Abstract
Vampire bats are notorious for being the sole mammals that strictly feed on fresh blood for their survival. While their saliva has been historically associated with anticoagulants, only one antihemostatic (plasminogen activator) has been molecularly and functionally characterized. Here, RNAs from both principal submandibular and accessory glands of Desmodus rotundus were extracted, and ~ 200 million reads were sequenced by Illumina. The principal gland was enriched with plasminogen activators with fibrinolytic properties, members of lipocalin and secretoglobin families, which bind prohemostatic prostaglandins, and endonucleases, which cleave neutrophil-derived procoagulant NETs. Anticoagulant (tissue factor pathway inhibitor, TFPI), vasodilators (PACAP and C-natriuretic peptide), and metalloproteases (ADAMTS-1) were also abundantly expressed. Members of the TSG-6 (anti-inflammatory), antigen 5/CRISP, and CCL28-like (antimicrobial) protein families were also sequenced. Apyrases (which remove platelet agonist ADP), phosphatases (which degrade procoagulant polyphosphates), and sphingomyelinase were found at lower transcriptional levels. Accessory glands were enriched with antimicrobials (lysozyme, defensin, lactotransferrin) and protease inhibitors (TIL-domain, cystatin, Kazal). Mucins, heme-oxygenase, and IgG chains were present in both glands. Proteome analysis by nano LC-MS/MS confirmed that several transcripts are expressed in the glands. The database presented herein is accessible online at http://exon.niaid.nih.gov/transcriptome/D_rotundus/Supplemental-web.xlsx. These results reveal that bat saliva emerges as a novel source of modulators of vascular biology.
Keywords: Vampire bat, Desmodus rotundus, hematophagy, bat ecology, proteome, Illumina, bioinformatics, bloodsucking, desmoteplase, rabies, sialogenin, sialome
INTRODUCTION
There are approximately 1100 species of bats worldwide, constituting 23% of all mammalian species. Only three species have adapted for blood feeding: Desmodus rotundus, Diphylla ecaudata, and Diaemus youngii. D. rotundus, known as the “common vampire bat,” bites its victims with its sharp incisor teeth, leaving a characteristic wound [1, 2]. While this bat may feed on snakes, lizards, turtles, reptiles, amphibians, ocelots, opossums, skunks, and other small mammals, it may also feed on cattle, horses, mules, goats, swine, poultry, sheep, and humans. The other two species feed mostly on birds [3–5].
One vampire bat drinks 15–25 ml in one blood meal, and an animal could be visited by several bats at night. The notable ability of bats to feed on blood indicate that an evolutionary process took place in the salivary glands (SGs) of these animals in which genes were recruited or evolved to produce biologically active peptides and proteins that, when secreted in the saliva, interfere with the hemostatic system of the host [1, 2, 6]. In this respect, the major SGs of the vampire bat associated with hematophagy are the principal submaxillary (also known as submandibular) and the accessory glands [7]. The glands secrete factors that inhibit clot formation and dissolve already formed clots, thus keeping blood flowing freely in a bite wound and enabling bats to drink their meal [8, 9]. Accordingly, saliva of vampire bats has been described to contain an uncharacterized platelet aggregation inhibitor [8, 10] and one anticoagulant (draculin) targeting FXa [11, 12]. Surprisingly, the only antihemostatic agent characterized thus far at the molecular level is a family of plasminogen activators: Desmodus salivary plasminogen activator (DSPA) α1 (DSPAα1, Desmoteplase), DSPAα2, DSPAβ, and DSPAγ [13–16]. In addition, a database search with the term “Desmodus rotundus” or “vampire bat” and “salivary gland” in mid-October 2012 retrieved only 16 sequences related to DSPA. These numbers are obviously an underestimation of the complexity of the genes expressed in the SG of D. rotundus.
SGs of bats are also known to be important reservoirs for rabies virus [3, 4]. In view of the long history of this disease in vampires and its growing incidence in bats, and our limited understanding of the molecular nature of antihemostatics found in the saliva, additional studies are needed to understand the composition of the SGs of these animals. With this goal, we have isolated the mRNA of the principal submaxillary and accessory SGs of D. rotundus [7] and sequenced almost 200 million reads using Illumina technology. The data were treated with several bioinformatics tools, which allowed us to comprehensively organize the sequences in a table that displays a remarkably large number of families. This table may be regarded as a database accessible online as a hyperlinked worksheet and displays biochemical, taxonomic, and gene ontology aspects for each family of protein. This report will improve our understanding of how bats feed on blood and how they transmit diseases that are of public health and veterinary importance.
MATERIALS AND METHODS
Collection of a Vampire Bat and mRNA Extraction
A female specimen of D. rotundus was collected by the authors under federal license issued for one of us (W.U.) by the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais—IBAMA (Brazilian Institute of Environment and Renewable Natural Resources) with nets, near the city of Botucatu in São Paulo State, Brazil [17, 18]. The collection and the aims of this study were also in agreement with Resolution number 21 (08/31/2006) by the Conselho de Gestão do Patrimônio Genético (Council for Management of Genetic Inheritance) based on the Medida Provisória (Provisional Authorization) n°. 186-16 (08/23/2001) and Federal Decree n°. 3.945 (09/28/2001) issued by the Brazilian Ministry of Environment. The bat was euthanatized according to the protocol, which agrees with the ethical principles in animal research adopted by the Brazilian College of Animal Experimentation (COBEA), and was approved by the Bioscience Institute/UNESP (Universidade Estadual de São Paulo) Ethics Committee on Use of Animals (CEUA). The principal and accessory glands were identified based on the anatomical studies by Disanto [19] and experience of one of us (E.C.V.) with bat anatomy [20]. After a ventral incision, the glands were removed, carefully cleaned from surrounding tissues, cut into two pieces, and immediately placed in RNAlater (Ambion, Austin, TX). After 2 days at 4°C, the glands were removed and placed in a petri dish. Then, a scalpel was used to remove two fragments from the right anterior principal gland—one from the central part (named 7M; not used) and one from the periphery (7L) of the gland. Two other fragments from the right posterior principal gland—one from the central part (8M) and one from the peripheral part (8L)—were also taken. One fragment was obtained from the center of the right accessory gland (AC). mRNA was isolated essentially as described [21] using a Micro-FastTrack 2.0 mRNA isolation kit (Invitrogen, San Diego, CA). The mRNA content was estimated by HT RNA Pico Sensitivity Reagent Kit (760635) on the Caliper LabChip GX (Perkin Elmer, Hopkinton, MA) according to the manufacturer’s instructions. The mRNA concentrations were as follows: 7L, 55.95 ng/ml; 7M, 17.18 ng/ml; 8L, 24.35 ng/ml; 8M, 29.96 ng/ml, and AC, 28.57 ng/ml.
mRNA Libraries and Sequencing
Four mRNA libraries (7L, 8L, 8M, and AC) were constructed from 10–400 ng mRNA using the TruSeq RNA Sample Prep Kit, version 2 (Illumina Inc., San Diego, CA). The resulting cDNA was fragmented using a Covaris E210 (Covaris, Woburn, MA). Library amplification was performed using eight cycles to minimize the risk of over-amplification. Unique barcode adapters were applied to each library. Individual libraries were quantitated by qPCR and then pooled in an equimolar ratio before sequencing on a HiSeq 2000 (Illumina) with ver. 3 flow cells and sequencing reagents. One lane of the HiSeq machine was used for the four libraries, yielding a total 191 090 454 reads of 101 nt in length, 148 804 634 being from the principal gland and the remaining from the AC. A paired-end protocol was used for all libraries. The peak quality value (varying from 0–40) was 37 in all reads, with less than 3% of the reads having a quality from 1 to 10. Raw data were processed using RTA 1.12.4.2 and CASAVA 1.8.2. mRNA library construction, and sequencing was done by the NIH Intramural Sequencing Center (NISC).
Sequence Assembly and Coding Sequence Extraction
Reads of all four libraries were assembled together with the ABySS software (Genome Sciences Centre, Vancouver, BC, Canada) [22, 23] in paired-end mode using various kmer (k) values (every even number from 50 to 96). Because the ABySS assembler tends to miss highly expressed transcripts [24], the Trinity assembler [25] was also used. The resulting assemblies were joined by an iterative BLAST and cap3 assembler [26]. Coding sequences (CDS) were extracted using an automated pipeline, based on similarities to known proteins, or by obtaining coding sequences containing a signal peptide [27]. Coding and their protein sequences were mapped into a hyperlinked Excel spreadsheet (presented as Supplemental File 1). Signal peptide, transmembrane domains, furin cleavage sites, and mucin-type glycosylation were determined with software from the Center for Biological Sequence Analysis (Technical University of Denmark, Lyngby, Denmark) [27–30]. The reads from the various libraries were mapped into the contigs using blastn [31] with a word size of 25, masking homonucleotide decamers, and allowing mapping to up to three different CDS if the BLAST results had the same score values. Mapping of the reads was also included in the Excel spreadsheet. Automated annotation of proteins was based on a vocabulary of nearly 250 words found in matches to various databases, including Swissprot, Gene Ontology, KOG, PFAM, and SMART, and a subset of the non-redundant protein database of the National Center for Biotechnology Information (NCBI) containing proteins from vertebrates. Further manual annotation was done as required. Detailed bioinformatics analysis of our pipeline can be found in our previous publication [26].
Sequence alignments were done with the ClustalX software package [32]. Phylogenetic analysis and statistical neighbor-joining bootstrap tests of the phylogenies were done with the Mega package [33]. In some cases, multiple alignment with CLUSTAL 5 or construction of phylogenetic trees was done using the websites http://pbil.univ-lyon1.fr/ and http://www.phylogeny.fr.
Mass Spectrometry and Protein Identification
A sample of the principal submaxillary (7L) or accessory glands (AC) were solubilized in NU/PAGE sample buffer containing DTT and loaded into a 4–12 % NU-PAGE gel with MES buffer (Invitrogen. Identification was performed on reduced and alkylated, trypsin-digested samples prepared by standard mass spectrometry (MS) protocols. The supernatant and two washes (5% formic acid in 50% acetonitrile) of the gel digests were pooled and concentrated by speed vac (Labconco, Kansas City, MO) to dryness directly in 200-μl polypropylene auto-sampler vials (Sun Sri, Rockwood, TN). Recovered peptides were resuspended in 5 μl of solvent A (0.1% formic acid, 2% acetonitrile, and 97.9% water).
Prior to MS analysis, the resuspended peptides were chromatographed directly on the column without trap clean-up. Bound peptides were separated at 500 nl/min generating 80–120 Bar pressure using an AQ C18 reversed-phase media (3-μ particle size and 200-μ pore) packed in pulled-tip, nanochromatography column (0.100 mm ID 150 mm L; Precision Capillary Columns, San Clemente, CA). Chromatography was performed in-line with an LTQ-Velos Orbitrap mass spectrometer (ThermoFisher Scientific, West Palm Beach, FL) and the mobile phase consisted of a linear gradient prepared from solvent A and solvent B (0.1% formic acid, 2% water, and 97.9% acetonitrile) at room temperature. Nano LC-MS (LC-MS/MS) was performed with a ProXeon Easy-nLC II multidimensional liquid chromatograph and temperature controlled Ion Max Nanospray source (ThermoFisher Scientific) in-line with the LTQ-Velos Orbitrap mass spectrometer.
Computer-controlled data-dependent automated switching to MS/MS by Xcalibur 2.1 software was used for data acquisition and provided the peptide sequence information. Data processing and databank searching were performed with Proteome Discoverer 1.2 and SEQUEST (Thermo Scientific, San José, CA). The data were searched against protein sequences in the database Bat-pep-S2-ext FASTA file provided (47594 sequences). Parsimony analysis was performed using ProteoIQ software (www.nusep.com; NuSep, Athens, GA), and protein probabilities were calculated using the ProteinProphet algorithm as deployed in the program [34]. All proteins were required to have at least two peptides and two spectra per peptide and a probability of 95% in at least one gel lane. MS/MS results were mapped into the Excel spreadsheet with indication of the slices containing the most abundant ions in sorted order, among other information. Statistics and assignments of the MS/MS-derived data is presented as table (supplementary material), listing the m/z, z, and assigned amino acid sequence of parent ions and matching score.
Statistical analysis
The number of the reads originating from each of the four libraries were mapped into the CDS (Supplemental excel file, columns AY-BU). These numbers were compared among themselves by a X2 test. Because we observed no significant differences among the reads originating from different regions of the principal submandibular gland, we pooled these reads and compared them with those originating from the accessory glands. Significant results (P < 0.05 and minimum expected value > 10 are shown in column CC with a “Y”, followed by the normalized ratio of the reads calculated as A × K/(B +1) where A and B are the actual number of reads and K is a constant derived from the division of the total reads for the B library divided by the total reads of the A library. The denominator of the ratio is added of 1 to prevent division by zero.
Public data access
All results were assembled as a database that was comprehensively organized in an Excel table accessible online as a hyperlinked supplemental worksheet available from http://exon.niaid.nih.gov/transcriptome/D_rotundus/Supplemental-web.xlsx. Public access of the data can be found at the National Center for Biotechnology Information (NCBI) under bioproject PRJNA178123. Raw illumina reads can be downloaded from the short read archives (SRA) of the NCBI under numbers SRR606899, SRR606902, SRR606908 and SRR606911. Over 8,000 protein sequences were deposited in GenBank under accession GABZ00000000 of the transcriptome shotgun annotation (TSA) portal.
RESULTS AND DISCUSSION
Bat SG Dissection
To isolate the mRNA from one vampire bat, one healthy specimen was captured alive using appropriate nets placed in a rural area located close to the city of Botucatu in the countryside of São Paulo State, Brazil. Figure 1A shows a female D. rotundus used in this study. After euthanizing the specimen according to an approved ethics protocol, the animal was placed in a dashboard and a ventral incision was performed (Figure 1B and 1C) using the anatomic indications provided by Disanto (1960) and the expertise of one us to dissect bats [20]. In a close-up view, Figure 1D shows two single ACs that occupy a medial position close to the trachea. The principal submaxillary glands (PS) are located laterally to the accessory glands and consist of two lobes, one anterior (aPS), and one posterior (pPS). Lymph nodes are also present superiorly to the principal submaxillary glands and are indicated as LN. The glands were carefully dissected, cut into two pieces, and placed in a RNAlater, which was used to preserve the tissue mRNA. Then, a fragment of approximately 0.5 cm was taken, and the mRNA was isolated and quantified using standard techniques [21].
Figure 1.
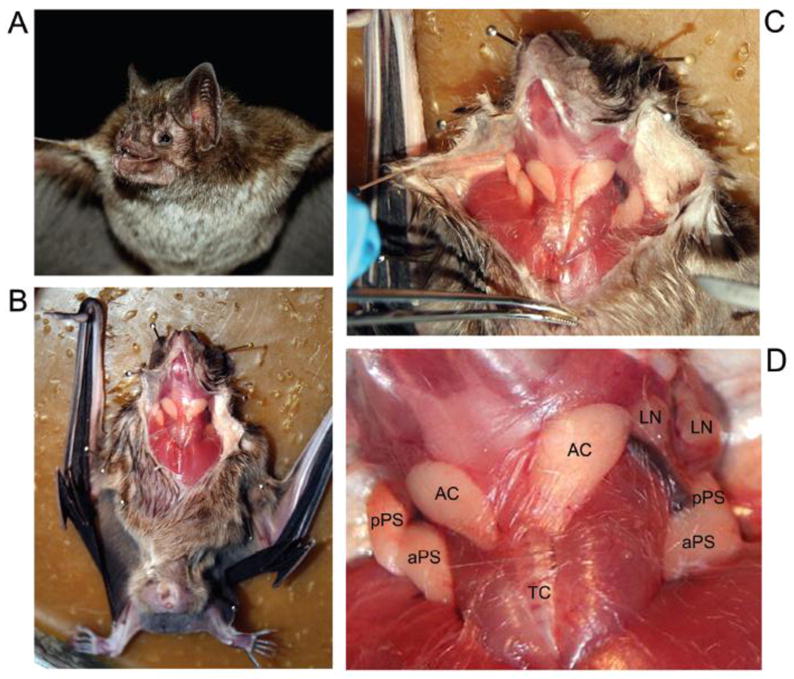
Dissection of Desmodus rotundus glands associated with hematophagy. (A) Female specimen of D. rotundus used in this study. (B) and (C) Ventral incision of the bat demonstrates the anatomic relationships of the glands. (D) A closer picture illustrates the disposition of the glands in regard to other tissues. AC, accessory gland; aPS, anterior principal submaxillary gland; pPS, posterior principal submaxillary gland; LN, lymph node; TC, trachea.
Preliminary Characterization of the Salivary Proteome of D. rotundus
To obtain information on protein expression in the SGs of D. rotundus, we used a proteomic approach using high-density 1D gel electrophoresis. The gel was sliced into 20 parts, and protein content was followed by tryptic digestion and reversed-phase HPLC/MS/MS. The results obtained were blasted against a database constructed with mRNA from the glands (see below). Results of this experiment using the principal submaxillary gland lead to abundant identification of polypeptides (Figure 2A). According to our analysis, a semaphorin was the most expressed protein in the principal gland (855 ions in fraction 8) with virtual no expression in the AC. Semaphorins are involved in neurogenesis and vascular growth [35, 36]. Plasminogen activator (DSPAγ, slice 13) was highly expressed (597 ions) followed by members of the lipocalin (163 ions), TSG-6 (101 ions), secretoglobin (65 ions) and antigen-5/CRISP families (57). We also found evidence for expression of ADAMTS-1 (46 ions), vasodilator (pituitary adenylate cyclase activating peptide; PACAP) (14 ions), tissue factor pathway inhibitor (TFPI, 9 ions), and sphingomyelinase (10 ions). Ions compatible with apyrase, DNAse, and dipeptidyl peptidase were also recovered. These proteins were generally found in close agreement with their estimated molecular masses, and their function is discussed in detail in the next section. Our analysis also demonstrated the presence of several other proteins in the principal submaxillary and AC glands, as well, including matrix proteins (collagen), proteinase inhibitors (protease C inhibitor, α-macroglobulin), components of the complement pathway, galectins, lysozyme, lipases, N-arginine dibasic convertase, and antigens of T cells.
Figure 2.
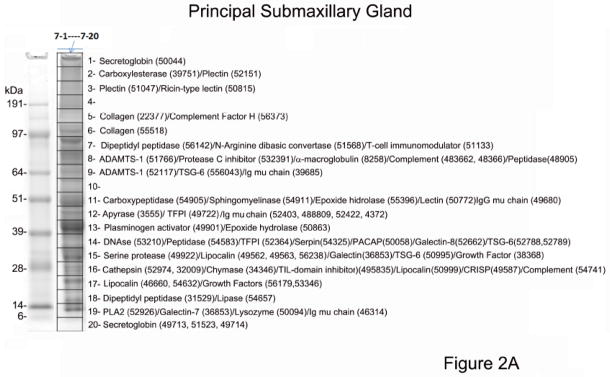
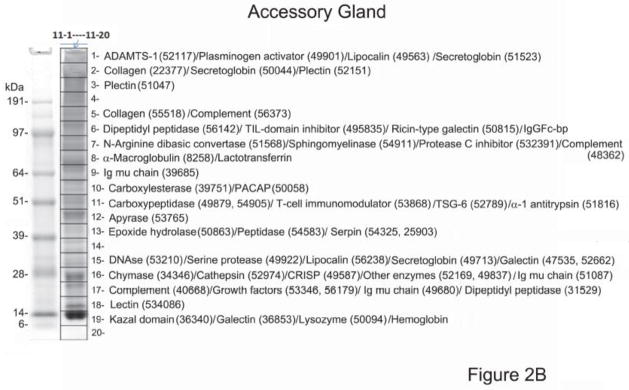
Proteome of Desmodus rotundus salivary gland (SG) homogenates. (A) Principal submaxillary gland. (B) Accessory gland. The gel lanes (“7” for the principal gland and “11” for the accessory gland) show the separation of ~50–100 μg SG proteins. The grid (1–20) represents gel slices submitted for tryptic digest and MS/MS identification. Numbers is parenthesis indicate the corresponding transcripts present in Table 4. Numbers at the left indicate the MW in kD of the protein standards. For a complete list of the proteome see supplemental spreadsheet which displays complete results, including ion abundances and statistical analysis.
The AC was found to express several proteins found in the principal submaxillary gland, although at much lower expression levels (Figure 2B). These proteins were identified as apyrases, DNAses, ADAMTS-1, secretoglobins, lipocalins, plasminogen activators, PACAP, chymases, antigen-5/CRISP, lectins, and other families. The AC were enriched with Kazal-domain containing proteins and, in particular, for trypsin inhibitor-like (TIL) domain-containing proteins (410 ions in the AC, and 4 ions in the principal gland), and lysozyme (351 ions in the AC and 17 in the principal gland) which have antimicrobial properties. In addition, two members of the bactericidal/permeability-increasing protein, a potent antimicrobial protein [37, 38] were identified predominantly in the AC, with 605 and 390 ions on slices 16 and 15, respectively and less than 10 ions in the principal gland. Lactotransferrin (or lactoferrin) was found abundantly represented in the AC with 1,085 ions in fraction 8, and only 19 ions in fraction 1 of the principal gland. Lactotransferrin is a well-known antimicrobial [39, 40]. Of note, bat saliva inhibits Factor Xa and the N-terminus of the purified component named draculin [11] has been identifiable as lactotransferrin. However, it remains to be confirmed whether recombinant bat salivary lactotransferrin display anticoagulant activity.
Hemoglobin subunits (on slice 19) are among the most abundantly detected proteins from the accessory gland, and were also found in the principal gland proteome. Hemoglobin could be considered a finding based on tissue blood content, but the target protein sequence for this study was obtained from the “de novo” assembly of the salivary transcriptome, indicating there are transcripts in the gland coding for these blood proteins. IgGFc-binding protein (slice 6, 410 ions, normalized spectra count – NPC – of 176) was abundantly found in the AC, but not well represented in the principal gland (slice 16, 1 ion, NPC=0.25). This protein has multiple von Willebrand domains (VWD) and Trypsin-inhibitor like cysteine rich domains in tandem. Its function is unknown. Several housekeeping proteins were also identified. All proteome results are displayed in the supplemental spreadsheet found at http://exon.niaid.nih.gov/transcriptome/D_rotundus/Supplemental-web.xlsx.
The results under the column “Slice information” (in magenta – columns AO and AX) are summarized as, for example, 11→18| 12→18| 13→2|. This indicates that 18 fragments were found in Fraction 11, while 18 and 2 peptides were found in fractions 12 and 13, respectively. Furthermore, this summary included protein identification only when two or more peptide matches to the protein were obtained from the same gel slice.
mRNA Sequencing
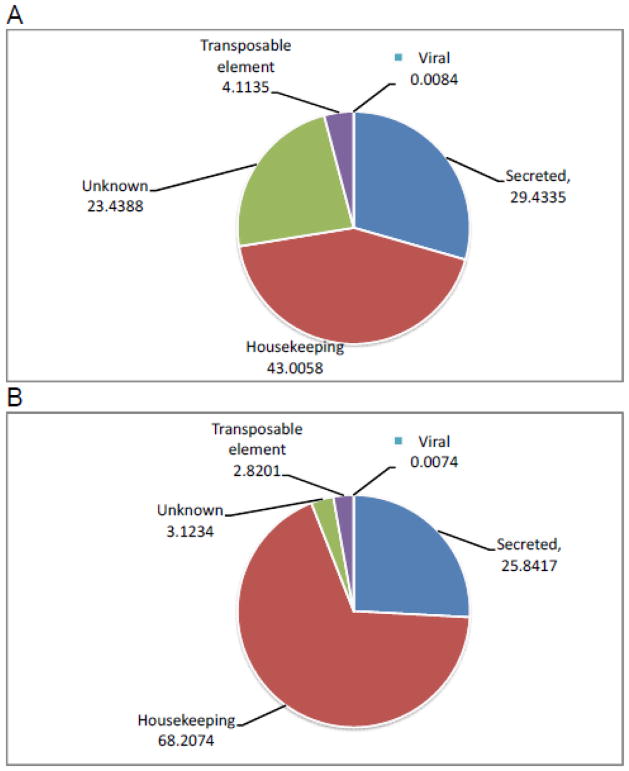
The 191 090 454 reads were assembled into 565 517 contigs larger than 120 nt, of which 219 561 were larger than 500 nt. The larger open reading frames from each contig containing a signal peptide were extracted from the 565 517 contigs, as well as those open reading frames matching at least 60% of the length of proteins on the Swissprot, Gene Ontology, and a subset of the non-redundant database containing vertebrate sequences. These two predicted protein sets were combined and their redundancy removed by discarding equal sequences or the smaller sequence matching a larger one. This subset comprised a total of 47 575 protein sequences that mapped 188 588 838 reads (these reads can be redundantly mapped into up to three CDS; see Materials and Methods), which can be used to evaluate relative expression of different functional classes of messages. Accordingly, 68 % of the reads originated from putative housekeeping (H) products, 26 % from genes coding for putative secreted (S) proteins, while 3.1% were of unknown (U) function, 2.8 derived from transposable elements (TE), and 0.01% from viruses. On the other hand, if we compare the percentage of the number of CDS in the several functional categories, we obtain 43 % of the CDS for the H category, 29 for S, 23 for Un, and 4 and 0.008% for TEs and viruses (Figure 3A and 3B and Table 1). The most striking change between the two measurements is the increase of the U class from 3% of the total of reads to 23% of the total of CDS, which is a consequence of the low level of reads/CDS in this class (528 as compared with 3,480 and 6,286 in the S and H classes).
Figure 3.
Functional classification of the extracted coding sequences from the principal and accessory salivary glands transcriptomes of Desmodus rotundus based on (A) the number of coding sequences or (B) the number of reads. The numbers represent the percentage of the total.
Table 1.
Functional Classification of Coding Sequences (CDS) Extracted from the Salivary Glands of Desmodus rotundusa
| Class | Number of CDS | % of Total | Number of Reads | % of Total | Reads/CDS |
|---|---|---|---|---|---|
| S | 14 003 | 29.4335 | 48 734 568 | 25.8417 | 3480 |
| H | 20 460 | 43.0058 | 128 631 603 | 68.2074 | 6287 |
| U | 11 151 | 23.4388 | 5 890 440 | 3.1234 | 528 |
| TE | 1957 | 4.1135 | 5 318 340 | 2.8201 | 2718 |
| V | 4 | 0.0084 | 13 887 | 0.0074 | 3472 |
|
| |||||
| Total | 47 575 | 100 | 188 588 838 | 100 | |
CDS, coding sequence; H, housekeeping; S, secreted; U, unknown; TE, transposable element; V, viral.
The H class was further subdivided into several functional categories (Table 2): cytoskeleton protein, detoxification, oxidant metabolism, extracellular matrix, immunity, metabolism (aminoacid, carbohydrate, energy, intermediate, lipid and nucleotide) nuclear export, nuclear regulation, protein export, protein modification, proteasome machinery, protein synthesis machinery, signal transduction and viral product. Not surprisingly, transcripts associated with protein synthesis machinery, signal transduction, and transcription machinery rank in the three top categories. The “unknown conserved” category ranks 4th in the list, representing our ignorance in basic cellular functions [41]. CDS associated with secretory functions were further subdivided into several categories according to Table 3. Among the most expressed CDS are plasminogen activators (2.8% of the S class reads), Kunitz domain-containing proteins (1.8%), lipocalins and other lipid carriers (20%), lipophilin and secretoglobins (18%), lysozyme (3.4%), and mucins (19%). Of note, an expanded family of small peptides with no matches to known proteins was found, accounting for 4.4% of the reads. No proteome results were found indicating whether they are expressed, but our acrylamide gel did not resolve their small size. They remain as public peptide targets for future proteomic experiments. We also found many products that are normally associated with the lymphoid tissue including immunoglobulin chains and other immune-related proteins. Notably, Ig μ chains accounted for 4.2% of the total reads of the S class. Finally, 9,978 other putative unclassified peptides accounted for 17% of the S class reads.
Table 2.
Functional Classification of the Coding Sequences (CDS) Associated with a Housekeeping Function from the Sialotranscriptome of Desmodus rotundus
| Class | Number of CDS | % of Total in Class | Number of Reads | % of Total | Reads/CDS |
|---|---|---|---|---|---|
| Protein synthesis machinery | 1009 | 29.7063 | 38 211 726 | 29.7063 | 37 871 |
| Signal Transduction | 4189 | 12.0478 | 15 497 297 | 12.0478 | 3700 |
| Transcription Machinery | 2478 | 11.3367 | 14 582 559 | 11.3367 | 5885 |
| Unknown Conserved | 3133 | 6.9203 | 8 901 678 | 6.9203 | 2841 |
| Immunity | 237 | 5.9781 | 7 689 713 | 5.9781 | 32 446 |
| Protein Export | 1348 | 5.4833 | 7 053 262 | 5.4833 | 5232 |
| Extracellular Matrix | 647 | 4.8787 | 6 275 600 | 4.8787 | 9700 |
| Protein Modification | 1060 | 3.4420 | 4 427 517 | 3.4420 | 4177 |
| Transporters and Channels | 1018 | 3.1183 | 4 011 094 | 3.1183 | 3940 |
| Nuclear Regulation | 878 | 2.6363 | 3 391 136 | 2.6363 | 3862 |
| Cytoskeletal Proteins | 657 | 2.4969 | 3 211 847 | 2.4969 | 4889 |
| Proteasome Machinery | 691 | 2.3213 | 2 985 883 | 2.3213 | 4321 |
| Lipid Metabolism | 569 | 2.0583 | 2 647 596 | 2.0583 | 4653 |
| Transcription Factor | 537 | 1.4466 | 1 860 827 | 1.4466 | 3465 |
| Carbohydrate Metabolism | 488 | 1.3102 | 1 685 353 | 1.3102 | 3454 |
| Energy Metabolism | 405 | 1.0735 | 1 380 859 | 1.0735 | 3410 |
| Signal Transduction/Apoptosis | 284 | 0.8062 | 1 037 026 | 0.8062 | 3652 |
| Amino Acid Metabolism | 197 | 0.6997 | 900 062 | 0.6997 | 4569 |
| Oxidant Metabolism/Detoxification | 82 | 0.6966 | 896 011 | 0.6966 | 10 927 |
| Nucleotide Metabolism | 212 | 0.6566 | 844 633 | 0.6566 | 3984 |
| Intermediary Metabolism | 127 | 0.3754 | 482 826 | 0.3754 | 3802 |
| Nuclear Export | 84 | 0.2516 | 323 594 | 0.2516 | 3852 |
| Detoxification | 123 | 0.2337 | 300 574 | 0.2337 | 2444 |
| Storage | 7 | 0.0256 | 32 930 | 0.0256 | 4704 |
|
| |||||
| Total | 20 460 | 100 | 128 631 603 | 100 | |
Table 3.
Functional Classification of the Coding Sequences (CDS) Associated with Putative Secreted Proteins from the Sialotranscriptome of Desmodus rotundus
| Class | Number of CDS | % of Total in Class | Number of Reads | % of Total | Reads/CDS |
|---|---|---|---|---|---|
| Enzymes | |||||
|
| |||||
| Phosphatase | 1 | 0.0071 | 1780 | 0.0037 | 1780 |
| Apyrase | 3 | 0.0214 | 9484 | 0.0195 | 3161 |
| DNAse | 3 | 0.0214 | 22 891 | 0.0470 | 7630 |
| Metalloprotease | 12 | 0.0857 | 66 638 | 0.1367 | 5553 |
| Plasminogen activator | 3 | 0.0214 | 1 401 038 | 2.8748 | 467, 13 |
| Other serine proteases | 6 | 0.0428 | 8987 | 0.0184 | 1498 |
| Clotting pathways serine proteases | 3 | 0.0214 | 65 441 | 0.1343 | 21 814 |
| Dipeptidyl and tripeptidyl peptidases | 3 | 0.0214 | 3893 | 0.0080 | 1298 |
| Legumains and cathepsins (may be lysosomal) | 26 | 0.1857 | 103 695 | 0.2128 | 3988 |
| Other peptidases | 41 | 0.2928 | 117 640 | 0.2414 | 2869 |
| Lipases | 15 | 0.1071 | 21 871 | 0.0449 | 1458 |
| Epoxide hydrolase | 4 | 0.0286 | 49 142 | 0.1008 | 12 286 |
| Other enzymes | 4 | 0.0286 | 3322 | 0.0068 | 831 |
|
| |||||
| Protease inhibitor domains | |||||
|
| |||||
| Kunitz domain | 15 | 0.1071 | 883 734 | 1.8134 | 58 916 |
| Amyloid Kunitz protein | 26 | 0.1857 | 101 236 | 0.2077 | 3894 |
| Collagen with VWB and Kunitz domains | 2 | 0.0143 | 13 211 | 0.0271 | 6606 |
| Cystatin | 1 | 0.0071 | 1 314 | 0.0027 | 1314 |
| Kazal domain | 11 | 0.0786 | 353 596 | 0.7256 | 32 145 |
| Serpins | 10 | 0.0714 | 21 337 | 0.0438 | 2134 |
| TIL domain containing protein | 2 | 0.0143 | 414 128 | 0.8498 | 207 064 |
| Metalloproteinase inhibitor domain | 2 | 0.0143 | 1287 | 0.0026 | 644 |
| Other protease inhibitors | 4 | 0.0286 | 2317 | 0.0048 | 579 |
|
| |||||
| Lipocalin and other lipid carriers | 11 | 0.0786 | 9 541 325 | 19.5781 | 867 393 |
|
| |||||
| Lipophilin/secretoglobin precursors | 15 | 0.1071 | 8 651 572 | 17.7524 | 576 771 |
|
| |||||
| Antigen 5/CRISP family | 1 | 0.0071 | 448 565 | 0.9204 | 448 565 |
|
| |||||
| β2-microglobulin/Class I MHC | 2 | 0.0143 | 10 913 | 0.0224 | 5457 |
|
| |||||
| Neuropeptide/hormones | 6 | 0.0428 | 712 298 | 1.4616 | 118 716 |
|
| |||||
| Extracellular matrix components | 14 | 0.1000 | 69 270 | 0.1421 | 4948 |
|
| |||||
| Lectins | 17 | 0.1214 | 275 140 | 0.5646 | 16 185 |
|
| |||||
| Antimicrobial peptides | 0.0000 | ||||
|
| |||||
| Defensin | 1 | 0.0071 | 112 | 0.0002 | 112 |
| Lysozyme | 3 | 0.0214 | 1 674 385 | 3.4357 | 558 128 |
| Other antimicrobial agents | 5 | 0.0357 | 22 364 | 0.0459 | 4473 |
| Complement and regulators | 11 | 0.0786 | 9147 | 0.0188 | 832 |
| Growth factors and immune regulators | 18 | 0.1285 | 11 680 | 0.0240 | 649 |
|
| |||||
| Immunity related products | |||||
|
| |||||
| C-C motif chemokine 7 | 5 | 0.0357 | 5330 | 0.0109 | 1066 |
| Chemokine (C-X-C motif) ligand 2 | 5 | 0.0357 | 3324 | 0.0068 | 665 |
| T cell immunomodulatory protein | 15 | 0.1071 | 80 958 | 0.1661 | 5397 |
| Other immunity related products | 15 | 0.1071 | 897 747 | 1.8421 | 59 850 |
| MHC class I and II antigen | 4 | 0.0286 | 1505 | 0.0031 | 376 |
| Ig μ chain | 68 | 0.4856 | 2 063 496 | 4.2342 | 30 346 |
| Ig γ chain | 30 | 0.2142 | 267 529 | 0.5490 | 8918 |
| Other Ig chains | 4 | 0.0286 | 7188 | 0.0147 | 1797 |
|
| |||||
| Mucins | 344 | 2.4566 | 9 343 275 | 19.1718 | 27 161 |
|
| |||||
| Expanded family 40-1 | 3104 | 22.1667 | 2 146 926 | 4.4053 | 692 |
|
| |||||
| Other putative secreted peptides | 9978 | 71.2562 | 8 391 175 | 17.2181 | 841 |
|
| |||||
| Conserved secreted protein | 130 | 0.9284 | 431 362 | 0.8851 | 3 318 |
|
| |||||
| Total | 14,003 | 100 | 48 734 568 | 100 | |
All results are placed in the supplemental table found at http://exon.niaid.nih.gov/transcriptome/D_rotundus/Supplemental-web.xlsx which describes the mol wt, pI, biochemical, taxonomic, putative function, matches to several databases, clusterization steps, relative proportion in the principal maxillary and accessory glands and several other specifics for each protein family, secreted or housekeeping, which has been sequenced in this project.
To minimize redundancy and highlight our most relevant data, results from the supplemental table were filtered to display only sequences coding for secreted proteins likely associated with hematophagy, but not exclusively. A condensed version of the supplemental table is presented as Table 4 (http://exon.niaid.nih.gov/transcriptome/D_rotundus/Table4-web.xlsx), which is a printable version (hyperlinked in the electronic version). Table 4 summarizes our transcriptome and proteome results for selected sequences found in the PS and AC of the vampire bat D. rotundus.
Table 4.
Transcriptional and proteoma analysis of D. rotundus Principal Submaxillary and Accessory glands.
| Peptide link | Total reads | M | * | SP | MW | pI | p | A/P | P/A | Comments/Target | 11 P | 11 S | 7 P | 7 S |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Putative secreted proteins | ||||||||||||||
|
| ||||||||||||||
| Enzymes | ||||||||||||||
|
| ||||||||||||||
| Phosphatase | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-508800 | 1780 | M | * | SIG | 55.22 | 9.19 | Y | 0.42 | 2.36 | Platelet function | ||||
|
| ||||||||||||||
| Apyrase | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-537653 | 3555 | M | * | SIG | 51.74 | 5.66 | Y | 0.67 | 1.49 | Platelet function | 1 | 12 | 1 | 12 |
|
| ||||||||||||||
| DNAse (Deoxyribonuclease) | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-499100 | 11477 | M | * | SIG | 33.91 | 6.06 | Y | 0.01 | 69.51 | Neutrophil function | ||||
| BatTrinityAbyss-532109 | 949 | G | * | BL | 36.22 | 5.81 | N | 1.18 | 0.84 | Neutrophil function | 1 | 15 | 1 | 14 |
|
| ||||||||||||||
| Metalloprotease (ADAMTS-1) | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-517665 | 10097 | M | * | SIG | 105.26 | 7.45 | Y | 0.01 | 74.75 | Angiogenesis | 0 | 1 | 8 | |
| BatTrinityAbyss-521171 | 6380 | M | * | SIG | 90.73 | 7.71 | Y | 2.34 | 0.43 | Angiogenesis | 1 | 1 | ### | 9 |
|
| ||||||||||||||
| Serine protease | ||||||||||||||
|
| ||||||||||||||
| Plasminogen activator | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-499018 | 1400766 | M | * | SIG | 44.42 | 8.78 | Y | 0 | 791.49 | Fibrinolytic | 1 | 1 | 1 | 13 |
|
| ||||||||||||||
| Other serine proteases | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-34346 | 381 | S | * | SIG | 27.93 | 9.24 | Y | 0.45 | 2.06 | Chymase | 1 | 16 | 1 | 16 |
|
| ||||||||||||||
| Clotting pathways serine proteases | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-499228 | 21684 | N | * | CYT | 16.93 | 6.15 | Y | 0.13 | 7.97 | Protease | ### | 15 | 1 | 15 |
| BatTrinityAbyss-489254 | 42256 | M | * | SIG | 30.95 | 5.8 | Y | 0.11 | 8.85 | Protease | ||||
|
| ||||||||||||||
| Dipeptidyl and tripeptidyl peptidases | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-561424 | 2794 | M | * | BL | 87.86 | 5.65 | Y | 0.4 | 2.45 | Dipeptidyl peptidases | 1 | 6 | 1 | 7 |
| BatTrinityAbyss-31529 | 825 | M | * | SIG | 52.1 | 6.32 | 1.13 | 0.87 | Dipeptidyl peptidases | 1 | 17 | 1 | 18 | |
|
| ||||||||||||||
| Legumains and cathepsins (may be lysosomal) | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-529744 | 6521 | M | * | SIG | 44.33 | 6.19 | Y | 0.65 | 1.53 | Cathepsin D | 1 | 16 | 1 | 16 |
| BatTrinityAbyss-32009 | 558 | V | * | SIG | 38.17 | 7.65 | 0.7 | 1.37 | Cathepsin S | 1 | 17 | 1 | 16 | |
|
| ||||||||||||||
| Other peptidases | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-515689 | 12851 | M | * | SIG | 117.08 | 5.93 | Y | 0.53 | 1.89 | Arginine convertase | 1 | 7 | 1 | 7 |
| BatTrinityAbyss-489052 | 2950 | M | * | BL | 107.07 | 5.73 | Y | 0.83 | 1.2 | Aminopeptidase | 1 | 7 | 1 | 8 |
| BatTrinityAbyss-549050 | 2294 | M | * | SIG | 50.21 | 6.44 | Y | 0.25 | 3.96 | Carboxy peptidases | 1 | 11 | 1 | 11 |
| BatTrinityAbyss-498793 | 4217 | M | * | SIG | 51.92 | 5.36 | Y | 0.87 | 1.14 | Carboxy peptidases | 1 | 11 | 1 | 11 |
| BatTrinityAbyss-545838 | 1041 | M | * | SIG | 37.83 | 5.22 | 0.93 | 1.05 | Prohibitin | 1 | 13 | 1 | 14 | |
|
| ||||||||||||||
| Lipases | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-549118 | 1863 | M | * | SIG | 51.34 | 5.79 | Y | 0.13 | 7.25 | Sphingomyelinase | ### | 7 | 1 | 11 |
| BatTrinityAbyss-39751 | 2505 | M | * | SIG | 62.08 | 6.1 | Y | 0.73 | 1.36 | Carboxylesterase | 1 | 10 | 1 | 2 |
|
| ||||||||||||||
| Epoxide hydrolase | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-508632 | 16840 | M | * | SIG | 52.4 | 6.79 | 1.04 | 0.96 | Epoxide hydrolase | 1 | 13 | 1 | 13 | |
|
| ||||||||||||||
| Other enzymes | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-521692 | 816 | M | * | CYT | 33.42 | 7.77 | 0.87 | 1.12 | Abhydrolase | 1 | 16 | 1 | 16 | |
| BatTrinityAbyss-498378 | 2194 | R | * | SIG | 33.27 | 9.01 | Y | 0.78 | 1.26 | Secreted protein | 1 | 16 | 1 | 16 |
|
| ||||||||||||||
| Protease inhibitor domains | ||||||||||||||
|
| ||||||||||||||
| Kunitz domain (TFPI-like) | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-523646 | 386161 | E | * | SIG | 19.32 | 9.45 | Y | 0.03 | 36.3 | Anticoagulant | 0 | 1 | 14 | |
|
| ||||||||||||||
| Kunitz domain (Kunitz-type protease inhibitor) | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-541822 | 3016 | M | * | SIG | 28.02 | 7.89 | 0.96 | 1.04 | Kunitz inhibitor | |||||
|
| ||||||||||||||
| Amyloid Kunitz protein | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-507602 | 1896 | M | * | SIG | 85.08 | 4.69 | Y | 0.2 | 4.8 | Mucin | ||||
| BatTrinityAbyss-507612 | 13922 | M | * | SIG | 78.72 | 4.72 | Y | 0.2 | 4.98 | Mucin | ||||
|
| ||||||||||||||
| Collagen with VWB and Kunitz domains | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-22377 | 4540 | M | * | SIG | 293.14 | 5.88 | Y | 0.87 | 1.15 | Collagen | 1 | 2 | 1 | 5 |
| BatTrinityAbyss-555181 | 8671 | M | * | SIG | 118.27 | 7.75 | Y | 0.75 | 1.33 | Collagen | 1 | 5 | 1 | 6 |
|
| ||||||||||||||
| Cystatin | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-41885 | 1314 | M | * | SIG | 16.48 | 8.9 | Y | 2.26 | 0.44 | Cysteine-type inhibitor | ||||
|
| ||||||||||||||
| Kazal domain | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-36340 | 45997 | M | * | SIG | 12.01 | 9.48 | Y | 4.91 | 0.2 | Protease inhibitor | 1 | 19 | 0 | |
|
| ||||||||||||||
| Serpins | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-548577 | 2192 | M | * | SIG | 46.37 | 4.79 | Y | 0.61 | 1.61 | Neuraserpin | ||||
| BatTrinityAbyss-518161 | 1976 | M | * | SIG | 46.8 | 6.68 | Y | 0.33 | 2.93 | α-1 antitrypsin | 1 | 11 | 0 | |
| DrSigp-SigP-532391 | 2305 | M | * | SIG | 61.63 | 4.97 | Y | 0.72 | 1.38 | Protease C1 inh. | 1 | 7 | 1 | 8 |
| BatTrinityAbyss-543253 | 3481 | R | * | CYT | 43.22 | 5.78 | Y | 1.86 | 0.54 | Serpin | 1 | 13 | 1 | 14 |
| BatTrinityAbyss-25903 | 909 | M | * | CYT | 42.7 | 6.15 | Y | 0.71 | 1.38 | Serpin | 1 | 13 | 1 | 14 |
|
| ||||||||||||||
| TIL domain containing protein | ||||||||||||||
|
| ||||||||||||||
| DrSigp-SigP-495835 | 342175 | M | * | SIG | 275.17 | 5.24 | Y | 4.98 | 0.2 | Protease inhibitor | 1 | 6 | ### | 16 |
|
| ||||||||||||||
| Metalloproteinase inhibitor domain | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-534229 | 862 | M | * | SIG | 24.18 | 9.01 | 0.84 | 1.16 | Protease Inhibitor | |||||
| BatTrinityAbyss-37180 | 425 | M | * | SIG | 23.04 | 8.58 | 0.74 | 1.29 | Protease Inhibitor | |||||
|
| ||||||||||||||
| Other protease inhibitors | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-8258 | 1034 | A | * | CYT | 92.78 | 5.61 | Y | 1.39 | 0.71 | α-Macroglobulin | 1 | 8 | 1 | 8 |
| DrSigp-SigP-210264 | 130 | M | SIG | 18.17 | 8.73 | 1.24 | 0.73 | Endopeptidase inhibitor | ||||||
| BatTrinityAbyss-321620 | 107 | M | SIG | 25.5 | 9.51 | 1.36 | 0.65 | Inter-α-trypsin inhibitor | ||||||
| DrSigp-SigP-36965 | 1046 | M | * | SIG | 15.45 | 7.76 | Y | 4.06 | 0.25 | Peptidase inhibitor | ||||
|
| ||||||||||||||
| Lipocalin and other lipid carriers | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-466603 | 212065 | P | * | SIG | 21.18 | 5.09 | Y | 0 | 523.66 | Lipocalin | 0 | 1 | 17 | |
| BatTrinityAbyss-495622 | 1585466 | M | * | SIG | 24.32 | 7 | Y | 0 | 919.23 | Lipocalin | ||||
| BatTrinityAbyss-496761 | 1873290 | M | * | SIG | 24.35 | 7.04 | Y | 0 | 885.83 | Lipocalin | ||||
| BatTrinityAbyss-495626 | 2899256 | K | * | SIG | 23.08 | 8.92 | Y | 0 | 1104.63 | Lipocalin | 0 | 1 | 15 | |
| BatTrinityAbyss-495631 | 964371 | M | * | SIG | 22.44 | 4.92 | Y | 0 | 1089.84 | Lipocalin | ||||
| BatTrinityAbyss-495624 | 316695 | M | * | SIG | 23.89 | 4.74 | Y | 0 | 844.63 | Lipocalin | 0 | 1 | 15 | |
| BatTrinityAbyss-495634 | 268861 | G | CYT | 18.05 | 9.71 | Y | 0 | 663.92 | Lipocalin | ### | 1 | 1 | 15 | |
| BatTrinityAbyss-546328 | 16850 | S | * | CYT | 18.3 | 4.78 | Y | 0.01 | 66.03 | Lipocalin | 0 | ### | 17 | |
| BatTrinityAbyss-562380 | 1146 | M | * | SIG | 29 | 9.67 | 0.84 | 1.17 | Lipocalin | 1 | 15 | 1 | 15 | |
| BatTrinityAbyss-500440 | 1402119 | M | * | SIG | 6.3 | 3.76 | Y | 0 | 1214.13 | Lipocalin | ||||
| BatTrinityAbyss-509991 | 1206 | P | * | CYT | 29.41 | 8.86 | Y | 0.61 | 1.61 | Lipocalin | 1 | 16 | 1 | 16 |
|
| ||||||||||||||
| Lipophilin/secretoglobin precursors | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-500441 | 1614499 | M | * | SIG | 6.7 | 5.97 | Y | 0 | 1435.33 | Antiinflammatory | ||||
| BatTrinityAbyss-500442 | 2063350 | M | * | SIG | 8.98 | 4.08 | Y | 0 | 1273.86 | Antiinflammatory | 0 | 1 | 1 | |
| BatTrinityAbyss-515986 | 1309024 | M | * | SIG | 10.32 | 8.34 | Y | 0 | 484.86 | Antiinflammatory | 1 | 2 | 1 | 20 |
| BatTrinityAbyss-497146 | 627793 | L | * | SIG | 11.05 | 8.68 | Y | 0 | 697.62 | Antiinflammatory | ||||
| DrSigp-SigP-515987 | 1090871 | M | * | SIG | 10.35 | 8.45 | Y | 0 | 451.73 | Antiinflammatory | ||||
| BatTrinityAbyss-515989 | 681163 | M | * | SIG | 5.76 | 5 | Y | 0 | 354.78 | Antiinflammatory | ||||
| BatTrinityAbyss-497133 | 163306 | F | * | CYT | 9.66 | 9.55 | Y | 0.02 | 46.27 | Antiinflammatory | ### | 15 | 1 | 20 |
| BatTrinityAbyss-497140 | 202043 | S | * | CYT | 12.08 | 9.69 | Y | 0.15 | 6.47 | Antiinflammatory | 1 | 15 | 1 | 20 |
| DrSigp-SigP-515229 | 345027 | M | * | SIG | 6.26 | 6.03 | Y | 0 | 657.26 | Antiinflammatory | ||||
|
| ||||||||||||||
| Antigen 5/CRISP family | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-495870 | 448565 | M | * | SIG | 27.39 | 8.79 | Y | 0.04 | 26.5 | CRISP | 1 | 16 | 1 | 16 |
|
| ||||||||||||||
| Beta2-microglobulin/Class I major histocompatibility complex | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-527040 | 10572 | M | * | SIG | 13.37 | 8.57 | Y | 0.71 | 1.41 | MHC | ||||
| BatTrinityAbyss-37556 | 341 | S | * | CYT | 19.81 | 6.27 | Y | 1.6 | 0.6 | MHC | ||||
|
| ||||||||||||||
| Neuropeptide/hormones | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-500584 | 357219 | M | * | SIG | 20.58 | 6.44 | Y | 0 | 486.05 | Vasodilator (PACA) | ### | 10 | 1 | 14 |
| BatTrinityAbyss-538594 | 23548 | M | * | SIG | 12.78 | ### | Y | 0 | 224.26 | Vasodilator (CNP) | ||||
|
| ||||||||||||||
| Extracellular matrix components | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-41612 | 1466 | R | * | SIG | 20.25 | 9.16 | Y | 4.9 | 0.2 | ECM | 1 | 17 | 0 | |
| BatTrinityAbyss-67131 | 188 | M | SIG | 19.38 | 5.65 | 0.58 | 1.51 | ECM | ||||||
| BatTrinityAbyss-510471 | 1914 | A | CYT | 86.72 | 6.29 | 0.98 | 1.02 | ECM | 1 | 3 | 1 | 3 | ||
| BatTrinityAbyss-521514 | 4436 | A | CYT | 330.47 | 5.58 | Y | 0.62 | 1.61 | ECM | ### | 2 | 1 | 2 | |
|
| ||||||||||||||
| Lectins | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-475358 | 289 | N | * | CYT | 26.53 | 8.89 | 1.17 | 0.81 | Galectin-3 | 1 | 15 | 1 | 15 | |
| BatTrinityAbyss-36853 | 411 | E | * | CYT | 20.77 | 6.25 | 0.73 | 1.31 | Galectin-7 | 1 | 19 | 1 | 19 | |
| BatTrinityAbyss-526622 | 3056 | R | * | CYT | 40.44 | 9 | Y | 0.8 | 1.25 | Galectin-8 | 1 | 15 | 1 | 14 |
| DrSigp-SigP-526623 | 1453 | M | * | SIG | 21.88 | 8.38 | Y | 0.69 | 1.43 | Galectin-8 | ||||
| BatTrinityAbyss-467016 | 231 | M | * | CYT | 18.95 | 5.43 | 1.42 | 0.67 | Galectin | |||||
| DrSigp-SigP-534086 | 59576 | M | * | SIG | 26.66 | 7.77 | Y | 4.57 | 0.22 | Lectin | 1 | 18 | 1 | 18 |
| BatTrinityAbyss-508153 | 13694 | V | * | CYT | 289.55 | 5.06 | Y | 0.35 | 2.87 | Lectin | ### | 6 | 1 | 3 |
|
| ||||||||||||||
| Antimicrobial peptides | ||||||||||||||
|
| ||||||||||||||
| Defensin | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-401005 | 112 | M | * | SIG | 7.52 | 9.05 | 1.78 | 0.51 | β-defensin | |||||
|
| ||||||||||||||
| Lysozyme | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-500942 | 1442243 | M | * | SIG | 16.63 | 9.34 | Y | 4.53 | 0.22 | Lysozyme | 1 | 19 | 1 | 19 |
|
| ||||||||||||||
| Other antimicrobial agents | ||||||||||||||
|
| ||||||||||||||
| DrSigp-SigP-429618 | 105 | M | * | SIG | 12.51 | 5.65 | 0.59 | 1.35 | BPI/LBP/CETP far | |||||
| BatTrinityAbyss-86412 | 163 | M | * | SIG | 22.15 | 9.03 | Y | 1.73 | 0.54 | lymphotoxin | ||||
|
| ||||||||||||||
| Complement and regulators | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-547414 | 1280 | A | * | CYT | 25.65 | 4.53 | Y | 0.51 | 1.92 | Complement | 1 | 16 | 1 | 16 |
| DrSigp-SigP-483662 | 454 | M | SIG | 43.51 | 6.21 | 0.95 | 1.02 | Complement | ### | 7 | ### | 8 | ||
| BatTrinityAbyss-483662 | 436 | N | SIG | 43.03 | 6.44 | 0.87 | 1.1 | Complement | ### | 7 | #### | 8 | ||
| BatTrinityAbyss-563735 | 2587 | R | * | SIG | 129.72 | 6.1 | Y | 1.26 | 0.79 | Complement | 1 | 5 | 1 | 5 |
| BatTrinityAbyss-40668 | 724 | M | * | SIG | 25.54 | 5.85 | Y | 0.4 | 2.35 | Complement | 1 | 17 | 1 | 16 |
|
| ||||||||||||||
| Growth factors and immune regulators | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-533465 | 1694 | M | * | SIG | 23.64 | 6.48 | Y | 0.3 | 3.26 | Stromal cell factor | 1 | 17 | 1 | 17 |
| DrSigp-SigP-38368 | 1290 | M | * | SIG | 28.1 | 4.57 | Y | 1.23 | 0.8 | hypothetical proteins | 1 | 15 | 1 | 15 |
|
| ||||||||||||||
| Immunity related products | ||||||||||||||
|
| ||||||||||||||
| C-C motif chemokine (CCL28-like) | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-506850 | 4563 | M | * | SIG | 14.37 | ### | Y | 0.03 | 30.37 | Chemokine | ||||
|
| ||||||||||||||
| T cell immunomodullatory protein | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-511328 | 3228 | M | * | BL | 90.89 | 5.26 | Y | 0.65 | 1.52 | T-cell antigen | ||||
| BatTrinityAbyss-511332 | 23206 | L | * | CYT | 88.87 | 5.18 | Y | 0.8 | 1.25 | T-cell antigen | ||||
| BatTrinityAbyss-511331 | 23214 | L | * | CYT | 87.9 | 5.22 | Y | 0.8 | 1.25 | T-cell antigen | 1 | 7 | 1 | 7 |
| BatTrinityAbyss-550694 | 2918 | M | * | SIG | 25.21 | 8.65 | Y | 3.99 | 0.25 | Insulin secretion | ||||
| BatTrinityAbyss-533680 | 338 | M | * | SIG | 49.25 | 7.76 | Y | 4.98 | 0.2 | BPI/LBP/CETP | 1 | 11 | 0 | |
|
| ||||||||||||||
| Other immunity related products (TSG-6) | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-527888 | 8330 | M | * | SIG | 31.29 | 7.6 | Y | 0.02 | 42.67 | TNF-inducible gene | 0 | 1 | 14 | |
| BatTrinityAbyss-509950 | 844810 | M | * | SIG | 27.15 | 5.4 | Y | 0 | 345.53 | TNF-inducible gene | 1 | 15 | 1 | 15 |
|
| ||||||||||||||
| MHC class I and II antigen | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-507167 | 456 | M | SIG | 31.63 | 5.76 | Y | 0.62 | 1.54 | MHC class I B | |||||
|
| ||||||||||||||
| Ig mu chain | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-39685 | 1674 | S | * | CYT | 49.76 | 5.81 | Y | 1.57 | 0.63 | IgG chain | 1 | 9 | 1 | 9 |
| BatTrinityAbyss-496807 | 479 | M | * | SIG | 15.15 | 9.12 | Y | 0.13 | 6.34 | IgG chain | ### | 17 | 1 | 11 |
| DrSigp-SigP-488712 | 579 | M | SIG | 15.98 | 8.47 | Y | 0.65 | 1.48 | IgG chain | 1 | 11 | 1 | 11 | |
| BatTrinityAbyss-510560 | 570247 | V | * | BL | 51.47 | 5.46 | Y | 0.12 | 8.15 | IgG chain | 1 | 11 | 1 | 12 |
| BatTrinityAbyss-138350 | 42 | V | CYT | 13.54 | 9.43 | 0.36 | 1.37 | IgG chain | 1 | 10 | 1 | 11 | ||
| BatTrinityAbyss-522329 | 586 | V | CYT | 14.08 | 7.94 | Y | 0.67 | 1.44 | IgG chain | 1 | 11 | 1 | 11 | |
|
| ||||||||||||||
| Ig lambda chain | ||||||||||||||
|
| ||||||||||||||
| BatTrinityAbyss-505962 | 35933 | L | * | SIG | 24.36 | 7.04 | Y | 0.15 | 6.51 | IgG chain | 1 | 17 | 1 | 16 |
| BatTrinityAbyss-46124 | 571 | M | SIG | 15.93 | 5.21 | Y | 2.19 | 0.45 | IgG chain | |||||
| BatTrinityAbyss-521576 | 399 | M | SIG | 15.31 | 5.07 | Y | 0.35 | 2.6 | IgG chain | |||||
| BatTrinityAbyss-505979 | 40645 | T | * | SIG | 26 | 8.17 | Y | 0.11 | 9 | IgG chain | 1 | 17 | 1 | 16 |
|
| ||||||||||||||
| Mucins (hundreds not shown) | ||||||||||||||
|
| ||||||||||||||
| DrSigp-SigP-493041 | 6272756 | M | * | SIG | 182.75 | 5.29 | Y | 5 | 0.2 | Mucin | 1 | 2 | 1 | 1 |
| BatTrinityAbyss-497247 | 1991177 | M | * | SIG | 13.25 | 4.55 | Y | 5 | 0.2 | Mucin | 1 | 15 | 1 | 18 |
M, indicates the presence of methionine.
SigP, presence of signal peptide.
MW, molecular weight of the the mature protein.
pI, isoelectric point.
, indicates the presence of stop codon.
A/P, ratio of transcripts present in the Acessory/Principal gland.
P/A, ratio of transcripts present in the Principal/Acessory gland.
p, p<0.05 for difference in expression levels for Principal vs Accessory glands.
11 P (Accessory Gland), 7 P (Principal Gland). Predict, predictive value for identification (1, maximum).
11 S (Accessory Gland), 7 S (Principal Gland). Slice from the 1D-gel (Figures 2A and 2B) where the protein was found in the proteome.
Inh., inhibitor. ECM, extracellular matrix.
SIG, signal peptide; CYT, cytosolic; BL, borderline prediction.
Principal Submaxillary (PS) Gland
Genes possibly associated with hematophagy
Plasminogen Activators
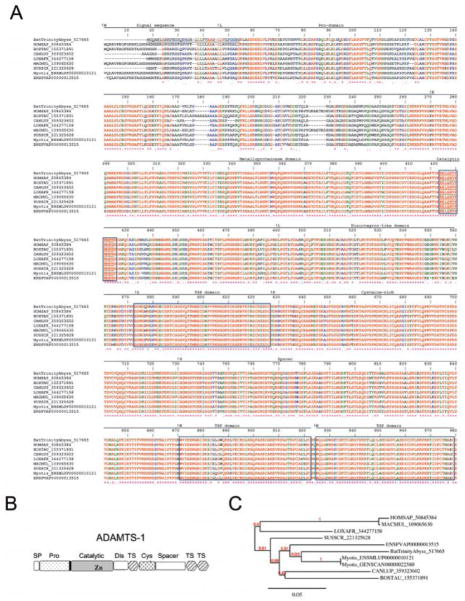
An activity compatible with fibrinolysis in bat saliva was reported in 1932 [42] and 1966 [8]. Only decades after the enzyme responsible for this activity was molecularly cloned [13, 14]. Desmodus salivary plasminogen activator (DSPA), or Desmoteplase (DSPA-α1), is the ortholog of human t-PA; however, it also exhibits several important biochemical properties and structural aspects that explain its mechanism of action. Plasminogen activators (e.g., human t-PA) display five domains: a fibronectin (or finger) domain (F), an EGF domain (E), two Kringle (K1, K2) domains, and a protease domain (P). These domains mediate t-PA binding to its cofactors and cell receptors, interact with inhibitors, and also catalytically cleave plasminogen with formation of plasmin, which digests fibrin [16]. Plasminogen activators are also involved in inflammation and tissue remodeling [43, 44].
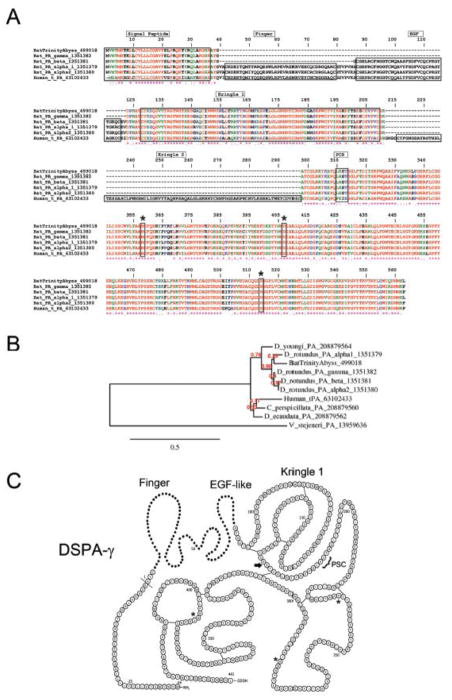
Four DSPA have been identified, and each variant exhibits high sequence homology to t-PA, although with complete domain deletions. For example, DSPAα1 and DSPAα2 harbor the same domains as human t-PA except for K2. Lack of this domains enables DSPA to interact with fibrin and interferes with inhibition of its function by plasminogen-activator inhibitor-1, the main physiologic inhibitor of fibrinolysis [43]. It has also been demonstrated that DSPA variants lack the plasmin-sensitive processing site (PSC); therefore, this proteins is less susceptible to cleavage into a two-chain form t-PA. This implies that all forms of DSPA are expressed as stable single-chain plasminogen activators. Notably, plasminogen activators from bats, such as DSPA-α1, are not inherently potent as fibrinolytic enzymes, but their activity is greatly enhanced in the presence of fibrin, a feature that distinguishes DSPA from other plasminogen activators [6, 15]. In fact, the activity of DSPAα1 is 105,000 times higher in the presence of fibrin than in its absence, while for t-PA, the factor is only 550 [15]. Despite its distinct sequence similarity, DSPA and t-PA share structural similarities that have been revealed by X-ray crystallography [45]. The enhanced specificity of DSPA for plasminogen in the presence of fibrin—and the longer half-life in plasma—are the basis for its potential clinical use in the treatment of stroke [16]. In our transcriptome analysis, we identified expression levels 791 times higher of DSPAγ in the PS gland than in the AC gland (Table 4), with over 1.4 million reads mapping to its CDS. The average coverage per base of the CDS was over 51,000. Remarkably, the abyss assembly did not retrieve this CDS, but rather many shorter fragments that appeared to contain retained introns. Only the trinity assembler was able to retrieve this CDS. However, abyss was able to retrieve many other CDS not retrieved by trinity (not shown). Figure 4A shows the Clustal alignment of DSPAγ sequenced in this project, human t-PA, and other DSPA deposited in the database, including DSPAγ, DSPAβ, DSPAα1, and DSPAα2. The phylogeny among vampire bat DSPA, human t-PA, or plasminogen activators from non-hematophagous bats (Carollia perspicillata) and snake venom Viridovipera stejeneri [46] is presented in Figure 4B. Predicted secondary structure of DSPAγ shows that it is a truncated form displaying only the K1, and protease domains (Figure 4C). Proteome studies presented in Figure 2A identified DSPAγ among the most expressed proteins. The function of this family of protein has been related to the fibrinolytic properties of saliva [16].
Figure 4.
Desmodus rotundus salivary plasminogen activator (DSPA). (A) Alignment of DSPAγ sequenced here and other plasminogen activator from D. rotundus or human t-PA. Accession numbers are indicated in the figure. The signal peptide, finger domain, EGF-like domain, Kringle 1, Kringle 2, and plasmin consensus sequence (PCS) are boxed for comparison. The catalytic triad (His, Asp, and Ser) are boxed and marked with an asterisk. Note that PSC (FRIK sequence) and Kringle 2 are present in t-PA only. (B) Phylogeny of plasminogen activators from vampire bats (D. rotundus, Diaemus youngi, and Diphylla ecaudata), non-hematophagous bats (Carollia perspicillata), and snake venom (Viridovipera stejeneri). (C) Predicted secondary structure for DSPAγ. The Finger and EGF-like domains present in DSPA-γ are shown as dots. Present in t-PA but not present in any DSPA are: PSC, plasmin sensitive processing site, and an additional Kringle (arrow). Asterisks indicate the amino acids that are part of the catalytic triad. Modified from [14].
Lipocalins
The lipocalin structure consists of an eight-stranded, antiparallel β-barrel forming a central hydrophobic cavity. Usually, lipocalins act by binding a small-molecule ligand, but they can also act by binding proteins in solution or receptors [47]. Transcriptome analysis indicates that at least four conserved members of the lipocalin superfamily are abundantly expressed in the principal gland, reaching 500–1000 higher levels than observed with the AC (Table 4). Notably, some unrelated bloodsucking arthropod orders (ticks and kissing bugs) studied so far express several members of the lipocalin family, which exemplifies typical cases of convergent evolution. Accordingly, lipocalins from hematophagous sources have been shown to interact and release vasodilator nitric oxide to the host [48] or to scavenge mast cell histamine, which increases permeability [49]. Other lipocalins have been shown to bind vasoconstrictor serotonin [50] or to mop pro-aggregatory molecule ADP [51, 52] and TXA2 [53, 54]. Still others interact with leukotrienes (e.g., LTB4) [55, 56] or macromolecules such as thrombin [57] or FIX/IXa [58]. Figure 5A shows the alignment of vampire bat lipocalins and some selected counterparts from vertebrata. Figure 5B is a phylogenetic tree that included hundreds of lipocalins sequenced from the SGs of hematophagous sources. Notably, vampire bat lipocalins do not clade with other lipocalins, suggesting divergence of function. Actually, lipocalins are notorious for their plasticity and reportedly exhibit distinct binding specificity despite high sequence homology among its members [47]. It is important to recognize that the high levels of expression of these proteins are necessary because biogenic amines and prostaglandins accumulate to near micromolar levels during inflammation and hemostasis [59]. In fact, lipocalins were among the most abundant ions retrieved in our proteome analysis of the PS gland (Figure 2A). It is possible that bat salivary lipocalins interact with pro-hemostatic molecules described above, explaining inhibition of platelet aggregation observed before with saliva [10]. Conceivably, bat salivary lipocalins diminish the pro-aggregatory tonus at site of incision, as reported for RPAI-1, moubatin, pallidipin, dipetalidipin, and triplatin isolated from blood-sucking arthropods [60].
Figure 5.
Desmodus rotundus salivary lipocalins. (A) Clustal alignment of vampire bat lipocalins sequenced here and other members from Vertebrata. Accession numbers are indicated in the figure. BOSTAU, Bos taurus; CANLUP, Canis lupu; EQUCAB, Equus caballus; SUSSCR; Sus scrofa. (B) Phylogenetic tree of lipocalins from D. rotundus and other hematophagous sources. Sequences derived from the nonredundant protein database of the National Center for Biotechnology Information are represented by six letters followed by the NCBI gi| accession number. The six letters derive from the first three letters of the genus and the first three letters from the species name. Protein sequences were aligned by the Clustal program, and the dendrogram was obtained with the Mega package after 10 000 bootstrap replications using the neighbor-joining algorithm, poisson model. The bar represents 20% amino acid divergence in the sequences. The color in the branches indicates the groups: red, vampire bat; blue, triatominae; green, tick; purple, mammalia.
Lipophilin/secretoglobin members
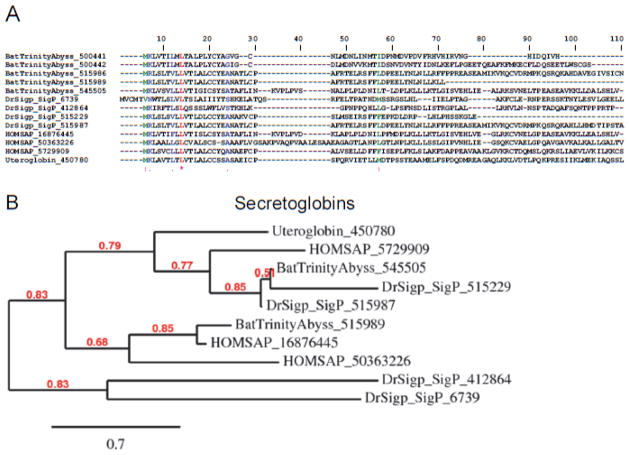
Secretoglobins are a family of secreted proteins found in mammals. They are found at high levels in many secretions including uterine, prostatic, pulmonary, lacrimal, and SGs. Secretoglobins are small (~10 kD) proteins with multiple functions that include immune modulation and inhibition of inflammation. The most studied member is uteroglobin, a steroid-inducible immunomodulatory protein that forms dimers containing internal hydrophobic cavities located at the interface between the two subunits. This is the location of binding of hydrophobic ligands, various eicosanoid mediators of inflammation—including PGD2, which is produced by mast cells and plays a major role in allergic reactions—and PGF2α a potent vasoconstrictor. Indirect evidence also suggests that uteroglobin binds leukotrienes [61]. Notably, we found high levels of expression for several bat salivary secretoglobins, with members of this family being expressed more than 1000 times in the PS gland in comparison to the AC (Table 4). Strikingly, a significant divergence in size and cysteine content was found among the members, suggesting divergence in function. The Clustal alignment of bat saliva secretoglobin and other members from Vertebrata are shown in Figure 6A, and their phylogenetic relationships in Figure 6B. The presence of abundantly expressed secretoglobins was confirmed by proteome analysis (Figure 2A). Conceivably, these proteins operate to remove pro-hemostatic mediators of inflammation, which affect platelet aggregation, leukocyte biology, or the vessel tonus. Of note, no member of the secretoglobin family have been sequenced in the SG of bloodsucking arthropods, indicating that recruitment of this family of proteins in the bat SG is, at present, a unique phenomenon among hematophagous animals. The specificity of bat secretoglobins remains to be proven through recombinant expression.
Figure 6.
Desmodus rotundus salivary secretoglobins. (A) Clustal alignment of vampire bat secretoglobins sequenced here and other members from humans. Accession numbers are indicated in the figure. HOMSAP, Home sapiens. (B) Phylogeny for the sequences depicted in (A).
Metalloproteases (ADAMTS-1)
ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) is a member of a group of secreted proteases with 19 members [62] in humans. ADAMTS is a branch of the ADAM (a disintegrin and metalloproteinase-like) subfamily of metalloproteinase found in snake venom metalloproteinases (reprolysins) [63, 64]. ADAMTSs are secreted molecules, some of which bind to the extracellular matrix. Structurally, ADAMTSs are synthesized as pre-proenzymes, contain a furin-recognition site, and comprise several domains. These include a signal peptide, a pro-domain, and a metalloproteinase catalytic domain with a zinc-binding motif (HEXXHXXG/N/SXXHD). Although ADAMTS also exhibits a disintegrin-like domain, no tripeptide (e.g., RGD) sequence has been found. Moreover, there is no evidence that ADAMTS associates with integrins, as reported for venom ADAMs and disintegrins from hematophagous sources [63–65]. Other domains include a central thrombospondin type I-like, a cysteine-rich domain, a spacer region, and a variable number of C-terminal thrombospondin type I-like repeats [62]. According to our analyses, the PS gland expresses a member of the ADAMTS family, which was identified as ADAMTS-1 ~ 70 times more than does the AC gland (Table 4). Figure 7A shows an alignment of bat saliva ADAMTS-1 and other members of this family form Vertebrata. It harbors all the domains present in ADAMTS-1 (Figure 7B) including the catalytic site, indicating that this molecule is enzymatically active. The phylogenetic association among ADAMTS-1 members is presented in Figure 7C. Further, high levels of expression were found for ADAMTS-1 in the PS gland as opposed to the AC gland (Figure 2A). Functionally, ADAMTS-1 cleaves aggregacan, versican [66], and interacts with the extracellular matrix, supporting involvement for the TSP domains [67, 68]. ADAMTS-1 has also been found to display anti-angiogenesis activity through inhibition of FGF-2-induced vascularization in the cornea and VEGF-induced angiogenesis in the chorioallantoic membrane assays [69]. Perhaps ADAMTS-1 in bat saliva affects angiogenesis, as previously reported for tick saliva [70, 71].
Figure 7.
Desmodus rotundus salivary metalloprotease (ADAMTS-1). (A) Clustal alignment of vampire bat ADAMTS-1 sequenced here and other members from Vertebrata. Accession numbers are indicated in the figure. HOMSAP, Homo sapiens; BOSTAU, Bos Taurus; CANLUP, Canis lupu; LOXAFR, Loxodonta Africana; MACMUL, Macaca mulatta; SUSSCR; Sus scrofa. The signal peptide (SP), pro-domain (Pro), metalloprotease domain catalytic site (boxed), disintegrin (DIS)-like domain, thrombospondin (TSP or TS) domains (boxed), cysteine-rich and spacer domain are based on human ADAMTS-1. Modified from [67, 68, 142, 143]. The predicted signal peptide for bat ADAMTS-1 is underlined (prediction by Signal P). (B) Modular gene organization of ADAMTS-1 members [68]. (C) Phylogeny of ADAMTS-1 from Vertebrata.
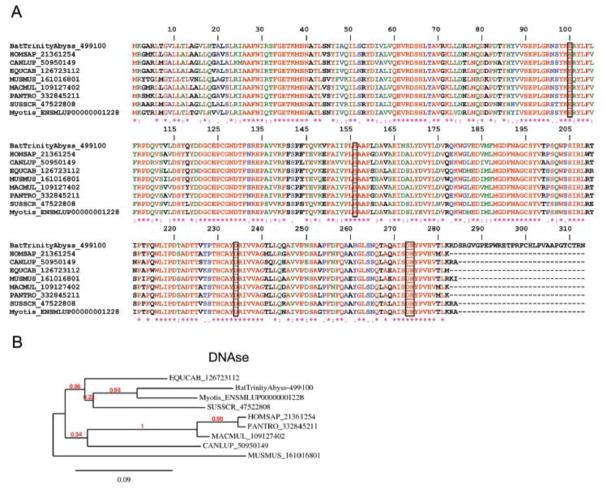
Endonucleases (deoxyribonucleases, DNAses)
Deoxyribonucleases are enzymes capable of hydrolyzing nucleic acids. They belong to the phosphodiesterase family and are capable of cleaving phosphodiester internal bonds within double-stranded (ds) or single-stranded (ss) DNA and RNA substrates [72]. Endonucleases from both eukaryotes and prokaryotes require magnesium for their activity. The function of endonucleases is associated with repair, recombination, transposition, and degradation of DNA; however, nucleases are also involved in cell defense and in promoting degradation of foreign nucleic acids. Recently, neutrophils, basophils, and mast cells were found to release neutrophil extracellular traps (NETs), which are extracellular DNA fibers comprising histones and neutrophil antimicrobial protein [73]. NETs are formed by a cell-death program that proceeds from dissolution of internal membranes followed by chromatin decondensation and cytolysis (NETosis). NETs induce coagulation through activation of FXII [74], stimulate platelet adhesion, and intercalate with a fibrin clot to generate a t-PA resistance scaffold [75]. Notably, bat SGs exhibit a high content of DNAse, being the eighth most abundant transcripts in the PS gland (Table 4). This signifies that very high levels of endonuclease activity are present in the saliva. In fact, this enzyme was identified as highly expressed in the PS gland according to our proteome analysis (Figure 2A). Of note, bat salivary DNAse is highly homologous to DNAse-I from several vertebrates (Figure 8A) and exhibits all conserved domains of this family including a catalytic site, DNA binding site, and metal-binding site, in addition to phosphate and actin-binding sites [76]. In contrast, bat DNAse exhibits a ~ 30-amino acid (aa) long residue in the C-terminus that is not present in other DNAses. Phylogenetic associations among DNAses from Vertebrata are shown in Figure 8B. Conceivably, bat DNAses plays a major role in preventing NET-associated procoagulant events and in promoting an environment appropriate for plasminogen activation by DSPA. Endonucleases are not specific for bat SG, as they have been found in mosquitoes, sand flies, and in simulidae [77] and snake venoms [78]. Consistent with a critical role of endonucleases as negative modulators of vascular biology, supplementation of DNAse-I effectively prevents thrombus formation in vitro and in vivo [74, 79].
Figure 8.
Desmodus rotundus salivary DNAses. (A) Clustal alignment of vampire bat DNAses sequenced here and other members from Vertebrata. Accession numbers are indicated in the figure. Residues involved in catalysis are boxed. HOMSAP, Homo sapiens; CANLUP, Canis lupu; EQUCAB, Equus caballus; MUSMUS, Mus musculus; MACMUL, Macaca mulatta; PANTRO, Pan troglodytes; SUSSCR; Sus scrofa. (B) Phylogeny for the sequences depicted in (A).
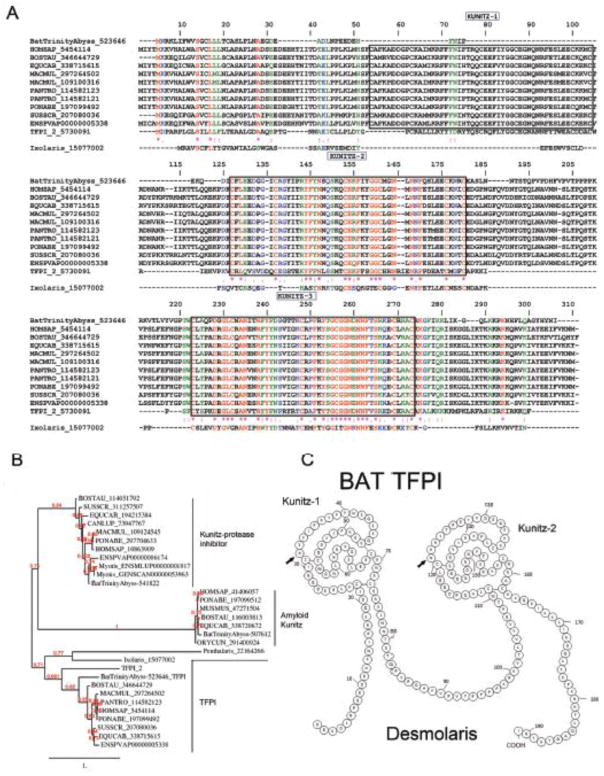
Kunitz domain-containing proteins (TFPI)
Kunitz proteins are known to inhibit blood coagulation such as through blockade of tissue factor, or other enzymes involved in inflammation [80]. The Kunitz fold can also perform functions beyond protease inhibition, such as ion channel inhibition. They are also abundantly expressed in snake venom, where they account for the neurotoxic activity of some venoms though K+ channel blockade [81]. From the transcriptome of D. rotundus, we found three distinct families of Kunitz-containing proteins (protease Kunitz inhibitor, amyloid Kunitz, and TFPI), which vary in length, cysteine content, and number of Kunitz. Of relevance for hematophagy, one family of Kunitz was found to be expressed 30–50 times more in the PS gland as opposed to the AC gland (Table 4). Members of this family contain two-Kunitz domains and display high sequence similarity to the anticoagulant TFPI-1, as depicted by a Clustal alignment (Figure 9A) and phylogenetic tree (Figure 9B). Of note, proteome studies (Figure 2A) identified TFPI as a relatively abundant protein in the PS gland. This discovery is important because TFPI controls the initiation of the coagulation cascade by the FVIIa/TF complex. TFPI is a 34- to 43-kD multidomain protein with an acidic amino terminus, three typical Kunitz-type inhibitor domains, and a basic carboxy terminus. The second Kunitz domain binds and inhibits FXa, and the first Kunitz domain binds the FVIIa/TF complex through formation of a final quaternary inhibitory complex consisting of FVIIa/TF/TFPI/FXa [82]. Interaction of TFPI-1 with FVIIa and FXa is mediated by arginine present in the P1 position of K1 and K2, respectively [80]. The third Kunitz plays a role in cell surface binding [83]. Another three- Kunitz inhibitor, named TFPI-2, exhibits arginine at the P1 position, but a glutamic acid and a serine are present in K2 and K3, respectively. TFPI-2 is a weak inhibitor of FVIIa/TF but strongly inhibits trypsin, chymotrypsin, plasma kallikrein, FXIa, and plasmin [84]. Inhibition of plasmin and the interaction of TFPI-2 with the matrix suggest that it plays an important role in regulation of extracellular matrix degradation and remodeling. Bat TFPI, herein named Desmolaris, resembles human TFPI-1 and TFPI-2 with respect to an arginine occupying the P1 position in the K1. This domain likely participates in inhibition of serine protease with specificity that remains to be determined. In contrast, the P1-position for K2 in Desmolaris is the polar amino acid asparagine. Of note, bovine and equine TFPI-1 displays a polar residue glutamine in the P1 position of K3. Similarly to TFPI-1 and TFPI-2, Desmolaris displays acidic residues in the N-terminus and basic residues in the C-terminus, which is involved with interaction with heparin [80]. Nevertheless, it remains to be determined whether Desmolaris works as an inhibitor of the FVIIa/TF complex and PAR2 signaling, such as Ixolaris [85, 86] or Penthalaris [87]. Alternatively, it might target other serine proteases such as FXa [88], which is inhibited by a molecule present in saliva (named draculin) and putatively identified as lactotransferrin [11]. Figure 9C shows the secondary structure prediction for Desmolaris, based on human TFPI [80].
Figure 9.
Desmodus rotundus salivary tissue factor pathway inhibitor (TFPI). (A) Alignment of vampire bat TFPI sequenced here and Vertebrata TFPI-1, and TFPI-2. Accession numbers are indicated in the figure. HOMSAP, Homo sapiens; BOSTAU, Bos taurus; EQUCAB, Equus caballus; SUSSCR; Sus scrofa; MACMUL, Macaca mulatta; PANTRO, Pan troglodytes. PONABE, Pongo abelii. Kunitz domains are boxed (numbering based on human TFPI). Ixolaris, a TFPI from the tick Ixoldes scapularis is also aligned [85]. (B) Phylogeny of Kunitz inhibitors (e.g., TFPI, Kunitz protease inhibitor, and amyloid Kunitz protein) from vampire bats and other members from Vertebrata. (C) Predicted secondary structure for two-Kunitz vampire bat TFPI, named Desmolaris. Numbering is based on the two Kunitz domains of Desmolaris. Arrows indicate the P1′ positions. Modified from [80].
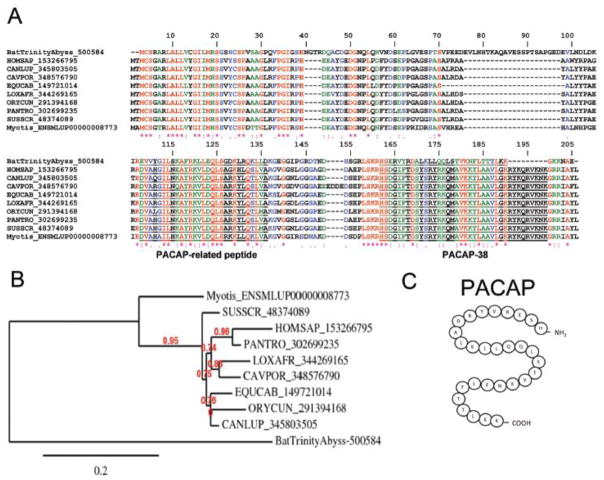
Pituitary adenylate cyclase activating peptide (PACAP)
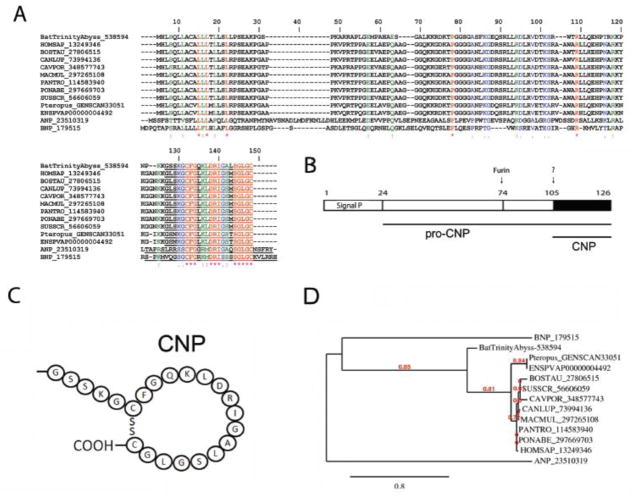
The critical importance of vasodilation in hematophagy is illustrated by the different chemical entities found in several SGs. To promote vasodilation, ticks use prostaglandins (PGE2, PGF2α, and PGI2), triatomines produce a gas (NO)-carrying molecule, and mosquitoes such as Aedes secrete tachykinin peptide [77, 89]. Notably, sandflies express maxadilan, a 61-aa peptide that mimics, without sharing sequence similarities, effects of endogenous PACAP [90]. Functionally, PACAP is 38-aa potent neuropeptide originally isolated from the hippocampus and derives— along with PACAP-related peptide—from cleavage of the 176-aa pre-pro-PACAP molecule. PACAP is a vasodilator that belongs to the secretin/glucagon/VIP/PACAP family [91, 92]. Activation of the PACAP receptor was also shown to inhibit lymphocyte activation and to have an immunosuppressive effect [93, 94]. Members of this family mediate their effects through G protein-coupled receptors VPAC1, VPAC2, and PAC1. These receptors transduce signaling through adenyl cyclase and phospholipase C pathways [95]. VPAC1 and VPAC2 receptors have essentially equivalent high affinities for both PACAP and VIP. PAC1 receptors have been described on neurons and smooth muscle [96] and have high affinity for PACAP and maxadilan. Notably, the PS gland from D. rotundus abundantly expresses a transcript to which over 330,000 reads were mapped that codes for a molecule with high similarity to other vertebrate PACAP (Table 4), as depicted by a Clustal alignment (Figure 10A), phylogenetic tree (Figure 10B), and proteome analysis (Figure 2A). In the bat version of PACAP, one insertion of approximately 20 aa at position 76 was observed. The secondary predicted structure of mature PACAP is shown in Figure 10C. The function of the bat PACAP peptide family is most probably related to vasodilation.
Figure 10.
Desmodus rotundus salivary pituitary adenyl cyclase activating peptide. (PACAP). (A) Alignment of vampire bat PACAP sequenced here and Vertebrata PACAP. HOMSAP, Homo sapiens; CANLUP, Canis lupu; CAVPOR, Cavia porcellus; EQUCAB, Equus caballus; LOXAFR, Loxodonta africana; ORYCUN, Oryctolagus cuniculus; PANTRO, Pan trogloditis; SUSSCR; Sus scrofa. (B) Phylogeny of PACAP sequences depicted in (A). (C) Predicted secondary structure for vampire bat mature PACAP [96].
C-type natriuretic peptide (CNP)
In mammals, the natriuretic peptide family consists of highly homologous polypeptide cardiac hormones: ANP, BNP, and C-type natriuretic peptide (CNP). Both ANP and BNP have analogous endocrine actions and exert both peripheral (natriuresis, vasodilatation, inhibition of aldosterone synthesis, and anti-mitogenic) and central (thirst suppression, inhibition of vasopressin adrenocorticotropic hormone release, and anti-sympathetic) effects to lower blood volume and pressure. Additional members of the natriuretic peptide family include urodilatin, a 32-residue elongated ANP that acts exclusively in the renal circulation, and the snake-derived Dendroaspis natriuretic peptide (DNP) [97]. CNP was the third member of the natriuretic peptide family to be discovered. It is present at high concentrations in the cardiovascular system and other peripheral tissues, most notably in vascular endothelial cells. Like ANP and BNP, CNP is expressed and stored as a pro-hormone and converted to the active peptide by a multistep process that involves a ubiquitous “pro-protein convertase” furin. CNP is a potent relaxant of arterial and venous smooth muscle in most mammalian species in vitro and is thought to act locally as a paracrine/autoregulator, as it is cleared rapidly from the circulation and is present at very low concentrations in plasma. Nevertheless, infusion of CNP lowers blood pressure in several species, including humans. Indeed, CNP has also been characterized as an endothelium-derived hyperpolarizing factor. CNP interacts with two subtypes of natriuretic peptide receptor, namely NPR-B and NPR-C. These receptors contain at the C-terminus a guanylate cyclase functional domain that generates the second messenger cGMP for signaling purposes (e.g., vasodilation) [98]. The bat PS SGs express high transcriptional levels of CNP (Table 4). Figure 11A show a Clustal alignment and phylogeny of bat salivary PACAP and its vertebrate counterparts. Notably, bat and human CNP share a similar primary sequence and domain structure (Figure 11B) except for residues in positions 2, 9, 16 and 17. A predicted secondary structure for mature bat salivary PACAP is shown in Figure 11C, and a phylogenetic tree for PACAP-related members is presented in Figure 11D. The function of bat CNP is related to vasodilation.
Figure 11.
Desmodus rotundus salivary C-type natriuretic peptide (CNP). (A) Alignment of vampire bat CNP sequenced here and Vertebrata CNP. HOMSAP, Homo sapiens; BOSTAU, Bos taurus; CANLUP, Canis lupu; CAVPOR, Cavia porcellus; MACMUL, Macaca mulatta; PANTRO, Pan troglodytes. PONABE, Pongo abelii; SUSSCR; Sus scrofa. (B) Modular gene organization of CNP members. (C) Predicted secondary structure for vampire bat mature CNP. (D) Phylogeny of CNP sequences depicted in (A).
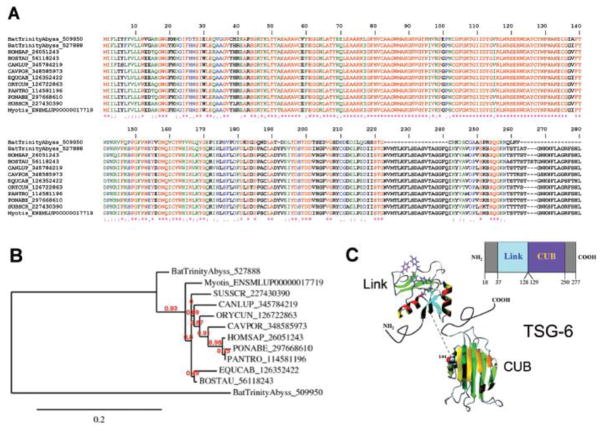
TNF-α-stimulated gene 6 (TSG-6)
TNF-α-stimulated gene 6 (TSG-6) was initially cloned from TNF-treated fibroblasts [99]. Structurally, TSG-6 is composed of two distinct homology regions. The N-terminal domain of TSG-6 represents a structural motif known as the Link module, which confers affinity for hyaluronan. Its presence defines a family of hyaluronan binding proteins designated as hyalectans. The C terminal half of TSG-6 forms a CUB (complement subcomponents C1r/C1s, Uegf, BMP-1) domain whose function remains unidentified [100]. Notably, TSG-6 has been found to potentiate the antiplasmin activity of inter-α-inhibitor (IαI) as the result of an interaction between the Link module domain of TSG-6 and the bikunin chain of IαI [101]. TSG-6 also inhibits neutrophil migration to interact with macrophage CD44 and modulate NF-κB signaling. TSG-6 inhibits inflammation, according to several in vivo models [100, 102]. Bat saliva expresses at least two members of the TSG-6 family (Table 4) whose Clustal alignment is presented in Figure 12A. One member displays two deletions at the C-terminal. Figure 12B shows the phylogeny among Vertebrata TSG-6 members. Of note, expression of TSG-6 was confirmed by the proteome of the PS gland (Figure 2A). Figure 12C depicts a modular representation of the gene that composes the Link and CUB domains and the structure of mature TSG-6. In bat saliva, this protein may function as an anti-inflammatory molecule.
Figure 12.
Desmodus rotundus salivary TSG-6. (A) Alignment of vampire bat TSG-6 sequenced here and Vertebrata TSG-6. HOMSAP, Homo sapiens; BOSTAU, Bos taurus; CANLUP, Canis lupu; CAVPOR, Cavia porcellus; EQUCAB, Equus caballus; ORYCUN, Oryctolagus cuniculus; PANTRO, Pan troglodytes; PONABE, Pongo abelii; SUSSCR; Sus scrofa. (B) Phylogeny of TSG-6 sequences depicted in (A). (C) Modular gene organization and structure of TSG-6. Modified from [100].
Chemokine CCL28
CCL28 is a chemokine signaling via CCR10 and CCR3 that is selectively expressed in certain mucosal tissues such as exocrine glands, trachea, and colon. CCL28 is particularly abundant in SGs and plays an important role in mucosal immunity as a chemoattractant for IgA-producing plasma cells into the mucosal lamina propria [103]. Notably, the C terminus of human CCL28 has a significant sequence similarity to histatin-5, a histidine-rich candidacidal peptide in human saliva [104]. More recently, it has been shown that CCL28 had a potent antimicrobial activity against Candida albicans, Gram-negative bacteria, and Gram-positive bacteria. Mechanistically, CCL28 exerts its antimicrobial activity under low-salt conditions and rapidly induces membrane permeability in target microbes [105]. In our analysis, bat salivary CCL28 was found to be abundant at the transcriptional (Table 4) and proteome (Figure 2A) levels. It is highly homologous to the human counterpart according to a Clustal analysis (Figure 13A) and phylogenetic tree (Figure 13B). CCL28 may function as a broad-spectrum antimicrobial protein in the saliva.
Figure 13.
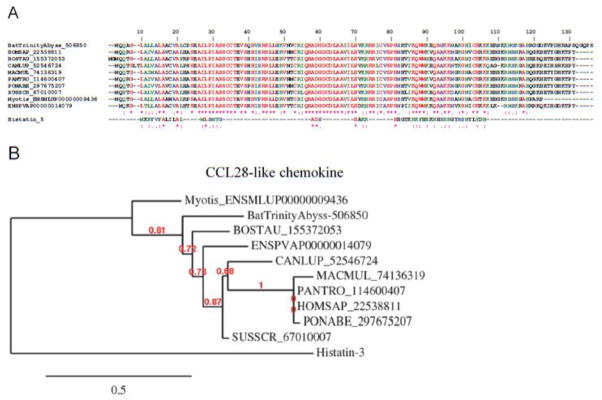
Desmodus rotundus salivary CCL28. (A) Alignment of vampire bat CCL28 sequenced here and other members from Vertebrata. HOMSAP, Homo sapiens; BOSTAU, Bos taurus; CANLUP, Canis lupu; MACMUL, Macaca mulatta; PANTRO, Pan troglodytes; PONABE, Pongo abelii; SUSSCR; Sus scrofa. (B) Phylogeny of TSG-6 sequences depicted in (A).
Apyrases, phosphatases, Antigen-5/CRISP, sphingomyelinase, epoxide hydrolase, and dipeptidyl peptidases
The PS gland also contains a number of transcripts that are expressed at levels 1.5–25 higher than those found in the AC gland (Table 4). Several transcripts were confirmed to be expressed according to our proteome analysis (Figure 2B). The protein families coded by these transcripts will be briefly covered here. For example, apyrases are Ca2+-dependent nucleotidases that play an important role in degrading pro-aggregatory ADP [106], which releases from red blood cells and other cell types [107]. Interestingly, a phosphatase was slightly enriched in the PS gland. The role of this enzyme is likely associated with removal of polyphosphates, which have been implicated in activation of coagulation FXII and FXI. FXIa activates the coagulation cascade leading to FXa, and thrombin formation, which activates PARs. FXIIa activates pre-kallikrein to kallikrein, resulting in bradykinin formation. Bradykinin, a major inducer of pain triggers increase in vessel permeability leading to edema formation [108]. Dipeptidyl peptidases are also found in the sialotranscriptome of D. rotundus, with higher expression in the PS gland indicated by both transcriptome and proteomic analysis. Of note, dipeptidyl peptidases were shown to have high specificity in destroying plasma bradykinin in tick saliva[109]. Several secretory carboxypeptidases were also found overexpressed in the PS gland (by both transcriptome and proteome analysis), and these may also be involved in degradation of peptidic mediators of inflammation such as bradykinin and anaphylatoxins [110, 111]. Acid sphingomyelinase was detected at higher expression in the PS gland, in concordance with the proteomic result (Table 4 and Figure 2A). This enzyme may be related to immunomodulation of the host [112]. We also found Antigen-5 family members, which belong to the larger CAP family [113] found in mammals, snakes, plants [113, 114] and are among the most ubiquitous proteins in the SG of blood sucking arthropods [115]. With few exceptions [59, 116], their function is unknown. Mucins were found at high levels and may help to lubricate mouthparts. Finally Ig chains were detected according to transcriptome and proteome analysis of the gland. Evidently, the function of these proteins in bat saliva can only be assigned after recombinant expression.
Accessory Gland
Genes associated with antimicrobial peptide (defensin, lysozyme, and lactotransferrin) and protease inhibitors (cystatins, serpins, Kazal, and TIL domain-containing proteins)
Transcripts coding for the full-length sequence for an antimicrobial peptide of the defensin—and particularly lysozyme—families were expressed at higher levels in the AC gland, as indicated by both transcriptome and proteome analysis (Figure 2B). Antimicrobial peptides are a regular finding in sialotranscriptomes of hematophagous insects and ticks. These peptides, when ingested with the blood meal, help control bacterial growth in the gut and may also protect their host-feeding lesions from infection [117]. Cystatins are cysteine protease inhibitors of nearly 100 aa. Tick saliva is particularly rich in cystatins, which have been shown to target cathepsins L and S, to block inflammation, and to suppress dendritic cell maturation [118]. In the bat, one member of the cystatin family was identified in the AC gland. It exhibits a high degree of homology to vertebrate cystatin, suggesting a similar function. Serpins are a ubiquitous protein family associated with the function of serine protease inhibition, from which the family name derives [119]. Bat salivary serpins found in both transcriptome and proteome analysis may target several proteases, with specificity that remains to be determined. We also found sequences homologous to neuroserpin, a serpin that inhibits tPA, uPA, and plasmin, suggesting that it may modulate the activation of fibrinolysis triggered by DSPA [120]. Low levels of α1-antitrypsin, an inhibitor of proinflammatory elastase, were also sequenced. AC glands also express a protease C1 inhibitor that blocks C1-esterase, inhibits chymotrypsin and kallikrein, and is a very efficient inhibitor of FXIIa [121]. The canonical TIL domain is found in many protease inhibitors and exerts antimicrobial function. Members of this family have been found ubiquitously in blood-feeding insect and tick sialomes, but very few have been characterized [77, 89, 115, 122, 123]. Its presence was confirmed in the proteome studies (Figure 2B). We also discovered Kazal domain-containing transcripts and proteins, which are conceivably associated with serine protease inhibition and antimicrobial activity [124].
Epoxide hydrolases were found expressed in both glands to a similar level, by both transcriptome and proteome analysis. If expressed in saliva, this enzyme could decay the levels of epoxyeicosatrienoic acids in the host; however these lipids are vasodilatory [125] and should have a beneficial effect for vampire bat feeding. The role of these enzymes in feeding is not apparent. Several other proteins were detected in our results of the proteome study for AC gland shown in Figure 2B, including galectin, sphingomyelinase, lectin, PACAP, TSG-6, apyrase, IgGFc-binding protein, lactotransferrin, heme-oxygenase (gi 417398396) and others.
Vampire bat SG proteins: notable modulators of vascular biology
Bloodsucking animals such as ticks, mosquitoes, sand flies, hookworms, leeches, and bats are a notable source of bioactive molecules that counteract the host response to injury through a number of distinct mechanisms [77, 89, 115, 122, 123, 126, 127]. These animals also operate as vectors of several infectious diseases, either by serving as a vehicle to deliver the infectious agent or by mechanism that exacerbate the immune response at bite sites. Therefore, study of hematophagous secretions has been of particular interest to fully understand the pharmacologic armamentarium of saliva on one hand, and the vectorial capacity of a given bloodsucking animal on the other. In the case of vampire bats, the SGs have functionally evolved to face the exigencies posed by an obligate blood diet and to counteract the host response to injury caused by the skin incision.
Understanding of the composition of saliva and related substances has largely relied on the massive sequencing and bioinformatics analysis of the cDNAs obtained through library constructions. These results have often been complemented with a proteomic approach aimed to confirm the presence of a certain family of proteins in the tissue. To fully characterize the sequences that compose the genes of D. rotundus SGs involved with hematophagy, we have employed Illumina technology, which greatly enhances the coverage of transcripts compared with cDNA libraries used thus far. Illumina uses RNA obtained directly from the tissue with very few amplification steps, excluding the limitations of potential overamplification associated with cDNA library construction. Sequencing of millions of reads indicates that the complexity of the PS gland is much larger and not limited to activities previously found in the saliva [8, 10, 13–16].
On a technical note, as indicated above regarding transcript abundance and their assembly, the abyss assembler produced more and longer transcripts than the trinity assembler, but failed to retrieve the most abundant transcripts possibly because it is a genome assembler where the expected read abundance should be even, while trinity was optimized for transcriptome assembly and takes into consideration transcript abundance to join the reads. Transcripts not retrieved by Abyss (or retrieved in very fragmented way) included those of the endonucleases, plasminogen activator, PACAP, antigen 5 and many others. These considerations were elaborated before on a comparative study of RNAseq assemblers [24] and will not be further discussed here.
Notably, the most abundant genes recruited by the bat PS gland have been characterized in bloodsucking arthropods—or in human physiology—as fibrinolytic enzymes (e.g., t-PA and DSPA), vasodilators (e.g., PACAP and CNP), or putative inhibitors of platelet aggregation (e.g., secretoglobins and lipocalin), blood coagulation (e.g., TFPI), neutrophil function (e.g., DNAses), macrophage activity (e.g., TSP-6), and angiogenesis (e.g., ADAMTS-1). When compared with those for bloodsucking arthropods, our results do not show a large degree of redundancy or family expansions among members of these protein families with the exception, perhaps, of the secretoglobins. It appears that the relatively short evolutionary history of bats—known to feed on blood for about 2 million years—may be responsible for this level of complexity of transcripts that is not comparable, for instance, with ticks or venomous animals [89, 128–132]. Nevertheless, the adaptive roles of this diversity in some families are explained at least in part by a gene-duplication phenomenon (e.g., Kunitz-type inhibitors such as TFPI) or deletions (e.g., desmoteplase)[133–135]. The functions of several bat sequences described here are unknown. Cloning and expressing select genes will help in characterizing their targets. It may also provide tools to better understand vascular biology and the immune system. A diagram with the targets of the most abundant bat salivary proteins present in the principal submaxillary and how they may affect vascular biology is shown in Figure 14. The inset shows the relative abundance of the most abundant transcript in the principal submaxillary vs the AC gland.
Figure 14.
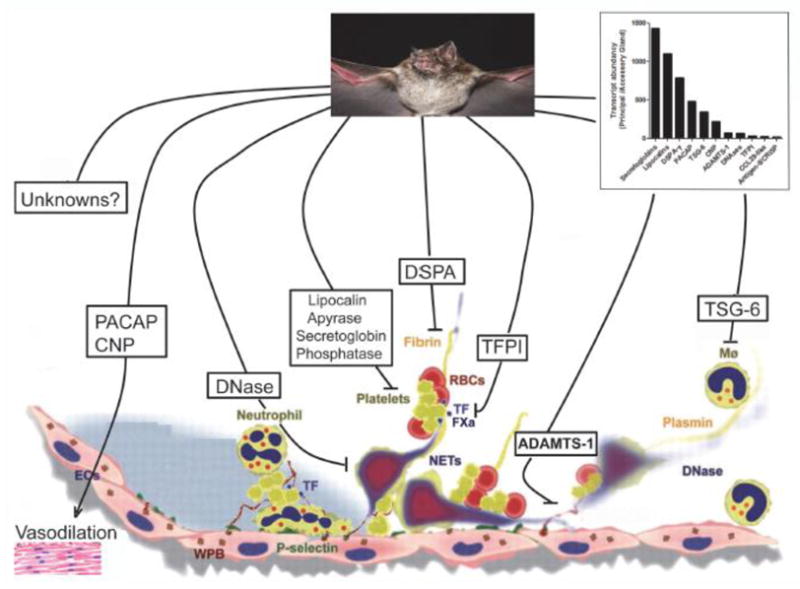
Desmodus rotundus saliva negatively modulates vascular biology. Vascular injury is accompanied by exposure of collagen, which promotes platelet adhesion and activation. Activated platelets release ADP and TXA2, which recruit other platelets to the site of injury [106]. Monocytes, activated platelets, and endothelium generate or recruit tissue factor–containing microparticles, leading to thrombin formation [74, 79]. Vascular injury also results in activation of endothelial cells that interact with platelets and neutrophils [144], resulting in NET formation, which supports platelet and red blood cell adhesion and facilitates fibrin generation [145]. Bat saliva may down-modulate several of these processes through intense production of plasmin by DSAP [16], blockade of initiation of the coagulation cascade by TFPI [80], and inhibition of NET-mediated coagulation activation by DNAse [145]. Platelet aggregation and vasoconstriction are likely blocked by lipocalins, secretoglobins [61], apyrases [107], and phosphatases [108]. Neutrophil and macrophages are also affected by TSG-6 [100], while angiogenesis might be modulated by ADAMTS-1 [62]. Finally, vasodilation is promoted by PACAP [91] and CNP [97]. Inset, relative abundance of transcripts in the principal submaxillary vs accessory glands. The transcripts correspond to the sequences (in parentheses) found in Table 4: Secretoglobin (500441), lipocalin (495626), plasminogen activator (499018), PACAP (500584), TSG-6 (509950), C-type natriuretic peptide (CNP, 538594), ADAMTS-1 (517665), DNAses (499100), TFPI (523646), CCL-28 (506850), antigen-5/CRISP (495870). For details, see text and Table 4 at http://exon.niaid.nih.gov/transcriptome/D_rotundus/Table4-web.xlsx).
Finally, bats are well known vectors of rabies [3, 4]. While the role of saliva in virus transmission has been documented in tick-borne diseases [136, 137], it is remarkable to note that components identified at high transcriptional levels in the SG of D. rotundus exhibit immunomodulatory properties and could be important for virus transmission. For example, PACAP affects dendritic cell production of inflammatory cytokines and displays immunomodulatory properties in vivo [138, 139]. Immunomodulation might also occur through inhibition of NET-derived neutrophil function through DNAses [140] or blockade of the coagulation cascade by TFPI [141], among other mechanisms [141].
CONCLUSION
The catalog of transcripts and proteins reported here will allow an in-depth investigation of the repertoire of antihemostatics and immunomodulators present in vampire bat saliva. It might also provide leads to understand whether and how salivary components contribute to host infection by rabies virus and other infectious agents transmitted by D. rotundus.
Supplementary Material
Significance.
Vampire bat saliva emerges as a novel source of antihemostatics which modulate several aspects of vascular biology.
Highlights.
Vampire bats are strict blood feeders.
Only one antihemostatic (plasminogen activator, desmoteplase) has been molecularly characterized.
200 million reads from the salivary glands of Desmodus rotundus were sequence by Illumina
Several novel families of proteins affecting hemostasis and the immune system were identified
Transcript products were confirmed by proteome analysis.
Bat saliva emerges as a novel source of modulator of Vascular Biology.
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (USA). We thank Drs. Glenn Nardone, Renee Olano, Carl Hammer, and Ming Zhao (Research Technology Branch, NIAID, NIH) for support and continuous assistance with the proteome study. We are thankful to Brenda Rae Marshall (DPSS, NIAID) for editorial assistance.
ABBREVIATIONS
- AC
accessory gland
- aPS
anterior PS
- pPS
posterior PS
- CDS
coding sequence
- CNP
C-type natriuretic peptide
- DPSA
Desmodus salivary plasminogen activator
- H
housekeeping class of function
- LN
lymph node
- MS
mass spectrometry
- NET
neutrophil extracellular trap
- PACAP
pituitary adenylate cyclase activating peptide
- pPS
posterior PS
- PS
principal submaxillary gland
- PSC
plasmin-sensitive processing site
- S
secreted class of function
- SG
salivary gland
- TE
transposable element
- TIL
trypsin inhibitor-like
- TSG-6
TNF-α-stimulated gene 6
- U
unknown class of function
Footnotes
The authors declare no competing financial interest.
Because I.M.B.F. and J.M.C.R. are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kunz TH, Fenton MB, editors. Bat Ecology. Chicago: University of Chicago Press; 2003. pp. 1–775. [Google Scholar]
- 2.Bredt A, Uieda W, Pedro WA, editors. Plantas e morcegos, na recuperação de áreas degradadas e na paisagem urbana. Brasília: Rede de Sementes do Cerrado; 2012. p. 273. [Google Scholar]
- 3.Arellano-Sota C. Biology, ecology, and control of the vampire bat. Rev Infect Dis. 1988;10 (Suppl 4):S615–9. doi: 10.1093/clinids/10.supplement_4.s615. [DOI] [PubMed] [Google Scholar]
- 4.Mayen F. Haematophagous bats in Brazil, their role in rabies transmission, impact on public health, livestock industry and alternatives to an indiscriminate reduction of bat population. Journal of veterinary medicine B, Infectious diseases and veterinary public health. 2003;50:469–72. doi: 10.1046/j.1439-0450.2003.00713.x. [DOI] [PubMed] [Google Scholar]
- 5.Greenghall AM, Schmidt U. Feeding behavior Natural History of Vampire Bats. 1988:111–31. [Google Scholar]
- 6.Schleuning WD. Vampire bat plasminogen activator DSPA-alpha-1 (desmoteplase): a thrombolytic drug optimized by natural selection. Haemostasis. 2001;31:118–22. doi: 10.1159/000048054. [DOI] [PubMed] [Google Scholar]
- 7.Tandler B, Toyoshima K, Phillips CJ. Ultrastructure of the principal and accessory submandibular glands of the common vampire bat. The American journal of anatomy. 1990;189:303–15. doi: 10.1002/aja.1001890403. [DOI] [PubMed] [Google Scholar]
- 8.Hawkey C. Plasminogen activator in saliva of the vampire bat Desmodus rotundus. Nature. 1966;211:434–5. doi: 10.1038/211434c0. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright T. The plasminogen activator of vampire bat saliva. Blood. 1974;43:317–26. [PubMed] [Google Scholar]
- 10.Hawkey C. Inhibitor of platelet aggregation present in saliva of the vampire bat Desmodus rotundus. Br J Haematol. 1967;13:1014–20. doi: 10.1111/j.1365-2141.1967.tb08870.x. [DOI] [PubMed] [Google Scholar]
- 11.Apitz-Castro R, Beguin S, Tablante A, Bartoli F, Holt JC, Hemker HC. Purification and partial characterization of draculin, the anticoagulant factor present in the saliva of vampire bats (Desmodus rotundus) Thromb Haemost. 1995;73:94–100. [PubMed] [Google Scholar]
- 12.Fernandez AZ, Tablante A, Beguin S, Hemker HC, Apitz-Castro R. Draculin, the anticoagulant factor in vampire bat saliva, is a tight-binding, noncompetitive inhibitor of activated factor X. Biochim Biophys Acta. 1999;1434:135–42. doi: 10.1016/s0167-4838(99)00160-0. [DOI] [PubMed] [Google Scholar]
- 13.Gardell SJ, Duong LT, Diehl RE, York JD, Hare TR, Register RB, et al. Isolation, characterization, and cDNA cloning of a vampire bat salivary plasminogen activator. J Biol Chem. 1989;264:17947–52. [PubMed] [Google Scholar]
- 14.Kratzschmar J, Haendler B, Langer G, Boidol W, Bringmann P, Alagon A, et al. The plasminogen activator family from the salivary gland of the vampire bat Desmodus rotundus: cloning and expression. Gene. 1991;105:229–37. doi: 10.1016/0378-1119(91)90155-5. [DOI] [PubMed] [Google Scholar]
- 15.Bringmann P, Gruber D, Liese A, Toschi L, Kratzchmar J, Schleuning WD, et al. Structural features mediating fibrin selectivity of vampire bat plasminogen activators. J Biol Chem. 1995;270:25596–603. doi: 10.1074/jbc.270.43.25596. [DOI] [PubMed] [Google Scholar]
- 16.Medcalf RL. Desmoteplase: discovery, insights and opportunities for ischaemic stroke. Br J Pharmacol. 2012;165:75–89. doi: 10.1111/j.1476-5381.2011.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uieda W, Chaves ME. Bats from Botucatu region, State of São Paulo, Southeastern Brazil. Chiroptera Neotropical. 2005;11:224–6. [Google Scholar]
- 18.Reis NRPA, Pedro WA, Lima IP. Morcegos do Brasil. Universidade Estadual de Londrina; 2007. pp. 1–253. [Google Scholar]
- 19.Disanto PE. Anatomy and histochemistry of the salivary glands of the vampire bat, Desmodus rotundus murinus. Journal of morphology. 1960;106:301–35. doi: 10.1002/jmor.1051060305. [DOI] [PubMed] [Google Scholar]
- 20.Vicente EC, Emiliani VC, Oliveira LF, Favero S. Análise morfométrica e anatômica do útero em famílias de quirópteros (mammalia) que ocorrem no Brazil. Ensaios e Ciencia. 2006;10:63–73. [Google Scholar]
- 21.Francischetti IM, My Pham V, Mans BJ, Andersen JF, Mather TN, Lane RS, et al. The transcriptome of the salivary glands of the female western black-legged tick Ixodes pacificus (Acari: Ixodidae) Insect Biochem Mol Biol. 2005;35:1142–61. doi: 10.1016/j.ibmb.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, et al. De novo transcriptome assembly with ABySS. Bioinformatics (Oxford, England) 2009;25:2872–7. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 23.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome research. 2009;19:1117–23. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC bioinformatics. 2011;12 (Suppl 14):S2. doi: 10.1186/1471-2105-12-S14-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS One. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein engineering. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 28.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–12. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 29.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 30.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 34.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 35.Sakurai A, Doci CL, Gutkind JS. Semaphorin signaling in angiogenesis, lymphangiogenesis and cancer. Cell research. 2012;22:23–32. doi: 10.1038/cr.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vadasz Z, Attias D, Kessel A, Toubi E. Neuropilins and semaphorins - from angiogenesis to autoimmunity. Autoimmunity reviews. 2010;9:825–9. doi: 10.1016/j.autrev.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochemical Society transactions. 2003;31:785–90. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 38.Beamer LJ, Carroll SF, Eisenberg D. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science (New York, NY. 1997;276:1861–4. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 39.Ho S, Pothoulakis C, Koon HW. Antimicrobial Peptides and Colitis. Curr Pharm Des. 2012 doi: 10.2174/13816128130108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–7. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 41.Galperin MY, Koonin EV. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic acids research. 2004;32:5452–63. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bier OE. Action anticoagulante et fibrinolytique de l’extract des glands salivaires d’une chauve-souris hematophage (Desmodus rotundus) C R Soc Biol. 1932;110:129–31. [Google Scholar]
- 43.Loscalzo J. An overview of thrombolytic agents. Chest. 1990;97:117S–23S. doi: 10.1378/chest.97.4_supplement.117s. [DOI] [PubMed] [Google Scholar]
- 44.Medcalf RL. Fibrinolysis, inflammation, and regulation of the plasminogen activating system. J Thromb Haemost. 2007;5 (Suppl 1):132–42. doi: 10.1111/j.1538-7836.2007.02464.x. [DOI] [PubMed] [Google Scholar]
- 45.Renatus M, Stubbs MT, Huber R, Bringmann P, Donner P, Schleuning WD, et al. Catalytic domain structure of vampire bat plasminogen activator: a molecular paradigm for proteolysis without activation cleavage. Biochemistry. 1997;36:13483–93. doi: 10.1021/bi971129x. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Wisner A, Xiong Y, Bon C. A novel plasminogen activator from snake venom. Purification, characterization, and molecular cloning. J Biol Chem. 1995;270:10246–55. doi: 10.1074/jbc.270.17.10246. [DOI] [PubMed] [Google Scholar]
- 47.Andersen JF, Gudderra NP, Francischetti IM, Ribeiro JM. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Arch Insect Biochem Physiol. 2005;58:97–105. doi: 10.1002/arch.20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribeiro JM, Hazzard JM, Nussenzveig RH, Champagne DE, Walker FA. Reversible binding of nitric oxide by a salivary heme protein from a bloodsucking insect. Science. 1993;260:539–41. doi: 10.1126/science.8386393. [DOI] [PubMed] [Google Scholar]
- 49.Paesen GC, Adams PL, Harlos K, Nuttall PA, Stuart DI. Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure. Mol Cell. 1999;3:661–71. doi: 10.1016/s1097-2765(00)80359-7. [DOI] [PubMed] [Google Scholar]
- 50.Andersen JF, Francischetti IM, Valenzuela JG, Schuck P, Ribeiro JM. Inhibition of hemostasis by a high affinity biogenic amine-binding protein from the saliva of a blood-feeding insect. The Journal of biological chemistry. 2003;278:4611–7. doi: 10.1074/jbc.M211438200. [DOI] [PubMed] [Google Scholar]
- 51.Francischetti IM, Andersen JF, Ribeiro JM. Biochemical and functional characterization of recombinant Rhodnius prolixus platelet aggregation inhibitor 1 as a novel lipocalin with high affinity for adenosine diphosphate and other adenine nucleotides. Biochemistry. 2002;41:3810–8. doi: 10.1021/bi011015s. [DOI] [PubMed] [Google Scholar]
- 52.Francischetti IM, Ribeiro JM, Champagne D, Andersen J. Purification, cloning, expression, and mechanism of action of a novel platelet aggregation inhibitor from the salivary gland of the blood-sucking bug, Rhodnius prolixus. The Journal of biological chemistry. 2000;275:12639–50. doi: 10.1074/jbc.275.17.12639. [DOI] [PubMed] [Google Scholar]
- 53.Assumpcao TC, Alvarenga PH, Ribeiro JM, Andersen JF, Francischetti IM. Dipetalodipin, a novel multifunctional salivary lipocalin that inhibits platelet aggregation, vasoconstriction, and angiogenesis through unique binding specificity for TXA2, PGF2alpha, and 15(S)-HETE. The Journal of biological chemistry. 2010;285:39001–12. doi: 10.1074/jbc.M110.152835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma D, Assumpcao TC, Li Y, Andersen JF, Ribeiro J, Francischetti IM. Triplatin, a platelet aggregation inhibitor from the salivary gland of the triatomine vector of Chagas disease, binds to TXA(2) but does not interact with glycoprotein PVI. Thromb Haemost. 2012;107:111–23. doi: 10.1160/TH11-10-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mans BJ, Ribeiro JM. A novel clade of cysteinyl leukotriene scavengers in soft ticks. Insect Biochem Mol Biol. 2008;38:862–70. doi: 10.1016/j.ibmb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaufays J, Adam B, Menten-Dedoyart C, Fievez L, Grosjean A, Decrem Y, et al. Ir-LBP, an ixodes ricinus tick salivary LTB4-binding lipocalin, interferes with host neutrophil function. PLoS One. 2008;3:e3987. doi: 10.1371/journal.pone.0003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noeske-Jungblut C, Haendler B, Donner P, Alagon A, Possani L, Schleuning WD. Triabin, a highly potent exosite inhibitor of thrombin. The Journal of biological chemistry. 1995;270:28629–34. doi: 10.1074/jbc.270.48.28629. [DOI] [PubMed] [Google Scholar]
- 58.Mizurini DM, Francischetti IM, Andersen JF, Monteiro RQ. Nitrophorin 2, a factor IX(a)-directed anticoagulant, inhibits arterial thrombosis without impairing haemostasis. Thromb Haemost. 2010;104:1116–23. doi: 10.1160/TH10.1160/TH10-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma D, Xu X, An S, Liu H, Yang X, Andersen JF, et al. A novel family of RGD-containing disintegrins (Tablysin-15) from the salivary gland of the horsefly Tabanus yao targets alphaIIbbeta3 or alphaVbeta3 and inhibits platelet aggregation and angiogenesis. Thromb Haemost. 2011;105:1032–45. doi: 10.1160/TH11-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Francischetti IM. Platelet aggregation inhibitors from hematophagous animals. Toxicon. 2010;56:1130–44. doi: 10.1016/j.toxicon.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee AB, Zhang Z, Chilton BS. Uteroglobin: a steroid-inducible immunomodulatory protein that founded the Secretoglobin superfamily. Endocrine reviews. 2007;28:707–25. doi: 10.1210/er.2007-0018. [DOI] [PubMed] [Google Scholar]
- 62.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox JW, Serrano SM. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275:3016–30. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- 64.Kini RM, Evans HJ. Structural domains in venom proteins: evidence that metalloproteinases and nonenzymatic platelet aggregation inhibitors (disintegrins) from snake venoms are derived by proteolysis from a common precursor. Toxicon. 1992;30:265–93. doi: 10.1016/0041-0101(92)90869-7. [DOI] [PubMed] [Google Scholar]
- 65.Assumpcao TC, Ribeiro JM, Francischetti IM. Disintegrins from hematophagous sources. Toxins. 2012;4:296–322. doi: 10.3390/toxins4050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stanton H, Melrose J, Little CB, Fosang AJ. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim Biophys Acta. 2011;1812:1616–29. doi: 10.1016/j.bbadis.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 67.Kuno K, Terashima Y, Matsushima K. ADAMTS-1 is an active metalloproteinase associated with the extracellular matrix. J Biol Chem. 1999;274:18821–6. doi: 10.1074/jbc.274.26.18821. [DOI] [PubMed] [Google Scholar]
- 68.Nagase H, Kashiwagi M. Aggrecanases and cartilage matrix degradation. Arthritis Res Ther. 2003;5:94–103. doi: 10.1186/ar630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vazquez F, Hastings G, Ortega MA, Lane TF, Oikemus S, Lombardo M, et al. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J Biol Chem. 1999;274:23349–57. doi: 10.1074/jbc.274.33.23349. [DOI] [PubMed] [Google Scholar]
- 70.Francischetti IM, Mather TN, Ribeiro JM. Tick saliva is a potent inhibitor of endothelial cell proliferation and angiogenesis. Thromb Haemost. 2005;94:167–74. doi: 10.1267/THRO05010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Francischetti IM, Mather TN, Ribeiro JM. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem Biophys Res Commun. 2003;305:869–75. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukae N, Enari M, Sakahira H, Fukuda Y, Inazawa J, Toh H, et al. Molecular cloning and characterization of human caspase-activated DNase. Proc Natl Acad Sci U S A. 1998;95:9123–8. doi: 10.1073/pnas.95.16.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–5. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 74.von Bruhl ML, Stark K, Steinhart A, Chandraratne S, Konrad I, Lorenz M, et al. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J Exp Med. 2012;209:819–35. doi: 10.1084/jem.20112322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuchs TA, Brill A, Duerschmied D, Schatzberg D, Monestier M, Myers DD, Jr, et al. Extracellular DNA traps promote thrombosis. Proc Natl Acad Sci U S A. 2010;107:15880–5. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, et al. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–58. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ribeiro JM, Mans BJ, Arca B. An insight into the sialome of blood-feeding Nematocera. Insect Biochem Mol Biol. 2010;40:767–84. doi: 10.1016/j.ibmb.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fox JW. A brief review of the scientific history of several lesser-known snake venom proteins: l-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon. 2012 doi: 10.1016/j.toxicon.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 79.Darbousset R, Thomas GM, Mezouar S, Frere C, Bonier R, Mackman N, et al. Tissue factor-positive neutrophils bind to injured endothelial wall and initiate thrombus formation. Blood. 2012;120:2133–43. doi: 10.1182/blood-2012-06-437772. [DOI] [PubMed] [Google Scholar]
- 80.Broze GJ, Jr, Girard TJ. Tissue factor pathway inhibitor: structure-function. Front Biosci. 2012;17:262–80. doi: 10.2741/3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harvey AL. Twenty years of dendrotoxins. Toxicon. 2001;39:15–26. doi: 10.1016/s0041-0101(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 82.Broze GJ, Jr, Warren LA, Novotny WF, Higuchi DA, Girard JJ, Miletich JP. The lipoprotein-associated coagulation inhibitor that inhibits the factor VII-tissue factor complex also inhibits factor Xa: insight into its possible mechanism of action. Blood. 1988;71:335–43. [PubMed] [Google Scholar]
- 83.Piro O, Broze GJ., Jr Role for the Kunitz-3 domain of tissue factor pathway inhibitor-alpha in cell surface binding. Circulation. 2004;110:3567–72. doi: 10.1161/01.CIR.0000148778.76917.89. [DOI] [PubMed] [Google Scholar]
- 84.Chand HS, Foster DC, Kisiel W. Structure, function and biology of tissue factor pathway inhibitor-2. Thromb Haemost. 2005;94:1122–30. doi: 10.1160/TH05-07-0509. [DOI] [PubMed] [Google Scholar]
- 85.Francischetti IM, Valenzuela JG, Andersen JF, Mather TN, Ribeiro JM. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002;99:3602–12. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- 86.Carneiro-Lobo TC, Schaffner F, Disse J, Ostergaard H, Francischetti IM, Monteiro RQ, et al. The tick-derived inhibitor Ixolaris prevents tissue factor signaling on tumor cells. J Thromb Haemost. 2012;10:1849–58. doi: 10.1111/j.1538-7836.2012.04864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Francischetti IM, Mather TN, Ribeiro JM. Penthalaris, a novel recombinant five-Kunitz tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick vector of Lyme disease, Ixodes scapularis. Thromb Haemost. 2004;91:886–98. doi: 10.1160/TH03-11-0715. [DOI] [PubMed] [Google Scholar]
- 88.Corral-Rodriguez MA, Macedo-Ribeiro S, Barbosa Pereira PJ, Fuentes-Prior P. Tick-derived Kunitz-type inhibitors as antihemostatic factors. Insect Biochem Mol Biol. 2009;39:579–95. doi: 10.1016/j.ibmb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 89.Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci. 2009;14:2051–88. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lerner EA, Iuga AO, Reddy VB. Maxadilan, a PAC1 receptor agonist from sand flies. Peptides. 2007;28:1651–4. doi: 10.1016/j.peptides.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson JM, Kellogg DL., Jr Thermoregulatory and thermal control in the human cutaneous circulation. Front Biosci (Schol Ed) 2010;2:825–53. doi: 10.2741/s105. [DOI] [PubMed] [Google Scholar]
- 92.Kellogg DL, Jr, Zhao JL, Wu Y, Johnson JM. VIP/PACAP receptor mediation of cutaneous active vasodilation during heat stress in humans. J Appl Physiol. 2010;109:95–100. doi: 10.1152/japplphysiol.01187.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qureshi AAAA, Ohnuma MTM, Granstein RD, Lerner EA. Immunomodulatory properties of maxadilan, the vasodilator peptide from sand fly salivary gland extracts. Am J Trop Med Hyg. 1996;54:665–71. doi: 10.4269/ajtmh.1996.54.665. [DOI] [PubMed] [Google Scholar]
- 94.Delgado M, Abad C, Martinez C, Juarranz MG, Leceta J, Ganea D, et al. PACAP in immunity and inflammation. Annals of the New York Academy of Sciences. 2003;992:141–57. doi: 10.1111/j.1749-6632.2003.tb03145.x. [DOI] [PubMed] [Google Scholar]
- 95.Conconi MT, Spinazzi R, Nussdorfer GG. Endogenous ligands of PACAP/VIP receptors in the autocrine-paracrine regulation of the adrenal gland. Int Rev Cytol. 2006;249:1–51. doi: 10.1016/S0074-7696(06)49001-X. [DOI] [PubMed] [Google Scholar]
- 96.Harmar AJ, Fahrenkrug J, Gozes I, Laburthe M, May V, Pisegna JR, et al. Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide: IUPHAR review 1. Br J Pharmacol. 2012;166:4–17. doi: 10.1111/j.1476-5381.2012.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ahluwalia A, Hobbs AJ. Endothelium-derived C-type natriuretic peptide: more than just a hyperpolarizing factor. Trends Pharmacol Sci. 2005;26:162–7. doi: 10.1016/j.tips.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 98.Baxter GF. The natriuretic peptides. Basic Res Cardiol. 2004;99:71–5. doi: 10.1007/s00395-004-0457-8. [DOI] [PubMed] [Google Scholar]
- 99.Wisniewski HG, Vilcek J. TSG-6: an IL-1/TNF-inducible protein with anti-inflammatory activity. Cytokine Growth Factor Rev. 1997;8:143–56. doi: 10.1016/s1359-6101(97)00008-7. [DOI] [PubMed] [Google Scholar]
- 100.Milner CM, Day AJ. TSG-6: a multifunctional protein associated with inflammation. J Cell Sci. 2003;116:1863–73. doi: 10.1242/jcs.00407. [DOI] [PubMed] [Google Scholar]
- 101.Mahoney DJ, Mulloy B, Forster MJ, Blundell CD, Fries E, Milner CM, et al. Characterization of the interaction between tumor necrosis factor-stimulated gene-6 and heparin: implications for the inhibition of plasmin in extracellular matrix microenvironments. J Biol Chem. 2005;280:27044–55. doi: 10.1074/jbc.M502068200. [DOI] [PubMed] [Google Scholar]
- 102.Tuo J, Cao X, Shen D, Wang Y, Zhang J, Oh JY, et al. Anti-inflammatory recombinant TSG-6 stabilizes the progression of focal retinal degeneration in a murine model. Journal of neuroinflammation. 2012;9:59. doi: 10.1186/1742-2094-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kunkel EJ, Kim CH, Lazarus NH, Vierra MA, Soler D, Bowman EP, et al. CCR10 expression is a common feature of circulating and mucosal epithelial tissue IgA Ab-secreting cells. J Clin Invest. 2003;111:1001–10. doi: 10.1172/JCI17244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kavanagh K, Dowd S. Histatins: antimicrobial peptides with therapeutic potential. J Pharm Pharmacol. 2004;56:285–9. doi: 10.1211/0022357022971. [DOI] [PubMed] [Google Scholar]
- 105.Hieshima K, Ohtani H, Shibano M, Izawa D, Nakayama T, Kawasaki Y, et al. CCL28 has dual roles in mucosal immunity as a chemokine with broad-spectrum antimicrobial activity. J Immunol. 2003;170:1452–61. doi: 10.4049/jimmunol.170.3.1452. [DOI] [PubMed] [Google Scholar]
- 106.Watson SP, Auger JM, McCarty OJ, Pearce AC. GPVI and integrin alphaIIb beta3 signaling in platelets. J Thromb Haemost. 2005;3:1752–62. doi: 10.1111/j.1538-7836.2005.01429.x. [DOI] [PubMed] [Google Scholar]
- 107.Atkinson B, Dwyer K, Enjyoji K, Robson SC. Ecto-nucleotidases of the CD39/NTPDase family modulate platelet activation and thrombus formation: Potential as therapeutic targets. Blood cells, molecules & diseases. 2006;36:217–22. doi: 10.1016/j.bcmd.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 108.Muller F, Mutch NJ, Schenk WA, Smith SA, Esterl L, Spronk HM, et al. Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell. 2009;139:1143–56. doi: 10.1016/j.cell.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ribeiro JM, Mather TN. Ixodes scapularis: salivary kininase activity is a metallo dipeptidyl carboxypeptidase. Exp Parasitol. 1998;89:213–21. doi: 10.1006/expr.1998.4296. [DOI] [PubMed] [Google Scholar]
- 110.Oshima G, Kato J, Erdos EG. Subunits of human plasma carboxypeptidase N (Kininase I; anaphylatoxin inactivator) BBa. 1974;365:344–8. [Google Scholar]
- 111.Ribeiro JMC, Spielman A. Ixodes dammini: Salivary anaphylatoxin-inactivating activity. Exp Parasitol. 1986;62:292–7. doi: 10.1016/0014-4894(86)90034-2. [DOI] [PubMed] [Google Scholar]
- 112.Utermohlen O, Herz J, Schramm M, Kronke M. Fusogenicity of membranes: the impact of acid sphingomyelinase on innate immune responses. Immunobiology. 2008;213:307–14. doi: 10.1016/j.imbio.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 113.Gibbs GM, Roelants K, O’Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins--roles in reproduction, cancer, and immune defense. Endocr Rev. 2008;29:865–97. doi: 10.1210/er.2008-0032. [DOI] [PubMed] [Google Scholar]
- 114.Wang YL, Kuo JH, Lee SC, Liu JS, Hsieh YC, Shih YT, et al. Cobra CRISP functions as an inflammatory modulator via a novel Zn2+- and heparan sulfate-dependent transcriptional regulation of endothelial cell adhesion molecules. The Journal of biological chemistry. 2010;285:37872–83. doi: 10.1074/jbc.M110.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mans BJ, Francischetti IMB. Sialomic perspectives on the evolution of blood-feeding behavior in arthropods: future therapeutics by natural design. In: Kini RM, Clemetson KJ, Markland FS, McLane MA, Morita T, editors. Toxins and Hemostasis From bench to bedside. Springer; New York: 2010. pp. 21–44. [Google Scholar]
- 116.Ma D, Gao L, An S, Song Y, Wu J, Xu X, et al. A horsefly saliva antigen 5-like protein containing RTS motif is an angiogenesis inhibitor. Toxicon. 2010;55:45–51. doi: 10.1016/j.toxicon.2009.06.038. [DOI] [PubMed] [Google Scholar]
- 117.da Silva BR, de Freitas VA, Nascimento-Neto LG, Carneiro VA, Arruda FV, de Aguiar AS, et al. Antimicrobial peptide control of pathogenic microorganisms of the oral cavity: a review of the literature. Peptides. 2012;36:315–21. doi: 10.1016/j.peptides.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 118.Schwarz A, Valdes JJ, Kotsyfakis M. The role of cystatins in tick physiology and blood feeding. Ticks and tick-borne diseases. 2012;3:117–27. doi: 10.1016/j.ttbdis.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Olson ST, Gettins PG. Regulation of proteases by protein inhibitors of the serpin superfamily. Prog Mol Biol Transl Sci. 2011;99:185–240. doi: 10.1016/B978-0-12-385504-6.00005-1. [DOI] [PubMed] [Google Scholar]
- 120.Galliciotti G, Sonderegger P. Neuroserpin. Front Biosci. 2006;11:33–45. doi: 10.2741/1778. [DOI] [PubMed] [Google Scholar]
- 121.Davis AE, 3rd, Mejia P, Lu F. Biological activities of C1 inhibitor. Mol Immunol. 2008;45:4057–63. doi: 10.1016/j.molimm.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JD, King GF, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- 123.Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 124.Rimphanitchayakit V, Tassanakajon A. Structure and function of invertebrate Kazal-type serine proteinase inhibitors. Dev Comp Immunol. 2010;34:377–86. doi: 10.1016/j.dci.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 125.Marino JP., Jr Soluble epoxide hydrolase, a target with multiple opportunities for cardiovascular drug discovery. Curr Top Med Chem. 2009;9:452–63. doi: 10.2174/156802609788340805. [DOI] [PubMed] [Google Scholar]
- 126.Takac P, Tsujimoto H, Champagne DE. Hypotensive proteins from hematophagous animals. Toxins and Hemostasis From bench to bedside. 2010 [Google Scholar]
- 127.Maritz-Olivier C, Stutzer C, Jongejan F, Neitz AW, Gaspar AR. Tick anti-hemostatics: targets for future vaccines and therapeutics. Trends Parasitol. 2007;23:397–407. doi: 10.1016/j.pt.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 128.Mans BJ, Neitz AW. Adaptation of ticks to a blood-feeding environment: evolution from a functional perspective. Insect Biochem Mol Biol. 2004;34:1–17. doi: 10.1016/j.ibmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 129.Fry BG. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–20. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, Juarez P, et al. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–74. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 131.Ligabue-Braun R, Verli H, Carlini CR. Venomous mammals: a review. Toxicon. 2012;59:680–95. doi: 10.1016/j.toxicon.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 132.Kini RM. Toxins in thrombosis and haemostasis: potential beyond imagination. J Thromb Haemost. 2011;9 (Suppl 1):195–208. doi: 10.1111/j.1538-7836.2011.04279.x. [DOI] [PubMed] [Google Scholar]
- 133.Tellgren-Roth A, Dittmar K, Massey SE, Kemi C, Tellgren-Roth C, Savolainen P, et al. Keeping the blood flowing-plasminogen activator genes and feeding behavior in vampire bats. Die Naturwissenschaften. 2009;96:39–47. doi: 10.1007/s00114-008-0446-0. [DOI] [PubMed] [Google Scholar]
- 134.Louw E, van der Merwe NA, Neitz AW, Maritz-Olivier C. Evolution of the tissue factor pathway inhibitor-like Kunitz domain-containing protein family in Rhipicephalus microplus. Int J Parasitol. 2012 doi: 10.1016/j.ijpara.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 135.Dai SX, Zhang AD, Huang JF. Evolution, expansion and expression of the Kunitz/BPTI gene family associated with long-term blood feeding in Ixodes Scapularis. BMC Evol Biol. 2012;12:4. doi: 10.1186/1471-2148-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nuttall PA. Molecular characterization of tick-virus interactions. Front Biosci. 2009;14:2466–83. doi: 10.2741/3390. [DOI] [PubMed] [Google Scholar]
- 137.Hubalek Z, Rudolf I. Tick-borne viruses in Europe. Parasitol Res. 2012;111:9–36. doi: 10.1007/s00436-012-2910-1. [DOI] [PubMed] [Google Scholar]
- 138.Delgado M, Gonzalez-Rey E, Ganea D. VIP/PACAP preferentially attract Th2 effectors through differential regulation of chemokine production by dendritic cells. FASEB J. 2004;18:1453–5. doi: 10.1096/fj.04-1548fje. [DOI] [PubMed] [Google Scholar]
- 139.Delgado M, Pozo D, Ganea D. The significance of vasoactive intestinal peptide in immunomodulation. Pharmacol Rev. 2004;56:249–90. doi: 10.1124/pr.56.2.7. [DOI] [PubMed] [Google Scholar]
- 140.Phillipson M, Kubes P. The neutrophil in vascular inflammation. Nat Med. 2011;17:1381–90. doi: 10.1038/nm.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Opal SM, Esmon CT. Bench-to-bedside review: functional relationships between coagulation and the innate immune response and their respective roles in the pathogenesis of sepsis. Crit Care. 2003;7:23–38. doi: 10.1186/cc1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boerboom D, Russell DL, Richards JS, Sirois J. Regulation of transcripts encoding ADAMTS-1 (a disintegrin and metalloproteinase with thrombospondin-like motifs-1) and progesterone receptor by human chorionic gonadotropin in equine preovulatory follicles. Journal of molecular endocrinology. 2003;31:473–85. doi: 10.1677/jme.0.0310473. [DOI] [PubMed] [Google Scholar]
- 143.Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R, et al. Purification and cloning of aggrecanase-1: a member of the ADAMTS family of proteins. Science. 1999;284:1664–6. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- 144.Furie B, Furie BC. Mechanisms of thrombus formation. N Engl J Med. 2008;359:938–49. doi: 10.1056/NEJMra0801082. [DOI] [PubMed] [Google Scholar]
- 145.Fuchs TA, Brill A, Wagner DD. Neutrophil extracellular trap (NET) impact on deep vein thrombosis. Arterioscler Thromb Vasc Biol. 2012;32:1777–83. doi: 10.1161/ATVBAHA.111.242859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.