Abstract
Over 2 billion people are infected with M. tuberculosis (M.tb); however, only 5–10% of those infected will develop active disease. Recent data suggest that containment is controlled locally at the level of the granuloma and that granuloma architecture may differ even within a single infected individual. Formation of a granuloma likely requires exposure to mycobacterial components released from infected macrophages but the mechanism of their release is still unclear. We hypothesize that exosomes, which are small membrane vesicles containing mycobacterial components released from infected macrophages, could promote cellular recruitment during granuloma formation. In support of this hypothesis, we found that C57BL/6 mouse-derived bone marrow macrophages treated with exosomes released from M.tb-infected RAW264.7 cells secrete significant levels of chemokines and can induce migration of CFSE-labeled macrophages and splenocytes. Exosomes isolated from the serum of M. bovis BCG infected mice could also stimulate macrophage production of chemokines and cytokines ex vivo but the level and type differed during the course of a 60 day infection. Interestingly, the exosome concentration in serum correlated strongly with mouse bacterial load, suggesting some role in immune regulation. Finally, hollow fiber-based experiments indicated that macrophages treated with exosomes released from Mtb-infected cells could promote macrophage recruitment in vivo. Exosomes injected intranasally could also recruit CD11b+ cells into the lung. Overall, our study suggests that exosomes may play an important role in recruiting and regulating host cells during an M. tuberculosis infection.
Introduction
Mycobacterium tuberculosis (M.tb) is an intracellular pathogen and the causative agent of tuberculosis (TB). An estimated 1.7 million individuals died from TB in 2009 and another eight million were infected (1). Although over 2 billion people have been infected with Mtb, only 5–10% of those infected will develop active disease (2). This percentage increases dramatically in HIV/Mtb co-infected patients. Evidence suggests a robust immune response in both individuals who control as well as succumb to infection; however, the molecular and immunological mechanisms that dictate control or susceptibility remain mostly undefined. Nevertheless, recent data suggests that containment or lack thereof is controlled locally at the level of the granuloma (3, 4, 5). In humans these dynamic structures consist of a caseated center surrounded by activated macrophages, fibrotic tissue and lymphocytes (6). The formation of these granulomas requires cytokines such as TNF-α as well as chemotactic factors to promote recruitment and activation of macrophages and later T cells (7). Infected macrophages are considered the primary producers of these mediators but published data also suggest that mycobacterial components can facilitate granuloma formation (8,9). However, the mechanism by which these mycobacterial components gain access to the extracellular milieu has not been defined. We hypothesized that exosomes released from infected macrophages in vivo might be the carriers of these mycobacterial components.
Exosomes are 30–100nm microvesicles of endocytic origin, which are secreted by hematopoietic and non-hematopoietic cells (10,11). They are known to function in intercellular communication, including antigen presentation and T cell activation (12), as well as immune suppression (13). Exosomes isolated from macrophages infected with M.tb in vitro contain mycobacterial components including ManLAM and other mycobacterial lipids (14) and can activate macrophages and antigen-specific T cells in vitro and in vivo (15). However, these previous studies did not address the production of exosomes during an in vivo infection, the activity of these in vivo-derived exosomes, or whether exosomes could promote cellular recruitment. To address these questions, we defined the chemokine response and cell recruitment activity of non-infected macrophages after treatment with different exosome populations. We also isolated exosomes from mice infected with M. bovis BCG and analyzed their biological activity using a cytokine membrane array. Finally, we evaluated the exosome-treated macrophages for recruitment of cells in vivo using a hollow fiber implantation model (16). The use of a PVDF hollow fiber allows for movement of soluble components but restricts cellular exchange between inside and outside the fiber. The hollow fiber can be removed and analyzed for cells attached to the external surface. We found that exosome concentration in serum correlated strongly with bacterial load in M. bovis BCG-infected mice and that these exosomes could stimulate macrophage chemokine production ex vivo. We also found that macrophages treated with exosomes released from infected cells are chemotactic for leukocytes both in vitro and in vivo. Our study suggests that exosomes released from infected cells during a mycobacterial infection could facilitate cellular recruitment as well as other immune functions.
Materials and Methods
Ethics Statement
The University of Notre Dame is credited through the Animal Welfare Assurance (#A3093–01). All animal studies were conducted according to the Institutional Animal Care and Use Committee (IACUC) guidelines.
Macrophage culture and bacterial strains
Bone marrow-derived macrophages (BMM) were isolated from 6–8 week old female C57BL/6 mice and cultured in vitro as previously described (17). The mouse macrophage cell line RAW 264.7 was maintained in RPMI supplemented with 10% fetal bovine serum, 10 mM sodium pyruvate and 25 mM HEPES. M.tb H37Rv, M. smegmatis and M. bovis BCG were each grown in Middlebrook 7H9 broth supplemented with OADC until mid logarithmic growth phase and frozen down as stocks in growth media plus 15% glycerol. Prior to use, the bacterial stocks were thawed and the mycobacteria were de-clumped by a brief sonication and passed through syringe fitted with 27 gauge needle at least ten times.
Isolation of exosomes from cell culture supernatants
Confluent monolayers of RAW 264.7 mouse macrophage cell line (1.5 × 108 cells) were infected with Mtb, M. smegmatis or left uninfected as controls. Before infection, the bacterial cultures were opsonized by incubation for 2 hours with normal horse serum and an uptake assay was undertaken to obtain approximately 80% infectivity, as described (17). The RAW264.7 macrophages were infected with bacteria for 4 hours followed by washes with 1X PBS. The cells were cultured in RPMI containing exosome-free FBS (10% final concentration) and exosomes were isolated from the culture supernatants of infected and uninfected RAW 264.7 cells (Un exosomes) after 72 hours and purified on linear sucrose gradient, as previously described (18). The exosomes formed a distinct ring based on their density (1.13 and 1.18 g/ml) and were carefully recovered from the gradient. In absence of a distinct ring, gradient fractions 5, 6 and 7 were collected, pooled and washed in PBS (1X). Exosomes were also purified using ExoQuick purification (System BioSciences, CA) for different experiments. On average, 20μg of purified exosomes were obtained from 10 million cells.
Mouse infections and CFU determination
C57BL/6 mice were infected retro-orbitally with M. bovis BCG (106 bacilli per mouse) or injected with equal volume of PBS. Mice were sacrificed at different time points (4 mice per group) and serum and spleens were collected. Exosomes were isolated from mouse serum by precipitating in ExoQuick solution overnight following manufacturer’s instructions. Exosomes isolated from M. bovis BCG-infected or PBS-treated mice are referred to as BCG exosomes or PBS exosomes respectively. Spleens from infected mice were homogenized and passed through a 70μm cell strainer. The cell suspension was treated with RBC lysis buffer followed by washes in 1X PBS. The suspension was serially diluted in PBS+Tween 80 (0.05% v/v) and plated on 7H10 agar supplemented with OADC. Colony Forming Units (CFU) were determined after 4 weeks.
Antibody array and ELISA for chemokine levels
Primary bone marrow macrophages (1 × 106 cells) were stimulated with exosomes at 40 μg/ml (Un and Rv exosomes) or 500 μg/ml (BCG and PBS exosomes) for 16 hours or left untreated. The cell culture supernatants were harvested and particulates were removed by centrifugation. The supernatants were tested immediately for cytokine/chemokine levels using Mouse Cytokine Array panel A array kit (R&D Systems) following manufacturer’s instructions. Briefly, cell culture supernatants were mixed with a cocktail of biotinylated detection antibodies and incubated with the nitrocellulose membrane that contains 40 different anti-cytokine/chemokine capture antibodies spotted in duplicate. Any cytokine/antibody complex formed is bound to the immobilized capture antibody. The membranes were incubated with chemiluminiscent substrate (Pierce) and exposed for to X-ray film. In separate experiments, the cell culture supernatants were also tested for MIP-1α and RANTES levels by ELISA (Biosource, Camarillo, CA and R&D Systems, Minneapolis, MN respectively) following manufacturer’s instructions.
In vitro transwell cell migration assay
BMM were seeded in the bottom chamber of the transwell plate (Costar, Corning. NY, 8.0 μm pore size polycarbonate membrane filter) and stimulated with exosomes as described previously (19). BMM or splenocytes were labeled with 2.5 μM CFSE (Molecular Probes Inc, Eugene, OR.). The labeled cells were seeded in the top chamber of the transwell and allowed to migrate for 2 hours. The filters were rinsed with 1X PBS to remove unattached macrophages and placed on a slide mounted with a coverslip. Fluorescent macrophages which migrated to the bottom surface of the transwell filter were counted in 10 randomly selected fields using Zeiss Observer fluorescent microscope and transmigration index was calculated as the ratio of migration in presence of stimulation to migration in absence of stimulation. Splenic cells that migrated to the bottom chamber of the transwell plate were counted on a hemacytometer and stained for flow cytometry analysis.
Flow cytometry
The cells were rinsed with DPBS and gently scraped and counted on hemacytometer using trypan blue to assess viability. The cells were washed in FACS buffer and blocked with 10% mouse serum and stained with PE conjugated anti-mouse Ly6G/6E, (BD Pharmingen, San Diego, CA), FITC conjugated F4/80, (eBiosciences, San Diego, CA), PE-Cy5 conjugated anti-mouse TCR beta, (BD Pharmingen, San Diego, CA) or PE conjugated CD11b, (BD Pharmingen, San Diego, CA) using isotype antibodies as controls. Cells were analyzed for protein surface expression using a Beckman Coulter flow cytometer and percent positive cells calculated.
Electron Microscopy & Nanosight
Exosomes were resuspended in 150mM Ammonium biocarbonate buffer and 10 μl was loaded onto a Carbon-formvar-coated copper grid. The sample was left on the grid for 5 minutes to form a monolayer and the remaining sample was wiped off using a clean filter paper. The grids were allowed to dry at room temperature. The sample was stained with 5μl 2% filtered uranyl acetate solution for 2 minutes and dried. The grids were loaded onto TITAN 80–300 Scanning Transmission Electron Microscope (STEM) with an accelerating voltage set to 80 kV and images were taken with Fei’s Tecnai software. The exosomes were also characterized for size distribution and quantitated by NanoSight LM10 using light scatter from the 635 nm red laser and using NTA 2.2 Analytical software.
Western blots
Exosomes (40μg) were resuspended in RIPA buffer (25mM Tris-HCl pH 7.6, 150mM NaCl,1% NP-40, 1% sodium deoxycholate & 0.1% SDS) with protease inhibitors. The suspension was mixed with Laemmli buffer, heated at 95°C for 5 minutes and chilled on ice for 5 minutes before loading onto SDS-gel. Rabbit anti-human CD63 and CD9 antibodies (System Biosciences) were used as primary and goat anti-Rabbit IgG HRP was used as secondary antibody.
In vivo cellular recruitment around hollow fiber implant and in mouse lungs
Cellular recruitment was studied in 6–8 week old female hairless, immunocompetent SKH1 mice obtained from Charles River Laboratories and further bred in the Freiman Animal facility. BMMs were left untreated or were treated with exosomes or infected with M. bovis BCG for 16 hours and the cell suspension was placed in the lumen of PVDF hollow fibers (molecular mass cutoff 500 kD; Spectrum Laboratories). The hollow fibers were heat-sealed at both ends with a final length of approximately 2 cm (16). A small incision was made in the SKH1 mice on the dorsal surface beneath the neck and two fibers were implanted subcutaneously into the mouse, one fiber on each side of the body cavity. The incision in the skin was sealed using liquivet tissue adhesive and lidocaine was applied at the site of the incision. The fibers were recovered from mice after 16 days and resuspended in 10% formalin solution for histological studies or in 1X DPBS+10 mM EDTA followed by vortexing to detach the cells for flow cytometry.
To study cellular recruitment in lungs, C57BL6 mice (4–5 mice per group) were treated with PBS, 25 μg of exosomes or infected with M. bovis BCG at 1× 106 CFU per mouse intranasally. After 5 days mice were sacrificed and lungs from 4–5 mice per group were pooled. Total lung cells were treated with Collagenase for 2 hours followed by treatment with Ack lysis buffer (150mM NH4Cl, 1mM KHCO3 and 0.1mM EDTA) to lyse RBCs. Cells were stained with FITC-conjugated anti-mouse F4/80, PE-conjugated anti-mouse CD11b or PE-conjugated anti-mouse Ly6G using isotype-matched antibodies as background control.
Histology
The fibers were fixed in 10% Neutral Buffered Formalin overnight and then transferred to 70% EtOH. The samples were processed using a Shandon Citadell 2000 automated tissue processor. After fixation, samples were dehydrated, embedded in paraffin, and sectioned at 4 μm using a Leica RM 2155 automated microtome. Sections were stained with hematoxylin and eosin (H&E) to examine general tissue and cellular morphology.
Statistical analyses
Data was analyzed by a one-tailed or paired Student’s t test. Statistical significance was assumed at p ≤ 0.05.
Results
Vesicles purified by ExoQuick™ were of similar activity and size compared to sucrose gradient purified vesicles
Purifying exosomes from mouse serum and other sources by sucrose gradient is cumbersome, and time-consuming, with relatively low yield (20% to 40%). Therefore, alternative purification methods are needed. ExoQuick, which uses a proprietary precipitation technique to purify exosomes from serum, media and other sources, has been shown to give exosomes of high enough quality for functional and RNA/protein characterization studies (http://www.systembio.com/exoquick) (20). Therefore, we evaluated ExoQuick-purified vesicles for size and shape using transmission electron microscopy. As shown in Figure 1A, the purified vesicles were of the expected size of exosomes (50 to 100 nm by TEM) and appeared to have minimal vesicles outside this size range. To look more carefully at the size distribution of the ExoQuick-purified exosomes and to compare this with the “gold-standard” sucrose gradient-purified exosomes, we evaluated the exosomes using a NanoSight LM10 which allows for a determination of vesicle size between 30 to 1000 nm. As shown in figure 1B, there was no clear difference in the vesicle size distribution between the ExoQuick- and sucrose gradient-purified exosomes. Interestingly, the mean size for exosomes using this instrument, which tracks the movement of a particle and calculates its size based on Brownian motion, was 128 nm and 144 nm for the ExoQuick- and sucrose gradient-purified exosomes, respectively. This is larger then what size is calculated from the TEM indicating that how exosomes are processed or observed will affect the size measurement. Finally, we observed that exosomes purified from M. smegmatis-infected RAW 264.7 cells by ExoQuick or sucrose gradient showed similar ability to stimulate TNF-α production when added to mouse bone marrow-derived macrophages (Fig. 1C). However, ExoQuick may precipitate other vesicle populations or protein aggregates that we could not detect with the present methods.
Fig. 1. Comparison of exosomes purified on a linear sucrose gradient vs. ExoQuick™.
Exosomes were isolated from cell culture supernatants of M. smegmatis infected or uninfected RAW264.7 macrophages. Exosomes were purified using the ExoQuick precipitation solution or on a linear sucrose gradient (0.25M–2.5M). (A) The exosomes were characterized morphologically by transmission electron microscopy. (B) The exosomes were characterized for size distribution and quantitated by NanoSight LM10 using light scatter from the 635 nm red laser. (C) RAW264.7 macrophages were seeded in a 6-well plate and were left untreated or treated with exosomes at 40 μg/ml for 16 hours. The cell culture supernatants were tested for TNF-α levels by ELISA.
Macrophages treated with exosomes isolated from Mtb-infected cells secrete chemokines and promote macrophage migration in vitro
Exosomes purified from cell culture supernatants of Mtb-infected or uninfected RAW 264.7 cells. Bone marrow-derived macrophages (BMM) were treated with exosomes at 40 μg/ml (equivalent to ~ 2.3 × 1010 exosomes/ml based on NanoSight LM10 measurements) for sixteen hours or left untreated and the cell culture supernatants were harvested. Supernatants were assayed for cytokine/chemokine levels using a mouse cytokine array kit (Fig. 2A). Pixel densities for the different array proteins from three experiments were defined and plotted (Fig.2B). Resting bone marrow macrophages released measurable levels of TIMP1, CXCL1, M-CSF, CCL2, MIP-2, IP-10 and Il-1ra. Treatment of macrophages with exosomes derived from uninfected RAW 264.7 cells (Un-exosomes) induced higher expression of sICAM1, IL-1ra and MIP-2 in comparison to resting cells. However, treatment of macrophages with exosomes released from Mtb infected cells (Rv-exosomes) induced not only measureable levels of TNF-α, MCP-5, MIP-1α, MIP-1β, RANTES and G-CSF but also relatively higher levels of sICAM-1, MIP-2 and IL-1ra compared to resting or Un-exosome-treated macrophages. For a complete list of antibodies spotted on the array see supplement table 1.
Fig. 2. Macrophages treated with exosomes released from mycobacterial-infected cells secrete cytokines and chemokines.
Exosomes from Mtb-infected, M. smegmatis-infected or uninfected RAW264.7 macrophages were purified by ExoQuick and used to treat BMM at 40 μg/ml for 16 hours. The supernatants were harvested and assayed for immune proteins using mouse cytokine arrays. Cytokine arrays were compared between untreated BMM and those treated with exosomes from uninfected or H37Rv-infected cells (A) or between BMM treated with exosomes from M. smegmatis- or H37Rv-infected cells (C). The pixel densities for each spot were calculated using ImajeJ software and plotted. Results are the mean of three separate experiments +SD and asterisk (*) indicates statistical difference when compared to (B) untreated/resting cells (RC) or (D) Rv exosome-treated cells (p value ≤ 0.05). The results obtained in antibody array analysis for Rv exosomes were confirmed for RANTES (E) and MIP-1α (F) by ELISA and the results are the mean of three separate experiments including SD. Asterisk (*) indicating statistical difference when compared to untreated/resting cells (RC) (p value ≤ 0.05). (A1,A2), (F1,F2) and (A23,A24) correspond to reference numbers for cytokine array (see supplement table 1 for arrangement of antibodies on the array).
Interestingly, although exosomes released from M.tb-infected macrophages induced chemokine/cytokine production, the exosomes from cells infected with M. smegmatis, a nonpathogenic mycobacteria, induced a much more robust macrophage response (Fig. 2C and D). Previous studies by our group and others indicated that macrophages infected with nonpathogenic mycobacteria induce macrophage activation to a greater extent compared to infection with pathogenic mycobacteria (17,21). This ability to induce a more robust macrophage response seems to extend to the exosomes released from cells infected with nonpathogenic compared to pathogenic mycobacteria and likely is due to differences in the mycobacterial components retained in/on the exosomes.
RANTES and MIP-1α are among the C-C chemokines known to induce the migration of monocytes and lymphocytes (22) and we confirmed their elevated expression in macrophages treated with exosomes released from H37Rv-infected RAW 264.7 cells (Fig. 2E and F). Since these chemokines were released from macrophages treated with Rv exosomes, we tested whether the treated macrophages could stimulate transmigration of macrophages using an in vitro transwell system. BMM seeded in the bottom chamber of a transwell were treated with sucrose gradient purified exosomes for 16 hours. CFSE-labeled BMM were added to the upper chamber and allowed to migrate for 2 hours. As shown in Figure 3A, we observed an increased number of CFSE-labeled macrophages which had migrated through the transwell filter when these cells were exposed to Rv exosomes compared to untreated or Un exosome-treated BMM. The number of labeled macrophages which migrated to the bottom of the transwell filter were quantified by counting 10 random fields for each treatment group (Fig. 3B).
Fig. 3. Exosomes from H37Rv-infected cells induce transmigration of CFSE-labeled bone marrow derived macrophages.
BMMs seeded in bottom chamber of 6-well transwell plate were left untreated or treated with sucrose-gradient purified exosomes at 40 μg/ml for 16 hours. CSFE-Labeled BMMs were added to the upper chamber and allowed to migrate for 2 hours. (A) Fluorescent cells which migrated to the bottom of the filter were visualized by fluorescent microscopy. (B) Cells in 10 random fields were counted and transmigration index was calculated as a ratio of migration in presence and absence of exosome stimulation. Results are the mean of three separate experiments +SD and asterisk (*) indicates statistical difference when compared to untreated/resting cells (RC) (p value ≤ 0.05).
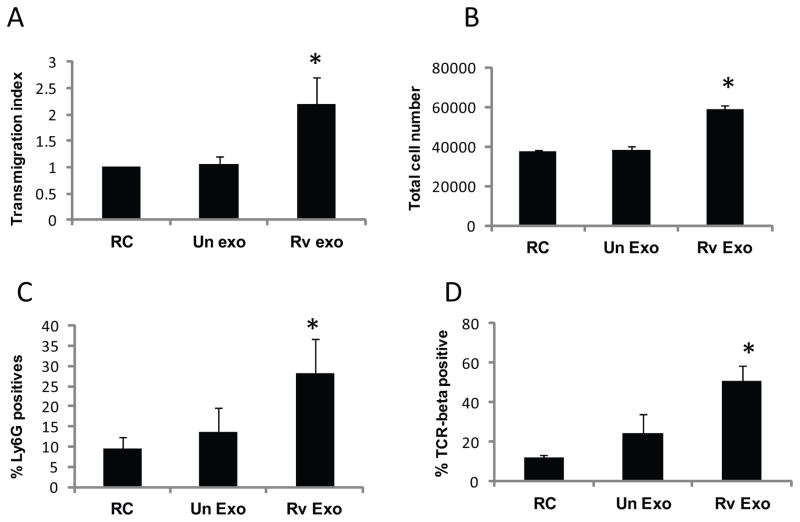
Macrophages treated with Rv exosomes induce transmigration of M. bovis BCG-sensitized splenocytes
Bronchoalveolar lavage fluid isolated from TB patients show increased levels of chemokines including RANTES (23). The production of these chemokines is required for the proper trafficking of leukocytes during the course of an Mtb infection (24). We tested splenocytes isolated from M. bovis BCG-sensitized mice for migratory activity following exposure to exosome-treated or untreated BMM. Splenocytes isolated from mice five weeks post-infection were labeled with CSFE and added to the top chamber of a transwell plate. The bottom chambers contained BMM that were stimulated with sucrose-purified exosomes or were left untreated. Splenic macrophages that migrated to the bottom surface of the transwell filter were counted in ten randomly selected fields. As shown in Figure 4A, an increased number of splenic macrophages migrated through the filter when cells were exposed to BMM treated with Rv-exosomes compared to BMM treated with exosomes from uninfected cells. Splenic cells that migrated into the bottom chamber of the transwell were also quantified and again were higher in wells containing Rv exosome-treated BMM (Fig. 4B). To look at the cell populations, flow cytometry was performed using PE-conjugated Ly6G/6E and Cy5-conjugated TCRβ antibodies. Macrophages treated with Rv exosomes significantly enhanced transmigration of neutrophils (Fig. 4C) and T cells (Fig. 4D) compared to untreated cells. Interestingly, splenocytes isolated from uninfected mice also showed increased migratory activity upon exposure to Rv exosome-treated BMM, suggesting that the migratory activity is not dependent upon prior activation following infection (data not shown).
Fig. 4. Exosomes from H37Rv-infected macrophages induce transmigration of M. bovis BCG sensitized splenocytes.
Splenocytes were isolated from mice 5 weeks after an infection with 1 × 106 CFU M. bovis BCG. BMM were seeded in lower chamber of transwell and stimulated with sucrose-gradient purified exosomes at 40 μg/ml for 16 hours. CFSE-labeled splenocytes were added to the top chamber of transwell and cells were allowed to migrate for 2 hours. (A) Splenic macrophages migrated to the bottom of the filters were counted in 10 random fields and results are represented as transmigration index. (B) Cells migrated to the bottom chamber were collected and counted on a hemacytometer. Additional cells were stained with PE-conjugated Ly6G (C) or Cy5-conjugated TCR-beta (D) to characterize the neutrophils and T cells respectively and analyzed by flow cytometry. Results are the mean of two separate experiments +SD and asterisk (*) indicates statistical difference when compared to untreated/resting cells (RC) (p value ≤ 0.05).
Elevated exosome concentration in serum from M. bovis BCG-infected mice
The initial experiments defined the chemotactic activity of exosomes isolated from cultured infected macrophages. To begin evaluating exosome production during the course of an in vivo infection, we isolated serum from M. bovis BCG-infected or naïve mice at different times post-infection and purified the exosomes using ExoQuick. To confirm that the vesicles purified by ExoQuick were primarily exosomes, samples isolated from day 10 infection were analyzed by TEM, NanoSight and by Western blot. As shown in Figure 5A, the purified vesicles observed by TEM had the expected size and shape of exosomes. The size distribution of the particles observed on the NanoSight LM10 further supports that the majority of vesicles were exosomes (Fig. 5B). Finally, the vesicles expressed the host exosomal markers CD63 and CD9 (Fig.5C).
Fig. 5. Isolation of exosomes from M. bovis BCG infected mice.

C57BL/6 mice were infected with 1 × 106 CFU of M. bovis BCG or were injected with PBS as control. Exosomes were isolated from mouse serum using ExoQuick and quantified by BCA. The exosomes were characterized morphologically by transmission electron microscopy (A) or by NanoSight (B). The exosome pellet resuspended in RIPA buffer was loaded on SDS-PAGE gel and probed for exosome markers CD63 and CD9 (C).
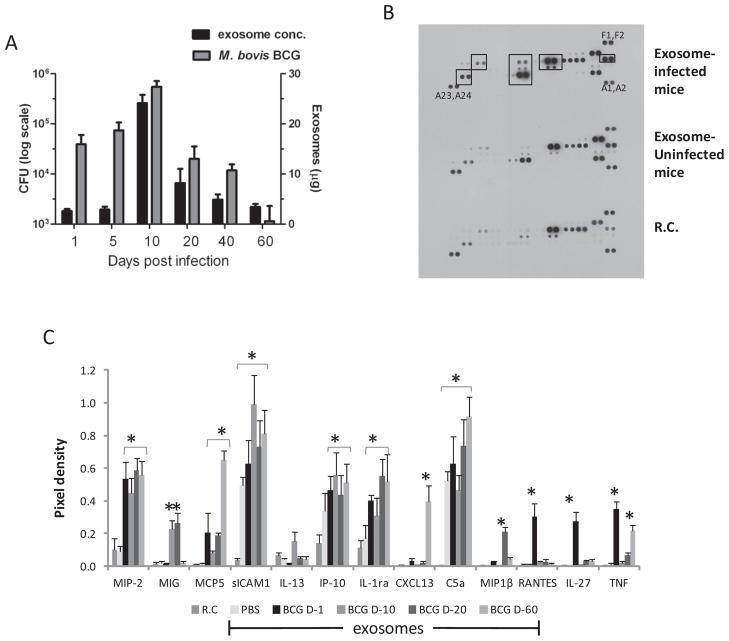
As shown in Figure 6A, the kinetics of bacterial load showed an initial increase that peaked at day 10 followed by a gradual decline through day 60. Interestingly, the exosome concentration showed similar kinetics, suggesting that infection induces exosome secretion correlating with bacterial burden. We subsequently tested whether exosomes from the serum of infected mice could promote a macrophage response ex vivo. We used exosomes isolated at different times post-infection. BMMs were treated with the serum-derived exosomes or were left untreated for 16 hours. Cell culture supernatants were harvested and assayed for cytokine/chemokine levels using the mouse cytokine array kit (Fig. 6B). Exposure of macrophages to exosomes from uninfected mice induced significantly higher expression of sICAM1 and C5a compared to resting cells alone. Treating macrophages with exosomes from infected mice resulted in significantly higher levels of MIP-2, MIG, MCP5, sICAM-1, IP-10, IL-1ra, CXCL13, C5a, MIP1β, RANTES, IL-27 and TNF-α compared to the resting cells (Fig. 6C). However, the expression profiles differed between treatments with exosomes isolated at different time points post-infection. For example, CXCL13 expression was only significantly higher using exosomes isolated at day 60. RANTES, TNF-α and IL-27 levels were only significantly higher using exosomes isolated at day 1. However, some cytokines, such as MIP-2, showed higher expression using exosomes isolated at any time point (Fig. 6C).
Fig. 6. Exosome production in M. bovis BCG infected mice correlates with bacterial load and show pro-inflammatory activity.
(A) C57BL/6 mice infected with 106 CFU M. bovis BCG were sacrificed at various time points and serum exosome concentration and CFU were determined. Results are representative of three independent experiments each with 4 mice per group. BMM were treated for 18 hours with 500 μg/ml of serum-derived exosomes isolated at day 1, 10, 20 and day 60 (D-1 through D-60) post M. bovis BCG infection or with exosomes from PBS-treated control mice. The culture supernatants of the treated BMM were analyzed for cytokines and chemokines using the mouse cytokine arrays according to the manufacturer’s instructions. (B) Shown is the array using exosomes from day 10 infected mice. (C) The pixel densities for each spot were calculated using ImajeJ software and plotted. Results are representative of three independent experiments with SD and asterisk (*) indicates statistical difference when compared to untreated/resting cells (RC) (p value ≤ 0.05). (A1,A2), (F1,F2) and (A23,A24) correspond to reference numbers for cytokine array (see supplement table 1 for arrangement of antibodies on the array).
Exosomes from Mtb-infected macrophages can induce cellular recruitment in vivo
To define whether exosomes can also recruit myeloid cells into the lungs, we intranasally administered ExoQuick-purified exosomes into mice and harvested the lung tissue 5 days post-treatment. As shown in figure 7, the lungs of mice treated with Rv exosomes or infected with M. bovis BCG had a significantly higher number of CD11b+ cells compared to mice treated with PBS or Un exosomes. There was also a moderate increase in the number of F4/80+ and Ly6G+ cells (macrophages and neutrophils respectively) in BCG-infected or Rv exosome-treated mice.
Fig. 7. Intranasal administration of Rv exosomes in C57BL/6 mice induces cellular recruitment in lungs.

C57BL/6 mice were intra-nasally administered PBS or 25 μg of exosomes in PBS or infected with 1× 106 CFU M. bovis BCG. Mice were sacrificed at day 5 and lungs from 4 mice per group were pooled. Total lung cells were treated with Collagenase for 2 hours and cells were stained with FITC-conjugated anti-mouse F4/80, PE-conjugated anti-mouse CD11b, PE-conjugated anti-mouse Ly6G or with isotype controls. Plotted is the number of cells which stained positive for the given markers. Results are representative of two independent experiments with SD. Asterisk (*) indicates statistical difference when compared to untreated/resting cells (RC) (p value ≤ 0.05).
Hollow fibers containing exosome-treated macrophages can induce macrophage recruitment in vivo
To study the effect of macrophages exposed to exosomes on inducing migration and recruitment of host immune cells in vivo, untreated, exosome-treated or M. bovis BCG-infected BMM were removed from plates and the cells were added to preconditioned PVDF hollow fibers. The sealed hollow fibers were implanted subcutaneously in immunocompetent SKH1 mice (Fig. 8A). The fibers have a 500-kDa molecular mass cut off which allows diffusion of soluble factors but prevents entry of host cells into the fibers. Fibers were recovered from mice after 16 days and assessed for cell recruitment by histology and flow cytometry. Hematoxylin-eosin staining showed increased cellular infiltration around the hollow fibers when these were filled with BMM treated with Rv exosomes compared to untreated BMM or cells treated with exosomes from uninfected cells (Fig. 8B). This suggests that macrophages treated with Rv exosomes secrete chemokines that diffuse from the hollow fibers and lead to enhanced recruitment of host immune cells. A similar recruitment of cells was observed when hollow fibers were filled with M. bovis BCG-infected BMM (Fig. 8B).
Fig. 8. Macrophages treated with Rv exosomes can induce cell recruitment in vivo.
(A) BMM stimulated with exosomes, infected with M. bovis BCG or left untreated were gently removed from plates and added into the lumen of PVDF hollow fibers which were sealed at both ends. The fibers were then implanted subcutaneously into SKHI mice and recovered after 16 days. (B) The fibers were fixed in 10% formalin solution and perifiber tissue was stained with hematoxylin and eosin. Results are representative of three independent experiments each with 4 mice per group. In a separate group of 4 mice, cells were removed from the fibers and stained for macrophages and neutrophils using FITC-conjugated F4/80 (C) and PE-conjugated Ly6G (D) respectively and analyzed by flow cytometry. Results are the mean of three separate experiments +SD and asterisk (*) indicates statistical difference when compared to untreated/resting cells (RC) (p value ≤ 0.05).
To define the cell populations recruited to the hollow fibers, cells were removed from the fibers, antibody-stained and analyzed by flow cytometry. As shown in Figure 8C, there was an enhanced recruitment of macrophages to the hollow fibers when these were filled with BMM treated with Rv exosomes but not with cells treated with exosomes from uninfected cells. However, no significant differences were observed in neutrophil recruitment between the fibers filled with exosome-treated or untreated BMM (Fig. 8D).
Discussion
Mtb is an intra-macrophage pathogen. Macrophages provide the primary niche for bacterial survival and replication but also provide a mechanism for control through T cell-mediated activation of uninfected macrophages. Following infection, mycobacteria are phagocytosed by alveolar macrophages in the lung and subsequently colonize the underlying epithelial layer, inducing an inflammatory response. This triggers recruitment of mononuclear cells from neighboring blood vessels as a reservoir of host cells for multiplying bacteria and the initiation of granuloma formation (25). As recruited macrophages differentiate into multinucleated giant cells, foamy macrophages and epitheloid macrophages, the phase of rapid bacterial multiplication is followed by a “containment” state that is characterized by active lymphocyte recruitment (26). However it is not clear whether chemokine production by infected macrophages are the only mediators of cellular recruitment since previous in vitro studies indicate that Mtb-infected macrophages show limited production of proinflammatory molecules relative to cells infected with less pathogenic mycobacteria (17,21). Other contributors to the cellular recruitment process may include Mtb cell wall components, which are known to be released into the extracellular milieu (27,28). Coupling some of these mycobacterial components onto beads induced cellular accumulation characteristic of granuloma formation in mice (8). Using a human granuloma model it was found that LAM and oxygenated mycolic acids induce human macrophages to differentiate into multinucleated giant cells and foamy macrophages, respectively (29). Interestingly, Mtb within these foamy macrophages shows differential trafficking and appear to be in a dormant non-replicative state, and therefore these mycobacteria could serve as the bacterial reservoir during latency (30). Moreover, foamy macrophages may play a critical role in disease transmission as the death of these cells through necrosis results in the release of cholesterol and other lipids within the granuloma promoting caseation (29,30). However, these previous studies with mycobacterial lipids have used artificial experimental systems (e.g., coating lipids on beads) and it has not been shown that granuloma formation can be promoted by mycobacterial components under more physiological conditions. We hypothesized that exosomes could function in vivo to transport mycobacterial lipids and proteins to the extracellular environment and promote granuloma formation.
Exosomes are 30–100 nm size microvesicles that are known to primarily function in intercellular communication (31). They are present in various bodily fluids including blood, bronchoalveolar lavage fluid, breast milk and urine, indicating that they may function both locally and systemically (32, 33). Their presence in tissue has been observed in EM studies using a sheep ileal Peyer’s patch model where it was shown that epithelial cells release 50 nm particles from their apical surface (34). Although the presence of exosomes in granulomas has not been observed directly, we have isolated exosomes containing mycobacterial components from the bronchoalveolar lavage (BAL) fluid of Mtb-infected mice, suggesting their release from infected macrophages within the granuloma (35). In recent studies by Qazi et al., they observed that patients with sarcoidosis, a granulomatous disease of the lung, secrete more exosomes into the BAL fluid compared to healthy controls and these exosomes had increased expression of MHC class I and II, CD9, CD63, and CD81, as well as increased pro-inflammatory activity (36).
Previous studies in our lab have demonstrated that exosomes released from Mtb-infected cells can induce a proinflammatory response when exposed to naïve macrophages (37), as well as suppress macrophage responses to IFN-γ (19). These exosomes contain mycobacterial components including over 40 mycobacterial proteins as well as glycolipids such as ManLAM (38) and can activate antigen specific CD4+ and CD8+ T cell responses in vitro and in vivo (15). Recently it has been shown that mycobacteria also release active membrane vesicles that may provide a mechanism for incorporation of mycobacterial components into exosomes (39). Since exosomes released from M.tb-infected cells can induce physiological changes in neighboring cells, we were interested in evaluating their ability to recruit cells that drive granuloma formation.
Unfortunately, at present it is difficult to define the contribution of exosomes to granuloma formation and infection control compared to cytokines and chemokines produced by infected macrophages since both are present during an in vivo infection. The most straightforward approach would be to use mice deficient in exosome production for the infection experiments. However, the cellular components involved in exosome biogenesis remain unknown. The few published studies have highlighted the role of Rab27a, Rab35 and other Rab proteins (40,41). However, these proteins are involved in many vesicular transport processes and the effect of modulating their expression would not be limited to exosome biogenesis. Therefore, we required the physical separation of exosomes from the infected macrophages for both the in vitro and in vivo experiments.
Our initial experiments focused on whether exosomes stimulated chemokine production since chemotactic factors play a key role in cellular recruitment and granuloma formation upon Mtb infection. We found that exposure of BMMs to exosomes released from infected cells but not from uninfected cells result in increased levels of various chemokines including RANTES and MIP-1α. This supports our general findings that exosomes released from Mtb-infected cells can promote a proinflammatory response in naïve macrophages (37). To determine if this chemokine production can promote cell migration, we used a transwell system with exosome-treated BMMs in the bottom and CSFE-labeled macrophages in the top chamber. These experiments show that exosomes released from Mtb-infected macrophages can promote macrophage chemotaxis. We also observed that the activity of exosomes to promote cell migration was not limited to macrophages as T cells present within splenocytes also showed marked migration when exposed to exosome-treated macrophages. Recent studies by Esser et al. indicated that exosomes from human APCs contain enzymes for leukotriene biosynthesis and can promote granulocyte migration and recruitment (42).
At present there have been only limited studies defining exosome production during the course of an in vivo infection. Studies have shown that exosomes released from herpes simplex virus-or Epstein-Barr virus-infected cells can modulate the activity of uninfected cells through the transfer of signaling molecules and functional microRNAs (43). In the present study we isolated exosomes from the serum of M. bovis-BCG-infected mice and analyzed the exosomes for biological activity ex vivo. Interestingly, we found that exosome concentration tightly correlated with bacterial load, suggesting that the presence of mycobacteria was driving the exosome production. At present we do not know the cellular source of the exosomes but it likely involves infected as well as uninfected cells. From the cytokine array it is clear that exosomes isolated from infected mice induce a different macrophage response compared to exosomes from uninfected mice. However, RANTES, MIP-2 and TNF-α expression was also elevated in macrophages treated with exosomes from Mtb-infected cells, suggesting some commonality between in vitro- and in vivo-derived exosomes. Of note, the exosomes isolated at different times post-infection induced different responses when incubated with macrophages ex vivo, suggesting that the exosomes change in composition during the course of an infection. How exosome composition changes in the context of both host and mycobacterial components await future investigation.
It is important to note that although our TEM and Nanosight data suggest that the vesicle population we isolated from serum by ExoQuick were primarily the size expected of exosomes, it is likely that other types of vesicles are also present in our preparation. ExoQuick purification is not specific for exosomes but it does allow for purification of vesicles from small volumes of serum. Further, although our previous experiments indicated that cultured macrophages infected with M. bovis BCG do not release annexin V stained vesicles (37), it is still possible that some vesicles released from our infected cells are not derived from fusion of the MVB. These differences in purification procedures may also account for the differences in mean vesicle size seen between ExoQuick purified and sucrose-purified samples.
To determine whether exosomes can induce cellular recruitment in vivo, we intra-nasally administered exosomes into the lung and measured cellular recruitment. However, this technique, while informative, since it measures the ability of exosomes to recruit cells into the lung, does not directly address whether exosome-treated macrophages may promote granuloma formation. To address this question, we used the hollow fiber implantation technique, which has been used previously to evaluate granuloma formation in vivo (16,44). The semi-diffusible PVDF hollow fibers allow diffusion of small soluble molecules including chemotactic factors, but prevent traffic of host immune cells across the fibers, thus facilitating the analysis of recruited cells in the perifiber area in vivo. The lumen of hollow fibers were filled with untreated, exosome-treated, or M. bovis BCG-infected macrophages and implanted subcutaneously into mice. We chose 16 days for isolation of hollow fibers to provide sufficient time for macrophage-induced cellular recruitment and to minimize foreign body inflammation associated with fiber implantation. Moreover, we were able to observe live macrophages within the hollow fibers after removal from the mice 16 days post-implantation (data not shown). Our results indicate that macrophages which are treated with exosomes containing mycobacterial components can promote macrophage recruitment in vivo. The general distribution of macrophages onto the hollow fibers suggests no specific foci along the membrane which is expected since the macrophages and the chemokines would be uniformly distributed within the fiber. Our flow cytometry data support our histological findings. However, although there were neutrophils in the perifiber tissue, we did not observe any significant difference in recruitment of neutrophils between the different treatment groups. This suggests that the neutrophil recruitment may be more associated with the tissue injury induced by the fiber implantation than from the BMM-induced recruitment.
In summary, our data support a role for exosomes in promoting macrophage release of chemotactic factors and in macrophage recruitment both in vitro and in vivo. What mycobacterial and/or host components are responsible for this activity await further investigation but likely involve lipids such as ManLAM, PIM and trehalose dimycolate, which have been shown individually to promote granuloma formation (9). Moreover, additional studies are needed to elucidate the role of exosomes in controlling Mtb infection, but such studies require a better understanding of exosome biogenesis and the availability of new methodology to specifically block their production.
Supplementary Material
Acknowledgments
This work was supported through the grant AI052439 from the National Institute of Allergy and Infectious Diseases.
We would like to thank Sarah Chapman, Michael Pinn, and LG Klinkenberg for their excellent technical assistance.
Abbreviations
- APCs
Antigen presenting cells
- BCG
Bacillus Calmette-Guerin
- BMM
Bone marrow derived macrophages
- CD4; CD8
cluster of differentiation 4; cluster of differentiation 8
- PBS
Phosphate Buffered Saline
- EM
Electron microscopy
- IFN-γ
Interferon-gamma
- Ly6G/6E
Lymphocyte antigen 6G/6E
- ManLAM
Mannosylated lipoarabinomannan
- MIP-1ά
macrophage inflammatory protein 1 alpha
- M.tb
Mycobacterium tuberculosis
- Nm
nanometer
- OADC
Oleic Albumin Dextrose Catalase
- PIM
Phosphatidylinositol Mannoside
- PVDF
polyvinylidene fluoride
- R.C
Resting cells
- TB
Tuberculosis
- WHO
World Health Organization
- RIPA
Radioimmunoprecipitation assay
Footnotes
Authorship
Prachi Singh performed most of the experiments and co-wrote the manuscript, Victoria Smith also performed some of the experiments, Petros Karakousis provided the expertise for the hollow fiber experiments and helped edit the manuscript, Jeff Schorey supervised the studies and co-wrote the manuscript.
References
- 1.WHO. WHO Report 2010. WHO; Geneva: 2010. Global tuberculosis control. [Google Scholar]
- 2.Flynn JL. Immunology of tuberculosis and implications in vaccine development. Tuberculosis. 2004;84:93–101. doi: 10.1016/j.tube.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Russel DG, Barry CE, 3rd, Flynn J. Tuberculosis: What we don’t know can and does hurt us. Science. 2010;328:852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bold TD, Ernst JD. Who Benefits from granulomas, mycobacteria or host? Cell. 2009;136:17–19. doi: 10.1016/j.cell.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev. 2011;240:252–268. doi: 10.1111/j.1600-065X.2010.00984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 8.Rhoades ER, Geisel RE, Butcher BA, McDonough S, Russell DG. Cell wall lipids from Mycobacterium bovis BCG are inflammatory when inoculated within a gel matrix: characterization of a new model of the granulomatous response to mycobacterial components. Tuberculosis. 2005;85:159–176. doi: 10.1016/j.tube.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Geisel RE, Sakamoto K, Russell DG, Rhoades ER. In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guerin is due principally to trehalose mycolates. J Immunol. 2005;174:5007–5015. doi: 10.4049/jimmunol.174.8.5007. [DOI] [PubMed] [Google Scholar]
- 10.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation: Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 11.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 12.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 13.Peche H, Heslan M, Usal C, Amigorena S, Cuturi MC. Presentation of donor major histocompatibility complex antigens by bone marrow dendritic cell-derived exosomes modulates allograft rejection. Transplantation. 2003;76:1503–1510. doi: 10.1097/01.TP.0000092494.75313.38. [DOI] [PubMed] [Google Scholar]
- 14.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giri PK, Schorey JS. Exosomes derived from M. bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PLoS One. 2008;3:e2461. doi: 10.1371/journal.pone.0002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med. 2004;200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roach SK, Schorey JS. Differential regulation of the mitogen-activated protein kinases by pathogenic and non pathogenic mycobacteria. Infect Immun. 2002;70:3040–3052. doi: 10.1128/IAI.70.6.3040-3052.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J Biol Chem. 2007;282:25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh PP, LeMaire C, Tan JC, Zeng E, Schorey JS. Exosomes released from M. tuberculosis infected cells can suppress IFN-γ mediated activation of naïve macrophages. PLoS One. 2011;6:e18564. doi: 10.1371/journal.pone.0018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor DD, Zacharis W, Gercel-Taylor C. Exosome isolation for proteomic analysis and RNA profiling. Methods Mol Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 21.Beltan E, Horgen L, Rastogi N. Secretion of cytokines by human macrophages upon infection by pathogenic and non-pathogenic mycobacteria. Microb Pathogenesis. 2000;28:313–318. doi: 10.1006/mpat.1999.0345. [DOI] [PubMed] [Google Scholar]
- 22.Algood HMS, Chan J, Flynn JL. Chemokines and tuberculosis. Cytokine & Growth Factor Rev. 2003;14:467–477. doi: 10.1016/s1359-6101(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 23.Kurashima K, Mukaida N, Fujimura M, Yasui M, Nakazumi Y, Matsuda T, Matsushima K. Elevated chemokine levels in bronchoalveolar lavage fluid of tuberculosis patients. Am J Respir Crit Care Med. 1997;155:1474–1477. doi: 10.1164/ajrccm.155.4.9105097. [DOI] [PubMed] [Google Scholar]
- 24.Sadek MI, Sada E, Toossi Z, Schwander SK, Rich EA. Chemokines induced by infection of mononuclear phagocytes with mycobacteria and present in lung alveoli during active pulmonary tuberculosis. Am J Respir Cell Mol Biol. 1998;19:513–521. doi: 10.1165/ajrcmb.19.3.2815. [DOI] [PubMed] [Google Scholar]
- 25.Flynn JL, Chan J, Pinn PL. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol. 2011;4:271–278. doi: 10.1038/mi.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulrichs T, Kaufmann SHE. New insights into the functions of granulomas in human tuberculosis. J Pathol. 2006;208:261–269. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- 27.Beatty WL, Ullrich HJ, Russel DG. Mycobacterial surface moieties are released from infected macrophages by a constitutive exocytic event. Eur J Cell Biol. 2001;80:31–40. doi: 10.1078/0171-9335-00131. [DOI] [PubMed] [Google Scholar]
- 28.Beatty WL, Rhoades ER, Ullrich HJ, Chatterjee D, Heuser JE, Russel DG. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic. 2000;1:235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 29.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffé M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons M, Raposo G. Exosomes – vesicular carriers for intercellular communication. Curr Opinion in Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Admyre C, Johansson SM, Qazi KR, Filén J, Lahesmaa R, Norman M, Neve EPA, Scheynius A, Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 33.Miranda KC, Bond DT, McKee M, Skog J, Păunescu TG, Da Silva N, Brown D, Russo LM. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landsverk T. The follicle-associated epithelium of the ileal Peyer’s patch in ruminants is distinguished by its shedding of 50 nm particles. Immunol Cell Biol. 1987;65:251–261. doi: 10.1038/icb.1987.28. [DOI] [PubMed] [Google Scholar]
- 35.Kruh NA, Schorey JS, Dobos KM. Mycobacterium tuberculosis/Book1. Tuberculosis Biomarkers: Prospects from the bench to the clinic. [Google Scholar]
- 36.Qazi KR, Toregrosa PP, Dahlberg B, Greenewald J, Eklund A, Gabriel S. Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis. Thorax. 2010;65:1016–1024. doi: 10.1136/thx.2009.132027. [DOI] [PubMed] [Google Scholar]
- 37.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110:3234–3244. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giri PK, Kruh NA, Dobos KM, Schorey JS. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics. 2010;10:3190–3202. doi: 10.1002/pmic.200900840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, Jimenez J, Glatman-Freedman A, Jacobs WR, Jr, Porcelli SA, Casadevall A. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest. 2011;121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 41.Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung SY, Lauterbach MA, Grønborg M, Bakhti M, Möbius W, Rhee JS, Barr FA, Simons M. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A–C. J Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esser J, Gehrmann U, D’Alexandri FL, Hidalgo-Estevez AM, Wheelock CE, Scheynius A, Gabrielsson S, Radmark O. Exosomes from human macrophages and dendritic cells contain enzymes for leukotrine biosynthesis and promote granulocyte migration. J Allergy Clin Immunol. 2010;126:1032–1040. doi: 10.1016/j.jaci.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 43.Meckes DG, Jr, Raab-Traub N. Microvesicles and viral infection. J Virol. 2011;85:12844–54. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klinkenberg LG, Sutherland LA, Bishai WR, Karakousis PC. Metronidazole lacks activity against Mycobacterium tuberculosis in an in vivo hypoxic granuloma model of latency. J Infect Dis. 2008;198:273–283. doi: 10.1086/589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.