Abstract
The precise regulation of morphogenesis is a key mechanism by which cells respond to a variety of stresses, including those encountered by microbial pathogens in the host. The polarity protein Cdc42 regulates cellular morphogenesis throughout eukaryotes, and we explore the role of Cdc42 proteins in the host survival of the human fungal pathogen Cryptococcus neoformans. Uniquely, C. neoformans has two functional Cdc42 paralogs, Cdc42 and Cdc420. Here we investigate the contribution of each paralog to resistance to host stress. In contrast to non-pathogenic model organisms, C. neoformans Cdc42 proteins are not required for viability under non-stress conditions. In the presence of cell stress, strains deleted for either paralog show defects in thermotolerance and morphogenesis, likely as a result of their roles in the organization of actin and septin structures during bud growth and cytokinesis. These proteins act downstream of C. neoformans Ras1 to regulate its morphogenesis subpathway, but not its effects on mating. Cdc42, and not Cdc420, is required for virulence in a murine model of cryptococcosis. The C. neoformans Cdc42 proteins likely perform complementary functions with other Rho-like GTPases to control cell polarity, septin organization, and hyphal transitions that allow survival in the environment and in the host.
Keywords: septins, pathogenesis, signal transduction, Ras1, stress
Introduction
In order to successfully inhabit diverse environments, microorganisms must rapidly adapt a complex life cycle to unexpected and unpredictable stress conditions. In the case of microbial pathogens, cells adapt to the unique stresses within the infected host, including variations in pH, temperature, and iron and nutrient availability. To survive in this environment, microorganisms often direct quite dramatic morphogenetic processes to maintain or alter their cell size, shape, or volume. For example, the human fungal pathogen Candida albicans must be able to transition between yeast-like morphology and hyphal states in order to cause disease (Leberer et al., 1997; Lo et al., 1997). In the case of thermally dimorphic fungi such as Histoplasma capsulatum and Blastomyces dermatitidis, host temperature induces the switch from a filamentous environmental morphology to a yeast-like pathogenic form(Maresca and Kobayashi, 2000). The role of thermotolerance in morphogenesis and pathogenesis is not limited to these dramatic cases: the human fungal pathogen Cryptococcus neoformans maintains a yeast-like morphology in the host despite exposure to conditions which may disrupt the establishment of signals for polarized growth and budding (Nichols et al., 2007; Price et al., 2008b).
Spores of the fungal pathogen C. neoformans are inhaled from the environment, initially establishing infection in the lungs. This opportunistic pathogen can disseminate to the central nervous system and result in lethal infection especially in immuno-compromised individuals. Previous investigations have identified several cellular factors required for C. neoformans survival under the host physiological temperature of 37°C, including trehalose-6-phosphate synthase (Tps1) activity, calcineurin signaling, and Ras proteins (Alspaugh et al., 2000; Odom, 1997; Petzold et al., 2006). Elements of the C. neoformans Ras pathway have been shown to control cytoskeletal architecture and cellular morphogenesis in a temperature-dependent manner (Alspaugh et al., 2000; Nichols et al., 2007).
Like many yeasts, C. neoformans responds to non-lethal cell stress with a transient depolarization of its actin cytoskeleton (Ho and Bretscher, 2001). In viable cells, actin is rapidly re-polarized to the site of new bud formation to facilitate subsequent cell division. In contrast, C. neoformans ras1Δ mutant cells display a marked delay in actin re-polarization at 37°C (Waugh et al., 2002a). This results in temperature-dependent morphological changes and growth arrest. The ras1Δ mutant strains are accordingly avirulent in animal models of cryptococcosis (Alspaugh et al., 2000). These results are consistent with accumulating data from other microorganisms in which Ras proteins are important upstream determinants of actin cytoskeletal integrity and cell stress response (Ho and Bretscher, 2001; Onken et al., 2006).
A growing body of evidence suggests that C. neoformans Ras1 cooperates with highly conserved Rho-GTPases such as Cdc42 and Rac1 to regulate the morphology of the yeast form under stress conditions, and to direct the morphological transition to filamentous growth during mating (Nichols et al., 2007; Price et al., 2008a; Vallim et al., 2005). Rho-GTPases, which are known to control morphogenetic signals in many species, act as molecular switches, cycling between inactive GPD-bound and active GTP-bound forms. Proteins that facilitate and regulate these transitions include Guanine nucleotide Exchange Factors (GEFs), GTPase Activating Proteins (GAPs), and Guanine nucleotide Dissociation Inhibitors (GDIs). In C. neoformans, the Cdc42-specific GEF Cdc24 and the GDI Rdi1 have been shown to control morphogenetic events in a Ras1-dependent manner (Nichols et al., 2007; Price et al., 2008a).
Cdc42 is a major component of the eukaryote polarisome and a key regulator of morphogenesis among yeast and filamentous fungi. In Saccharomyces cerevisiae, CDC42 is essential for growth (Johnson and Pringle, 1990; Johnson, 1999). Polar accumulation and activation of ScCdc42p at presumptive bud sites is followed by the recruitment and organization of the actin cytoskeleton, polarity machinery, and the septin ring, culminating in the formation and separation of a daughter cell (Ayscough et al., 1997; Douglas et al., 2005). In the absence of functional Cdc42, or in the absence of the GEF Cdc24 S. cerevisiae cells are apolar and arrest as large, unbudded cells (Adams et al., 1990; Johnson, 1999).
In addition to its role in polar growth, Cdc42 is a key element in morphological transitions required for pathogenicity. In C. albicans, activated CaCdc42p is required for the yeast-hyphal transition and pathogenesis (Bassilana et al., 2003; Su et al., 2007; Ushinsky et al., 2002; VandenBerg et al., 2004). The basidiomycetous corn smut Ustilago maydis requires UmCdc42p for organization of the actin contractile ring during cytokinesis and for virulence (Böhmer et al., 2008; Mahlert et al., 2006). In this and other filamentous fungi CDC42 is not essential, and coordination of the polarisome is shared with closely related Rac proteins (Bassilana and Arkowitz, 2006; Boyce et al., 2003; Gorfer et al., 2001; Hurtado et al., 2000; Mahlert et al., 2006). The degree to which functional overlap is present between these two closely related proteins is species specific.
In C. neoformans, Ras1 mediates its effect through a branched signal transduction cascade, and there is indirect evidence that Cdc42 and Rac provide specificity between these branches. Rac1 has been shown to be primarily involved in the formation of mating hyphae, while morphogenesis and thermotolerance in the yeast form appears to be dependent on components of traditional Cdc42 pathways (Nichols et al., 2007; Price et al., 2008a; Vallim et al., 2005). For example, C. neoformans cdc24Δ mutant cells arrest as large unbudded cells under stress conditions, similar to ras1Δ mutants. However, in contrast to ras1Δ strains, cdc24Δ mutants have no defect in filamentous growth or mating. Additionally, over-expression of RDI1, a negative regulator of Cdc42 function, results in mis-localization of Cdc42 and subsequent defects in yeast morphogenesis and cytokinesis (Price et al., 2008a). Also, consistent with a role for Cdc42 in Ras1-mediated signaling, over-expression of Cdc42 homologs in the ras1Δ background rescues the striking thermotolerance defects. In contrast to other fungal species, C. neoformans has two highly related Cdc42 paralogs, Cdc42 and Cdc420. To directly address the specific roles of these two paralogs in the Ras1 pathway, we examined the role of these two related GTPases in Ras-mediated morphogenesis and stress response.
Results
Two Cdc42 paralogs are non-essential
In contrast to other fungal species, the C. neoformans var. grubii genome encodes two closely related Cdc42 paralogs, Cdc42 and Cdc420 (Figure 1a). Both proteins contain predicted Rho-GTPase domains, including conserved GTP binding and hydrolysis domains, effector binding domains, and the C-terminal prenylation sites and lysine rich regions required for membrane localization (Johnson, 1999). Both paralogs maintain the canonical TQXD sequence at position 115, which distinguishes Cdc42 GTPases from the related Rac GTPases (TKXD) (Chen et al., 1993). Moreover, identification of a specific residue that differentiates Cdc42 (F56) from Rac (W56) clearly places both paralogs in the Cdc42 class (Gao et al., 2001). The two protein sequences share 84% similarity and diverge primarily in the Rho-insert domain (Figure 1a). The regions around the two genes are not syntenic, implying a single gene duplication, rather than a larger duplication event.
Figure 1. C. neoformans species maintain two Cdc42 paralogs.
A) Alignment of C. neoformans (Cn) Cdc42 and Cdc420 protein sequences with Cdc42 protein sequences from S. cerevisiae (Sc), S. pombe (Sp), and U. maydis (Um). Key domains are indicated and the C. neoformans Rac1 sequence is provided for comparison. Residue 56 distinguishes Cdc42(F) and Rac(W) proteins by providing specificity with GEFs (Hlubek et al., 2008). B) Phylogenetic tree showing the relatedness of Cdc42 protein sequences from three C. neoformans species, U. maydis C. cinerea, P. marneffei, S. pombe, Candida species, A. fumigatus, S. cerevisiae, and R. oryzae (rooted).
A survey of Cryptococcus species genomes revealed duplicated CDC42 genes in three closely related varieties, C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii. We chose to examine the genomes of a variety of fungi in basidiomycete, ascomycete, and zygomycete lineages for CDC42 duplication, including genomes for which CDC42 function has been well characterized, as well as those in which CDC42 function remains unclear. Examination of the partially completed Tremella mesenterica genome, representing the most closely related group of basidiomycetes to the Cryptococcus genus, identified a single CDC42 homolog, with higher similarity to C. neoformans CDC42 than to CDC420. The incomplete T. mesenterica genome project limits more definitive assessments at this time. In the more distantly related basidiomycetes Ustilago maydis and Coprinopsis cinerea no duplication is present. No duplicated CDC42 genes were identified among ascomycete fungi, including S. cerevisiae, and S. pombe, or in the pathogenic ascomycetes, including Candida species, P. marneffei, and A. fumigatus, in which functional CDC42 homologs are joined by functional RAC homologs (Momany, 2002). However, we did identify two as yet uncharacterized CDC42 paralogs, encoding all expected CDC42 domains, in the distantly related pathogenic zygomycete Rhizopus oryzae.
Cladograms were constructed for the CDC42 sequences using Bayesian inference and a maximum likelihood heuristic search (see materials and methods). Trees were rooted with Rhizopus oryzae, the most distantly related taxon to all of the other taxa included in the analyses. In both cases, there was significant statistical nodal support (MrBayes posterior probabilities values ≥ 95% and bootstrap values ≥ 70%) for each of the Cryptococcus CDC42 and CD420 clades, as well as for the clade consisting of both paralogs. Similar topography was generated using multiple techniques, and we present the maximum likelihood analysis (Figure 1b). CDC42 sequence from these collected species support a model in which Cryptococcus species maintain a CDC42-like copy and a CDC420-like copy.
It should be noted that the genes CDC42 and CDC420 were previously named DCH2 (Genbank ID: DQ991434) and CDC42 (Genbank ID: DQ991433) respectively (Nichols et al., 2007). These names were assigned based on initial similarity to S. cerevisiae and S. pombe sequences. Based on accumulating genome data and on the data presented here, we have determined that DCH2 is the major paralog, and have therefore chosen to rename it CDC42. The gene previously identified as CDC42 will now be known as CDC420 to reflect both its sequence divergence and its more limited role in C. neoformans biology.
To determine whether C. neoformans CDC42 paralogs are essential and to investigate function, we generated cdc42Δ and cdc420Δ null mutants in which the entire open reading frame of each gene was deleted (Table 1). Viable mutants were obtained for each allele in multiple, independent transformation events, indicating that, unlike in S. cerevisiae and S. pombe, these genes are not essential in C. neoformans (Johnson et al., 1990; Miller and Johnson, 1994). The mutant strains grew at rates comparable to wild type at 30°C, and they demonstrated no defects in capsule or melanin production, two virulence-associated phenotypes. ERB007 (cdc420Δ) and ERB010 (cdc42Δ) were selected as representative strains and are used for all phenotypic characterizations presented here.
Table 1.
| Strain | Genotype | Source/Reference |
|---|---|---|
| H99 | MAT α | (Perfect et al., 1980) |
| KN99a | MAT a | (Nielsen et al., 2003) |
| ERB002 | MAT α cdc420∷nat | this study |
| ERB005 | MAT α cdc42∷nat | this study |
| ERB007 | MAT a cdc420∷neo | this study |
| ERB010 | MAT a cdc42∷nat | this study |
| ERB011 | MAT α cdc42∷nat cdc420∷neo | this study |
| ERB012 | MAT α cdc420∷nat CDC420-neo | this study |
| ERB013 | MAT α cdc42∷nat CDC42-neo | this study |
| LK002 | MAT a CDC10-mcherry-neo | (Kozubowski and Heitman, 2009) |
| ERB018 | MAT a cdc420∷nat CDC10-mcherry- neo | this study |
| ERB023 | MAT α cdc42∷nat CDC10-mcherry-neo | this study |
| CSB40 | MAT α ste20∷ura | (Nichols et al., 2004) |
| ERB024 | MAT α ste20∷ura CDC10-mcherry-neo | this study |
| ERB025 | MAT α cdc42∷nat cdc420∷neo CDC42∷CDC420 hyg | this study |
| ERB026 | MAT α cdc42∷nat cdc420∷neo CDC420∷CDC42 hyg | this study |
A cdc42Δ cdc420Δ double mutant (ERB011) was obtained by mating and spore dissection of the single mutants. CDC42 and CDC420 are located on chromosomes 14 and 7 respectively and are therefore unlinked. Random spore analysis revealed that recombination occurred at the expected frequency between the single mutants, with appropriate segregation of genetic markers (mating type, CDC42∷nat allele, and CDC420∷neo allele). However, the cdc42Δ cdc420Δ double mutants were isolated at a lower than expected frequency (1:45), suggesting a synthetic phenotype with respect to growth or spore viability.
Cdc42 paralogs are implicated in thermotolerance and morphogenesis
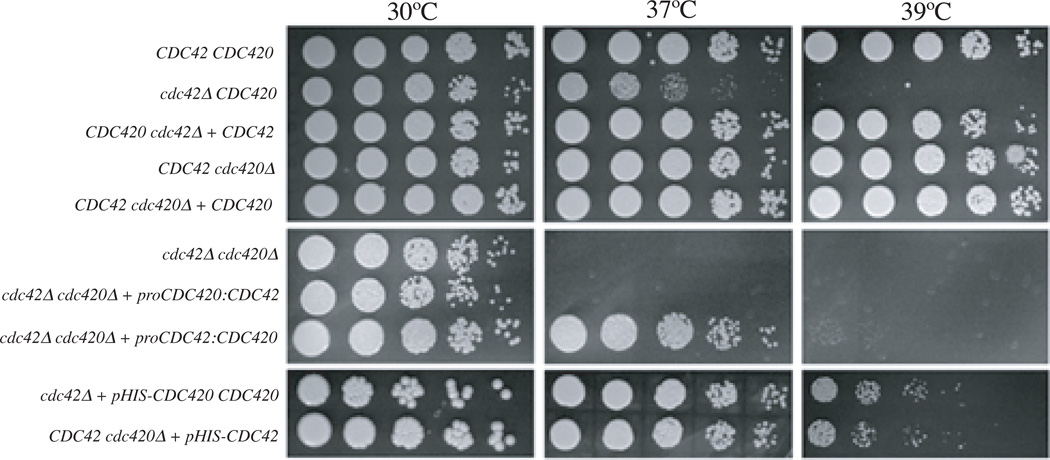
Previous data suggest that the Cdc42 paralogs may function downstream of Ras1 to control yeast cell morphogenesis and growth at elevated temperature. To characterize the specific contributions of the two paralogs to morphogenesis and thermotolerance, we examined the growth of single and double cdc42Δ and cdc420Δ deletion strains at 30°C, 37°C, and 39°C (Figure 2). Strains were spotted in five- fold serial dilutions and incubated on YPD medium for 48 hours at 30°C, 37°C, and 39°C. In this assay the cdc42Δ mutant, and not the cdc420Δ mutant, displayed a decrease in growth relative to wild type at 37°C. At 39°C this difference becomes more pronounced, consistent with ras1Δ and cdc24Δ defects in growth ((Nichols et al., 2007). Reintroduction of the CDC42 gene under its endogenous promoter restored wild type growth at 39°C to the cdc42Δ mutant strain. Similarly, over-expression of CDC420 restored wild type growth at 39°C to cdc42Δ cells. The loss of both CDC42 paralogs had a synthetic effect: the cdc42Δ cdc420Δ double mutant showed a slight decrease in growth at 30°C. At 37°C the double mutant displayed a more severe growth defect than either single mutant, and similar to the growth defect of the ras1Δ mutant at 37°C (Nichols et al., 2007).
Figure 2. Growth defects of cdc42Δ, cdc420Δ, and cdc42Δ cdc420Δ mutants.
Indicated strains were spotted in 5-fold serial dilutions onto solid YPD medium and incubated at 30°, 37°, or 39°C as indicated for 48 hours.
Cdc42 paralogs are required for cytokinesis
Growth defects at high temperature are consistent with previous results suggesting a role for these proteins in Ras1-mediated thermotolerance. Previous experiments demonstrated that over-expression of CDC42 and CDC420 suppressed the ras1Δ and cdc24Δ mutant growth defects at high temperature (Nichols et al., 2007). We confirmed that over-expression of either CDC42 or CDC420 restores growth at 37°C to the ras1Δ mutant (data not shown). Together these data suggest overlapping but non-redundant roles of the Cdc42 and Cdc420 GTPases in C. neoformans morphogenesis, thermotolerance, and Ras1 signaling.
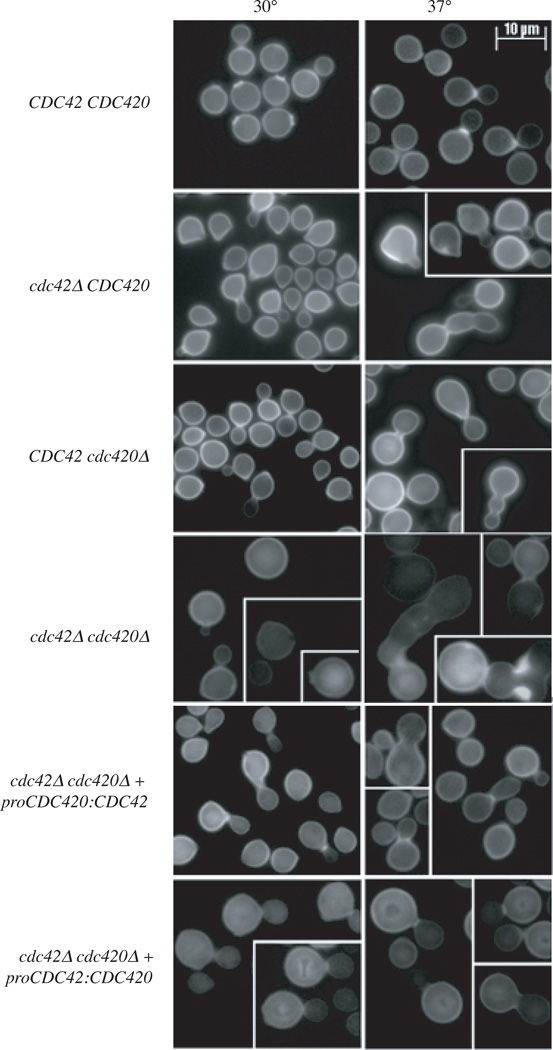
Upon exposure to temperature stress (37°C), ras1Δ and cdc24Δ cells increase in size compared to wild type, and arrest as round, unbudded cells. Similarly, cdc42Δ and cdc420Δ cells are larger than wild type cells when exposed to temperature stress (Figure 3). The cdc42Δ, cdc420Δ, and cdc42Δ cdc420Δ mutants do not simply arrest as unbudded cells. Instead these mutants have morphological defects including elongated buds at high temperature, and evidence of failed cytokinesis (cells with wide bud necks in the case of cdc420Δ mutants, and cells with multiple buds in the case of cdc42Δ and cdc42Δcdc420Δ mutants). Consistent with their more severe growth defects, cdc42Δ cdc420Δ cells are larger than the single mutants at 30°C, and this effect becomes more pronounced after incubation at 37°C (Figure 3).
Figure 3. Cdc42 paralogs are implicated in thermotolerance and morphogenesis.
A) Wild type, cdc42Δ cdc420Δ, cdc42Δ cdc420Δ, proCDC42:CDC420 and proCDC420:CDC42 strains were inoculated into YPD and grown to mid-log phase. Cultures were split and refreshed with media pre-warmed to 30° or 37°C and allowed to grow under the indicated conditions for 24 hours. Cells were fixed and stained with Calcofluor for imaging. (Bar = 10µm).
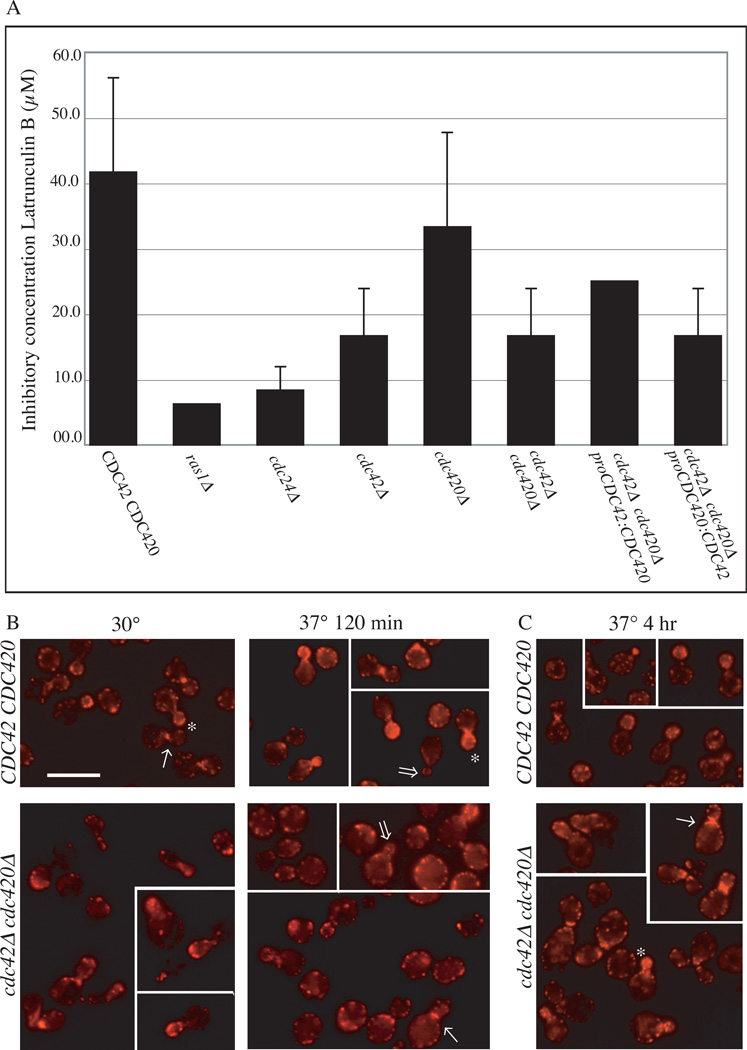
Ras1 pathway defects in high temperature growth and morphogenesis correlate with sensitivity to the actin inhibitory drug latrunculin B. Compared to wild type, ras1Δ and cdc24Δ cells are highly sensitive to latrunculin B when incubated at 30°C (Figure 4a). Consistent with their role as downstream elements of the Ras1 pathway for organization of the actin cytoskeleton, cells deficient for CDC42 or both CDC42 and CDC420 are also more susceptible than wild type cells to this drug.
Figure 4. CDC42 paralogs play a role in actin re-polarization after exposure to stress.
A) Indicated strains were examined in the presence of the actin inhibitor Latrunculin B in a 96 well format. Cells (106ml−1) were incubated in the presence of decreasing concentrations of Latruculin B (100µM to 0µM) for 48 hours (n=3). The concentration of drug that failed to inhibit growth was noted for each strain. B, C) Wild type and cdc42Δ cdc420Δ cells were grown to log phase, and cultures were split and refreshed with pre-warmed media at 30° and 37°C. Cultures were grown at 30° or 37°C. After 120 minutes (B) or 4 hours (C) aliquots were fixed, permeablized, and stained with rhodamine-conjugated phalloidin to reveal actin structures. Budding cells were categorized as polarized or apolar according to previously established criteria (Nichols et al., 2007; Waugh et al., 2002b). (Bar = 10µm)
CDC42 is induced during high temperature growth
The phenotypic changes associated with loss of both CDC42 and CDC420 in the double mutant strongly argue that both genes are expressed and functional. We used semi-quantitative - reverse-transcriptase-PCR and Real-Time PCR to determine relative levels of expression for these genes in wild type and single mutant strains at 30° and 37°C. Although CDC42 and CDC420 share highly similar amino acid and nucleotide sequences, we were able to design primers that differentially amplify the two genes and their transcripts. As demonstrated by semi-quantitative RT-PRC in Figure 5a, the CDC42 gene is transcribed at a higher level than CDC420 in wild type cells growing at 30°C. Moreover, CDC42, but not CDC420, is significantly induced (6-fold) during growth at 37° compared to expression at 30°C (Figure 5b). Expression of CDC42 was only slightly increased in the cdc420Δ background. This up-regulation is consistent with a role for CDC42 in the response to stress. Conversely, CDC420 is not significantly induced at 37°C, and its expression is not up-regulated in the absence of CDC42. The low level of CDC420 expression under both permissive and stress conditions may explain the subtle phenotypes of the cdc420Δ mutant strain. Intriguingly, CDC420 is located near the telomere of chromosome 7, perhaps indicating a gene silencing effect (or trans effect) on its level of expression.
Figure 5. CDC42 paralogs are differentially expressed.
A) Primer pairs specific for CDC420 or CDC42 transcripts were identified and assessed by semi-quantitative RT-PCR in wild type cDNA. B) CDC42 and CDC420 transcripts were detected by real-time PCR in wild type (H99), cdc42Δ, and cdc420Δ strains at 30° and 37°C. Bars represent the fold change in expression between temperature conditions, relative to an internal control (GPD) (n=4). C) Promoter swap constructs were generated and introduced in the double mutant background.
Expression levels are insufficient to account for differences in Cdc42 paralog activity
We performed a “promoter swap” experiment to determine if the differences in CDC42 and CDC420 function are strictly due to different levels of gene expression. As demonstrated in the schematic in Figure 5c, we created constructs in which the CDC42 gene is expressed from the CDC420 promoter (proCDC420:CDC42) and in which the CDC420 gene is expressed from the CDC42 promoter (proCDC42:CDC420). The constructs were transformed into the cdc42Δcdc420Δ double mutant strain and several independent transformants were obtained. Expression levels were assessed by northern analysis and real-time PCR. We chose to study strains with a range of gene expression before focusing on strains in which the CDC42 and CDC420 promoters most closely approximated activity in the wild-type strains.
We first assessed the proCDC42:CDC420 strains for expression level using northern analysis with the CDC420 open reading frame as a probe. As observed in wild-type cells, transcription from the CDC42 promoter was induced at 37°C. A survey of 8 strains with various levels of expression showed that increased expression level correlated with increased rescue of high temperature growth (Supplemental Figure S1). Analysis by real-time PCR identified strain ERB025, in which CDC42 promoter activity was comparable to wild type at 30° and 37°C (less than 2 fold difference). Importantly proCDC42:CDC420 restored growth at 37°C, but was insufficient to restore growth at 39°C to the double mutant, in contrast to the cdc420Δ single mutant, in which CDC42 is intact (Figure 2). Resistance to latrunculin b was also slightly decreased in this strain relative to the cdc420Δ single mutant (Figure 4a). Finally, proCDC42:CDC420 was insufficient to rescue morphological defects of the cdc42Δ cdc420Δ double mutant (Figure 3). ERB025 cells suffered few apparent defects in cytokinesis, but were significantly larger than their cdc420Δ or cdc42Δ counterparts at 30° and 37°C.
Next we assessed the proCDC420:CDC42 strains for expression level by real-time PCR, probing for CDC42. Again we observed a range of expression levels which correlated with growth at high temperature (data not shown). We identified ERB026, in which CDC420 promoter activity was similar to wild type levels (less than two fold difference). Consistent with a model in which induction is required for high temperature growth, proCDC420:CDC42 (ERB026) failed to rescue growth at 37°C (Figure 2) and latrunculin b sensitivity was comparable to that of the double mutant (Figure 4a). However, ERB026 cells were morphologically similar to the cdc420Δ and cdc42Δ single mutants at 30° and 37°C.
It appears that CDC42 is the major allele in C. neoformans during yeast phase growth, while CDC420 encodes a functional paralog that provides a basal level of function sufficient for growth at permissive conditions. Upon exposure to stress conditions, CDC42 transcription is induced, and this up-regulation contributes to growth at high temperature: in the absence of the inducible promoter (cdc42Δ cells and proCDC420:CDC42 cells), C. neoformans is unable to grow under stress conditions. Together these results indicate that expression levels of these genes account for most but not all differences in gene function and suggest that both genes have undergone some degree of sub-functionalization. If differences in transcription level were sufficient to explain the differences in phenotype of the cdc42Δ and cdc420Δ strains, we would expect proCDC42:CDC420 to completely restore the temperature growth and morphology of the double mutant to that of the cdc420Δ mutant. Instead, this strain fails to grow at 39°C, and these cells are significantly larger than either of the single mutant strains.
Cdc42 paralogs aid actin re-polarization under stress conditions
In order to more fully explore the contribution of Cdc42 proteins to morphogenesis and thermotolerance in C. neoformans, we undertook a careful morphological characterization of the cdc42Δ cdc420Δ double deletion mutant. To investigate the organization of actin in these cells, we examined the actin cytoskeleton under stress conditions by staining with rhodamine-conjugated phalloidin. We previously established that the Ras1 protein likely mediates its effects on thermotolerance through its regulation of the actin cytoskeleton, and its latrunculin b sensitivity profile supports this model. As a result, cells deficient in RAS1 or CDC24 fail to efficiently re-polarize their cytoskeleton following exposure to 37°C temperature stress (Nichols et al., 2007; Waugh et al., 2002a). Similar to loss of RAS1 or CDC24, loss of both CDC42 paralogs resulted in defects in actin re-polarization during exposure to temperature stress (Figure 4b, (Nichols et al., 2007)). After 120 minutes of growth at 37°C in liquid culture, cdc42Δ cdc420Δ double mutants demonstrated defective actin polarization to the growing bud compared to wild type cells under the same conditions (25% in cdc42Δ cdc420Δ cells vs 91% of wild type cells, n=100).
Defects in actin polarization do not completely account for defects in morphogenesis of the cdc42Δ cdc420Δ mutant. Four hours after exposure to 37°C, a small but observable fraction of cdc42Δ cdc420Δ mutant cells have polarized actin to the bud tip, and have trans-located actin to contractile rings at bud necks (Figure 4c). Even after 24 hours at 37°C a sub-population of these mutants with significant morphology defects, including multiple unseparated multinucleate buds, will maintain actin structures consistent with budding and cytokinesis (Supplemental Figure 2). Moreover, the cdc42Δ cdc420Δ double mutant is able to propagate at 30°C, indicating that proteins other than Cdc42 are able to direct cell polarity and morphogenesis events under non-stress growth conditions. Despite this actin polarization and recruitment to the bud neck, cells deficient for Cdc42 paralogs demonstrate persistent failures in cytokinesis, the major defect of cdc42Δ cdc420Δ double mutant cells.
CDC42 is required for normal septin localization
Septins are a highly conserved family of proteins shown to play key structural roles in cytokinesis. S. cerevisiae Cdc42 is essential for the assembly of septin rings at the bud neck (Cid et al., 2001). Similarly, C. albicans Cdc42 activity is required for normal septin organization during hyphal development (Cid et al., 2001; Court and Sudbery, 2007). Recently, the C. neoformans septin proteins Cdc3, Cdc11, and Cdc12 have been demonstrated to be required for normal morphogenesis at 30°C and 37°C (Kozubowski and Heitman, 2009). Given the cdc42Δ cdc420Δ double mutant defect in cytokinesis, and the important role septins play in this process, we examined septin organization in the absence of Cdc42 paralogs using a fluorescently tagged Cdc10 septin protein.
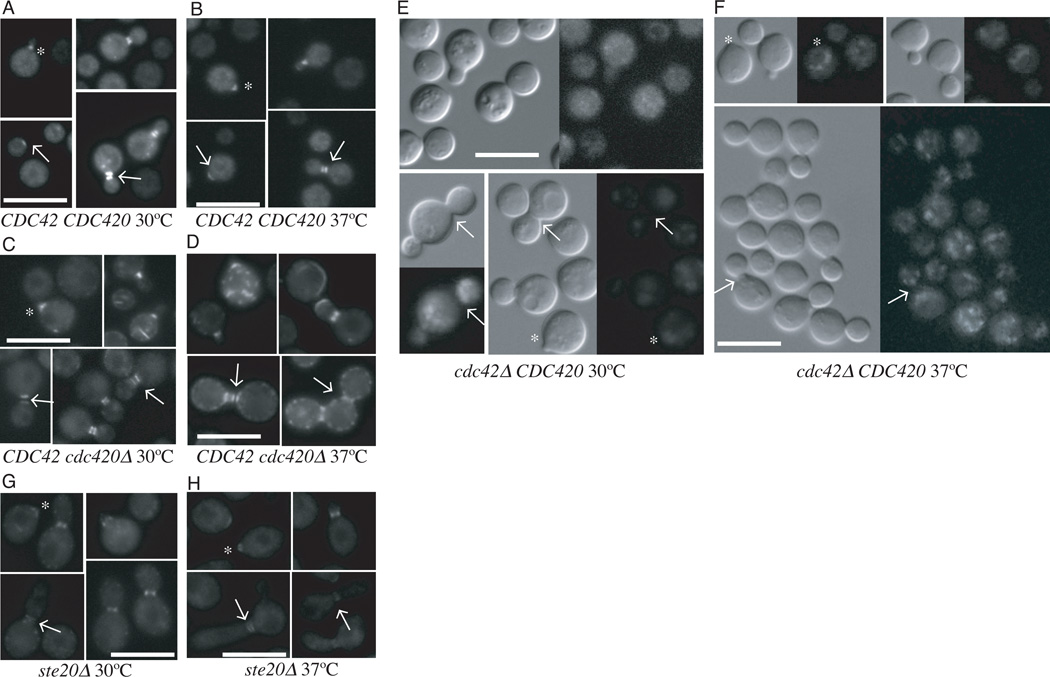
C. neoformans septin proteins Cdc3, Cdc10 and Cdc11, most likely in association with Cdc12, are recruited to the presumptive bud site, where they form filaments and are organized into a ring structure (Kozubowski and Heitman, 2009). We confirmed that at 30°C Cdc10-mCherry localized to the bud neck in wild type cells (Figure 6a). During bud growth, the septin filaments undergo a series of structural rearrangements, forming first an hourglass shaped collar around the neck, and then two rings on either side of the dividing neck.Wildtype cells are able to maintain this program of septin recruitment and reorganization at 37 °C (Figure 6b).
Figure 6. Cdc42 paralogs are involved in septin organization.
The localization of a Cdc10-mCherry construct was examined in wild type (A, B) cdc420Δ (C, D) cdc42Δ (E, F) and ste20Δ (G, H) cells grown at 30° or 37°C respectively. Cells were inoculated in to YPD and grown to log phase. Cultures were split, refreshed with pre-warmed media, and allowed to grow for 4 hours. Cells were gently fixed before imaging as described. (Bar = 10µm)
In the absence of CDC42, there is a complete failure in septin localization (Figure 6e). The Cdc10-mCherry fusion protein fails to localize to nascent buds and contractile necks in cdc42Δ mutant cells, remaining as globular patches throughout the cell cycle at 30°C. Similar defects in septin protein localization are present at 37°C (Figure 6f).
Unlike cdc42Δ mutant cells, and consistent with their more subtle morphological defects, the majority of cdc420ΔC cells have septin localization similar to wild type cells at 30°C (Figure 6c). Cdc10-mCherry localizes to the nascent bud and forms contractile rings at 30°C. A minority of cdc420Δ cells show the aberrant accumulation of Cdc10 aggregates (Figure 6c). At 37°C, the frequency of aggregates increases, and in some cells with normal Cdc10-mCherry localization, septin collars are elongated across the neck (Figure 6d). These results are consistent with a model in which Cdc42 paralogs facilitate cell growth and proliferation by recruiting septins to the nascent bud site and directing their reorganization during budding and cytokinesis.
Cdc42 paralogs do not organize septins through Ste20, the Cla4 homolog
Cdc42 GTPases interact with their targets via CRIB binding domains. It has previously been shown in yeast-2-hybrid assays that the Cla4 homolog CnSte20 interacts with Cdc42 and Cdc420 via this domain (Nichols et al., 2007; Wang et al., 2002). Cells deficient in STE20 are temperature sensitive and exhibit a characteristic defect in cytokinesis at 37°C. In S. cerevisiae, Cla4 has been shown to act in complex with Cdc42 during the initial assembly of the septin ring (Kadota et al., 2004). Moreover, Cla4 plays a role in Rac-mediated septation in a number of filamentous fungi (Leveleki et al., 2004; Rolke and Tudzynski, 2008). We therefore asked if ste20Δ cells have a defect in septin organization similar to cdc42Δ (Figure 6g, h). Surprisingly, these cells show normal septin structures at bud tips and at bud necks at 30°C and 37°C, suggesting that cytokinesis defects of the ste20Δ strain are not due to the same septin organization or localization defects observed in the cdc42Δ or cdc420Δ mutants.
Cdc42 plays a role in sporulation
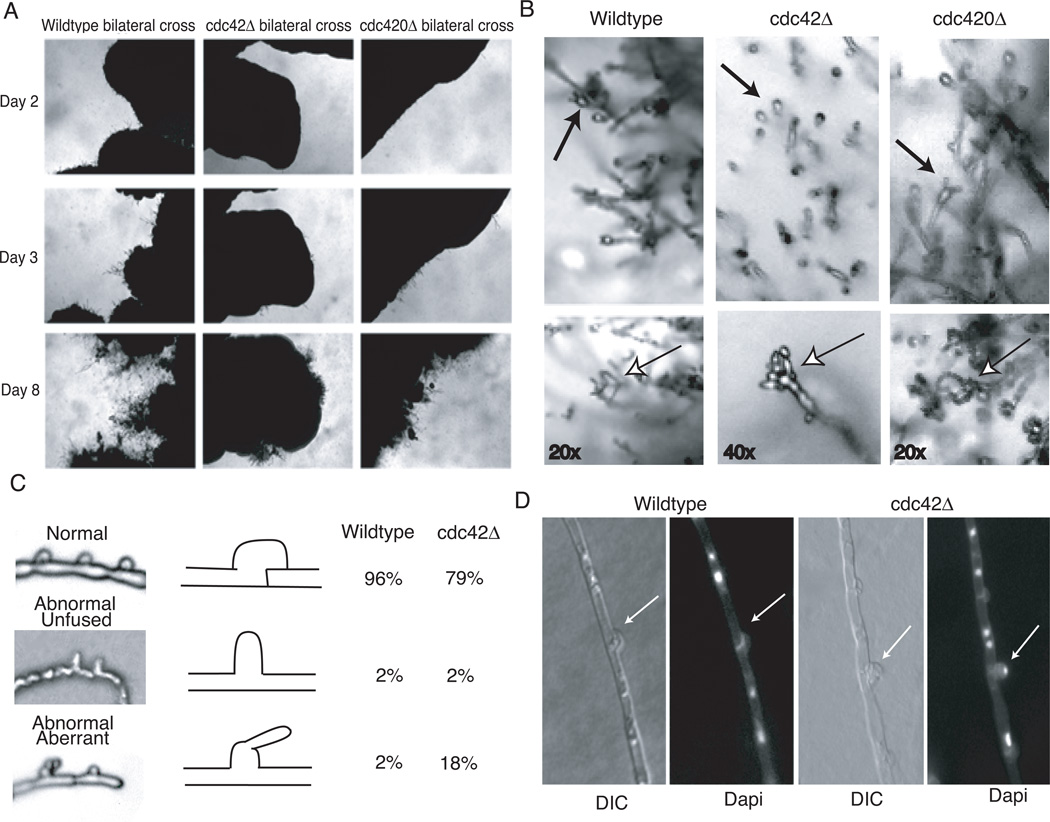
To test the role of CDC42 paralogs in C. neoformans mating, we examined the effects of cdc420Δ and cdc42Δ mutations in sexual crosses (Figure 7). There were no significant defects in unilateral matings, when single deletion mutants were crossed with the congenic wild type strain of the opposite mating type (data not shown). In bilateral crosses of MATα cdc420Δ × MATa cdc420Δ, we observed no significant defects in fusion, filamentation, or the formation of fruiting structures (Figure 7a, b). Interestingly, mating reactions in which both parents were cdc42Δ mutants exhibited multiple defects in the mating process. First, initial filamentation was delayed (Figure 7a). Mating filaments in wild type crosses typically appear within the first 24–48 hours of incubation of V8 mating media, but the mating hyphae of the cdc42Δ bilateral cross were not evident until approximately 72 hours. Even after these initial filaments had emerged, the foci of mating filamentation were less robust than those of wild type crosses. Also, sexual basidiospores were readily observed in wild type crosses by day eight (Figure 7b); however in the cdc42Δ bilateral mutant cross basidia heads were aberrant and basidiospores chains failed to form, even after 3 weeks of incubation. Moreover, although fused clamp connections were observed in bilateral cdc42Δ mating hyphae, there was a significant increase in the incidence of abnormal morphology among clamp cells (Figure 7c).
Figure 7. Cdc42 but not Cdc420 is required for clamp cell formation and sporulation.
A) Wild type × wild type, cdc42Δ × cdc42Δ, and cdc420Δ × cdc420Δ cells were co-cultured on V8 mating medium and allowed to incubate in the dark at room temperature. The same location on each plate was photographed every 24 hours. B) Basidia heads characteristic of each bilateral cross were identified and spore chains were photographed in situ. C) Slide squashes of mating filaments were prepared as described. Clamps were categorized as normal, abnormal unfused, or abnormal aberrant. 100 clamps were counted for each strain. D) Mating plugs were fixed, permeablized, and stained with Dapi to reveal nuclei.
In order to quantify this clamp defect, three classifications of clamp cell were established: normal, abnormal--unfused, and abnormal with aberrant growth. Schematics of the three classes, as well as representative samples, can be seen in Figure 7c. Clamp connections were counted and classified in wild type and cdc42Δ bilateral mating reactions that had incubated for two weeks under normal mating conditions. Clamp connections in cdc42Δ bilateral matings were more likely to be aberrant than in wild type matings (20% in cdc42Δ bilateral vs. 4% in wt cross). Intact fused clamp connections are required for normal nuclear migration along the growing mating hypha, and defects in clamp cell formation might be expected to result in alterations in nuclear delivery to the terminal basidia, the site of nuclear fusion, meiosis, and sporulation. The high percentage of aberrant clamp connections in the cdc42Δ bilateral mating reactions might therefore explain the failure of sexual basidiospore formation.
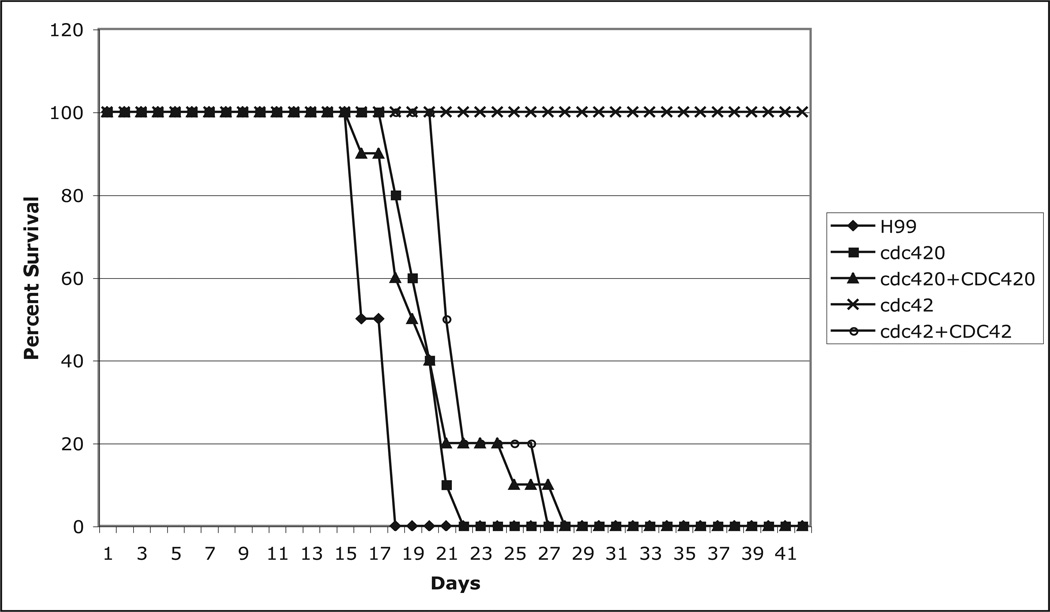
Cdc42, but not Cdc420, is required for C. neoformans pathogenicity in a mouse model of infection
Thermotolerance is essential for the pathogenicity of C. neoformans, and several temperature-sensitive mutants display reduced virulence in animal models of cryptococcal infection (Alspaugh et al., 2000; Odom, 1997). We therefore tested the role of CDC42 paralogs in C. neoformans pathogenicity using the murine inhalation model of cryptococcosis (Cox et al., 2003). A/Jr mice (10 mice per strain) were inoculated by nasal inhalation (5×106 cells/mouse) with the wild type, cdc42Δ, cdc42Δ+CDC42 reconstituted, cdc420Δ, or cdc420Δ +CDC420 reconstituted strain. By 28 days after infection, all mice infected with wild type or either of the reconstituted strains had succumbed to the infection (Figure 8). Mice infected with cdc420Δ mutant strains died at the same rate as mice infected with the wild type strain, indicating that the subtle defect in high temperature growth in this strain did not effect its virulence in this model. By contrast, mice infected with the cdc42Δ mutant strain survived to the completion of the experiment. Mice in this group appeared well throughout the assay, and gained weight at the same rate as mice in the uninfected control group, indicating that CDC42, like RAS1 and CDC24, is essential for virulence in C. neoformans.
Figure 8. Cdc42 but not Cdc420 is required for virulence.
A/Jcr mice (10 per strain) were inoculated intra-nasally with the indicated strains (5 × 105 cells). Mice were monitored daily for weight loss and neurological symptoms and sacrificed at predetermined clinical endpoints, which correlate with an imminent lethal infection.
Discussion
The C. neoformans genome encodes two functional Cdc42 paralogs
We have shown here that both CDC42 and CDC420 encode true Cdc42 homologs. The two paralogs contain all predicted Cdc42 domains, and share the canonical TQXD motif at position 115–118 which distinguishes them from Rac homologs (Chen et al., 1993; Johnson, 1999; Vallim et al., 2005). Loss of each Cdc42 paralog individually leads to distinct phenotypes, although the cdc420Δ phenotype is subtle, and loss of both paralogs generates an additive phenotype. Our analysis of CDC42 and CDC420 expression suggests that this difference in phenotype is partially related to gene dosage, with CDC42 expressed at much higher levels than than CDC420, and further induced in response to high temperature stress. Moreover, promoter swap experiments demonstrated that expression of CDC42 at CDC420 levels rescues morphological defects at 30°C, but is insufficient to rescue defects at 37°C, indicating that this induction is required for high temperature growth.
In addition to differences in expression level, the Cdc42 paralogs may have begun to undergo sub-functionalization: The two Cdc42 proteins maintain conserved residues in key regulatory domains and clearly control similar events in actin polarization and morphogenesis. However, there are differences between the two sequences, including alterations within the membrane localization domain and the Rho-insert/Rdi1-binding domain (Johnson, 1999). These differences in primary structure may contribute to differences in protein function. For example, differences in septin defects in the single mutants point to different roles for Cdc42 and Cdc420 in septin recruitment and organization. Additionally, the promoter swap experiments indicate that proCDC42:CDC420 does not fully complement CDC42 mutant defects.
Interestingly, the CDC42 gene tree reflects morphology and Cdc42 function rather than species relationship (Figure 1b). Cdc42 proteins of yeast phase fungi group together, while filamentous phase ascomycetes and the fission yeast form a clade with basidiomycetes. This may reflect an emerging theme in the role of Rho-GTPases in fungal morphogenesis and certainly merits further investigation (Momany, 2002).
C. neoformans is unusual among eukaryotes in that it has duplicated many elements of the Ras pathway, including Ras1/Ras2, Rac1/Rac2, Cdc42/Cdc420, and Ste20/Pak1 (Alspaugh et al., 2000; Nichols et al., 2007; Vallim et al., 2005; Wang et al., 2002; Waugh et al., 2002a). It is interesting to note that CDC420 is located in a subtelomeric region likely to be regulated by epigenetic silencing (Barton and Kaback, 2006). Although epigenetic regulation has not yet been described in C. neoformans, we anticipate that it will play a role in the transcriptional regulation of other duplicated paralogs in C. neoformans.
C. neoformans Cdc42 paralogs participate in Ras1-mediated response to stress
We recently demonstrated that the elevated temperature encountered within the infected host (37°C) results in a temporary depolarization of the C. neoformans actin cytoskeleton (Nichols et al., 2007). Following depolarization, wild type cells rapidly repolarize actin and continue budding. In contrast, defects in the Ras1 pathway, including loss of Ras1 or the Cdc42-GEF Cdc24, result in impaired actin re-polarization at high temperature. However, the precise mechanism by which Ras1 helps to establish yeast polarity in the presence of cell stress remains unclear. Additionally, the role of C. neoformans Ras1 in cell polarity is further suggested by the observation that this protein is required for the growth of this fungus as hyphae, a process requiring continual re-establishment of cell polarity (Waugh et al., 2002a). In a number of fungi, including S. pombe, Colletotrichum trifoli, and P. marneffei, Ras proteins act upstream of the Rho-GTPases, providing a plausible mechanism linking Ras and cell polarity (Boyce et al., 2005b; Chang et al., 1994; Chen et al., 2006; Li and Chang, 2003a). Consistent with its role as a global regulator acting through the Rho-GTPases, C. neoformans ras1Δ morphogenesis defects can be complemented by over-expression of Cdc24, Cdc42 paralogs, or Rac1(Nichols et al., 2007; Vallim et al., 2005). Similar to the Ras1 and Cdc24 proteins, the C. neoformans Cdc42 paralogs are not required for growth in the absence of stress. However, the ras1Δ, cdc24Δ, and cdc42Δ cdc420Δ mutants display similar defects in growth at elevated temperature and susceptibility to the actin inhibitor latrunculin b. Together these data support a model in which Ras proteins control cell morphology through the highly conserved Cdc24/Cdc42 signaling pathway.
Cdc42 is required for septin organization and functions independently of the Cla4 homolog Ste20
Morphological defects of the cdc42Δ cdc420Δ mutant include elongated buds, aberrant bud necks, and failure to complete cytokinesis. Consistent with work in S. cerevisiae linking defects in bud growth (including aberrant neck morphology and elongated bud tips) to defects in septin organization, (Caviston et al., 2003; Gladfelter et al., 2002; Gladfelter et al., 2005; Iwase et al., 2006) we observed defects in septin assembly in the C. neoformans cdc42Δ and cdc420Δ mutants. Defects in septin organization may also explain the sporulation defects we observed in cdc42Δ bilateral mating. Kozubowski and Heitman (2009) have shown that septins are localized to areas of spore emergence in the basidia head during sporulation and that loss of septins results in sporulation defects.
In S. cerevisiae and in a number of filamentous fungi, the PAK kinase Cla4 has been identified as a downstream target of Cdc42/Rac GTPases responsible for cytokinesis. ScCla4p phosphorylation of the septins Cdc10 and Cdc3 is thought to stabilize the septin collar (Versele and Thorner, 2004). C. purpurea Cla4 interacts specifically with activated Rac, and its loss has adverse effects on invasive polarized growth and septation, similar to defects found in CpracΔ mutants (Rolke and Tudzynski, 2008).
The C. neoformans Cla4 homolog Ste20 plays a role in polarized growth and cytokinesis and interacts with Cdc42 and Cdc420 in yeast-two-hybrid assays (Nichols et al., 2007; Wang et al., 2002). Ste20 cells also have mating defects related to mis-sorting of nuclei, similar to our proposed mechanism for cdc42Δ defects in sporulation (Nichols et al., 2004). We therefore asked whether ste20Δ cells had defects in septin localization. Surprisingly, we found no defects in Cdc10 localization under any conditions tested, indicating that Cdc42 paralogs do not mediate septin organization via Ste20, although it remains likely that Cdc42 requires the activity of Ste20 for some other function related to cytokinesis.
Cdc42 and Rac1 share overlapping and distinct roles related to polar growth and septation
In S. cerevisiae and S. pombe Cdc42 is the center of polarity and is essential for both the organization of septins and the polarization of actin (Ayscough et al., 1997; Gladfelter et al., 2002; Irazoqui et al., 2003). In other eukaryotes, these two functions are shared between Cdc42 and the Rho-GTPase, Rac. The division of labor by these two highly related proteins has been adapted to different purposes in different filamentous fungi (Bassilana and Arkowitz, 2006; Boyce et al., 2005b; Gorfer et al., 2001; Hurtado et al., 2000; Mahlert et al., 2006; Rolke and Tudzynski, 2008; Virag et al., 2007). During hyphal growth in the dimorphic ascomycete P. marneffei, the Cdc42 homolog co-ordinates actin polarization while the Rac1 homolog co-ordinates septation events (Boyce et al., 2005a). In contrast, in the basidiomycete U. maydis Cdc42 plays the predominant role in cytokinesis, and Rac1 co-ordinates polar growth (Mahlert et al., 2006). Similarly, C. neoformans Rac1 has been previously implicated as the major regulator of hyphal growth (Vallim et al., 2005).
In contrast to the ascomycetes, C. neoformans Cdc42 is not absolutely required for actin polarization. Our results have shown that in the absence of Cdc42 paralogs, C. neoformans is able to polarize actin to buds under permissive conditions, and to some degree is able to form normal actin structures even under stress conditions. However, cells deficient for both paralogs have a significant defect in actin re-polarization after exposure to stress. It is possible that Cdc42 paralogs play a role in actin organization during budding under non-stress conditions, and that Rac1 rescues defects in actin localization in the absence of Cdc42 paralogs. This may be a non-native function of Rac1, particularly given the observation that the rac1Δ strain is able to bud normally (Vallim et al., 2005). Alternatively, it may be that in rac1Δ cells Cdc42 paralogs are able to assume non-native functions and coordinate the actin cytoskeleton to rescue defects in polar growth. The principle that Rac and Cdc42 proteins share overlapping roles in microbial morphogenesis is not without precedent. For example in P. marneffei, where Rac proteins play a predominant role in hyphal septation, dominant active Cdc42 is able direct septation in the absence of Rac1 during hyphal growth (Boyce et al., 2005b). However, a second level of complexity in C. neoformans may offer an alternate explanation: C. neoformans has a putative Rac1 paralog Rac2 (CNAG_05998.2), whose role in morphogenesis has yet to be investigated. Interestingly, Rac2 is also located in a subtelomeric region.
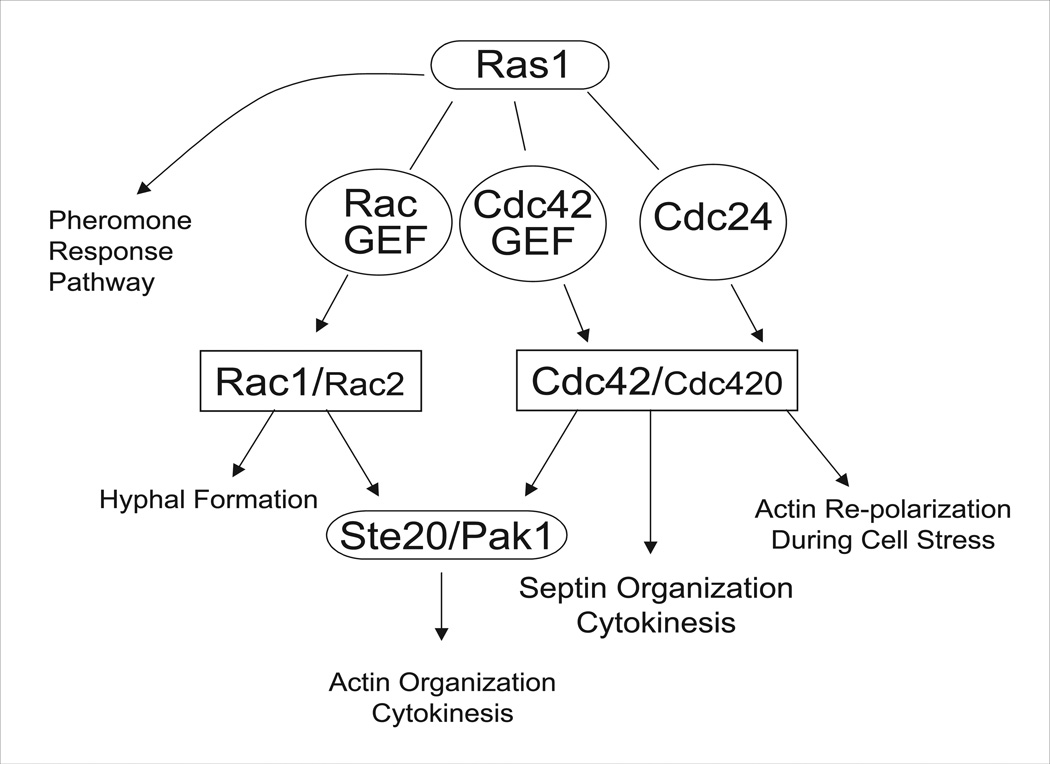
We therefore propose a model in which Cdc42 and Rac proteins coordinately regulate polar growth, and Cdc42 paralogs direct the organization of septins in C. neoformans (Figure 9). The components of this proposed signaling pathway appear to work together to mediate morphological events in response to cell stress. However, it is evident that the phenotypes of the cdc24Δ and cdc42Δ cdc420Δ strains are not identical. The cdc24Δ mutant, like the ras1Δ mutant, arrests at high temperature as single unbudded enlarged cells with depolarized actin. In contrast, the cdc42Δ and the cdc42Δ cdc420Δ mutants have severe morphological defects at high temperature, including cytokinesis defects. Two possible models may explain this observation. In the first, Cdc24 is required to enhance the activity of Cdc42 paralogs to a level sufficient to rescue morphological defects at 39°C, and differences in phenotype between the cdc24Δ and cdc42Δ cdc420Δ strains reflect baseline activity of the Cdc42 paralogs in the absence of upstream activation in the cdc24Δ strain. In the second model Cdc24 acts as a Cdc42 GEF specifically to regulate actin polarization in response to stress, and that other Cdc42-specific GEFs direct Cdc42’s role in cytokinesis. Consistent with this model, C. neoformans encodes multiple putative Cdc42 GEFs, which may facilitate such specificity (Nichols et al., 2007). In S. pombe two Cdc42 GEFs, Scd1 and Gef1, play distinct and overlapping roles in morphogenesis, cell cycle, and cytokinesis, and specify Cdc42 activation of downstream targets (Chang et al., 1994; Coll et al., 2003; Li and Chang, 2003b).
Figure 9. Proposed C. neoformans Ras1 pathway for growth and thermotolerance.
Ras1 regulates mating, morphogenesis, and thermotolerance via a branched signaling pathway. In this work, we have investigated the contribution of Cdc42 paralogs to morphogenesis and thermotolerance. Based on data presented here, as well as previously published work, we propose a pathway in which Ras1 signals through multiple Rho-GTPases: Cdc42, Cdc420, Rac1, and Rac2; with Cdc42 and Rac1 playing the major roles, and Cdc420, and possibly Rac2, playing minor roles. Rac1 primarily is required for hyphal morphogenesis, and may act through Ste20 and Pak1 to organize the actin cytoskeleton during cytokinesis. Cdc42, and to a lesser extent Cdc420, organize the septins during cytokinesis, and play a role in actin organization during growth under stress conditions. We additionally propose the existence of multiple as yet uncharacterized GEFs, which may provide specificity to Ras1 Rho-GTPase-mediated signaling.
Additionally, it is possible that the Ras1, Cdc24, and Cdc42 proteins do not act in a simple linear pathway, and that increasing layers of complexity facilitate different functions of the interacting signaling elements. There is growing evidence in other species to suggest that Cdc24 mediates its effect on polarity by facilitating the formation of complexes between Cdc42 and downstream effectors. For example, in S. pombe morphology and cytokinesis are regulated by a multi-protein complex composted of Ras1, Cdc42, the Cdc42-GEF Scd1, the alternate GEF Scd2, and the Ste20 homolog Shk1 (Chang et al., 1999; Chang et al., 1994; Hlubek et al., 2008; Li and Chang, 2003b). In U. maydis Rac proteins that have been engineered with Cdc42-like localization can rescue Cdc42 defects, pointing to a role for localization in function (Hlubek et al., 2008). Finally, although Cdc42 function has classically been understood to be Ras-like, switching from an off to an on state upon binding of GTP, recent work in S. cerevisiae suggests a model in which Cdc42 activity is analogous to that of the translation elongation factor Ef-Tu (Gladfelter et al., 2002; Irazoqui et al., 2003). In this model, GTP-bound Cdc42 forms a complex with its downstream effectors, and GTP hydrolysis results in a conformational change in the entire complex. In C. neoformans it is possible that the diversity of GEFs and GEF targets (Cdc42 and Cdc420, as well as Rac1 and Rac2), coupled with similar alternate modes of action, provide this organism with previously unrecognized flexibility in directing the effects of key polarity and cytoskeletal organization proteins under diverse growth conditions. Here we show that C. neoformans cells deficient for both Cdc42 paralogs (cdc42Δ cdc420Δ) display a number of morphological defects, including increased cell size, abnormal bud morphology, and a failure to complete cytokinesis. Consistent with these morphological defects, these cells failed to grow at host physiological temperatures, These phenotypic changes are consistent with known functions of Cdc42 proteins, including the establishment of a new bud site, and the direction of septin localization in other species (Etienne-Manneville, 2004). The accentuation of mutant phenotypes at elevated temperature is also consistent with prior observations in Ras signaling in S. cerevisiae and C. neoformans (Ho and Bretscher, 2001; Nichols et al., 2007). However, C. neoformans also demonstrates a unique interplay of multiple Cdc42 and Rac proteins, allowing flexibility of the signaling pathways that coordinate morphogenesis and stress response in this human pathogen.
Experimental Procedures
Nomenclature
The genes CDC42 and CDC420 were previously named DCH2 (Genbank ID: DQ991434) and CDC42 (Genbank ID: DQ991433) respectively (Nichols et al., 2007). As the major paralog, DCH2 has been renamed CDC42. The gene previously identified as CDC42 has been renamed CDC420 to reflect it more limited role in C. neoformans biology. Accession numbers for sequences used in the phylogenetic analysis are as follows: C. neoformans CDC42 XM_572208, CDC420 XM_571459; C. gattii CDC42 XM_767441, CDC420 XM_769683; U. maydis AF463452; C. cinerea XM_001833721; C. albicans XM_706814; C. glabrata XM_446201; P. marneffei AF330694; A. fumigatus NC_007195; S. pombe NC_003424; S. cerevisiae NC_001144.
Strains, media, and growth conditions
C. neoformans strains used in this study are listed in Table I. Strains were incubated on YPD medium (Sherman, 1991) or V8 mating medium (Kwon-Chung et al., 1992). ERB011 was obtained by mating ERB005 with ERB007 and dissecting the resulting spores. ERB018, ERB023, and ERB024 were generated by mating LK001 with ERB002, ERB010, or CSB40 respectively, and dissecting spores.. Mating assays were performed by co-culturing strains of opposite mating type on V8 mating medium in the dark at 25°C. For morphogenesis experiments, cells were inoculated into liquid YPD and grown to mid-log phase at 30°C, shaking at 150 RPM. Cultures were split and refreshed with media pre-warmed to 30° or 37°C, then incubated at the indicated temperature at 150 RPM.
Phylogeny
Genbank accession numbers for sequences used in the phylogenetic analysis are as follows: C. neoformans CDC42 XM_572208, CDC420 XM_571459; C. gattii CDC42 XM_767441, CDC420 XM_769683; U. maydis AF463452; C. cinerea XM_001833721; P. marneffei AF330694; A. fumigatus NC_007195; C. albicans XM_706814; C. glabrata XM_446201; S. pombe NC_003424; S. cerevisiae NC_001144. Sequences were aligned using MUSCLE 3.7, and the alignment checked and manually edited using MEGA 4 (Edgar, 2004; Tamura et al., 2007). Bayesian inference analyses were carried out using MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003). Prior default settings and the model of nucleotide substitution estimated by MrModeltest 2.3 (SYM+I+G) were used. Run settings were 1.5 million generations, four chains, a burnin of 3750 generations and sampling frequency of every 100th generation (Nylander, 2004). A maximum likelihood heuristic search was carried out using PAUP, with 100 random addition sequence replicates, TBR branch swapping, and the likelihood settings for the best-fit model estimated by Modeltest (TrNef+I+G) (Posada and Crandall, 1998; Swofford, 2003). Nonparametric bootstrapping with 1000 pseudo-replicates was used to obtain statistical support values for each node.
Molecular biology
To generate cdc42Δ and cdc420Δ deletion mutants using dominant selectable markers, PCR overlap extension was used to replace the entire CDC42 and CDC420 open reading frames with the neomycin or nourseothricin resistance cassettes in the H99α or KN99a strains as described (Fraser et al., 2003; McDade and Cox, 2001). Genomic integration was performed using the biolistic transformation method as described (Davidson et al., 2000; Toffaletti et al., 1993). Deletion strains were confirmed by PCR, Southern blot analysis, and RT-PCR. Primers used for PCR overlap extension include the following (5’ to 3’, sequence for NAT gene underlined): CDC42 Left flank: AA1468: GCCAGGGGTTGCACCGGGA and AA1409: GTCATAGCTGTTTCCTGTGTCTGCATGGTTGGCTAGG Right flank AA1412: CTGGCCGTCGTTTTACGTGCCTCATCCTTTGAAGAC and AA1469: TCGCTGTACATCGTCGAATC; sequence for NAT gene for cdc42∷NAT allele: AA1410: CCTAGCCAACCATGCAGACACAGGAAACAGCTATGAC and AA1411: GTCTTCAAAGGATGAGGCACGTAAAACGACGGCCAG; CDC420 Left flank: AA1466: AGAGAGGGGGAGTGGAGGTA; and AA1405: GTCATAGCTGTTTCCTGTATCTCTTAAAGTTGCGGGG Right flank AA1408: CTGGCCGTCGTTTTACGCTTGATCCTCTAGTACAC and AA1467: TGCAATCCTCGAACACTACG; NAT gene for cdc420∷NAT allele: AA1406: CCCCGCAACTTTAAGAGATACAGGAAACAGCTATGAC and AA1407: GTGTACTAGAGGATCAAGCGTAAAACGACGGCCAG. Promoter swapped constructs were generating using the following primers and co-transformed with pHYG7KB1 carrying a Hygromycin resistance cassette (5’ to 3’, sequence for open reading frame underlined): proCDC420:CDC42 Left flank AA1466: AGAGAGGGGGAGTGGAGGTA and AA1840: CGACACACTTGATTGTCTGCATTATCTCTTAAAGTTGCGG; Right flank AA1842: GTGCCTCATCCTTTGATACACATATATTAGATTTTTTCTC and AA1467: TGCAATCCTCGAACACTACG; ORF AA1839: CCGCAACTTTAAGAGATAATGCAGACAATCAAGTGTGTCG and AA1841: GAGAAAAAATCTAATATATGTGTATCAAAGGATGAGGCAC. proCDC42:CDC420 Left flank AA1468: GCCAGGGGTTGCACCGGGA and AA1836: ACACATTTGATAGTCTGCATGGTTGGCTAGGAGGGTTGTGG; Right flank AA1837: CCAAAAAGTGCTTGATCCTCTAGACCCCCTATGGCGACTAAG and AA1469: TCGCTGTACATCGTCGAATC; ORF AA1835: CCACAACCCTCCTAGCCAACCATGCAGACTATCAAATGTGT and AA1838: CTTAGTCGCCATAGGGGGTCTAGAGGATCAAGCACTTTTTGG.
Microscopy
Differential interference microscopy (DIC) and fluorescent images were captured with a Zeiss Axio Imager A1 fluorescent microscope equipped with an AxioCam MRM digital camera. Cells were fixed with 9% microfiltered formaldehye for 10 minutes, washed three times with 1XPBS, permeabilized with 1% Triton-PBS for 10 minutes, and washed three times with 1XPBS. Cells were stained with Calcofluor or Rhodamine conjugated Phalloidin. For cells containing the mCherry construct, localization was verified in live cells before fixing. Invitrogen Slow-Fade was added to slides to preserve the signal. Agar plugs of mating filament were fixed with 9% microfiltered formaldehye for 30 minutes, washed three times with 1XPBS, permeabilized for 30 minutes in 1% Triton-PBS, and washed three times with 1XPBS. Filaments were visualized by slide squash.
RNA extraction, cDNA preparation, and RT-PCR
Strains were grown to mid-log phase in YPD at 30°C, split and refreshed with pre-warmed media, then grown at 30° or 37°C for 120 minutes. Cells were collected by centrifugation and flash frozen on dry ice. Total RNA was extracted from lyophilized cells using the Qiagen RNA extraction kit and the “Purification of total RNA from plant cells, tissue, and filamentous fungi” protocol (2006). cDNA was prepared using the Clonetech Advantage RT-for-PCR kit (2006). Primer specificity was verified by qPCR in wild type and deletion strains. RT-PCR was performed as described, with annealing at 50°C (Cramer et al., 2006). Primers used were as follows: CDC42: Forward: AA778: CGTCCCCGCACTTATTGTC; Reverse: AA782: AGTCGCCATAGGGGGTTCTAAT. CDC420 Forward: AA1768: TTTGAGGGATGATCCAAAGCA; Reverse: AA1769: CATTCTTCAACCCCTTTTGC. GPD Forward: AA301: AGTATGACTCCAACAATGGTCG; Reverse: AA302: AGACAAACATCGGAGCATCAGC.
Animal Experiments
Using the murine inhalation model of systemic cryptococcosis, female A/Jcr mice were inoculated intra-nasally with 5 × 105 C. neoformans cells, as previously described (Cox et al., 2000). Briefly, groups of 10 mice were inoculated with one of five strains: H99 (CDC42 CDC420 wild type), ERB002 (cdc420Δ), ERB005 (cdc42Δ), ERB012 (cdc420Δ + CDC420 reconstituted), or ERB013 (cdc42Δ +CDC42 reconstituted). Mice were observed daily for signs of infection. Animals were sacrificed at predetermined clinical endpoints correlating with an imminent lethal infection. The statistical significance in the difference between the survival curves of the animals inoculated with each strain was evaluated using the log-rank test (JMP software; SAS Institute, Cary, NC). All studies were performed in compliance with institutional guidelines for animal experimentation.
Supplementary Material
Acknowledgements
This work was supported by PHS grants AI063242 and AI050128. ERB is supported by the Duke University Program in Genetics and Genomics. We thank Joseph Heitman for advice and support. We also thank the Fungal Genomes Initiative at the Broad Institute for sequence data. We acknowledge Diana Norton for her contribution to this work.
References
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. Cdc42 and Cdc43, Two additional Genes Involved in Budding and Establishment of Cell Polarity in the Yeast Saccharomyces cerevisiae. J Cell Biology. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans . Mol Microbiol. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton AB, Kaback DB. Telomeric silencing of an open reading frame in Saccharomyces cerevisiae. Genetics. 2006;173:1169–1173. doi: 10.1534/genetics.106.058420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M, Blyth J, Arkowitz RA. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryotic Cell. 2003;2:9–18. doi: 10.1128/EC.2.1.9-18.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M, Arkowitz RA. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryotic Cell. 2006;5:321–329. doi: 10.1128/EC.5.2.321-329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmer C, Böhmer M, Bölker M, Sandrock B. Cdc42 and the Ste20-like kinase Don3 act independently in triggering cytokinesis in Ustilago maydis. J Cell Sci. 2008;121:143–148. doi: 10.1242/jcs.014449. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, Hynes MJ, Andrianopoulos A. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J Cell Sci. 2003;116:1249–1260. doi: 10.1242/jcs.00319. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, Chang H, D’Souza CA, Kronstad JW. An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize. Eukaryotic Cell. 2005a;4:2044–2056. doi: 10.1128/EC.4.12.2044-2056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce KJ, Hynes MJ, Andrianopoulos A. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Mol Microbiol. 2005b;55:1487–1501. doi: 10.1111/j.1365-2958.2005.04485.x. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Longtine MS, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Molecular Biology of the Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E, Bartholomeusz G, Pimental R, Chen J, Lai H, Wang L, Yang P, Marcus S. Direct binding and In vivo regulation of the fission yeast p21-activated kinase shk1 by the SH3 domain protein scd2. Mol Cell Biol. 1999;19:8066–8074. doi: 10.1128/mcb.19.12.8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EC, Barr M, Wang Y, Jung V, Xu HP, Wigler MH. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- Chen C, Ha YS, Min JY, Memmott SD, Dickman MB. Cdc42 is required for proper growth and development in the fungal pathogen Colletotrichum trifolii. Eukaryotic Cell. 2006;5:155–166. doi: 10.1128/EC.5.1.155-166.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lim HH, Lim L. The CDC42 homologue from Caenorhabditis elegans. Complementation of yeast mutation. J Biol Chem. 1993;268:13280–13285. [PubMed] [Google Scholar]
- Cid VJ, Adamiková L, Sanchez M, Molina M, Nombela C. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 2001;147:1437–1450. doi: 10.1099/00221287-147-6-1437. [DOI] [PubMed] [Google Scholar]
- Coll PM, Trillo Y, Ametzazurra A, Perez P. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol Biol Cell. 2003;14:313–323. doi: 10.1091/mbc.E02-07-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H, Sudbery P. Regulation of Cdc42 GTPase activity in the formation of hyphae in Candida albicans. Mol Biol Cell. 2007;18:265–281. doi: 10.1091/mbc.E06-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, Harrison T, McDade H, Taborda C, Heinrich G, Casadevall A, Perfect JR. Superoxide Dismutase Influences the Virulence of Cryptococcus neoformans by Affecting Growth within Macrophages. Infection and Immunity. 2003;71:173–180. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer KL, Gerrald Q, Nichols CB, Price MS, Alspaugh JA. Transcription factor Nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryotic Cell. 2006;5:1147–1156. doi: 10.1128/EC.00145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Moore TD, Odom AR, Heitman J. Characterization of the MFalpha pheromone of the human fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2000;38:1017–1026. doi: 10.1046/j.1365-2958.2000.02213.x. [DOI] [PubMed] [Google Scholar]
- Douglas LM, Alvarez FJ, McCreary C, Konopka JB. Septin function in yeast model systems and pathogenic fungi. Eukaryotic Cell. 2005;4:1503–1512. doi: 10.1128/EC.4.9.1503-1512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42--the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Findley K, Rodriguez-Carres M, Metin B, Kroiss J, Fonseca A, Vilgalys R, Heitman J. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryotic Cell. 2009 doi: 10.1128/EC.00373-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryotic Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Xing J, Leto TL, Zheng Y. Trp(56) of rac1 specifies interaction with a subset of guanine nucleotide exchange factors. J Biol Chem. 2001;276:47530–47541. doi: 10.1074/jbc.M108865200. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Bose I, Zyla TR, Bardes EG, Lew DJ. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156:315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Kozubowski L, Zyla TR, Lew DJ. Interplay between septin organization, cell cycle and cell shape in yeast. J Cell Sci. 2005;118:1617–1628. doi: 10.1242/jcs.02286. [DOI] [PubMed] [Google Scholar]
- Gorfer M, Tarkka MT, Hanif M, Pardo AG, Laitiainen E, Raudaskoski M. Characterization of small GTPases Cdc42 and Rac and the relationship between Cdc42 and actin cytoskeleton in vegetative and ectomycorrhizal hyphae of Suillus bovinus. Mol Plant Microbe Interact. 2001;14:135–144. doi: 10.1094/MPMI.2001.14.2.135. [DOI] [PubMed] [Google Scholar]
- Hlubek A, Schink K, Mahlert M, Sandrock B, Bölker M. Selective activation by the guanine nucleotide exchange factor Don1 is a main determinant of Cdc42 signalling specificity in Ustilago maydis. Mol Microbiol. 2008;68:615–623. doi: 10.1111/j.1365-2958.2008.06177.x. [DOI] [PubMed] [Google Scholar]
- Ho J, Bretscher A. Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Molecular Biology of the Cell. 2001;12:1541–1555. doi: 10.1091/mbc.12.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado CA, Beckerich JM, Gaillardin C, Rachubinski RA. A rac homolog is required for induction of hyphal growth in the dimorphic yeast Yarrowia lipolytica. J Bacteriol. 2000;182:2376–2386. doi: 10.1128/jb.182.9.2376-2386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Iwase M, Luo J, Nagaraj S, Longtine MS, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in …. Journal of Cell Biology. 1990 doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Molecular Biology of the Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Heitman J. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of C. neoformans in print. 2009 doi: 10.1111/j.1365-2958.2009.06983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Wickes BL, Stockman L, Roberts GD, Ellis D, Howard DH. Virulence, serotype, and molecular characteristics of environmental strains of Cryptococcus neoformans var. gattii. Infect Immun. 1992;60:1869–1874. doi: 10.1128/iai.60.5.1869-1874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarga A, Valentin F, Andersson M, Lopez R. Web Services at the European Bioinformatics Institute. Nucleic Acids Research Web Services Issue. 2007 doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas DY. Virulence and hyphal formation of Candida albicans require the Ste20p–like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- Leveleki L, Mahlert M, Sandrock B, Bölker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol Microbiol. 2004;54:396–406. doi: 10.1111/j.1365-2958.2004.04296.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Chang E. Schizosaccharomyces pombe Ras1 effector, Scd1, interacts with Klp5 and Klp6 kinesins to mediate cytokinesis. Genetics. 2003a;165:477–488. doi: 10.1093/genetics/165.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chang EC. Schizosaccharomyces pombe Ras1 effector, Scd1, interacts with Klp5 and Klp6 kinesins to mediate cytokinesis. Genetics. 2003b;165:477–488. doi: 10.1093/genetics/165.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Mahlert M, Leveleki L, Hlubek A, Sandrock B, Bölker M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol Microbiol. 2006;59:567–578. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- Maresca B, Kobayashi G. Dimorphism in Histoplasma capsulatum and Blastomyces dermatitidis. Contrib Microbiol. 2000;5:201–216. doi: 10.1159/000060346. [DOI] [PubMed] [Google Scholar]
- McDade HC, Cox GM. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Johnson DI. Cdc42p GTPase Is Involved in Controlling Polarized Cell Growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany M. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr Opin Microbiol. 2002;5:580–585. doi: 10.1016/s1369-5274(02)00368-5. [DOI] [PubMed] [Google Scholar]
- Nichols CB, Fraser JA, Heitman J. PAK Kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans . Mol Biol Cell. 2004;15:4476–4489. doi: 10.1091/mbc.E04-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CB, Perfect ZH, Alspaugh JA. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2007;63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti D, Perfect JR, Heitman J. Sexual Cycle of Cryptococcus neoformans var. grubii and Virulence of Congenic a and Infection and Immunity. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken B, Wiener H, Philips MR, Chang EC. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci USA. 2006;103:9045–9050. doi: 10.1073/pnas.0603318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 1980;101:177–194. [PMC free article] [PubMed] [Google Scholar]
- Perrière G, Gouy M. WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Petzold EW, Himmelreich U, Mylonakis E, Rude T, Toffaletti D, Cox GM, Miller JL, Perfect JR. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect Immun. 2006;74:5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Nichols CB, Alspaugh JA. Cryptococcus neoformans Rho-GDP dissociation inhibitor mediates intracellular survival and virulence. Infect Immun. 2008a:5729–5737. doi: 10.1128/IAI.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Nichols CB, Alspaugh JA. Cryptococcus neoformans Rho-GDP dissociation inhibitor mediates intracellular survival and virulence. Infect Immun. 2008b doi: 10.1128/IAI.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolke Y, Tudzynski P. The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Mol Microbiol. 2008;68:405–423. doi: 10.1111/j.1365-2958.2008.06159.x. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods in Enzymology. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Su Z, Li H, Li Y, Ni F. Inhibition of the pathogenically related morphologic transition in Candida albicans by disrupting Cdc42 binding to its effectors. Chem Biol. 2007;14:1273–1282. doi: 10.1016/j.chembiol.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushinsky SC, Harcus D, Ash J, Dignard D, Marcil A. CDC42 Is Required for Polarized Growth in Human Pathogen Candida albicans. Eukaryotic Cell. 2002;1:95–104. doi: 10.1128/EC.1.1.95-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallim MA, Nichols CB, Fernandes L, Cramer KL, Alspaugh JA. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryotic Cell. 2005;4:1066–1078. doi: 10.1128/EC.4.6.1066-1078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBerg AL, Ibrahim AS, Edwards JE, Toenjes KA, Johnson DI. Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans. Eukaryotic Cell. 2004;3:724–734. doi: 10.1128/EC.3.3.724-734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol. 2004;164:701–716. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag A, Lee MP, Si H, Harris SD. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol Microbiol. 2007;66:1579–1596. doi: 10.1111/j.1365-2958.2007.06021.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Nichols CB, Lengeler KB, Cardenas ME, Cox GM, Perfect JR, Heitman J. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryotic Cell. 2002;1:257–272. doi: 10.1128/EC.1.2.257-272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh MS, Nichols CB, DeCesare CM, Cox GM, Heitman J, Alspaugh JA. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology (Reading, Engl) 2002a;148:191–201. doi: 10.1099/00221287-148-1-191. [DOI] [PubMed] [Google Scholar]
- Waugh MS, Nichols CB, DeCesare CM, Cox GM, Heitman J, Alspaugh JA. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans . Microbiology. 2002b;148:191–201. doi: 10.1099/00221287-148-1-191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.