Abstract

1-Deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) in the nonmevalonate isoprene biosynthesis pathway is a target for developing antimalarial drugs. Fosmidomycin, a potent DXR inhibitor, showed safety as well as efficacy against Plasmodium falciparum malaria in clinical trials. On the basis of our previous quantitative structure–activity relationship (QSAR) and crystallographic studies, several novel pyridine-containing fosmidomycin derivatives were designed, synthesized, and found to be highly potent inhibitors of P. falciparum DXR (PfDXR) having Ki values of 1.9–13 nM, with the best one being ∼11× more active than fosmidomycin. These compounds also potently block the proliferation of multidrug resistant P. falciparum with EC50 values as low as 170 nM. A 2.3 Å crystal structure of PfDXR in complex with one of the inhibitors is reported, showing that the flexible loop of the protein undergoes conformational changes upon ligand binding and a hydrogen bond and favorable hydrophobic interactions between the pyridine group and the PfDXR account for the enhanced activity.
Keywords: 1-deoxy-d-xylulose-5-phosphate reductoisomerase, antimalarial activity, X-ray protein crystallography, inhibitor design
Eukaryotic parasite Plasmodium spp. are the causative agents of malaria, among which Plasmodium falciparum produces the most severe form of human malaria and is responsible for the vast majority of deaths of malaria patients (∼1 million/year). In addition, this dreadful number could be rising because of the increased drug resistance of P. falciparum to inexpensive drugs such as chloroquine. Since 2001, the World Health Organization (WHO) has strongly recommended the use of artemisinin-based combination therapies to treat malaria.1 However, Plasmodium parasites are known to be able to develop drug resistance. After 10 years, P. falciparum strains that are resistant to these new drug combinations have been observed.1 There is therefore a pressing need to develop new antimalarial drugs.
1-Deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) is the second enzyme in the nonmevalonate isoprene biosynthesis pathway, catalyzing the reductive isomerization of 1-deoxy-d-xylulose-5-phosphate (DXP) to 2-methyl-d-erythritol-4-phosphate (MEP) using Mg2+ (or Mn2+) and NADPH as the cofactors, as schematically illustrated in Figure 1.2 This is used by most bacteria, as well as apicomplexan parasites such as Plasmodium spp., to make isopentenyl diphosphate and dimethylallyl diphosphate. These two diphosphate compounds are the only precursors for the biosynthesis of all isoprenoids and terpenoids, including important substances such as C15–C55 isoprenyl (e.g., farnesyl and undecaprenyl) diphosphates for anchoring proteins to the cell membrane. DXR is essential for the growth of all of these species including P. falciparum. In addition, humans and animals use the mevalonate pathway to synthesize IPP and DMAPP, making DXR an attractive target for discovering novel antimalarial drugs.3,4
Figure 1.

Reaction catalyzed by DXR.
Fosmidomycin (1, Chart 1) and its close analogue FR900098 (2) were found to be potent DXR inhibitors5 and possess antibacterial and -malarial activities.6 Particularly, several recent clinical trials showed that in combination with clindamycin, 1 is safe for human use and effective against P. falciparum malaria.7,8 However, because of a short half-life in plasma (∼1 h) as well as poor oral availability of 1, considerable interest has therefore been generated to develop potent, lipophilic DXR inhibitors with the hope that these compounds could have broad anti-infective activities as well as improved pharmacokinetic properties.9−20
Chart 1. Structures of DXR Inhibitors.
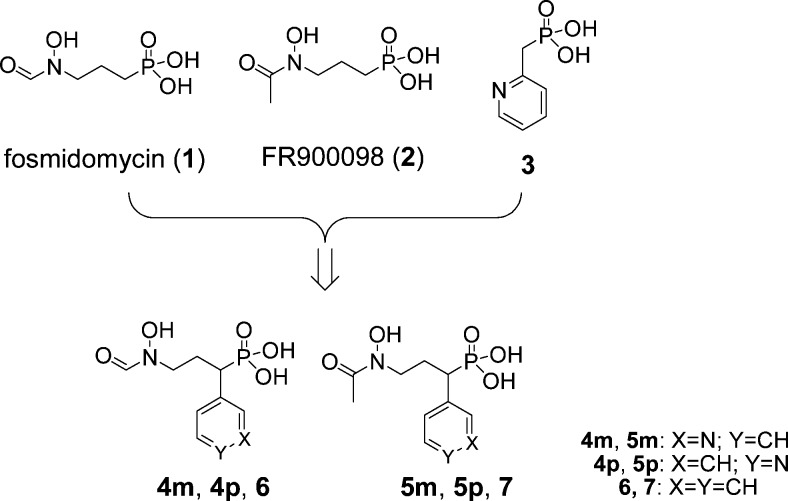
In our early work,19,20 a series of lipophilic, pyridine- or quinoline-containing phosphonate compounds, such as compound 3 in Chart 1, were found to be a new class of DXR inhibitors. Our structure–activity relationship (SAR) and quantitative SAR (QSAR) studies all showed the importance of the presence of an electron-deficient aromatic ring, such as a pyridine, at the α-position of the phosphonate group, as shown in Figure S1a in the Supporting Information. In addition, superposition of the crystal structures of Escherichia coli DXR (EcDXR) in complex with 1 and 3 (Figure S1b in the Supporting Information) suggests fosmidomycin derivatives with an α-pyridine substituent, such as compounds 4m, 5m, 4p, and 5p (m stands for meta-substituted pyridinyl compound and p stands for para-substituted pyridinyl) shown in Chart 1, could be particularly active, since these compounds likely have all favorable interactions with the protein as found for both 1 and 3. Compounds 6 and 7 (Chart 1) with a more electron-rich phenyl ring were included in the study as a comparison.
The general method for synthesizing compounds 4–7 is shown in Scheme 1. Pyridine-3-carboxaldehyde was reduced and converted to 3-picolinyl chloride, which was reacted with sodium salt of diethylphosphite to give 8. It was alkylated with allyl bromide, followed by ozonization to give aldehyde 9. Upon reductive amination with O-benzyl-hydroxylamine and NaBH3CN, the resulting hydroxylamine 10 was formylated or acetylated, followed by hydrolysis using bromotrimethylsilane (TMSBr) and hydrogenation, to give 4m or 5m. Compounds 4p and 5p were similarly prepared, starting from pyridine-4-carboxaldehyde. However, analogous compounds with an ortho-substituted pyridinyl cannot be synthesized from pyridine-2-carboxaldehyde, due to instability of the corresponding aldehyde 9 (or its O-benzyl-hydroxylamine oxime). Phenyl-substituted 6 and 7 were prepared similarly, starting from benzaldehyde.
Scheme 1. General Synthesis for Compounds 4–7.
Reagents and conditions: (i) NaBH4, MeOH. (ii) SOCl2. (iii) HP(O)(OEt)2, NaH, THF, 43% for three steps. (iv) n-BuLi, then allyl bromide, −78 – 0 °C. (v) O3, −78 °C, then Me2S. (vi) NH2OBn. (vii) NaBH3CN, pH 3, MeOH, 40% from 8. (viii) For R = H, HCOOH/Ac2O; for R = Me, Ac2O. (ix) TMSBr. (x) H2, Pd/C, 76% from 10.
The enzyme inhibitory activities of the newly synthesized compounds 4–7 were first tested against recombinant EcDXR. As shown in Table 1, compounds 4p and 5m were found to exhibit potent activity with Ki values of 35 and 42 nM, respectively, being as active as 1 (Ki = 34 nM). Compounds 4m and 5p (Ki = 87 and 82 nM) have only slightly reduced inhibition against EcDXR. However, compounds 6 and 7 containing a phenyl substituent were found to be markedly less active with Ki values of 1.5 and 0.93 μM, respectively, showing an average of ∼20-fold activity reduction as compared to pyridine-containing compounds 4 and 5.
Table 1. Activity of DXR Inhibitors 1–7.
| enzyme Ki (μM) |
IC50 against P. falciparum proliferation (μM) |
|||
|---|---|---|---|---|
| EcDXR | PfDXR | 3D7 strain | Dd2 strain | |
| 1 | 0.034 | 0.021 | 1.17 | 0.44 |
| 4m | 0.087 | 0.013 | 0.34 | 0.18 |
| 4p | 0.035 | 0.0089 | 0.18 | 0.17 |
| 5m | 0.042 | 0.0019 | 0.44 | 0.31 |
| 5p | 0.082 | 0.013 | 0.63 | 0.46 |
| 6 | 1.5 | 0.48 | NTa | NTa |
| 7 | 0.93 | 0.085 | 3.5 | 1.7 |
| CQ | NT | NT | 0.021 | 0.12 |
NT, not tested.
E. coli DXR has been used in the vast majority of previous studies, although EcDXR inhibition may not be relevant and predictive in the context of developing antimalarial drugs. To this end, we next tested the inhibitory activities of these compounds against recombinant Plasmodium falciparum DXR (PfDXR) enzyme. In our previous work,211 was determined also to be a potent inhibitor of PfDXR with a Ki value of 21 nM. We were pleased to find that pyridine-containing compounds 4m,p and 5m,p possess considerably higher inhibitory activity against PfDXR, as can be seen in Table 1 and representatively shown in Figure S2 in the Supporting Information. The Ki values of these compounds range from 1.9 to 13 nM, with the best compound 5m (Ki = 1.9 nM) having an acetyl and a pyridin-3-yl group being ∼11× more active than 1. The phenyl-containing compounds 6 and 7 again exhibited significantly lowered inhibitory potencies with Ki values of 480 and 85 nM, showing that the pyridine groups in 4m,p and 5m,p are able to provide an average of ∼30-fold enhanced activity against PfDXR.
With these highly potent inhibitors of P. falciparum DXR in hand, we next investigated the antimalarial activities of these compounds against the proliferation of erythrocyte-stage P. falciparum. The 3D7 strain of P. falciparum is sensitive to current antimalarial drugs, while the Dd2 strain is multidrug resistant, including chloroquine (CQ), pyrimethamine, and mefloquine. Chloroquine and 1 were included in our studies as positive controls. As also shown in Table 1, 1 was found to have good activity against the growth of P. falciparum strains 3D7 and Dd2 with EC50 values of 1.2 and 0.44 μM, respectively, while chloroquine exhibits more activity against the 3D7 strain (EC50 = 21 nM) than it does against the drug-resistant Dd2 strain (EC50 = 120 nM). The antimalarial activities of 1 and chloroquine are similar to those reported previously.6 As compared to 1, the novel pyridine-containing DXR inhibitors were found to have enhanced antimalarial activities. Formyl compounds 4m and 4p exhibit potent antimalarial activity (EC50 = 340 and 180 nM, respectively) against the 3D7 strain. Moreover, regardless of the drug resistance of the Dd2 strain, these two compounds possess even more pronounced activity against this strain of the parasite with EC50 values of 180 and 170 nM. The acetyl-containing compounds 5m and 5p have, however, relatively lowered antimalarial activity, as compared to 4m and 4p with a formyl group. The EC50 values of 5m are 440 and 630 nM against 3D7 and Dd2, respectively, and those of 5p are 310 and 460 nM. Compound 7 with a phenyl group was found have a relatively weak antimalarial activity against P. falciparum strains 3D7 and Dd2 (EC50 = 3.5 and 1.7 μM, respectively).
It is noteworthy that all of these DXR inhibitors including 1 exhibit more activity against chloroquine-resistant P. falciparum Dd2 strain, demonstrating that these compounds working in a different mechanism have the potential to become a new class of antimalarials without cross-drug resistance. It is also interesting that compounds 4m and 4p possess enhanced activities against the proliferation of P. falciparum, although they are relatively less active against the recombinant enzyme than the acetyl-containing inhibitor 5m. This could be attributed to lowered cell membrane permeability of 5m, since these inhibitors need to pass both membranes of erythrocyte and the parasite to reach their cellular target PfDXR.
Potential toxicities of the newly synthesized compounds were assessed using human noncancerous fibroblast WI-38 cells. All of the pyridine-containing compounds 4 and 5 have essentially no cytotoxicity with EC50 values of >300 μM against the growth of WI-38 cells, showing a very high selectivity index of >1700-fold. These compounds therefore represent promising leads for further antimalarial drug development.
The next question of interest is how these pyridine-containing inhibitors bind to P. falciparum DXR. Prior structural studies were mostly focused on EcDXR.17,19,22−25 Currently, only two crystal structures of PfDXR in complex with 1 and 2 are available.26 However, it is known that the conformation of DXR, especially for the flexible loop at the active site, changes considerably when a ligand having a different property (e.g., hydrophobicity) from 1 and 2 binds.19 Therefore, crystallographic studies for the binding of these pyridine-containing inhibitors to PfDXR are needed.
We determined the crystal structure of a quaternary complex of PfDXR with compound 5p, NADPH, and a Mn2+, at 2.3 Å. Parameters for data processing and refinement are listed in Table S1 in the Supporting Information. As shown in Figure 2A, the protein crystallizes as a homodimer, with each monomer being almost identical to the other. The binding conformation of NADPH is very similar to that in previous reported structures. Each of the two Mn2+ ions is anchored to PfDXR via coordination to three carboxylate groups of the residues Asp231, Glu233, and Glu315 (Figure 2B). The electron density and omit maps of inhibitor 5p are shown in Figure S3 in the Supporting Information. In addition, the racemic form of 5p was used in the study, and at the 2.3 Å resolution, it is not unambiguous to assign the absolute configuration of the α-carbon atom of compound 5p (Figure S3 in the Supporting Information). Fifty percent occupancy for each enantiomer was therefore assigned in the final structure.
Figure 2.

(A) Overall structure of PfDXR in complex with Mn2+ (brown sphere), 5p, and NADPH. (B) Close-up view of the active site of PfDXR:5p. (C) Active sites of the aligned structures of PfDXR:5p (in green) and PfDXR:1 (in orange) complexes. For clarity, only the flexible loops containing residues 290–299 of the two structures are shown as curved lines. (D) Aligned structures of PfDXR:5p (in green) and EcDXR:3 (in purple) complexes. The colored curved lines represent the flexible loops of the proteins.
Figures 2B and S4 in the Supporting Information show the close-up view of the PfDXR active site and detailed protein–ligand interactions. As always can be seen in other DXR–hydroxamate inhibitor complexes,22−25 the two O atoms of the hydroxamate group of 5p chelate the central Mn2+, with their distances being 2.1 and 2.4 Å. The acetyl O atom also forms a hydrogen bond with the side chain −OH of Ser232. The negatively charged phosphonate group of compound 5p is recognized by a polar environment, forming five hydrogen bonds as well as electrostatic interactions with Lys312, Asn311, Ser306, Ser270, and Ser269. The pyridine group of 5p was found to be favorably located in a mainly hydrophobic pocket defined by residues His293, Trp296, Met298, Cys338, and Pro358, which is not present in the reported crystal structure of the PfDXR:1 complex.26 In addition, a hydrogen bond between the N of the pyridine ring of 5p and the −SH of Cys338 was observed, with the distance of N···H–S being 3.4 Å. Because the structurally similar compound 7 cannot form a hydrogen bond with Cys338, the interaction could therefore contribute to the observed ∼6× more inhibitory potency of 5p. Moreover, this hydrogen bond, together with the favorable hydrophobic interactions described above, could be responsible for the enhanced activity of 5p against PfDXR as compared to 1. It is also remarkable that the methyl group of 5p has favorable hydrophobic interactions with Met298 and Trp296. This could explain that the acetyl-containing DXR inhibitors (e.g., 5m and 7) in general possess increased enzyme activity than the corresponding compounds having a formyl group.
Alignment of the structures of PfDXR:5p and PfDXR:1 was performed to find the conformational differences of the protein. As shown in Figures S5a in the Supporting Information, these two structures can be aligned very well with a root-mean-square (rms) deviation for the protein backbone being 0.5 Å. The only notable difference is the conformation of the flexible loop consisting of residues 290–299, which is part of the active site of PfDXR. As shown in Figure 2C, the loop is located much closer to the ligand-binding site in the PfDXR:1 structure, with the big, hydrophobic indole ring of Trp296 being right beneath the carbon skeleton of 1. This conformation allows the active site to be largely separated from the solvent, which is considered to be important for the enzyme activity or for the tight binding of polar inhibitor 1. For example, a previous mutagenesis study showed that replacing the corresponding Trp204 in Synechocystis DXR with an Ala, Val, or Leu residue renders the enzyme completely inactive.27 The only weakly active mutation is Trp204Phe having a smaller, aromatic phenyl side chain. In the PfDXR:5p structure, the flexible loop was observed to move up to 4.5 Å away from the center of the active site to hold the pyridine of 5p and allow the Trp296 indole and several other residues to have favorable hydrophobic interactions with the inhibitor.
We next compared the binding of inhibitor 5p to PfDXR with that of another pyridine-containing inhibitor 3 to EcDXR.19 Protein alignment indicated that these two structures have a rms deviation for the protein backbone of 1.9 Å (Figure S5b in the Supporting Information). The close-up view of the active site is shown in Figure 2D, in which the two phosphonate groups of 5p and 3 are very close to each other, with the distance between the two P atoms being 0.5 Å. The two pyridine rings were found also to be located in the same hydrophobic pocket. Relatively large differences can be observed with respect to the conformation of the flexible loops of these two enzymes. Not only the backbones of the loops exhibit an average distance for the Cα atoms of ∼4 Å between the two structures, but the orientations of the side chains of several residues are different as well. These conformational alterations could be driven mainly by two factors. First, compound 3 does not have a hydroxamate moiety. Second, the indole ring of Trp211 of EcDXR has a π–π interaction with the pyridine group of 3. The steric as well as π–π stacking interactions allow the EcDXR flexible loop to shift ∼4 Å on average toward the pyridine binding site. For the PfDXR:5p complex, the presence of the hydroxamate moiety seems not to favor the formation of a similar π–π interaction between Trp296 and the pyridine, due likely to the steric congestion between the hydroxamate moiety and the Trp296 indole group. Rather, the hydrophobic side chains of the flexible loop, such as Met298, Trp296, and His293, together with those of residues Pro358 and Cys338 from other chains, are orientated to create a generally hydrophobic environment to have favorable interactions with the pyridine ring as well as the methyl group of compound 5p.
In summary, four pyridine-containing compounds were designed, synthesized, and found to exhibit up to 11-fold enhanced inhibitory activity (Ki, 1.9–13 nM) against P. falciparum DXR as compared to 1. These noncytotoxic compounds also showed considerably improved antimalarial activity against the proliferation of multidrug-resistant P. falciparum with EC50 values as low as 170 nM, showing the promise to overcome the widespread drug resistance of the malaria parasites. The crystal structure of the PfDXR:5p complex reveals the exact binding pose of the highly potent inhibitor. The backbone of 5p binds to PfDXR in a similar fashion as 1. Conformational changes were observed for the flexible loop of PfDXR to form a mainly hydrophobic pocket that holds and has favorable interactions with the pyridine group of the inhibitor. In addition, the hydrogen bond between the pyridine N atom and the Cys338 should also contribute considerably to the increased potency.
Glossary
Abbreviations
- DXR
1-deoxy-d-xylulose-5-phosphate reductoisomerase
- EcDXR
Escherichia coli DXR
- PfDXR
Plasmodium falciparum DXR
Supporting Information Available
Supplementary Figures S1–S5, Supplementary Table, and Experimental Section. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The coordinates for the PfDXR:5p complex have been deposited in Protein Data Bank as entry 4GAE.
Author Present Address
‡ Department of Parasitology, Wuhan University School of Basic Medical Science, Wuhan 430071, China.
Author Contributions
† These authors contributed equally.
This work was supported by a grant (R21AI088123) from the National Institute of Health (NIH) to Y.S. We thank the staff of the X-ray Crystallography Facility at Baylor College of Medicine for assistance in data collection.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- The World Health Organization. Global Report on Antimalarial Efficacy and Drug Resistance: 2000–2010; WHO Press: Geneva, 2010. [Google Scholar]
- See a recent review and the references cited therein:Hunter W. N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [DOI] [PubMed] [Google Scholar]
- Obiol-Pardo C.; Rubio-Martinez J.; Imperial S. The methylerythritol phosphate (MEP) pathway for isoprenoid biosynthesis as a target for the development of new drugs against tuberculosis. Curr. Med. Chem. 2011, 18, 1325–1338. [DOI] [PubMed] [Google Scholar]
- Singh N.; Cheve G.; Avery M. A.; McCurdy C. R. Targeting the methyl erythritol phosphate (MEP) pathway for novel antimalarial, antibacterial and herbicidal drug discovery: Inhibition of 1-deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) enzyme. Curr. Pharm. Des. 2007, 13, 1161–1177. [DOI] [PubMed] [Google Scholar]
- Kuzuyama T.; Shimizu T.; Takahashi S.; Seto H. Fosmidomycin, a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 1998, 39, 7913–7916. [Google Scholar]
- Jomaa H.; Wiesner J.; Sanderbrand S.; Altincicek B.; Weidemeyer C.; Hintz M.; Turbachova I.; Eberl M.; Zeidler J.; Lichtenthaler H. K.; Soldati D.; Beck E. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 1999, 285, 1573–1576. [DOI] [PubMed] [Google Scholar]
- Missinou M. A.; Borrmann S.; Schindler A.; Issifou S.; Adegnika A. A.; Matsiegui P. B.; Binder R.; Lell B.; Wiesner J.; Baranek T.; Jomaa H.; Kremsner P. G. Fosmidomycin for malaria. Lancet 2002, 360, 1941–1942. [DOI] [PubMed] [Google Scholar]
- Oyakhirome S.; Issifou S.; Pongratz P.; Barondi F.; Ramharter M.; Kun J. F.; Missinou M. A.; Lell B.; Kremsner P. G. Randomized controlled trial of fosmidomycin-clindamycin versus sulfadoxine-pyrimethamine in the treatment of Plasmodium falciparum malaria. Antimicrob. Agents Chemother. 2007, 51, 1869–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz L.; Tritsch D.; Grosdemange-Billiard C.; Hemmerlin A.; Willem A.; Bach T. J.; Rohmer M. Isoprenoid biosynthesis as a target for antibacterial and antiparasitic drugs: phosphonohydroxamic acids as inhibitors of deoxyxylulose phosphate reducto-isomerase. Biochem. J. 2005, 386, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckle L.; de Andres-Gomez A.; Dick B.; Cox R. J.; Godfrey C. R. A fragment-based approach to understanding inhibition of 1-deoxy-d-xylulose-5-phosphate reductoisomerase. ChemBioChem 2005, 6, 1866–1874. [DOI] [PubMed] [Google Scholar]
- Munos J. W.; Pu X.; Liu H. W. Synthesis and analysis of a fluorinated product analogue as an inhibitor for 1-deoxy-d-xylulose 5-phosphate reductoisomerase. Bioorg. Med. Chem. Lett. 2008, 18, 3090–3094. [DOI] [PubMed] [Google Scholar]
- Silber K.; Heidler P.; Kurz T.; Klebe G. AFMoC enhances predictivity of 3D QSAR: A case study with DOXP-reductoisomerase. J. Med. Chem. 2005, 48, 3547–3563. [DOI] [PubMed] [Google Scholar]
- Kurz T.; Schlüter K.; Kaula U.; Bergmann B.; Walter R. D.; Geffken D. Synthesis and antimalarial activity of chain substituted pivaloyloxymethyl ester analogues of Fosmidomycin and FR900098. Bioorg. Med. Chem. 2006, 14, 5121–5135. [DOI] [PubMed] [Google Scholar]
- Haemers T.; Wiesner J.; Van Poecke S.; Goeman J.; Henschker D.; Beck E.; Jomaa H.; Van Calenbergh S. Synthesis of α-substituted fosmidomycin analogues as highly potent Plasmodium falciparum growth inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 1888–1891. [DOI] [PubMed] [Google Scholar]
- Woo Y. H.; Fernandes R. P.; Proteau P. J. Evaluation of fosmidomycin analogs as inhibitors of the Synechocystis sp. PCC6803 1-deoxy-d-xylulose 5-phosphate reductoisomerase. Bioorg. Med. Chem. 2006, 14, 2375–2385. [DOI] [PubMed] [Google Scholar]
- Kurz T.; Schlüter K.; Pein M.; Behrendt C.; Bergmann B.; Walter R. D. Conformationally restrained aromatic analogues of fosmidomycin and FR900098. Arch. Pharm. Chem. Life Sci. 2007, 340, 339–344. [DOI] [PubMed] [Google Scholar]
- Yajima S.; Hara K.; Sanders J. M.; Yin F.; Ohsawa K.; Wiesner J.; Jomaa H.; Oldfield E. Crystallographic structures of two bisphosphonate:1-deoxyxylulose-5-phosphate reductoisomerase complexes. J. Am. Chem. Soc. 2004, 126, 10824–10825. [DOI] [PubMed] [Google Scholar]
- Deng L.; Sundriyal S.; Rubio V.; Shi Z.; Song Y. Coordination chemistry based approach to lipophilic inhibitors of 1-deoxy-d-xylulose-5-phosphate reductoisomerase. J. Med. Chem. 2009, 52, 6539–6542. [DOI] [PubMed] [Google Scholar]
- Deng L.; Endo K.; Kato M.; Cheng G.; Yajima S.; Song Y. Structures of 1-deoxy-d-xylulose-5-phosphate reductoisomerase/lipophilic phosphonate complexes. ACS Med. Chem. Lett. 2011, 2, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L.; Diao J.; Chen P.; Pujari V.; Yao Y.; Cheng G.; Crick D. C.; Prasad B. V. V; Song Y. Inhibition of 1-deoxy-d-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies. J. Med. Chem. 2011, 54, 4721–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G.; Deng L.; Fryszczyn B. G.; Brown N. G.; Liu Z.; Jiang H.; Palzkill T.; Song Y. Thermodynamic investigation of inhibitor binding to 1-deoxy-d-xylulose-5-phosphate reductoisomerase. ACS Med. Chem. Lett. 2012, 3, 496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima S.; Nonaka T.; Kuzuyama T.; Seto H.; Ohsawa K. Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase complexed with cofactors: Implications of a flexible loop movement upon substrate binding. J. Biochem. 2002, 131, 313–317. [DOI] [PubMed] [Google Scholar]
- Steinbacher S.; Kaiser J.; Eisenreich W.; Huber R.; Bacher A.; Rohdich F. Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-d-erythritol 4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria drug development. J. Biol. Chem. 2003, 278, 18401. [DOI] [PubMed] [Google Scholar]
- Mac Sweeney A.; Lange R.; Fernandes R. P.; Schulz H.; Dale G. E.; Douangamath A.; Proteau P. J.; Oefner C. The crystal structure of E.coli 1-deoxy-d-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation. J. Mol. Biol. 2005, 345, 115–127. [DOI] [PubMed] [Google Scholar]
- Yajima S.; Hara K.; Iino D.; Sasaki Y.; Kuzuyama T.; Ohsawa K.; Seto H. Structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in a quaternary complex with a magnesium ion, NADPH and the antimalarial drug fosmidomycin. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2007, 63, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda T.; Tanaka N.; Kusakabe Y.; Nakanishi M.; Kitade Y.; Nakamura K. T.. Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum. Sci. Rep. 2011, 1, article number 9, DOI: 10.1038/srep00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes R. P.; Phaosiri C.; Proteau P. J. Mutation in the flexible loop of 1-deoxy-d-xylulose 5-phosphate reductoisomerase broadens substrate utilization. Arch. Biochem. Biophys. 2005, 444, 159–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.