Abstract
The serine/threonine kinase Akt/PKB promotes cancer cell growth and invasion through several downstream targets. Identification of novel substrates may provide new avenues for therapeutic intervention. Our study shows that Akt phosphorylates the cancer related transcription factor Runx2 resulting in stimulated DNA binding of the purified recombinant protein in vitro. Pharmacological inhibition of the PI3K/Akt pathway in breast cancer cells reduces DNA binding activity of Runx2 with concomitant reduction in the expression of metastasis related Runx2 target genes. Akt phosphorylates Runx2 at three critical residues within the runt DNA binding domain to enhance its in vivo genomic interactions with a target gene promoter, MMP13. Mutation of these three phosphorylation sites reduces Runx2 DNA binding activity, but does not interefere with CBFβ-Runx2 interactions. Consequently, expression of multiple metastasis-related genes is decreased and Runx2 mediated cell invasion is supressed. Thus, our work identifies Runx2 as a novel and important downstream mediator of the PI3K/Akt pathway that is linked to metastatic properties of breast cancer cells.
Keywords: PI3K, Akt, Runx2, Cell Signaling, Cancer, Transcription Factor, SUM159 breast cancer cells
INTRODUCTION
Breast cancer is one of the most common cancers, and the leading cause of mortality and morbidity among patients is the metastasis of the primary breast tumor to secondary organs that include brain, bone and lungs. Mechanistically, dysregulation of several signaling pathways and ectopic expression of oncogenic transcription factors contribute to the development and subsequent metastasis of breast cancer cells.
PI3K/Akt signaling is one of the most frequently deregulated pathways in breast cancer (Liu et al., 2007). Akt is activated in cells in response to various stimuli such as growth factors, hormones and extracellular matrix components. Activated Akt phosphorylates and regulates the activity of a host of proteins involved in cellular metabolism, proliferation, motility and apoptosis (Manning and Cantley, 2007). Interestingly, aggressive breast cancers frequently show amplification of Akt genes or mutations in other components of the signaling pathway that result in activation of Akt (Bellacosa et al., 1995; Dave et al., 2011; Dunlap et al., 2010). Although the PI3K/Akt pathway regulates the metastatic potential of human breast cancer cells (Qiao et al., 2007), only a handful of downstream effectors that mediate aberrant transcriptional programs in response to Akt signaling have been identified. For example, Akt enhances anchorage independent growth of breast cancer cells by direct phosphorylation of Y box binding protein-1 (YB- 1) (Sutherland et al., 2005). The actin binding protein girdin is another well-known Akt substrate that is required for IGF1 dependent cell movement of MDA-MB-231 breast cancer cells (Jiang et al., 2008).
Runt related transcription factors (Runx1, Runx2, Runx3) are lineage determining gene regulators involved in cell growth, proliferation and differentiation. Runx2 is a master regulator of osteoblast differentiation and bone formation (Lian and Stein, 2003), but it is also ectopically expressed in breast tumor cells where it contributes to metastasis of breast cancer to bone and formation of osteolytic lesions (Barnes et al., 2004; Barnes et al., 2003; Pratap et al., 2005; Pratap et al., 2006). High levels of Runx2 expression in breast cancer patients positively correlate with metastasis and poor clinical outcome of the disease (Das et al., 2009; Onodera et al., 2010). In osteoblasts Runx2 is a downstream effector of various signaling pathways, and several protein kinases have been shown to phosphorylate Runx2 and positively or negatively regulate its transcriptional activity during normal development) (Jonason et al., 2009). However, how Runx2 activity responds to signaling pathways that are associated with the onset and progression of breast cancer remains to be established. Here we show that Akt kinase directly phosphorylates Runx2 to regulate invasive properties of breast cancer cells. Our results indicate that Runx2 is an important downstream mediator of PI3K/Akt signaling in breast cancer.
EXPERIMENTAL PROCEDURES
Cell culture and treatments
The human breast cancer cell line SUM159 (a kind gift from Dr A. Mercurio, Department of Cancer Biology UMASS Medical School) was utilized for these studies due to high endogenous levels of both Runx2 and intact PI3K/Akt signaling. Cells were cultured in Ham’s F12 media (Hyclone) supplemented with 5% fetal bovine serum (FBS, Atlanta), 10 μg/ml insulin, 2 μg/ml hydrocortisone, 100U/ml penicillin, 100μg/ml streptomycin (Pen-Strep) and 2mM L-glutamine. MCF7 cells (which have minimal Runx2 levels and intact PI3K/Akt signaling) were cultured in DMEM supplemented with 10% FBS, Pen-Strep. 293T cells were cultured in DMEM supplemented with 10% FBS, Pen-Strep and 2mM L-glutamine. To block PI3K/Akt signaling, cells were treated with 20 μM LY294002 (Cell Signaling) or 20 μM Triciribine (Calbiochem). For transfection experiments, cells were transfected with various plasmids using Lipofectamine 2000 (Invitrogen).
MMTV-PyMT mice
Male FVB mice that were transgenic (+/−) for the PyV-MT antigen under the control of the mouse mammary tumor virus promoter (a kind gift from LM Shaw, Department of Cancer Biology, UMASS Medical School) were bred with female FVB/NJ mice (Jackson Labs), and female offspring positive for the transgene were saved for further analysis. Genotyping was performed by PCR as described previously for the PyV-MT transgene (Guy et al., 1992). Primary tumors as well as whole mammary glands from age matched controls were removed at indicated time points and whole cell lysates prepared for protein analyses.
Expression plasmids
GST tagged Runx2 pGEX bacterial expression plasmid was a kind gift from Dr M. Montecino (Universidad Andres Bello, Santiago, Chile). Constructs encoding for deletion mutants of GST-Runx2 were made by PCR amplification followed by ligation with pGEX vector (GE Health Care Life Sciences). The FLAG tagged Runx2 construct was created by ligating Runx2 cDNA into pCMV2 plasmid (Stratagene). Single and multiple point mutation constructs of Runx2 were synthesized using Site Directed Mutagenesis kits (Stratagene). Constitutively active (CA) and dominant negative (DN) mammalian expression constructs of Akt were purchased from Addgene (9008 and 9030) (Ramaswamy et al., 1999). CA Akt has an amino terminal src myristoylation sequence which targets Akt to the plasma membrane independent of PtdIns-3,4,5-P3 where it is phosphorylated by PDK1. Threonine 308 and serine 473, which are phosphorylated to activate Akt, are mutated to alanine in the DN Akt construct, and thus this mutant is kinase inactive. Wild type and mutant Runx2 cDNAs were cloned into pLenti-CMV-Blast-DEST vector using LR clonase (Invitrogen). Lentiviral particles were packaged in 293T cells as previously described (Pratap et al., 2009). Lentiviral particles were used to infect SUM159 and MCF7 cells in the presence of polybrene. Runx2 shRNA and control lentivirus used to make stable SUM19 cell line are described previously (Pratap et al., 2009).
Protein purification and in vitro kinase assay
GST fusion proteins were expressed in BL21 RP codon plus cells (Stratagene) and purified using glutathione agarose beads (Amersham Pharmacia). Protein quality was assessed by Coomassie staining. For in vitro kinase assays purified proteins were incubated for 30 min with 100 ng of purified Akt (upstate) in the presence of [γ-32P] ATP in kinase buffer (25 mM Tris pH 7.5, 10 mM MgCl2 and 5 mM β-glycerophosphate). The samples were resolved by 10% SDS-PAGE gel and bands viewed by autoradiograph.
Electrophoretic mobility shift assay
Runx consensus probe described previously (Zaidi et al., 2006) was labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England Biolabs) and purified using G-25 Sephadex column (Roche). Runx binding sites were mutated to generate corresponding probe for competition assays. DNA-protein binding reactions were carried out by incubating 10 fmol of radiolabeled probe with purified Runx2 or in vitro transcribed and translated (IVTT) Runx2 protein at room temperature for 20 min. Protein-DNA complexes were resolved on 4% nondenaturing polyacrylamide-Tris-borate-EDTA gel. For competition assays 100 fold excess of wild type or mutant probe was included in the reaction. Gels were dried and autoradiographed.
Chromatin immunoprecipitation (ChIP)
SUM159 cells overexpressing Runx2 were subjected to ChIP analysis as previously described (Pratap et al., 2005). Briefly, formaldehyde crosslinking was done followed by sonication to obtain DNA fragment with average size of 0.5 kb. Protein-DNA complexes were immunoprecipitated using Runx2 M70 antibody or IgG as a control. Purified DNA was amplified using specific primer sets and iTaq SYBR green supermix (Biorad) and fluorescence detected by ABI PRISM 700 system (Applied Biosystems). Primers flanking a Runx2 binding element in human MMP13 promoter region were used to assess the Runx2 occupancy. Primers amplifying promoter region of MARK3 gene were used as negative control (Supplemental Table S1).
RNA isolation, cDNA synthesis and Quantitative Real Time PCR (RT-QPCR)
RNA was isolated using Trizol reagent (Invitrogen) and trace DNA removed using DNA free RNA kit (Zymo). cDNA was synthesized using Superscript III (Invitrogen). cDNA was then amplified using gene-specific primers (Supplemental Table S1) and iTaq SYBR green supermix (BioRad) and fluorescence detected by ABI PRISM 700 system (Applied Biosystems).
Invasion assay
Cells were analyzed for invasion with Matrigel 24 well plates (BD Biosciences). Cells suspended in serum free medium were placed on each Matrigel insert and NIH3T3 conditioned medium was added to the bottom chambers of the plates. Cells were allowed to invade at 37°C. Non-invading cells were removed by scrubbing and cells were fixed and stained using HEMA 3 stain kit (Fisher) and counted manually under a light microscope.
Immunoprecipitation
FLAG-tagged Runx2 was overexpressed in 293T cells. Cells lysates in lysis buffer [20 mM HEPES, 10 μM ZnCl2, 1 mM MgCl2, 250 mM NaCl, 0.1% Triton X-100, supplemented with MG132, protease inhibitor cocktail tablets (Roche) and phosphatase inhibitor cocktails set 1&2 (Calbiochem)] were incubated with 50 μl of agarose-conjugated FLAG-M2 beads (Sigma) at 4°C for 2 h. Beads were washed in wash buffer (20 mM HEPES, 10 μM ZnCl2, 1 mM MgCl2, 150 mM NaCl, 0.1% Triton X-100), followed by SDS-PAGE and western blot analysis. Total Runx2 protein was detected using rabbit anti-FLAG antibody (Sigma) and phosphorylation determined by anti-phospho-serine/threonine antibody (BD Biosciences). In some experiments, immunoprecipitated samples were resolved by 12% SDS-PAGE and blots were probed with anti-Cbfβ antibody (abcam) to detect co-immunoprecipitation of Cbfβ with Runx2.
In Situ immunofluorescence microscopy
Cells grown on gelatin-coated cover slips were fixed using 3.7% formaldehyde and permeabilized in 0.1% Triton X-100. Cells were incubated with primary antibody at 37°C followed by incubation with FITC conjugated secondary antibody. DNA was visualized by DAPI (4′, 6-diamidino-2-phenylindole) staining. Images were recorded using an Epifluorescence Zeiss Axioplan 2 (Zeiss MicroImaging) microscope equipped with a charged coupled device. Images were captured and analyzed by MetaMorph Imaging Software (Molecular Devices).
Western blot
Cell lysates were prepared in direct lysis buffer (Zaidi et al., 2001) containing MG132 and protease inhibitor cocktail (Roche). Primary mammary tumors from MMTV-PyMT mice were removed along with mammary glands from mice negative for the transgene as controls at various time points. Proteins were prepared in RIPA buffer containing MG132, and protease inhibitor cocktail. Samples were resolved by SDS-PAGE followed by western blot analysis. Runx2 protein was detected using Runx2 monoclonal antibody or M70 antibody (Santa Cruz). pAkt and total Akt were detected using rabbit polyclonal antibodies (Cell Signaling). Blots were probed with B23 (Santa Cruz), Lamin (Santa Cruz) or βactin (Cell Signaling) antibodies for internal control.
Mass spectrometry
In vitro phosphorylated GST-Runx2 was resolved in an SDS-PAGE gel, silver stained and the Runx2 band excised. The sample was digested with trypsin and chymotrypsin and subjected to LC ESI MS/MS analysis. All spectra were analyzed with the Mascot searching algorithm.
RESULTS
Runx2 protein levels and Akt activity are upregulated in advanced mammary carcinomas
To examine the correlations among Runx2 expression, Akt activity and breast cancer progression, the MMTV-PyMT model of mammary cancer was utilized. In this transgenic mouse model, progression from premalignant to malignant to metastatic carcinoma is comparable with human breast cancer. Early malignant transformation in MMTV-PyMT mice occurs around 8 weeks of age and by 12 weeks the primary tumors in most mice progress to advanced carcinoma (Lin et al., 2003). Expression analysis of these mammary tumors revealed that Runx2 protein levels increased concomitantly with increased levels of phosphorylated Akt as tumors progressed (Fig. 1A). These results suggest that the biological activities of Runx2 and Akt contribute to late stage disease progression.
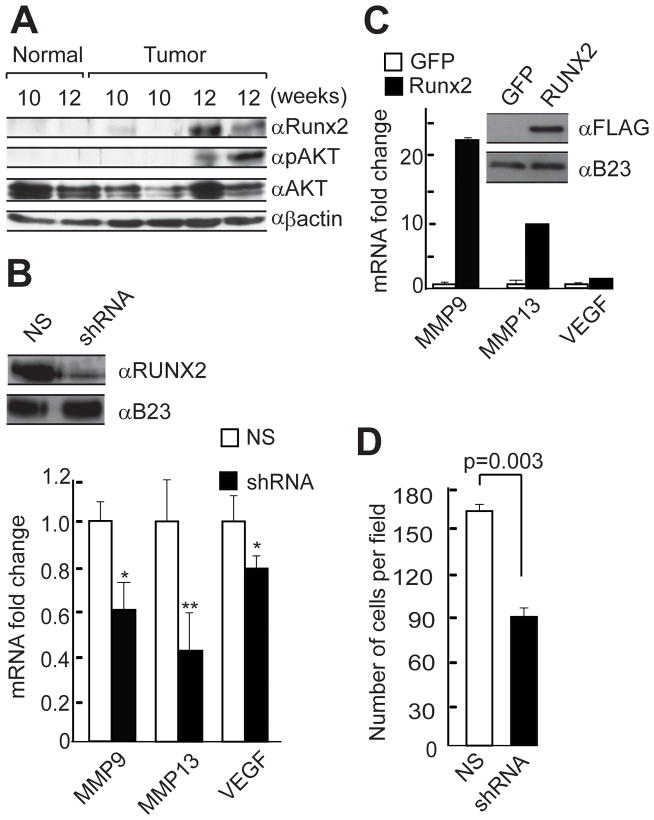
Figure 1. Runx2 regulates the expression of metastasis related genes and modulates the invasive potential of SUM159 breast cancer cells.
A) Primary mammary tumors from MMTV-PyMT mice at various stages of disease progression were removed along with mammary glands from mice negative for the transgene as controls at indicated time points, and whole cell lysates were prepared. Runx2, phosphorylated Akt (pAkt) and total Akt were detected by western blot analysis. B) Expression levels of metastasis related markers were monitored in SUM159 cells in which Runx2 protein was depleted using shRNA lentivirus. Runx2 protein was detected by western blotting using whole-cell lysates (top panel). B23 protein served as internal loading control. Endogenous mRNA levels of metastasis-related genes in response to depletion of Runx2 protein were measured by RT-QPCR analysis of total cellular RNA (bottom panel). Data were normalized to GAPDH mRNA levels. Error bars represent SD (n=4). p values calculated using Student’s t test are indicated. * p < 0.05, ** p < 0.01. C) Runx2 protein was over expressed in SUM159 cells using lentiviral vectors, and protein levels were monitored by western blot analysis of whole cell lysates. mRNA levels of metastasis-related genes were measured by RT-QPCR analysis of total cellular RNA. Experiments were performed using two independent biological samples with similar results, and one representative experiment is shown. D) The effect of Runx2 on cell invasion was monitored by Matrigel transwell assays using SUM159 cells that express non silencing (NS) or Runx2 shRNA. Cells were allowed to invade for 10 hours at 37°C. We counted two random fields in two wells each for three independent experiments. The data are presented as the number of invaded cells per field (p values based on Student’s t test are indicated).
Runx2 regulates metastasis-related genes and the invasive potential of breast cancer cells
We further evaluated the relationship between PI3K/Akt signaling and Runx2 activity in breast cancer cells. SUM159 cells express significant levels of ectopic Runx2 and have high basal activity of the PI3K/Akt pathway due to an activating mutation in the kinase domain of the PI3KCA gene (Ng et al., 2009). We initially examined the effect of modulating Runx2 activity on metastatic marker gene expression and invasion in SUM159 breast cancer cells. Western blot analysis of cell lysates showed that Runx2 is robustly expressed in SUM159 cells and its protein levels are nearly depleted by shRNA (Fig. 1B) and elevated upon forced expression (Fig. 1C). Reduction of Runx2 levels decreased and elevation increased expression of several metastasis related genes (MMP9, MMP13 and VEGF) (Fig. 1). The effect of Runx2 knockdown on the invasive potential of SUM159 breast cancer cells was investigated in Matrigel transwell assays. Depletion of Runx2 significantly diminished invasion of SUM159 cells (40–50% less invasion) (Fig. 1D). These results indicate that Runx2 regulates the expression of metastasis related proteins supporting ECM remodeling or angiogenesis, and modulates the invasive properties of SUM159 breast cancer cells.
PI3K/Akt signaling controls Runx2 responsive genes related to breast cancer cell invasion
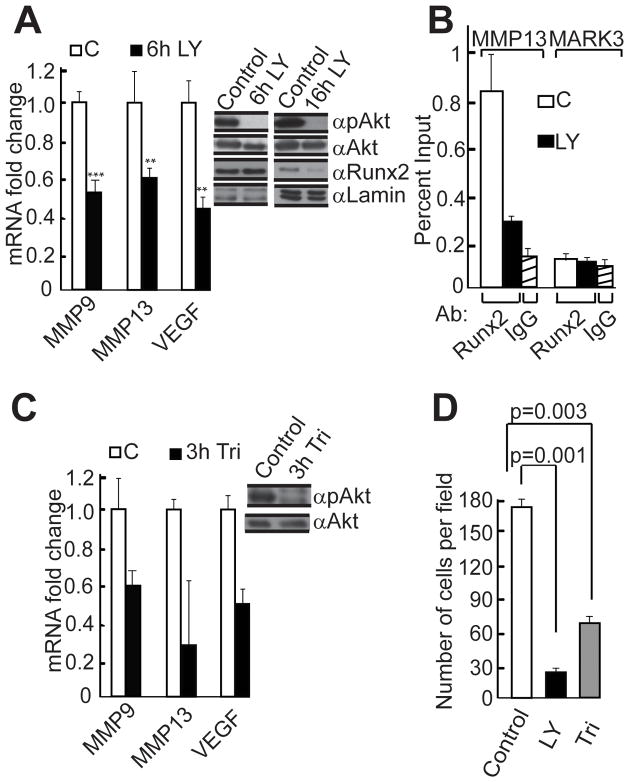
We next evaluated whether PI3K/Akt signaling modulates Runx2 activity in SUM159 breast cancer cells. We applied LY294002, a reversible inhibitor of PI3K and analyzed phospho-Akt (p-Akt) and total Akt levels by western blot to confirm that the inhibitor was indeed blocking Akt activation (Fig. 2A right panel). Inhibition of the PI3K/Akt pathway reduced expression of metastasis related Runx2 responsive genes (MMP9, MMP13 and VEGF) in SUM159 cells (Fig. 2A left panel). To address the mechanism by which the PI3K/Akt pathway regulates biological activity of Runx2, we examined how protein levels and DNA binding activity responded to modulation of PI3K/Akt signaling in breast cancer cells. Western blot analysis revealed that inhibition of the PI3K/Akt pathway by LY294002 in SUM159 cells for a short period (6 hr) did not change protein levels of Runx2; however, prolonged treatment (16 hr) with the inhibitor decreased the levels of Runx2 protein (Fig. 2A right panel). Importantly, chromatin immunoprecipitation (ChIP) analysis showed that inhibition of the PI3K/Akt pathway in SUM159 cells over-expressing Runx2 reduced genomic interactions of Runx2 with the MMP13 promoter (Fig. 2B). Inhibition of the PI3K/Akt pathway with triciribine, a cell-permeable tricyclic nucleoside that selectively inhibits Akt kinase activity, also reduced the expression of metastasis related Runx2 target genes (Fig. 2C). In complementary functional studies, inhibition of PI3Kinase/Akt signaling with either LY294002 or triciribine substantially reduced the invasive potential of SUM159 cells (Fig. 2D). Taken together, our results indicate that both PI3K and Runx2 contribute to breast cancer cell invasion and that PI3K/Akt signaling enhances the DNA binding potential of Runx2.
Figure 2. The PI3K/Akt pathway controls the levels of Runx2 target genes in SUM159 breast cancer cells.
A) Endogenous PI3K/Akt activity in SUM159 cells was blocked by treatment with 20μM LY294002 (LY), and total cellular protein and RNA samples were collected at indicated time points. The mRNA levels of Runx2 target genes were measured by RT-QPCR analysis. Data were normalized to GAPDH mRNA levels and results were quantified as described for Figure 1B. ** p < 0.01, *** p < 0.005. Runx2, phosphorylated Akt (pAkt) and total Akt were detected by western blot analysis. B) Genomic interactions of Runx2 were established by Chromatin Immunoprecipitation (ChIP) assay using Runx2 antibody. SUM159 cells over expressing WT Runx2 were treated with the PI3K/Akt inhibitor LY294002 for six hours. DNA recovered with the Runx2 antibody was subjected to QPCR analysis using MMP13 promoter primers. Normal rabbit IgG and primers amplifying the promoter region of the MARK3 gene were used as negative controls. Experiments were performed with two independent biological samples with similar results, and one representative experiment is shown. C) Endogenous Akt activity was blocked by treating SUM159 cells with 20μM Triciribine (Tri), and total cellular protein and RNA samples were collected after three hours. The mRNA levels of Runx2 target genes were measured by RT-QPCR analysis and data were normalized to GAPDH or HPRT mRNA levels. Experiments with two independent biological samples produced similar results and one representative experiment is shown. Phosphorylated Akt (pAkt) and total Akt were detected by western blot analysis. D) SUM159 breast cancer cells treated with LY294002 or triciribine or untreated control cells were analyzed in Matrigel invasion assays; results were quantified as described for Figure 1D.
Runx2 is a direct substrate of Akt kinase
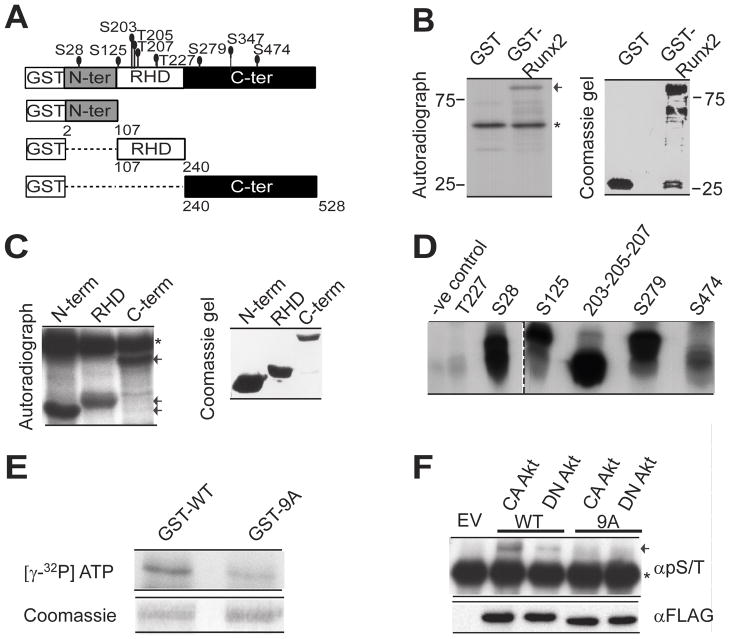
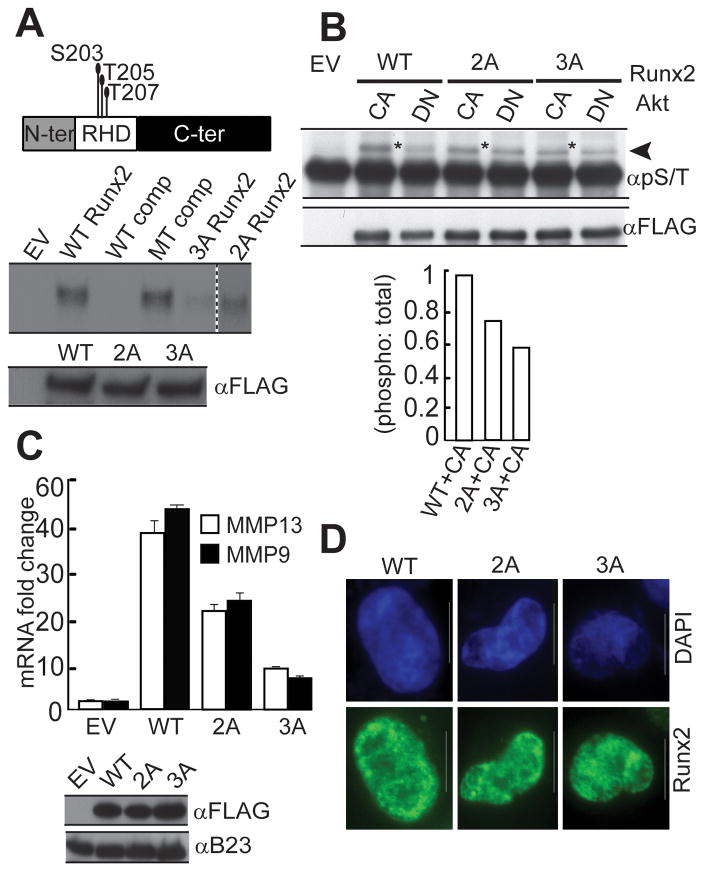
To delineate the mechanism by which the PI3K/Akt pathway affects the DNA binding potential of Runx2, we examined whether Akt kinase, which is the major kinase downstream of PI3 kinase, directly phosphorylates Runx2. In silico analysis (Songyang et al., 1994) identified nine serine or threonine residues in the runt homology domain (DNA binding domain) or in the N-terminal or C-terminal regions of Runx2 that may function as potential Akt phosphorylation sites (Fig. 3A and Supplemental Table S2). In vitro kinase assays using bacterially expressed proteins revealed that Akt strongly phosphorylates GST tagged WT Runx2 but not GST protein alone (Fig. 3B). In vitro phosphorylated full length Runx2 was further analyzed by mass spectrometry (MS) to identify phosphorylation sites. MS analysis identified a single phospho-peptide corresponding to the S28 site; however the analysis was technically limited and did not have coverage of the full protein (data not shown). When the S28 site was mutated to alanine in full length Runx2 (S28ARunx2), the mutant protein still showed strong phosphorylation by Akt in an in vitro kinase assay (Supplemental Fig. S1). To evaluate the remaining potential phosphorylation sites, three non-overlapping deletion mutants of Runx2 (N-terminus, Runt-domain, C-terminus) were subjected to phosphorylation by Akt in vitro (Fig. 3A). All three domains of Runx2 were robustly phosphorylated (Fig. 3C). In order to characterize the exact Akt phosphorylation sites, we next performed in vitro kinase assays with a panel of synthetic peptides spanning the putative phosphorylation sites (Supplemental Table S2). One of these peptides, encompassing S203/T205/T207, conforms to the most stringent criteria for a predicted Akt site. Akt strongly phosphorylated this peptide and three others (respectively spanning amino acids S28, S125 and S279), while very weakly phosphorylating two peptides (i.e., T227 and S474) (Fig. 3D). Akt did not significantly phosphorylate a mutant Runx2 protein (9ARunx2) in which all putative Akt phosphorylation residues were substituted by alanine (Fig. 3E). Taken together these findings demonstrate that Akt phosphorylates Runx2 at several sites in vitro.
Figure 3. Runx2 is a direct substrate of Akt kinase.
A) Schematic illustration of Runx2 protein showing putative Akt phosphorylation sites identified by the motif scanner prediction tool (http://scansite.mit.edu), as well as domains within Runx2 that were fused to GST for in vitro kinase assays [N-ter, Amino terminus (amino acids (aa) 2–107); RHD, RUNT homology domain (aa 107–241); C-ter, Carboxy terminus (aa 240–528)]. In vitro Akt kinase assays were performed with purified recombinant GST protein or GST fused to: B) full length Runx2, or C) selected deletion mutants. Proteins were resolved using 10% SDS PAGE and autoradiographed or stained using Coomassie. Arrows indicate full length or Runx2 deletions. Asterisks indicate autophosphorylation of Akt. D) Peptides corresponding to putative Akt phosphorylation sites in Runx2 were subjected to Akt kinase assay in vitro (see Supplemental Table S2). Samples were resolved in 20% Tris-tricine gels and autoradiographed. Technical difficulties prevented synthesis of peptide S347. E) Akt kinase assays were performed in vitro using [γ-32P] ATP with purified recombinant GST-fused full length WT Runx2 and 9ARunx2 mutant protein in which all nine predicted Akt sites were mutated to alanine. Proteins were resolved using 10% SDS PAGE and autoradiographed (top panel) or stained using Coomassie (bottom panel). F) In vivo phosphorylation of Runx2 was examined by immunoprecipitation assays using FLAG-tagged WT and 9ARunx2 in 293 T cells that co-expressed constitutively active (CA) or kinase inactive, dominant negative (DN) Akt. Runx2 was immunoprecipitated with FLAG beads and phosphorylation detected by western blot analysis with a general phospho serine/threonine antibody (pS/T) (top panel). The arrow indicates the phosphorylated Runx2 band. The asterisk indicates immuno-precipitated IgG. The blots were reprobed with FLAG antibody to detect total Runx2 (bottom panel).
To assess phosphorylation of Runx2 in mammalian cells, FLAG-tagged WT and 9ARunx2 were co-expressed with either constitutively active (CA) or kinase inactive, dominant negative (DN) Akt in 293T cells. Runx2 was then immunoprecipitated and its phosphorylation levels were analyzed using a general phospho-serine/threonine antibody (pS/T). Runx2 was strongly phosphorylated when co-expressed with CA Akt, whereas significantly reduced phosphorylation was observed in the presence of DN Akt (Fig. 3F). As expected, the 9ARunx2 mutant showed minimal Akt dependent phosphorylation. In addition, treatment of western blot membranes with lambda phosphatase eliminated pS/T antibody reactivity, thereby verifying the specificity of this antibody for phospho-proteins (Supplemental Fig. S2). Taken together, these data show that Runx2 is a direct substrate of Akt kinase in mammalian cells.
Akt increases DNA binding of Runx2 by direct phosphorylation of the runt homology domain without altering CBFβ interaction
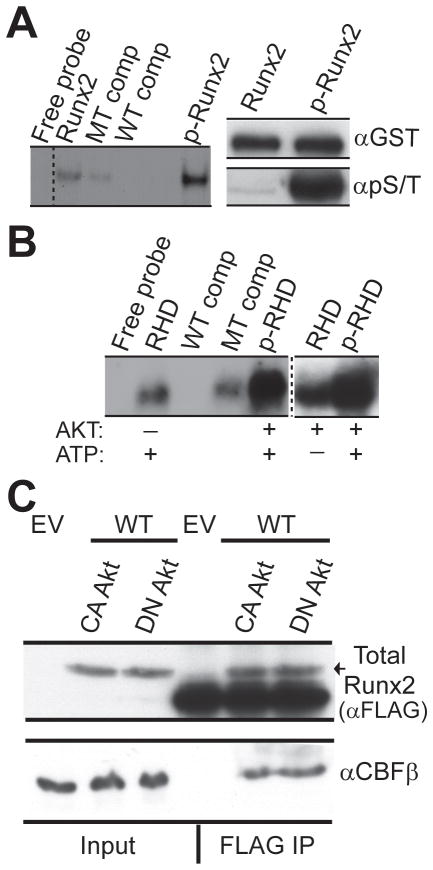
We next addressed whether phosphorylation by Akt can increase the DNA binding potential of Runx2 using GST fusion proteins and EMSA. GST-tagged full length Runx2 or the GST-tagged DNA binding domain both form complexes with the Runx consensus DNA oligonucleotide in the absence of Akt. However, phosphorylation by Akt dramatically increased the affinity of Runx2 for its cognate site (Fig. 4A). Akt phosphorylation also increased the binding of a deletion mutant spanning only the runt homology domain (Fig. 4B). These data show that Akt kinase enhances the DNA binding potential of Runx2 by direct phosphorylation of the runt homology domain. Thus, Akt phosphorylation directly impacts on the intrinsic DNA binding activity of Runx2.
Figure 4. Akt increases DNA binding of Runx2 by direct phosphorylation of the runt homology domain (RHD) without altering CBFβ interaction.
GST-tagged full length Runx2 was subjected to Akt kinase assay in vitro in the presence of non-radioactive ATP. The effect of Akt mediated phosphorylation on the DNA binding potential of full length Runx2 was measured by Electrophoretic Mobility Shift Assay (EMSA) using a Runx consensus oligonucleotide (left panel). Specificity of the Runx2 complex was established using binding assays with unlabeled WT or mutant (MT) competitor oligonucleotides. B) GST-tagged Runx2 RHD was subjected to Akt in vitro kinase assay in the presence of non-radioactive ATP and the effect on the DNA binding potential was measured by EMSA. Controls were carried out in the absence of either Akt or ATP, as indicated. C) FLAG-tagged WT Runx2 was co-expressed with constitutively active (CA) or kinase inactive, dominant negative (DN) Akt in 293T cells. Runx2 was immunoprecipitated with FLAG beads and samples were resolved using 8% and 12% SDS PAGE gels. Blots were probed with FLAG (top panel) and CBF β (bottom panel) antibodies.
The DNA binding affinity and stability of Runx2 are both increased by interaction with its cofactor CBFβ (Ito, 2008). Thus we further addressed whether phosphorylation status of Runx2 altered CBFβ interaction: FLAG-Runx2 was co-expressed with CA or DN Akt. Total Runx2 was isolated with FLAG-M2 beads (see Methods), shown in the upper panel of Fig. 4C. Analysis of this precipitate for the Runx2 binding partner CBFβ by western blot revealed that equivalent CBFβ was present in both the CA and DN Akt-transfected cells (Fig. 4C, lower panel). We conclude that formation of the Runx2-CBFβ complex is independent of Akt signaling
Akt phosphorylation deficient mutants of Runx2 show reduced DNA binding and transcriptional potential
We investigated the functional importance of Akt phosphorylation for Runx2 DNA binding and transcriptional activity by mutating candidate Akt sites. A panel of Runx2 mutants in which single, double or triple Akt sites in the runt homology domain were substituted by alanine was tested for DNA binding (Fig 5, Supplemental Table S3 and data not shown). None of the single alanine substitution mutants when produced by in vitro transcription and translation exhibited appreciable loss of DNA binding. However, two mutants in which either two (2ARunx2; S203/T205) or three residues (3ARunx2; S203/T205 and S207) were substituted by alanine showed reduced DNA binding (Fig. 5A). Comparison of the “CA lanes” (marked by asterisks in Fig. 5B) reveals a progressive decrease in the phosphorylation of Runx2 in the 2A and 3A mutants. Taken together, our data show that mutation of specific Akt phosphorylation sites within the runt homology domain decreases the DNA binding potential of Runx2.
Figure 5. Akt phosphorylation deficient mutants of Runx2 show reduced DNA binding and transcriptional potential.
A) DNA binding activity of in vitro transcribed and translated (IVTT) FLAG-tagged WT, 2A and 3A mutant Runx2 was measured by Electrophoretic Mobility Shift Assays (EMSA) using Runx consensus oligonucleotide. Specificity of the Runx2 complex was established using binding assays with unlabeled WT or mutant (MT) competitor oligonucleotides (middle panel). Proteins were resolved by 10% SDS PAGE gel and probed with FLAG antibody (bottom panel). B) Phosphorylation of WT and Runx2 mutants was examined by immunoprecipitation assays using FLAG-tagged Runx2 proteins expressed in 293T cells that co-express constitutively active (CA) or kinase inactive, dominant negative (DN) Akt. Runx2 phosphorylation was monitored with the pS/T antibody as described previously (top panel). * highlights Runx2 phosphorylation in WT and Runx2 mutants co-expressing CA Akt. The blots were reprobed with FLAG antibody to detect total Runx2 (middle panel). Bands were quantified using ImageJ software and the ratio of phosphorylated versus total protein is plotted (bottom panel). C) FLAG-tagged WT, 2A or 3A Runx2 proteins were expressed in SUM159 cells using lentiviral vectors. Endogenous mRNA levels of metastasis-related Runx2 responsive genes were measured by RT-QPCR analysis of total cellular RNA. Data were normalized to mCox mRNA levels. Two independent biological samples were analyzed, with similar results, and one representative experiment is shown. Runx2 was detected by western blot analysis of whole-cell lysates using FLAG antibody. B23 served as internal loading control. D) SUM159 cells expressing FLAG- tagged WT and mutant Runx2 proteins were processed for immunofluorescence microscopy using FLAG antibody. Images were captured using a Zeiss Axioplan digital microscope and Metamorph software was used for bio imaging.
We next analyzed the consequences of abrogating Akt mediated signaling through Runx2 on the expression of Runx2 responsive genes in breast cancer cells. Expression of the 2A or 3A Runx2 mutant in SUM159 cells reduced the mRNA levels of two key metastasis-related target genes relative to WT Runx2 (MMP9: 45% and 87% reduction, MMP13: 45% and 78% reduction, respectively, for 2A and 3A) (Fig. 5C). Furthermore, intra-nuclear localization of Runx2 is obligatory for its functional activity (Afzal et al., 2005; Javed et al., 2005). We observe that both mutant proteins retained proper sub-nuclear localization as determined by immunofluorescence microscopy (Fig. 5D). This result indicates that the demonstrated decrease in expression of genes is not related to mis-localization of Runx2 in the cytoplasm. Thus, abrogation of critical Akt phosphorylation sites directly impinges on the expression of metastasis related Runx2 responsive genes.
Runx2 is an important downstream mediator of the PI3K/Akt pathway for invasion of breast cancer cells
Our studies in highly metastatic SUM159 breast cancer cells demonstrate that the invasive potential of these cells is dependent on both aberrant Runx2 expression and activated PI3K/Akt signaling. To study the pathological role of Runx2 in invasion of breast cancer cells, we performed complementary experiments using WT and Akt site mutants of Runx2 in non-invasive MCF7 adenocarcinoma cells. These cells have an intact PI3K/Akt signaling axis (Fig. 6A) and barely detectable levels of endogenous Runx2. Forced expression of WT Runx2 increased invasion of MCF7 cells in culture (Fig. 6B), corroborating previous observations (Leong et al., 2010; Pratap et al., 2005). Mutation of the key Akt phosphorylation sites in Runx2 (i.e., S203, T205, T207; mutants 2A and 3A) reduced the Runx2 dependent invasive potential of MCF7. The extent to which invasion was decreased was consistent with the number of alanine substitutions (Fig. 6B), the phosphorylation status of the mutants (Fig. 5B) and DNA binding activity (Fig. 5A). Similar to results obtained with SUM159 cells, MCF7 cells expressing 2A or 3A mutant Runx2 protein had lower mRNA levels of MMP9 and MMP13 than cells expressing WT Runx2 (Fig. 6C). These results indicate that activation of the PI3K/Akt pathway and aberrant elevated expression of Runx2 both contribute to the increased invasive potential of breast cancer cells.
Figure 6. Runx2 is an important downstream mediator of the PI3K/Akt pathway for invasion of breast cancer cells.
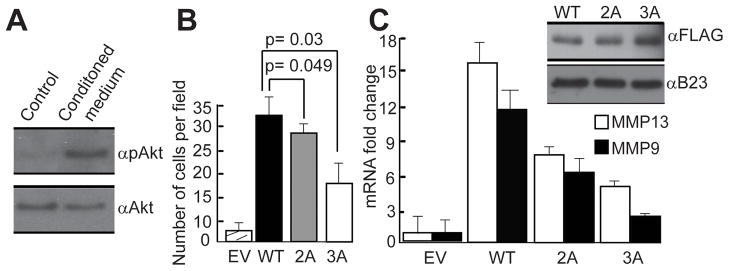
A) MCF7 cells have an intact PI3K/Akt signaling pathway. Cells were incubated with control or NIH3T3 conditioned medium for 10 minutes. Whole cell lysates were used to detect p-Akt and total Akt by western blot analysis. B) The effect of Akt mediated phosphorylation of Runx2 on cell invasion was monitored by Matrigel transwell assays using MCF7 cells that express WT, 2A or 3A Runx2. Cells were allowed to invade for 72 hours at 37°C and results were quantified as described in Figure 1. C) Protein and RNA samples were collected from MCF7 cells expressing FLAG-tagged WT, 2A or 3A Runx2. Runx2 was detected by western blot analysis using FLAG antibody. Endogenous mRNA levels of metastasis-related Runx2 responsive genes were measured by RT-QPCR analysis of total cellular RNA. Data were normalized to mCox mRNA levels. Experiments were performed in two independent biological samples with similar results, and one representative experiment is shown.
DISCUSSION
This study demonstrates that Runx2 is a novel substrate of Akt in breast cancer cells and an important downstream mediator of PI3K/Akt signaling that is pathologically associated with breast cancer metastasis. This finding is particularly important because the PI3K/Akt pathway is one of the major signaling pathways through which growth factors and other extracellular stimuli influence cellular growth, metabolism and invasion. In breast cancer cells, several mutations have been identified that hyperactivate PI3K/Akt signaling, which is essential to oncogenic transformation (Bellacosa et al., 1995; Da Silva et al., 2010; Dunlap et al., 2010; Gershtein et al., 2007). The identification of Runx2 as a key downstream target of the PI3K/Akt pathway in breast cancer cells provides a significant advance in understanding the role of oncogenic aberrations in control of cell growth and invasion.
Our study shows that inhibition of the PI3K/Akt pathway in breast cancer cells severely inhibits Runx2 DNA binding and expression of metastatic genes. Evidence presented to support this conclusion includes: several peptides spanning putative Akt sites throughout the Runx2 protein are directly phosphorylated by Akt; loss of Runx2 DNA binding activity occurs when critical Akt sites in the runt homology domain are mutated; Runx2-CBFβ interaction as well as the sub-nuclear organization of Runx2 foci are retained independently of Akt signaling. Inhibition of Akt signaling (either through pharmacological inhibition or genetic manipulations) prevents genomic interactions of Runx2 with a representative target promoter (i.e. MMP13) and reduces expression of a panel of Runx2 responsive genes that are involved in cancer metastasis. We conclude there is a significant contribution of Runx2 phosphorylation mediated by Akt signaling to the expression of Runx2-related metastatic genes.
Apart from the known pathological role of Runx2 in breast cancer (Pratap et al., 2011), Runx2 normally acts as a multifunctional osteogenic regulator of mesenchymal cells. In osteoblastic cells, Runx2 responds to multiple kinase pathways involving MAPK/ERK, PKA, PKC and Akt, with each having the ability to control its transcriptional activity (Ge et al., 2007; Ge et al., 2012). Moreover, previous studies have shown that Akt signaling promotes Runx2 mediated bone formation and osteoblast differentiation (Fujita et al., 2004; Kawamura et al., 2007; Ling et al., 2010; Yang et al., 2011). However, none of these studies investigated a ‘kinase/substrate’ mechanism for Akt stimulation of Runx2 activity. We find that Akt stimulates Runx2 DNA binding by direct phosphorylation at a principal motif in the central DNA binding domain to increase the invasive migration of breast cancer cells. This result is fully supported by the studies of Fujita and colleagues who showed that PI3K/Akt signaling enhances DNA binding, as well as Runx2 mediated differentiation and migration of chondrocytes and osteoblasts (Fujita et al., 2004). Nonetheless, the mechanisms by which PI3K/Akt signals to Runx2 can be cell context dependent and indirect, as evidenced in prostate cancer cells (Zhang et al., 2011).
Our demonstration that the cancer related activity of Runx2 does depend on the PI3K/Akt pathway indicates oncogenic convergence of two critical molecular mechanisms that contribute to the pathological properties of metastatic breast cancer cells. Our study shows that inhibiting total Akt activity in SUM159 cells leads to decreased cell invasion and that Runx2 is an important downstream mediator of this effect. There are three isoforms of Akt (Akt1, Akt2 and Akt3) encoded by three separate genes that appear to have distinct and opposite effects on breast cancer invasion and metastasis. Akt2 enhances invasion (Arboleda et al., 2003) whereas Akt1 has been shown to inhibit tumor invasion (Yoeli-Lerner et al., 2005). It will be interesting to examine if Runx2 is phosphorylated in an isoform specific manner by Akt in breast cancer cells.
In conclusion, this work clearly demonstrates the biochemical mechanism by which Runx2, a known contributor to progression of breast cancer metastasis, is directly regulated by the PI3K/Akt pathway. While much is known about the signals that activate the PI3K/Akt pathway, identification of Runx2 as a key downstream substrate of the PI3K/Akt pathway provides a molecular mechanism to control tumor cell invasion and enhances the mechanistic rationale for developing novel inhibitors of Akt and Runx2 as therapeutic targets in breast cancer.
Supplementary Material
Acknowledgments
Contract grant sponsor: National Institutes of Health. Grant numbers: P01 CA082834, P01 AR048818 and R01 AR039588.
Footnotes
This work was supported in part by National Institutes of Health grants P01 CA082834, P01 AR048818 and R01 AR039588. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204(1):63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, Danino M, Karlan BY, Slamon DJ. Overexpression of AKT2/protein kinase Bbeta leads to up-regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res. 2003;63(1):196–206. [PubMed] [Google Scholar]
- Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, Stein GS, Gerstenfeld LC. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases-associated osteolytic disease. Cancer Res. 2004;64(13):4506–4513. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- Barnes GL, Javed A, Waller SM, Kamal MH, Hebert KE, Hassan MQ, Bellahcene A, Van Wijnen AJ, Young MF, Lian JB, Stein GS, Gerstenfeld LC. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 2003;63(10):2631–2637. [PubMed] [Google Scholar]
- Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, Wan M, Dubeau L, Scambia G, Masciullo V, Ferrandina G, Benedetti Panici P, Mancuso S, Neri G, Testa JR. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64(4):280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- Da Silva L, Simpson PT, Smart CE, Cocciardi S, Waddell N, Lane A, Morrison BJ, Vargas AC, Healey S, Beesley J, Pakkiri P, Parry S, Kurniawan N, Reid L, Keith P, Faria P, Pereira E, Skalova A, Bilous M, Balleine RL, Do H, Dobrovic A, Fox S, Franco M, Reynolds B, Khanna KK, Cummings M, Chenevix-Trench G, Lakhani SR. HER3 and downstream pathways are involved in colonization of brain metastases from breast cancer. Breast Cancer Res. 2010;12(4):R46. doi: 10.1186/bcr2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Leong DT, Gupta A, Shen L, Putti T, Stein GS, van Wijnen AJ, Salto-Tellez M. Positive association between nuclear Runx2 and oestrogen-progesterone receptor gene expression characterises a biological subtype of breast cancer. Eur J Cancer. 2009;45(13):2239–2248. doi: 10.1016/j.ejca.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H, Narasanna A, Chakrabarty A, Hilsenbeck SG, Huang J, Rimawi M, Schiff R, Arteaga C, Osborne CK, Chang JC. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29(2):166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J, Le C, Shukla A, Patterson J, Presnell A, Heinrich MC, Corless CL, Troxell ML. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat. 2010;120(2):409–418. doi: 10.1007/s10549-009-0406-1. [DOI] [PubMed] [Google Scholar]
- Fujita T, Azuma Y, Fukuyama R, Hattori Y, Yoshida C, Koida M, Ogita K, Komori T. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol. 2004;166(1):85–95. doi: 10.1083/jcb.200401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176(5):709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Yang Q, Zhao G, Yu H, Kirkwood KL, Franceschi RT. Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J Bone Miner Res. 2012;27(3):538–551. doi: 10.1002/jbmr.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershtein ES, Scherbakov AM, Shatskaya VA, Kushlinsky NE, Krasil’nikov MA. Phosphatidylinositol 3-kinase/AKT signalling pathway components in human breast cancer: clinicopathological correlations. Anticancer Res. 2007;27(4A):1777–1782. [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci U S A. 2005;102(5):1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Enomoto A, Jijiwa M, Kato T, Hasegawa T, Ishida M, Sato T, Asai N, Murakumo Y, Takahashi M. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer Res. 2008;68(5):1310–1318. doi: 10.1158/0008-5472.CAN-07-5111. [DOI] [PubMed] [Google Scholar]
- Jonason JH, Xiao G, Zhang M, Xing L, Chen D. Post-translational Regulation of Runx2 in Bone and Cartilage. J Dent Res. 2009;88(8):693–703. doi: 10.1177/0022034509341629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N, Kugimiya F, Oshima Y, Ohba S, Ikeda T, Saito T, Shinoda Y, Kawasaki Y, Ogata N, Hoshi K, Akiyama T, Chen WS, Hay N, Tobe K, Kadowaki T, Azuma Y, Tanaka S, Nakamura K, Chung UI, Kawaguchi H. Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS One. 2007;2(10):e1058. doi: 10.1371/journal.pone.0001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong DT, Lim J, Goh X, Pratap J, Pereira BP, Kwok HS, Nathan SS, Dobson JR, Lian JB, Ito Y, Voorhoeve PM, Stein GS, Salto-Tellez M, Cool SM, van Wijnen AJ. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res. 2010;12(5):R89. doi: 10.1186/bcr2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS. Runx2/Cbfa1: a multifunctional regulator of bone formation. Curr Pharm Des. 2003;9(32):2677–2685. doi: 10.2174/1381612033453659. [DOI] [PubMed] [Google Scholar]
- Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163(5):2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L, Dombrowski C, Foong KM, Haupt LM, Stein GS, Nurcombe V, van Wijnen AJ, Cool SM. Synergism between Wnt3a and heparin enhances osteogenesis via a phosphoinositide 3-kinase/Akt/RUNX2 pathway. J Biol Chem. 2010;285(34):26233–26244. doi: 10.1074/jbc.M110.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Bagaitkar J, Watabe K. Roles of AKT signal in breast cancer. Front Biosci. 2007;12:4011–4019. doi: 10.2741/2367. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SS, Mahmoudi T, Danenberg E, Bejaoui I, de Lau W, Korswagen HC, Schutte M, Clevers H. Phosphatidylinositol 3-kinase signaling does not activate the wnt cascade. J Biol Chem. 2009;284(51):35308–35313. doi: 10.1074/jbc.M109.078261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Miki Y, Suzuki T, Takagi K, Akahira J, Sakyu T, Watanabe M, Inoue S, Ishida T, Ohuchi N, Sasano H. Runx2 in human breast carcinoma: its potential roles in cancer progression. Cancer Sci. 2010;101(12):2670–2675. doi: 10.1111/j.1349-7006.2010.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Imbalzano KM, Underwood JM, Cohet N, Gokul K, Akech J, van Wijnen AJ, Stein JL, Imbalzano AN, Nickerson JA, Lian JB, Stein GS. Ectopic runx2 expression in mammary epithelial cells disrupts formation of normal acini structure: implications for breast cancer progression. Cancer Res. 2009;69(17):6807–6814. doi: 10.1158/0008-5472.CAN-09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25(19):8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25(4):589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Pratap J, Lian JB, Stein GS. Metastatic bone disease: role of transcription factors and future targets. Bone. 2011;48(1):30–36. doi: 10.1016/j.bone.2010.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M, Iglehart JD, Pardee AB. Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res. 2007;67(11):5293–5299. doi: 10.1158/0008-5472.CAN-07-0877. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, Sellers WR. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1999;96(5):2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4(11):973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, Dedhar S, Nelson C, Pollak M, Leighton Grimes H, Miller K, Badve S, Huntsman D, Blake-Gilks C, Chen M, Pallen CJ, Dunn SE. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24(26):4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- Yang S, Xu H, Yu S, Cao H, Fan J, Ge C, Fransceschi RT, Dong HH, Xiao G. Foxo1 mediates insulin-like growth factor 1 (IGF1)/insulin regulation of osteocalcin expression by antagonizing Runx2 in osteoblasts. J Biol Chem. 2011;286(21):19149–19158. doi: 10.1074/jbc.M110.197905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20(4):539–550. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Choi JY, van Wijnen AJ, Stein JL, Lian JB, Stein GS. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114(Pt 17):3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Pratap J, Schroeder TM, JJW, Lian JB, van Wijnen AJ, Stein GS, Stein JL. Alterations in intranuclear localization of Runx2 affect biological activity. J Cell Physiol. 2006;209(3):935–942. doi: 10.1002/jcp.20791. [DOI] [PubMed] [Google Scholar]
- Zhang H, Pan Y, Zheng L, Choe C, Lindgren B, Jensen ED, Westendorf JJ, Cheng L, Huang H. FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer Res. 2011;71(9):3257–3267. doi: 10.1158/0008-5472.CAN-10-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.