DNA flaps function as intermediates in DNA metabolism. 3′-Flaps are generated as intermediates for homologous recombination. 5′-flaps, on the other hand, arise as necessary but dangerous DNA structures during DNA replication and DNA damage response. 5′-Flaps are generated either through strand displacement synthesis by a DNA polymerase or through the action of a DNA helicase, and generally, their degradation by 5′-nucleases is tightly coupled to their formation (Fig. 1A). Members of the family of 5′-flap endonucleases, as exemplified by FEN1, cut such 5′-flaps. Recently, however, another type of 5′-endonuclease, Dna2, has gained attention because of its involvement in multiple pathways involving the degradation of 5′-flaps. In the May 15th issue of Cell Cycle, Budd et al. showed that deletion of S. cerevisiae DNA2 leads to a permanent checkpoint arrest, resulting in a lethality that can be alleviated by the inactivation of key cell cycle checkpoint factors.1 In fact, it had been known for over a decade that the temperature sensitivity of dna2 hypomorphs could be suppressed by deletion of the checkpoint mediator RAD9.2 However, at that time, the mechanistic significance of this result was unclear, since the role of Dna2 nuclease in DNA metabolism had not yet been delineated.
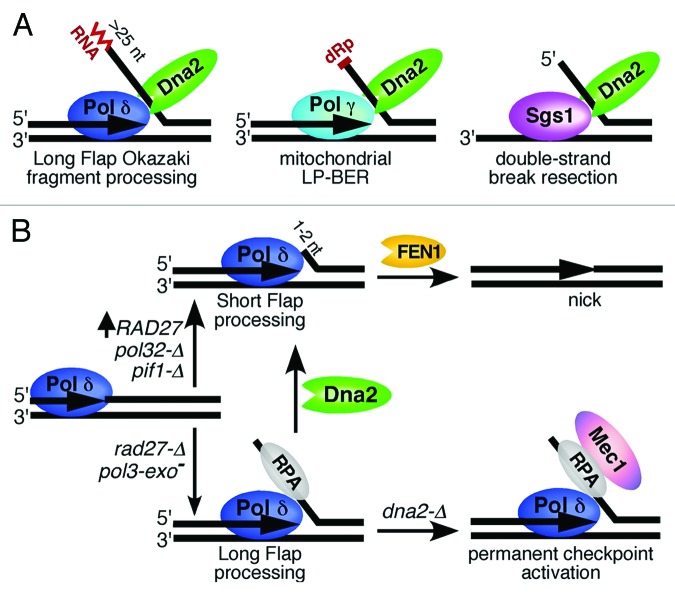
Figure 1. Dna2 nuclease in DNA metabolism. (A) Pathways in which Dna2 nuclease participates. (B) Balance of short vs. long flap distribution during Okazaki fragment maturation is affected by various mutations. RAD27 = FEN1. See text for details.
Dna2 was originally identified as a DNA helicase with an associated nuclease activity, but subsequent studies showed that its nuclease activity performs the primary function in the pathways studied and is essential for viability.3,4 The function of the helicase appears to be more ancillary in character. During Okazaki fragment maturation, 5′-flaps are generated through strand displacement synthesis by DNA polymerase δ.5 Most flaps remain small and undergo concomitant cleavage by FEN1, but some flaps escape cleavage and grow to a length of > 25 nt, which allows coating by the single-stranded binding protein RPA. These RPA-coated flaps are refractory to FEN1 cutting, but they become substrates for Dna2. Since Dna2 cutting is not precise but leaves a small 5′-flap of 2–6 nucleotides, further trimming by FEN1 or another flap endonuclease is still necessary (Fig. 1B). Similarly, during long-patch base excision repair in the mitochondria of mammalian cells, flaps generated by DNA polymerase γ are cut by FEN1 when they are small and by Dna2 when they are longer.6,7 Most recently, resection of the 5′-strand at a double-stranded break has been shown to be mediated by the coupled action of Sgs1 helicase and the Dna2 nuclease.8,9 This degradation process is unique in that FEN1 is apparently not involved. None of these pathways show a substantial dependence on the helicase function of Dna2.
Okazaki fragment maturation proceeds primarily through the generation of short flaps that are cut by FEN1, with the Dna2-dependent long flap pathway being a rare backup mechanism.10 Biochemical and genetic studies have shown that several factors can influence the balance of short vs. long flaps during Okazaki fragment maturation (Fig. 1B).5 This balance is an important consideration, because there is good evidence that the checkpoint-mediated lethality of a DNA2 deletion is caused specifically by the accumulation of long flaps during Okazaki fragment maturation. Lethality of dna2-Δ is suppressed by mutations and conditions that limit the accumulation of long flaps on the lagging strand of the replication fork; for instance, by the reduction in efficiency with which Pol δ carries out strand displacement synthesis (in pol32-Δ, that deletes its third subunit), or by deletion of PIF1, a helicase which actually promotes the generation of longer flaps.11 In addition, overexpression of RAD27 (encoding FEN1) increases the utilization of short flaps, thereby also limiting the accumulation of long flaps and thus suppressesing the lethality of dna2 hypomorphs. In contrast, the phenotypic defects caused by an increase in the generation of long flaps, either in a rad27-Δ mutant or in a proofreading-defective mutant of Pol δ, which results in increased strand displacement synthesis, can be rescued by overexpression of DNA2. Finally, the balance of short vs. long flaps can also be affected by post-translational modification of FEN1 and Dna2.12
Given the high importance assigned to the FEN1 pathway, one may wonder why a rad27-Δ mutant is viable but dna2-Δ is not. Budd et al. show that the answer lays in checkpoint activation.1 Rad27 deficiency is partially compensated by the action of another flap exonuclease, Exo1, and partially by Dna2 cutting of the longer flaps, which are generated more frequently in the rad27-Δ mutant. However, the rad27-Δ defect also leads to the generation of double-stranded breaks which require homologous recombination for repair, hence the lethality of rad27-Δ rad52-Δ double mutants. On the other hand, Dna2 deficiency leads to the persistent accumulation of long 5′-flaps, and these RPA-coated flaps likely serve as recruitment sites for the principal checkpoint kinase Mec1, initiating a continual checkpoint response with subsequent lethal consequences in G2/M. Remarkably, both the DNA damage checkpoint, which proceeds via the Rad9 mediator, and the replication checkpoint, which proceeds via the Mrc1 mediator, contribute to the terminal arrest of a DNA2 deletion, since maximal growth is recovered when the checkpoint functions of both mediators are eliminated. The latter observation is an indication that Okazaki fragment maturation is tightly coupled to its maturation by FEN1, Dna2 and DNA ligase, since failure in maturation leads to replication-fork stalling with associated activation of the Mrc1-dependent checkpoint pathway.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/15851
References
- 1.Budd ME, Antoshechkin IA, Reis C, Wold BJ, Campbell JL. Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint. Cell Cycle. 2011;10:1690–8. doi: 10.4161/cc.10.10.15643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Formosa T, Nittis T. Dna2 mutants reveal interactions with Dna polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–70. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–9. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 4.Lee KH, Kim DW, Bae SH, Kim JA, Ryu GH, Kwon YN, et al. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 2000;28:2873–81. doi: 10.1093/nar/28.15.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–36. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–82. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–6. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–11. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278:1618–25. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 11.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–80. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan L, Stewart J, Polaczek P, Campbell JL, Bambara RA. Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates. J Biol Chem. 2010;285:4398–404. doi: 10.1074/jbc.M109.086397. [DOI] [PMC free article] [PubMed] [Google Scholar]