bortezomib whereas treatment with the non-TR compound etoposide had no effect (Figure S2B-F). Similarly, the TR compounds were able to rescue cells from the histone deacetylase (HDAC) inhibitor vorinostat (Figure S2G), which kills cells via the induction of the pro-apoptotic proteins BMF and NOXA (Wiegmans et al., 2011).
MCL1 knockdown phenocopies TR compounds
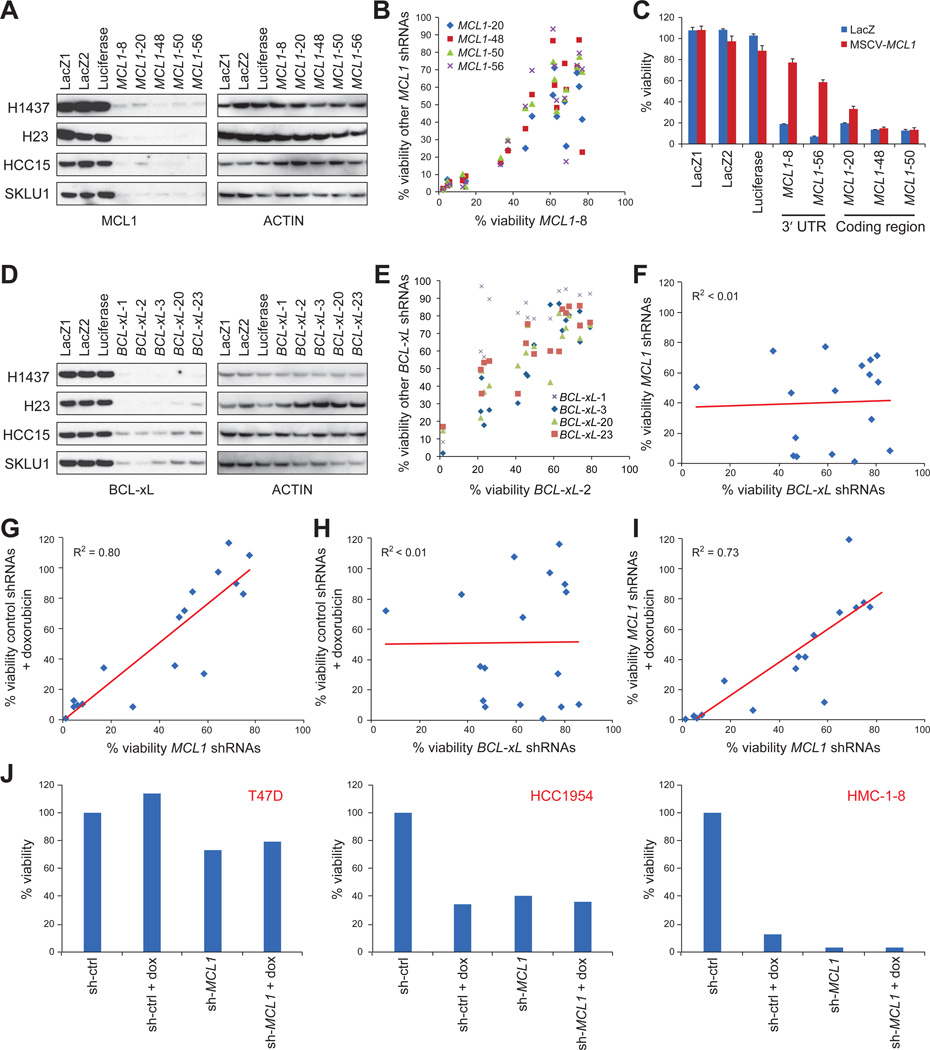
In order to determine whether MCL1 repression explains the activity of TR compounds, we tested whether their effects could be phenocopied by knockdown of MCL1. We treated 17 breast and 16 NSCLC cancer cell lines representing different levels of sensitivity to TR compounds with each of the five most effective shRNAs selected from a library of 60 anti-MCL1 shRNAs (Figure 3A). The response to the 5 MCL1 shRNAs was highly correlated (R2>0.64 for breast cell lines and R2>0.55 for NSCLC cell lines) (Figure 3B). Ectopic expression of MCL1 with a heterologous 3’ UTR at physiologically relevant levels was able to rescue cells from the 2 MCL1 shRNAs targeting the 3’ UTR of MCL1, but not the 3 MCL1 shRNAs targeting the coding region of MCL1 (Figure 3C), indicating that their cellular effects are most likely due to MCL1 repression as opposed to off-target effects.
Figure 3. MCL1 knockdown phenocopied TR compounds.
A. Knockdown efficiency of the five best MCL1 shRNAs as determined by Western blot.
B. Effects of one shRNA (MCL1-sh8) on the cell viability is plotted on the x-axis against the effects of the other four MCL1 shRNAs on the y-axis. Data were measured from 17 breast cancer cell lines.
C. Effect of expressing physiological levels of FLAG-tagged MCL1 on apoptosis induced by MCL1 shRNAs targeting the 3’ UTR of MCL1 (MCL1-8 and MCL1-56) or targeting the coding region of MCL1 (MCL1-20, MCL1-48 and MCL1-50) in HMC-1-8 cells. Error bars indicate standard deviation of duplicate measurements.
D. Five BCL-xL shRNAs effectively knocked down BCL-xL expression. H1437 and H23 cells had longer exposure time than HCC15 and SKLU1 cells for Western blots to show the shRNA effects.
E. Effects of one shRNA (BCL-xL-2) on cell viability is plotted on the x-axis against the effects of the other four BCL-xL shRNAs on the y-axis. Data were measured from 17 breast cancer cell lines.
F-I. Effects of MCL1 shRNAs (G, I) or BCL-xL shRNAs (F, H) on cell viability plotted on x-axis against sensitivity to MCL1 shRNAs (F), doxorubicin (G, H), or MCL1 shRNAs + doxorubicin (I) plotted on y-axis in 17 breast cancer cell lines. Cells were infected with viruses carrying shRNAs for MCL1 or BCL-xL for 3 days, or 2 days followed by treatment with 5µM doxorubicin for an additional 24 hours (median of 5 MCL1 shRNAs or BCL-xL shRNAs as indicated).
J. Examples of cell lines that were resistant (T47D), partially sensitive (HCC1954) or sensitive (HMC-1-8) to MCL1 inhibition. dox: doxorubicin.
See also Figure S3.
In addition, we generated shRNAs against BCL-xL to test whether MCL1-dependent cells were sensitive to knockdown of other anti-apoptotic genes. The responses to the 5 most effective BCL-xL shRNAs (out of the 24 shRNAs tested) were highly correlated (Figure 3D-E, S3A), but these responses did not correlate with response to the MCL1 shRNAs (R2=0.002) (Figure 3F, S3B).
Impaired viability induced by doxorubicin was strongly correlated with the effects of MCL1 shRNAs (R2=0.80 for breast cancer cells (Figure 3G) and R2=0.74 for NSCLC). Conversely, doxorubicin sensitivity did not correlate with the effects of shRNAs targeting BCL-xL (R2=0.0001 for breast cancer cell lines) (Figure 3H). Furthermore, doxorubicin did not induce additional significant cell death after MCL1 knockdown, consistent with MCL1 repression being a major effector of doxorubicin action (Figure 3I-J). Triptolide yielded similar results, suggesting that this is a general property of TR compounds (Figure S3C). Taken together, these results further support the notion that a subset of tumor cells are dependent upon MCL1 for survival, and that TR compounds act largely via MCL1 repression.
Discovering predictive biomarkers of MCL1 essentiality
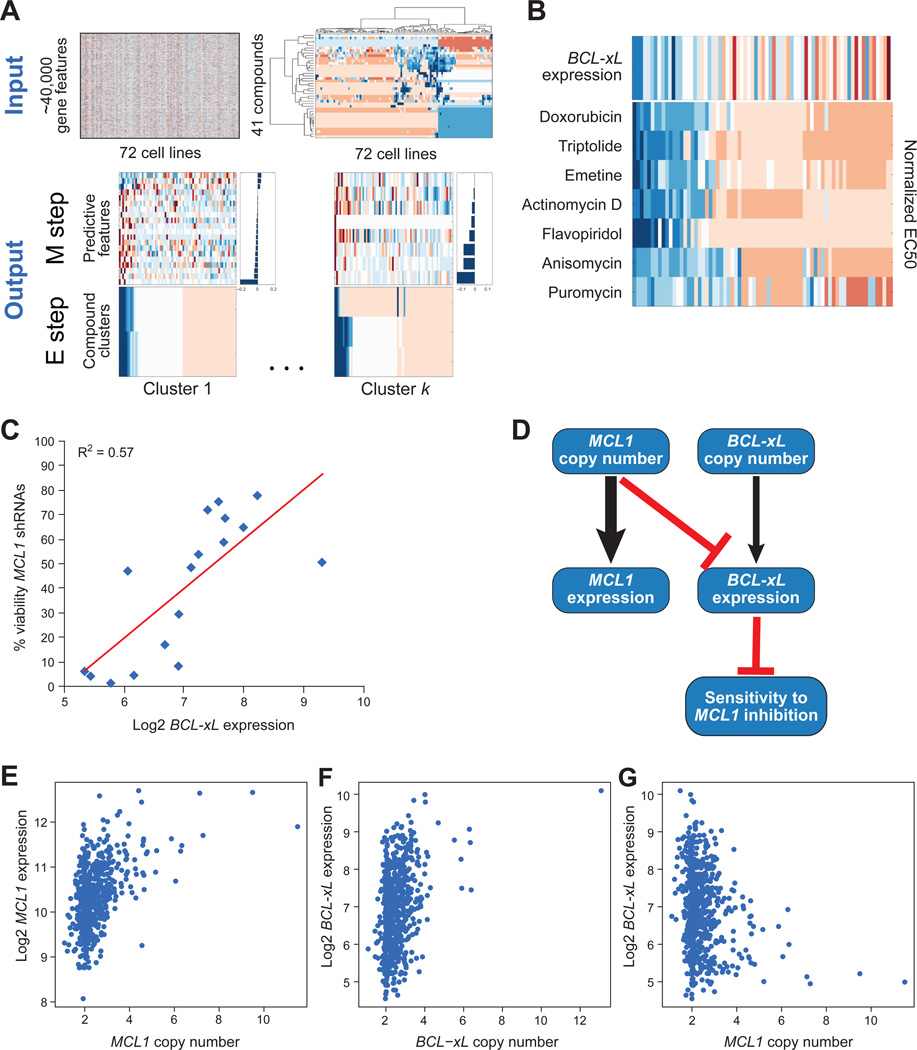
We next sought to discover biomarkers that are predictive of MCL1 essentiality by comparing TR compound sensitivities with genomic data. Such biomarkers would prove useful for the prediction of sensitivity to any present or future MCL1 inhibitors. We developed an analytical method to infer groups of compounds that induce sensitivity in similar cancer genetic subtypes and infer predictive biomarkers of sensitivity to each compound group. Briefly, the method uses an expectation-maximization (EM) algorithm and iterates until convergence between clustering groups of compounds based on the similarity of their response profiles and uses an elastic net algorithm to infer a predictive model for each group based on its genetic features (Lee et al., 2009). The method further employs a bootstrapping procedure to obtain a parsimonious model containing only robustly predictive features (Figure 4A, also see Supplemental Experimental Procedures for details).
Figure 4. Unbiased computational analysis identified TR compounds sensitivity predicted by low expression of BCL-xL.
A. Schematic of biomarker discovery algorithm (see Supplemental Experimental Procedures for details).
B. BCL-xL expression as a predictive biomarker of TR compound sensitivity. The Expectation Maximization algorithm identified one cluster that contained all of the TR compounds (bottom panel; blue indicates drug sensitivity, red indicates resistance), coupled to a single predictive feature, BCL-xL RNA expression (top panel; blue indicates low level of expression, red indicates high expression).
C. Correlation between expression level of BCL-xL (x-axis) with cell viability upon MCL1 knockdown (y-axis), represented as the percent viability relative to control shRNAs, averaged over 2 replicates treated for 3 days with 5 different shRNA constructs targeting MCL1 across a panel of 17 breast cancer cell lines.
D. The ARACNE reverse engineering algorithm was used to identify direct interactions between sensitivity to TR compounds and MCL1 and BCL-xL gene expression and gene copy number. Arrows represent inferred direct interactions with the weight of the edge proportional to the mutual information between the corresponding nodes. Arrows indicate the presumed direction of causality (e.g. copy number to gene expression). Black lines indicate positive correlations and red lines indicate negative correlations.
E-G. Comparisons of gene expression and copy number values of MCL1 and BCL-xL across 643 cancer cell lines. E) MCL1 copy number versus MCL1 expression. F) BCL-xL copy number versus BCL-xL expression. G) MCL1 copy number versus BCL-xL expression.
We examined the genetic features (copy number and expression data for >18,000 genes, and mutation data for 34 genes) across 72 cell lines for which we had TR compound sensitivity measurements. To ensure that our predicted biomarkers were specific to sensitivity induced by the TR compounds, we also performed dose-response measurements on 37 additional control compounds (Table S2). The algorithm identified a cluster of compounds consisting of all of the TR compounds (actinomycin D, doxorubicin, flavopiridol, and triptolide), as well as 3 additional compounds (puromycin, emetine, and anisomycin) that function as global repressors of protein translation (Figure 4B). Similar to MCL1 mRNA, the extremely short half-life of MCL1 protein likely explains the selective effects of protein translation inhibitors on MCL1 activity.
The predictive model of sensitivity to the group of transcriptional and translational repressors contained only a single feature, corresponding to mRNA expression of BCL-xL. Specifically, low expression of BCL-xL was associated with sensitivity and high expression of BCL-xL was associated with resistance to compounds that repress MCL1 expression. The half-life of BCL-xL protein is much longer than that of MCL1 (Figure S1C), consistent with its ability to prevent apoptosis induced by transcriptional and translational inhibitors. Also consistent with this observation, sensitivity to MCL1 shRNAs anti-correlated with BCL-xL mRNA levels in the 17 breast cancer cell lines (R2=0.57) (Figure 4C).
We next sought to derive a computational model for the causal interactions that explain how MCL1 and BCL-xL influence sensitivity to TR compounds. We applied the ARACNE reverse engineering algorithm (Basso et al., 2005; Margolin et al., 2006), which is designed to deconvolute direct and indirect interactions among a set of covariates, and derived a network of direct interactions among variables corresponding to gene expression and copy number of MCL1 and BCL-xL and sensitivity to TR compounds. We used as input to the algorithm a matrix of values across the panel of 72 cell lines, corresponding to normalized expression and copy number of MCL1 and BCL-xL, as well as sensitivity to the TR compounds, computed as the average of normalized IC50 values across all TR compounds. This approach yielded a model in which expression of BCL-xL was indeed the direct predictor of sensitivity to TRs (Figure 4D). As expected, gene expression of BCL-xL and MCL1 was directly influenced by the copy number of the respective genes (Figure 4E-F). Interestingly, the model indicated an epistatic relationship between MCL1 copy number and BCL-xL expression. MCL1 copy number was negatively correlated with BCL-xL expression (Figure 4G), suggesting that MCL1 amplification may decrease the selective pressure requiring BCL-xL for inhibition of apoptosis.
Sequestration of pro-apoptotic proteins by MCL1 and BCL-xL
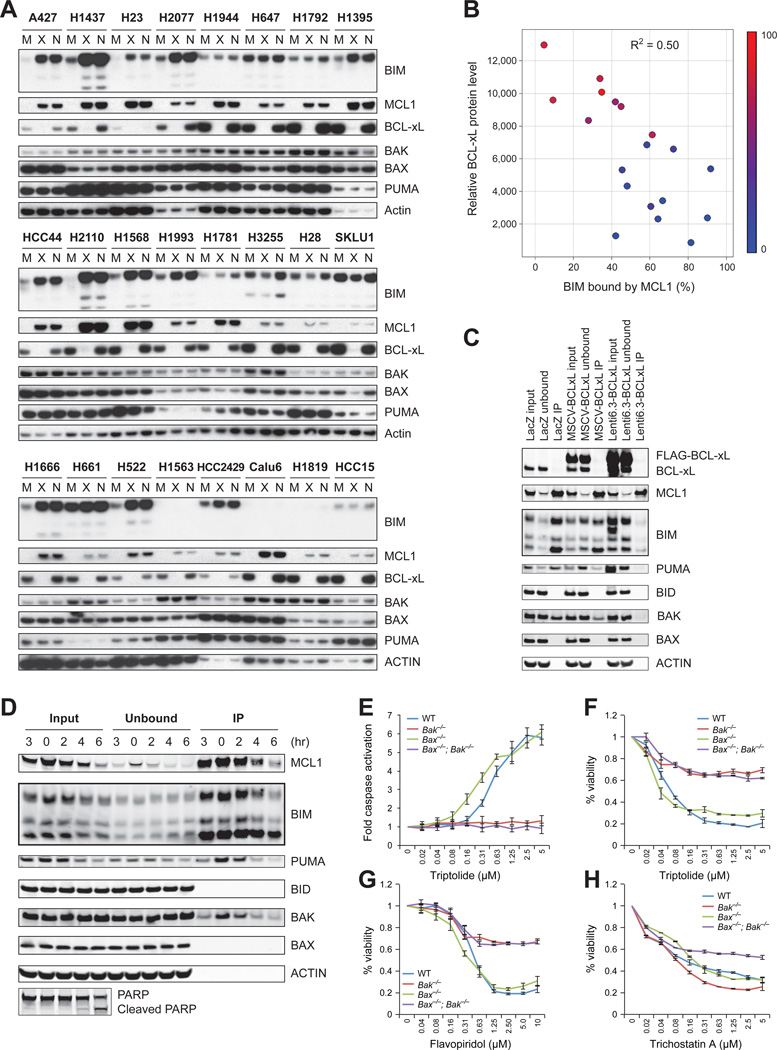
The above data suggested that breast and lung cancer cells with low expression of BCL-xL rely on MCL1 to sequester pro-apoptotic proteins. Upon repressing of MCL1 protein levels, pro-apoptotic proteins might be released from MCL1 and cause downstream caspase activation and apoptosis. BIM binds to all anti-apoptotic proteins (Merino et al., 2011). In a panel of 19 NSCLC cell lines, in cells expressing low levels of BCL-xL, depletion of MCL1 by immunoprecipitation resulted in depleting nearly the entirety of BIM (Figure 5A-B). In contrast, in cells expressing high levels of BCL-xL, only a small fraction of BIM was sequestered by MCL1 (Figure 5A-B). Furthermore, when BCL-xL was over-expressed in cells that normally have low levels of BCL-xL, the fraction of BIM bound by MCL1 decreased significantly (Figure 5C). These experiments demonstrate a shuttling of BIM sequestration between MCL1 and BCL-xL, depending on their relative expression levels. To explore whether the release of BIM from MCL1 explains the apoptotic effect of MCL1-repressing TR compounds, we repeated the MCL1-BIM co-immunoprecipitation experiments under conditions of TR treatment. Surprisingly, despite the TR compounds triptolide or flavopiridol significantly reducing MCL1 levels, the majority of BIM protein remained bound to the residual MCL1 (Figure S4A-B). In addition, BIM knockdown by shRNA did not abrogate the sensitivity to TR compounds (Figure S7C-G), although we cannot exclude the possibility that more complete BIM knock-down might have a more dramatic effect.
Figure 5. MCL1 is the major anti-apoptotic protein in cells with low BCL-xL expression.
A-B. Cell lysates of the indicated NSCLC cell lines were subjected to immunoprecipitation with an anti-MCL1 antibody (M), an anti-BCL-xL antibody (X) or no antibody control (N). The indicated proteins left in the post-immunoprecipitation fraction were detected using Western blotting (A). The fraction of unbound BIM and BIM input was quantified by Image J software and the color denotes cell viability after 24h of treatment with triptolide (B).
C. HMC-1-8 cells stably expressing FLAG-BCL-xL via the MSCV retroviral vector or the Lenti6.3 lentiviral vector, or LacZ control were subjected to immunoprecipitation using an anti-MCL1 antibody. Input, unbound and Immunoprecipitation (IP) fraction were analyzed by Western blotting. The IP fraction was 5-fold more concentrated than input and unbound.
D. HMC-1-8 cells were treated with 2µM triptolide for indicated time, and subjected to immunoprecipitation with an anti-MCL1 antibody conjugated to agarose beads, eluted with an MCL1 peptide, and Western blotted for pro-apoptotic proteins. The IP fraction was 5-fold more concentrated than input and unbound.
E-G. Wild type (WT), Bak−/−, Bax−/− and Bax−/−;Bak−/− MEFs were treated with TR compounds triptolide (E-F) or flavopiridol (G), or non-TR compound trichostatin A (H), at indicated concentrations for 7 hours (E, for caspase activation) or 24 hours (F-H, for viability). Error bars indicate standard deviation of duplicate measurements. See also Figure S4.
Because BIM seemed unlikely to be the principle pro-apoptotic mediator of MCL1 repression, we considered other candidate proteins. MCL1 co-immunoprecipitation experiments showed that while the majority of PUMA, BAK and BAX proteins were not bound by MCL1 (Figure 5A, S4A), significant amounts of PUMA and BAK were pulled down by MCL1, and overexpression of BCL-xL disrupted this interaction (Figure 5C,D). MCL1-bound PUMA decreased after triptolide-mediated MCL1 repression, but this result is best explained by triptolide’s concomitant repression of PUMA expression (Figure 5D). To test the possibility that BAK release from MCL1 explains the TR effect, we used Bak−/− MEFs to determine contribution of Bak in TR compound-induced apoptosis. Bak deletion nearly completely rescued cells from TRs but did not protect cells from the non-TR compound trichostatin A (Figure 5E-H). BAX and BAK are both multidomain pro-apoptotic BCL2 family proteins. However, BAK proved unique in that we did not detect MCL1-BAX interaction in co-immunoprecipitation experiments (Figure 5C,D), and Bax−/− cells were not rescued from TR compounds (Figure 5E-G). Taken together, our data suggest that MCL1 protects cells from cell death at least in part through sequestration of BAK, and this sequestration is diminished with TR compound-mediated MCL1 repression.
BCL-xL predicts MCL1 dependency in vivo
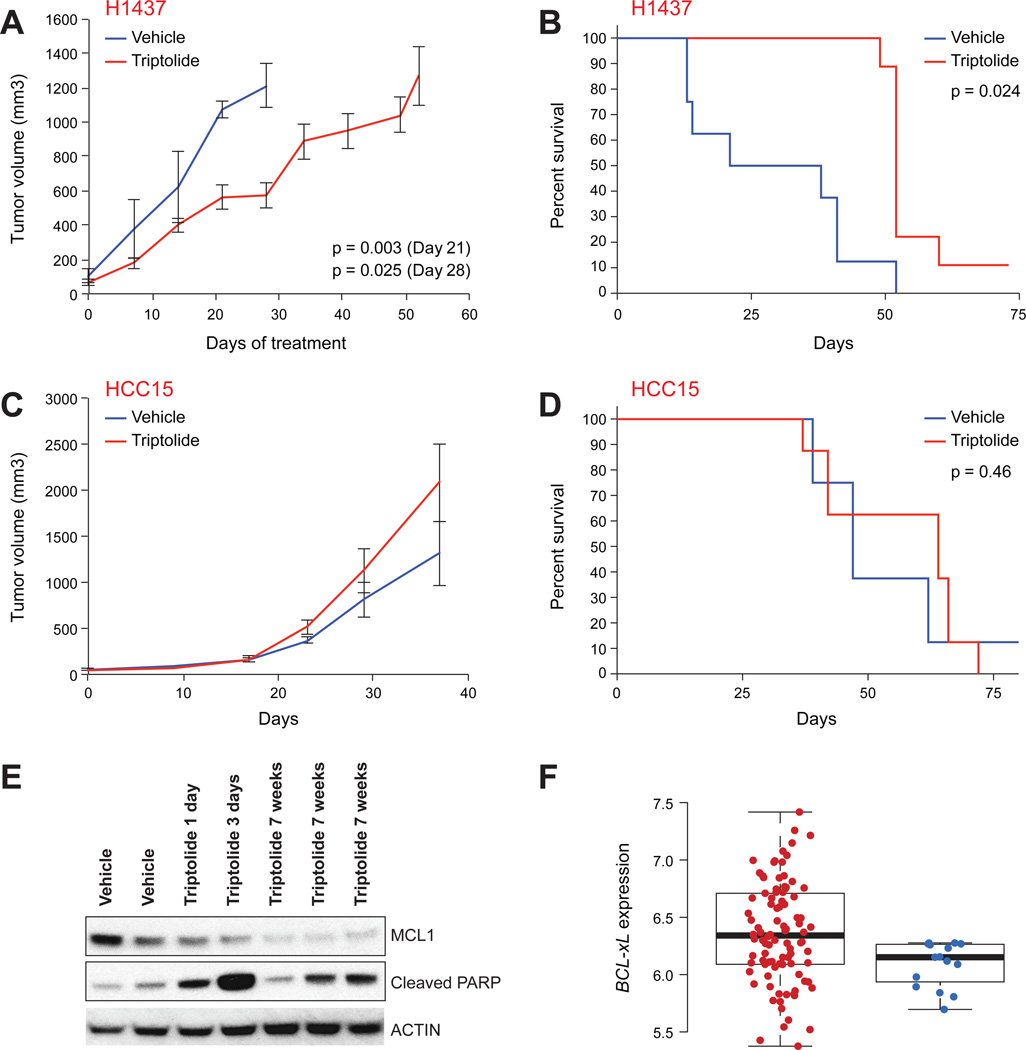
An important question in developing biomarkers of MCL1 dependency is whether resistance mechanisms observed in vitro hold in vivo, where tumor-microenvironment interactions are known to modulate apoptotic mechanisms (Bewry et al., 2008; Buggins et al., 2010). We therefore examined the in vivo response of two NSCLC cell lines grown as xenografts in NOD-SCID mice. H1437 cancer cells express low levels of BCL-xL and are sensitive to triptolide (as a prototype MCL1 repressor) in vitro. HCC15 cells, in contrast, express high levels of BCL-xL and are triptolide-resistant in vitro. This pattern of sensitivity persisted in vivo. Triptolide significantly attenuated the growth of the H1437 NSCLC cancer model (Figure 6A-B). By contrast, in the HCC15 xenograft model, triptolide did not significantly affect tumor volume or survival of the mice (Figure 6C-D). Western blotting of whole tumor lysates demonstrated that treatment with triptolide decreased MCL1 protein abundance and increased PARP cleavage in the H1437 xenograft model (Figure 6E), indicated that triptolide repressed MCL1 expression and induced apoptosis in vivo.
Figure 6. Cells expressing low levels of BCL-xL were sensitive to triptolide in vivo.
A-D. H1437 (low BCL-xL expression; A, B) and HCC15 (high BCL-xL expression; C, D) NSCLC cell lines were grown as xenograft in NOD-SCID mice and the mice treated with triptolide or vehicle as indicated. A,C. Tumor volume. B,D. Survival curves. Error bars indicate standard deviation of tumor volume of 8–9 mice.
E. Triptolide treatment in vivo reduced MCL1 expression and induced PARP cleavage, a marker of apoptosis in tumors. Mice were treated either with vehicle alone or triptolide for 7 weeks (lanes marked Vehicle and triptolide 7 weeks, respectively), or with vehicle for 7 weeks followed by 1–3 days of triptolide (middle lanes).
F. Correlation between BCL-xL expression levels and resistance to epirubicin in ER-negative breast cancer patients (GEO accession number GSE16446). The distribution of BCL-xL expression levels, averaged over all BCL-xL probesets on the Affymetrix U133 Plus 2 array is displayed as a box plot for patients who obtained (N=16, blue dots, right column) or did not obtain (N=98, red dots, left column) pathological complete response to single agent epirubicin treatment. Whiskers of box-plots represent 5%–95% data span. Box-plots present median, 25th, and 75th percentiles of data.
See also Figure S5.
Our model predicts that patients with high levels of BCL-xL expression are resistant to TRs. To test this hypothesis, we investigated the relationship between BCL-xL gene expression and clinical response to neoadjuvant treatment with the anthracycline epirubicin in 114 estrogen receptor-negative breast cancer patients for which it was determined whether a complete pathological response (pCR) was achieved (Desmedt et al., 2011). BCL-xL showed significant differential expression between patients who achieved pCR and those who did not (Figure 6F). As previously reported, expression of topoisomerase 2A (TOP2A) did not correlate with response to epirubicin (Figure S5), consistent with our finding that anthracyclines kill tumor cells via a transcriptional repressive mechanism rather than via a topoisomerase inhibitory mechanism as has been generally assumed.
BCL-xL is a functional determinant of MCL1 dependency
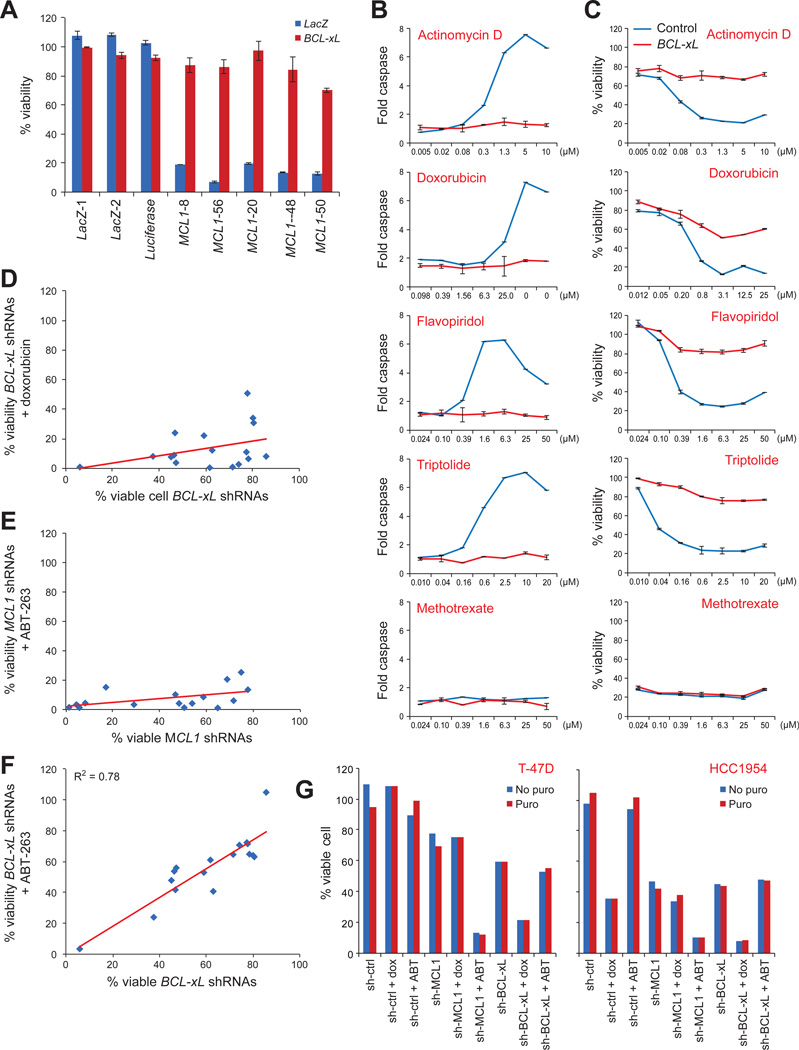
We next investigated whether BCL-xL was simply a marker of MCL1 dependency or it was a functional determinant of response. Overexpression of BCL-xL in MCL1-dependent lines protected them from apoptosis induced by MCL1 shRNAs or TR compounds (Figure 7A-C), but not by other cytotoxic agents such as methotrexate (Figure 7B-C), suggesting a specific effect for TR compounds. Conversely, BCL-xL knockdown conferred sensitivity in cell lines otherwise resistant to TR compounds. Cell lines resistant to treatment with TR compounds (using doxorubicin as a representative example) were sensitive to combined treatment with BCL-xL shRNAs (Figure 7D,G), and cell lines resistant to treatment with MCL1 shRNAs were sensitive to combined treatment with the BCL-xL inhibitor ABT-263 (Figure 7E,G). The viability of cells treated with BCL-xL shRNAs was highly correlated with viability after treatment with the BCL-xL inhibitor ABT-263, and combined treatment of cells with ABT-263 and BCL-xL shRNAs did not yield synergistic effects (Figure 7F-G).
Figure 7. BCL-xL expression determined sensitivity to TR compounds and MCL1 repression.
A. Effect of ectopic expression of BCL-xL on apoptosis induced by MCL1 shRNAs in HMC-1-8 cells. Error bars indicate standard deviation of duplicate measurements.
B-C. Ectopic expression of BCL-xL protected sensitive HMC-1-8 cells from TR compounds, but not methotrexate, as measured both by caspase activity (B) and cell viability (C). Error bars indicate standard deviation of duplicate measurements.
D-F. The effects of BCL-xL shRNAs (D,F) or MCL1 shRNAs (E) on cell viability plotted on x-axis against sensitivity to the combination of BCL-xL shRNAs (D,F) or MCL1 shRNAs (E) and doxorubicin (D) or ABT-263 (E,F) in 17 breast cancer cell lines. Cells were infected with viruses carrying shRNAs for MCL1 or BCL-xL for 3 days, or 2 days followed by treatment with 5µM doxorubicin or 1µM ABT-263 for an additional 24 hours (median of 5 MCL1 shRNAs or BCL-xL shRNAs as indicated).
G. Examples of combination treatment of MCL1 or BCL-xL shRNAs and doxorubicin (dox) or ABT-263 (ABT). puro: puromycin.
See also Figure S5.
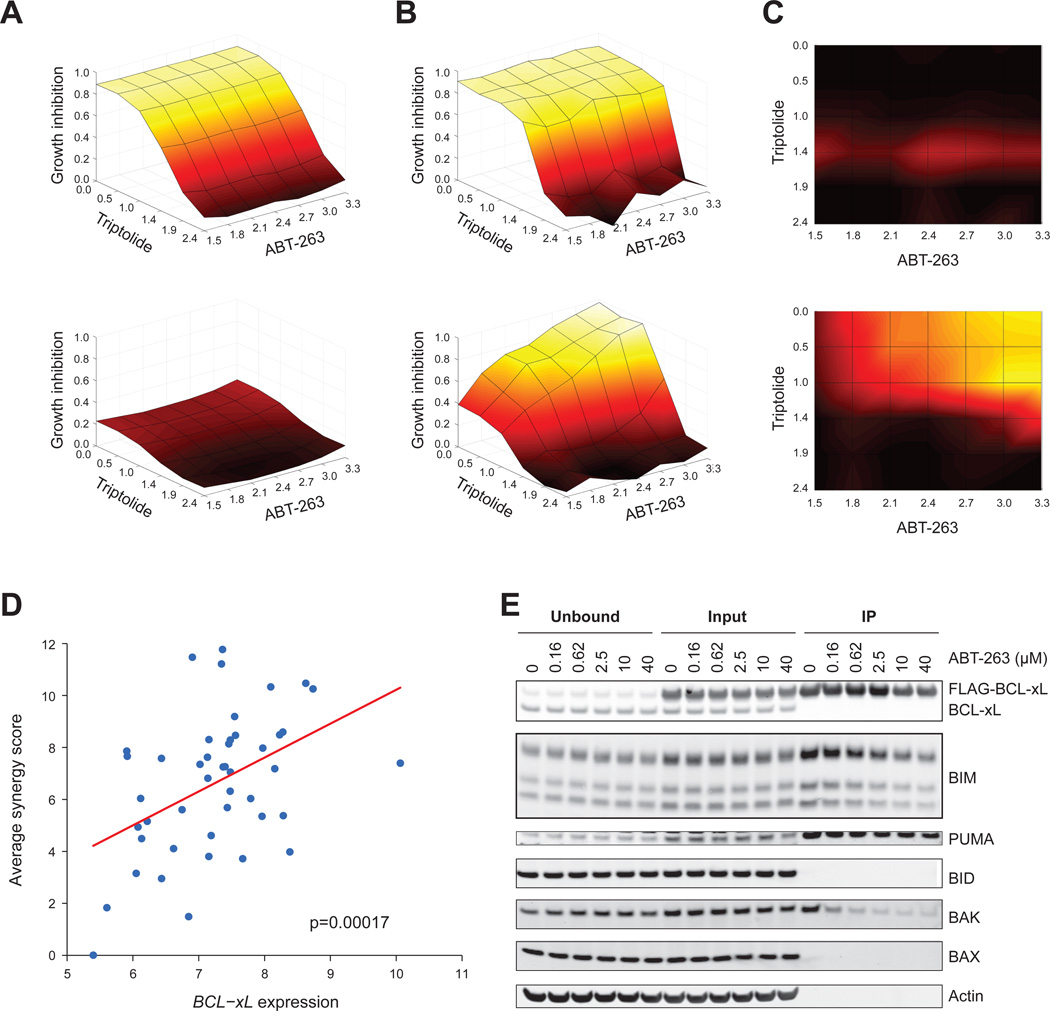
The above data suggest that TR compounds would exhibit a synergistic effect when used in combination with BCL-xL inhibitors. We treated a panel of 74 NSCLC cancer cell lines with a 42-point dose-response matrix (6 concentrations of triptolide or Actinomycin D, and 7 concentrations of ABT-263 or ABT-737). We examined the synergy between TR compounds and BCL-xL inhibitors for each cell line by computing the excess growth inhibition over the Bliss independence model for each combination of compound concentrations (Figure 8A-C). Cell lines that were highly sensitive to TR compounds showed no evidence of synergy when treated in combination with ABT-737. Cell lines that were resistant to TR compounds and to BCL-xL inhibitors were sensitive to the combination (Figure 8A-C).
Figure 8. TR compounds and BCL-xL inhibitor compounds synergistically killed cells with high BCL-xL expression.
A-C. Example of synergy calculation. Cell lines expressing BCL-xL at the lowest (NCI-H23, top panel) and highest (SKLU1, bottom panel) levels, across a panel of 74 NSCLC cell lines.
A. Bliss Independence model. Dose-response curves for single-agent treatment with ABT-263 and triptolide were used to compute the null-hypothesis surface of expected response to combination treatment with ABT-263 and triptolide using a Bliss independence model. The xand y-axis represent concentrations of ABT-263 and triptolide in log10(nM), and the z-axis (as well as the color scale) represents the expected percent growth inhibition for each combination of compound concentrations under the null hypothesis model.
B. Dose response matrix. Observed response surfaces for the combination experiments were plotted.
C. Excess over Bliss independence. Excess percent growth inhibition over the null hypothesis model was computed for each combination of compound concentrations by subtracting the growth inhibition values displayed in (A) from those in (B). Black represents no excess growth inhibition and yellow represents excess growth inhibition (scale 0 to 1).
D. BCL-xL expression level versus compound synergy across the panel of 74 cell lines. The x-axis displays the log2 expression level of BCL-xL. The y-axis displays the synergy score, computed by summing the excess over bliss independence values (as shown in C) over all combinations of compound concentrations. The synergy scores displayed in the plot represent the average synergy score over the 4 combination experiments, performed by pairing triptolide or actinomycin D with ABT-263 or ABT-737.
E. Cells stably expressing FLAG-tagged BCL-xL were treated with ABT-263 at indicated concentrations for 3 hours, and subjected to immunoprecipitation with an anti-FLAG antibody beads, and eluted with 3X FLAG peptides. The IP fraction was 5-fold more concentrated than input and unbound.
A synergy score was computed for each combination experiment in each of the 74 NSCLC cell lines by summing the excess over Bliss independence across all dose combinations. The synergy score was averaged over the 4 combination experiments, performed by pairing triptolide or actinomycin D with ABT-263 or ABT-737. This synergy score was highly correlated with expression of BCL-xL (Figure 8D), suggesting that high expression of BCL-xL determines the synergistic relationship between TR compounds and BCL-xL inhibitory compounds, and that resistance to TR compounds, caused by high expression of BCL-xL, can be overcome by treating in combination with BCL-xL inhibitors. Consistent with this notion, ABT-263 released BAK from BCL-xL (Figure 8E).
DISCUSSION
At an accelerating pace, the genomic characterization of human cancer is elucidating the molecular basis of the disease. Recent large-scale analyses of gene copy number in cancer demonstrated that the genes encoding the BCL2-family proteins MCL1 and BCL-xL are frequent targets of amplification. Low-level MCL1 amplification is particularly notable, representing one of the most common copy number abnormalities in all of human cancer (Beroukhim et al., 2010). In support of a functionally important role of MCL1, numerous studies have elucidated the critical role of MCL1 in preventing tumor cell death (Warr and Shore, 2008).
Using a multiplexed Luminex bead-based assay, we screened for compounds that reduced MCL1 expression while preserving the expression of proapoptotic genes. Although the compounds that emerged from this screen were general transcriptional repressor (TR) compounds (as opposed to specifically targeting the MCL1 locus), they preferentially repressed MCL1 because of the short half-life of MCL1 mRNA and protein. Multiple lines of evidence suggest that TR compounds induce apoptosis in cancer cells primarily through repression of MCL1 expression including 1) upon treatment with TR compounds, MCL1 protein levels decreased rapidly and preceded caspase activation; 2) ectopic expression of physiological levels of MCL1 rescued cancer cells from TR compounds, despite the expression of other genes still being repressed; 3) the pattern of TR compound sensitivity across a panel of cancer cell lines closely mirrored the pattern of sensitivity of those cell lines to MCL1 knock-down by RNAi; 4) of over 40,000 genomic features measured, the top feature that predicted sensitivity to TR compounds was the low expression of BCL-xL, which shares redundant function with MCL1; 5) Ectopic expression of BCL-xL rescued cancer cells from TR compounds; 6) MCL1 repression by TR compounds resulted in the release of, pro-apoptotic protein BAK protein from MCL1; and 7) Bak deficiency protected cells from TR compounds. These results suggest that the mechanism of cell death induced by TR compounds is best explained by MCL1 inhibition.
This indicated that some of the widely used chemotherapeutic drugs such as anthracyclines may preferentially repress MCL1 to induce apoptosis in tumor cells. Although the anti-tumor effect of anthracyclines has long been speculated to be related to the drug’s inhibition of DNA topoisomerase II (Desmedt et al., 2011; Moretti et al., 2009), and an association between low TOP2A expression and anthracycline response in ER-negative breast cancer patients has been reported (Martin, 2011), our data suggest that their activity may be largely explained by inhibition of transcription, leading most dramatically to the repression of short-lived MCL1 transcripts. While it is possible that multiple mechanisms of action explain the anti-tumor effects of anthracyclines, at least in the experimental cancer models studied here, anthracycline gene expression consequences most reflected transcriptional inhibition rather than DNA topoisomerase II inhibition. Furthermore, the similar pattern of sensitivity of cell lines to MCL1 knockdown compared to anthracycline treatment is also consistent with an MCL1-mediated transcriptional inhibitory effect. Lastly, our observation that BCL-xL expression is predictive of resistance to MCL1 repression both in model systems and in patients with breast cancer further strengthens the anthracycline-MCL1 connection. We note that the concentration of doxorubicin used in our experiments approximates that observed in human tumor tissues (1.9–24.4 µM) (Rossi et al., 1987). Doxorubicin stimulates topoisomerase II-mediated DNA cleavage only at low concentrations, whereas at doses greater than ~ 0.4 µM, topoisomerase II-mediated DNA cleavage is lost (Tewey et al., 1984). These data therefore suggest that at clinically relevant concentrations, anthracyclines act as transcriptional repressors, as opposed to DNA damaging agents.
The transcriptional inhibitory role of anthracyclines is also of importance when considering anthracycline-based combination therapies. The transcriptional induction of pro-apoptotic proteins has been reported to be crucial for the efficacy of many classes of anti-neoplastic agents including radiation (Jeffers et al., 2003; Villunger et al., 2003), the proteasome inhibitor bortezomib (Gomez-Bougie et al., 2007; Voortman et al., 2007), the HDAC inhibitor vorinostat (Wiegmans et al., 2011), and the kinase inhibitors imatinib (Kuroda et al., 2006) and erlotinib (Gong et al., 2007). Anthracyclines may block the induction of such pro-apoptotic proteins and counteract, rather than synergize, with those therapies. For example, we found that doxorubicin treatment actually rescues cancer cells from bortezomib- and vorinostat-induced killing (Figure S2). Such antagonistic actions may be preventable by adjusting the dosing schedule of combination therapies, but the results serve as a reminder that knowledge of mechanisms of action should ideally be considered in developing combination strategies.
Taken together, the results reported here elucidate a strategy for the development of MCL1 inhibitors as cancer therapeutics. The multiplexed, gene-expression-based high-throughput screening approach described here holds promise for the future discovery of specific inhibitors of MCL1 expression and for the use of chemical genomic approaches to elucidate small-molecule mechanisms of action. The study also highlights the power of genomically-characterized cell lines for the discovery of predictive biomarkers of drug response. Most immediately, the work suggests an approach to the clinical development of any MCL1 inhibitor in breast and NSCLC tumors, focusing on tumors expressing low levels of BCL-xL as a patient-selection strategy.
Experimental Procedures
Cell culture, caspase and viability assay
All human cell lines were part of the Broad-Novartis Cancer Cell Line Encyclopedia (CCLE) Project (http://www.broadinstitute.org/ccle/home) or gifts from Dr. Mathew Meyerson at the Broad Institute. Bak−/−, Bax−/− and Bax−/−;Bak−/− MEFs were gifts from Dr. Anthony Letai at the Dana Farber Cancer Institute. Caspase activity was measured by Caspase-Glo 3/7 kit and cell viability was measured by CellTiter-Glo (both from Promega, Madison, WI). cDNAs for ectopic expression were from Human ORFeome collection by Dr. Mark Vidal at Dana-Farber Cancer Institute. Compounds were purchased from commercial sources listed in the supplement table 2, or were synthesized at the Broad Institute.
High-throughput gene expression screen for proximal apoptosis genes and gene expression profiling by microarray
MCF7 cells growing in 384 well dishes were treated with 2,922 small molecules from small molecule libraries from the Broad Institute Chemical Biology Program for 8 hours before being lysed. mRNA in cell lysates was hybridized to dT20-conjugated plates (Qiagen, Valencia, CA) and then reverse transcribed by Superscript II (Promega, Madison WI). The resulting covalently attached cDNA was amplified by ligation-mediated amplification. For each gene to be assayed, upstream and downstream probes with unique barcode tag and universal primer sites were annealed to targeted cDNA and ligation by Taq DNA ligase (New England Biolabs, Ipswich, MA) generated a sequence complementary to the transcript. The ligation product was PCR amplified using biotin-conjugated universal primers. The PCR products were then captured by hybridization to probes complementary to the barcodes that were linked to uniquely colored polystyrene beads (Luminex, Austin, TX). The products were subsequently stained with streptavidin-phycoerythrin (SAPE) (Invitrogen, Carlsbad, CA). Each gene product was identified by the color of its capture bead and quantified using the associated SAPE fluorescence, as measured by a Luminex detector (Luminex, Austin, TX). MCF-7 cells were treated with 500 nM triptolide or 2.5 µM actinomycin D for 2, 4 or 6 hours, and gene expression was profiled using Affymetrix microarrays (GEO accession number GSE28662).
RNAi and ectopic expression
shRNAs viral infection was performed as previously described (Moffat et al., 2006). The targeted sequences for the best shRNAs for MCL1 and BCL-xL are listed in Supplemental Experimental Procedures. Cell viability following treatment with MCL1 or BCL-xL shRNAs was compared to results using three control shRNAs against luciferase or LacZ. For combination studies, cells infected with lentivirus carrying shRNAs were selected with or without puromycin (2.5µg/ml) for two days before small molecules were added. Cell viability was measured 24h after addition of small molecules. A FLAG tag was added to N-terminal of MCL1 or BCL-xL and FLAG-MCL1 or FLAG-BCL-xL was cloned into an Entry vector (Invitrogen, Valencia, CA) followed by recombination into an MSCV destination vector. BCL-xL entry clone was also cloned into the pLenti6.2 destination vector (Invitrogen, Valencia, CA) for Figure 7A-C.
Western blot and co-immunoprecipitation
Cells in figure 6A were lysed in cell lysis buffer (Cell Signaling Technology, Danvers, MA). Otherwise, cells were lysed in CHAPS lysis buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% CHAPS, 1mM EDTA, 1mM EGTA, protease inhibitors and Phosstop (Roche, Indianapolis, IN), and 20µM MG132). Protein lysates were incubated with antibody for MCL1 (Santa-Cruz Biotechnology Inc., Santa Cruz, CA) or BCL-xL (Cell Signaling Technology, Danvers, MA) overnight, then protein-A/G Plus beads (Santa-Cruz Biotechnology Inc., Santa Cruz, CA) were added and incubated for an additional 4 hours. Agarose beads conjugated with an MCL1 antibody and an MCL1 peptide was used in Figure 5D. Anti-FLAG beads and 3X FLAG peptides (both from Sigma, St Louis, MO) were used in Figure 8E. Antibodies for MCL1 Westerns were from Santa-Cruz Biotechnology Inc., (Santa Cruz, CA,) and BD Pharmigen (San Jose, CA), BCL-xL, BIM, PUMA, BAK and PARP antibodies were from Cell Signaling Technology, (Danvers, MA). BAX and ACTIN antibodies were from Millipore (Billerica, MA). Protein quantification was performed by Image J.
Animal studies
Mice were imaged 2 weeks after SQ injection of H1437-Luc-mCherry or HCC15-Luc-mCherry cells to identify mice with established tumor burden. Tumor measurements were taken twice weekly to track tumor volume. All mice had established tumors at 2 weeks and were entered into treatment groups each containing 8 or 9 mice, with all groups having around the same BLI average. Treatments were administered daily via IP injection and mice were measured weekly for six weeks. The animals had tumor measurements twice weekly. The time to sacrifice was determined by tumor volume reaching 1500 cm2, or tumor ulceration. The xenograft mice were generated, housed and bred in the Dana-Farber Cancer Institute (DFCI) animal facility. All animal experiments were approved by the Dana-Farber Cancer Institute IACUC.
Supplementary Material
Highlights.
Chemical genomics identified MCL1 repressors including anthracyclines.
MCL1 is a major downstream target of transcriptional inhibitors.
Expression level of BCL-xL is a resistance biomarker for MCL1 inhibitors.
BCL-xL is a major determinant of resistance to anthracyclines.
Acknowledgements
This work was supported by NIH grant P01 CA068484 and 5U54CA112962 to TRG. G.W. was supported by a postdoctoral fellowship from Damon Runyon Cancer Research Foundation. The authors thank the Broad Institute Chemical Biology Platform for assistance in small-molecule screening, the Broad Institute Genetic Analysis Platform for Affymetrix microarray profiling, the Broad-Novartis Cancer Cell Line Encyclopedia project for data sharing, Joshua Gould for assistance in data analysis, Jinyan Du, Xiaodong Lu and Emily Gray for technical assistance, and Leslie Gaffney and Lauren Solomon for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SIGNIFICANCE
Human tumors effectively escape cell death by activating anti-apoptotic mechanisms, one of which is amplification of MCL1. We describe here a chemical genomic approach to the discovery of repressors of MCL1 expression. We also identify high BCL-xL expression as a primary resistance mechanism to MCL1 inhibition, including resistance to anthracyclines, which we show act at least in part through an MCL1-inhibitory mechanism.
Supplemental information
Supplemental information includes five figures, two tables, and Supplemental Experimental Procedures and can be found with this article online.
References
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KW, Cooper GM. Rapid turnover of mcl-1 couples translation to cell survival and apoptosis. J Biol Chem. 2007;282:6192–6200. doi: 10.1074/jbc.M610643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewry NN, Nair RR, Emmons MF, Boulware D, Pinilla-Ibarz J, Hazlehurst LA. Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance. Mol Cancer Ther. 2008;7:3169–3175. doi: 10.1158/1535-7163.MCT-08-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggins AG, Pepper C, Patten PE, Hewamana S, Gohil S, Moorhead J, Folarin N, Yallop D, Thomas NS, Mufti GJ, et al. Interaction with vascular endothelium enhances survival in primary chronic lymphocytic leukemia cells via NF-kappaB activation and de novo gene transcription. Cancer Res. 2010;70:7523–7533. doi: 10.1158/0008-5472.CAN-10-1634. [DOI] [PubMed] [Google Scholar]
- Chen R, Keating MJ, Gandhi V, Plunkett W. Transcription inhibition by flavopiridol: mechanism of chronic lymphocytic leukemia cell death. Blood. 2005;106:2513–2519. doi: 10.1182/blood-2005-04-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Dai Y, Harada H, Dent P, Grant S. Mcl-1 down-regulation potentiates ABT-737 lethality by cooperatively inducing Bak activation and Bax translocation. Cancer Res. 2007;67:782–791. doi: 10.1158/0008-5472.CAN-06-3964. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Di Leo A, de Azambuja E, Larsimont D, Haibe-Kains B, Selleslags J, Delaloge S, Duhem C, Kains JP, Carly B, et al. Multifactorial approach to predicting resistance to anthracyclines. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- Gomez-Bougie P, Wuilleme-Toumi S, Menoret E, Trichet V, Robillard N, Philippe M, Bataille R, Amiot M. Noxa up-regulation and Mcl-1 cleavage are associated to apoptosis induction by bortezomib in multiple myeloma. Cancer research. 2007;67:5418–5424. doi: 10.1158/0008-5472.CAN-06-4322. [DOI] [PubMed] [Google Scholar]
- Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. Puma is an essential mediator of p53-dependent and - independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Keuling AM, Felton KE, Parker AA, Akbari M, Andrew SE, Tron VA. RNA silencing of Mcl-1 enhances ABT-737-mediated apoptosis in melanoma: role for a caspase-8-dependent pathway. PLoS One. 2009;4:e6651. doi: 10.1371/journal.pone.0006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska M, Fenoglio-Preiser CM, Krajewski S, Song K, Macdonald JS, Stemmerman G, Reed JC. Immunohistochemical analysis of Bcl-2 family proteins in adenocarcinomas of the stomach. Am J Pathol. 1996a;149:1449–1457. [PMC free article] [PubMed] [Google Scholar]
- Krajewska M, Krajewski S, Epstein JI, Shabaik A, Sauvageot J, Song K, Kitada S, Reed JC. Immunohistochemical analysis of bcl-2 bax bcl-X, and mcl-1 expression in prostate cancers. Am J Pathol. 1996b;148:1567–1576. [PMC free article] [PubMed] [Google Scholar]
- Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DC, Kimura S, Ottmann OG, Druker BJ, Villunger A, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14907–14912. doi: 10.1073/pnas.0606176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SI, Dudley AM, Drubin D, Silver PA, Krogan NJ, Pe'er D, Koller D. Learning a prior on regulatory potential from eQTL data. PLoS Genet. 2009;5:e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenroth SJ, Crews CM. Triptolide-induced transcriptional arrest is associated with changes in nuclear substructure. Cancer Res. 2008;68:5257–5266. doi: 10.1158/0008-5472.CAN-07-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Morgan-Lappe S, Huang X, Li L, Zakula DM, Vernetti LA, Fesik SW, Shen Y. 'Seed' analysis of off-target siRNAs reveals an essential role of Mcl-1 in resistance to the small-molecule Bcl-2/Bcl-X(L) inhibitor ABT-737. Oncogene. 2006 doi: 10.1038/sj.onc.1210166. [DOI] [PubMed] [Google Scholar]
- Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Romero A, Cheang MCU, Lopez Garcia-Asenjo JA, Garcia-Saenz JA, Oliva B, Roman JM, He X, Casado A, de la Torre J, Furio V, Puente J, Caldes T, Vidart JA, Lopez-Tarruella S, Diaz-Rubio E, Perou CM. Genomic predictors of response to doxorubicin versus docetaxel in primary breast cancer. Breast Cancer Res Treat. 2011;128:10. doi: 10.1007/s10549-011-1461-y. [DOI] [PubMed] [Google Scholar]
- Merino D, Strasser A, Bouillet P. Bim must be able to engage all pro-survival Bcl-2 family members for efficient tumor suppression. Oncogene. 2011 doi: 10.1038/onc.2011.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Moretti E, Oakman C, Di Leo A. Predicting anthracycline benefit: have we made any progress? Current opinion in oncology. 2009;21:507–515. doi: 10.1097/CCO.0b013e328331a501. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Peck D, Crawford ED, Ross KN, Stegmaier K, Golub TR, Lamb J. A method for high-throughput gene expression signature analysis. Genome Biol. 2006;7:R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Rossi C, Gasparini G, Canobbio L, Galligioni E, Volpe R, Candiani E, Toffoli G, D'Incalci M. Doxorubicin distribution in human breast cancer. Cancer Treat Rep. 1987;71:1221–1226. [PubMed] [Google Scholar]
- Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. XPB a subunit of TFIIH, is a target of the natural product triptolide. Nature chemical biology. 2011;7:182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer research. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A. p53- and drug-induced apoptotic responses mediated by BH3- only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- Voortman J, Checinska A, Giaccone G, Rodriguez JA, Kruyt FA. Bortezomib, but not cisplatin, induces mitochondria-dependent apoptosis accompanied by up-regulation of noxa in the non-small cell lung cancer cell line NCI-H460. Molecular cancer therapeutics. 2007;6:1046–1053. doi: 10.1158/1535-7163.MCT-06-0577. [DOI] [PubMed] [Google Scholar]
- Warr MR, Shore GC. Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med. 2008;8:138–147. doi: 10.2174/156652408783769580. [DOI] [PubMed] [Google Scholar]
- Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, Opferman JT, Sallan SE, den Boer ML, Pieters R, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmans AP, Alsop AE, Bots M, Cluse LA, Williams SP, Banks KM, Ralli R, Scott CL, Frenzel A, Villunger A, et al. Deciphering the molecular events necessary for synergistic tumor cell apoptosis mediated by the histone deacetylase inhibitor vorinostat and the BH3 mimetic ABT-737. Cancer research. 2011;71:3603–3615. doi: 10.1158/0008-5472.CAN-10-3289. [DOI] [PubMed] [Google Scholar]
- Zhou P, Levy NB, Xie H, Qian L, Lee CY, Gascoyne RD, Craig RW. MCL1 transgenic mice exhibit a high incidence of B-cell lymphoma manifested as a spectrum of histologic subtypes. Blood. 2001;97:3902–3909. doi: 10.1182/blood.v97.12.3902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.