Abstract
Studies suggest that mitochondrial DNA (mtDNA) haplogroups are associated with antiretroviral therapy (ART)-related metabolic complications and distal sensory polyneuropathy (DSP), but there have been few studies in persons of African descent. We explored such associations in South African adults. Clinical and laboratory data and DNA specimens from a cross-sectional study were used. Sequencing and Phylotree determined African mtDNA subhaplogroups. Wilcoxon and regression analyses determined associations between mtDNA subhaplogroups and ART-related complications. The 171 participants represented six major haplogroups: L0 (n=78), L1 (n=3), L2 (n=30), L3 (n=53), L4 (n=1), and L5 (n=6). Analyses were restricted to 161 participants representing L0, L2, and L3: 78% were female; the median age was 36 years. All had been exposed to thymidine analogues, 42% were on lopinavir/ritonavir (lopinavir/r), and 58% were on either efavirenz or nevirapine. Median (IQR) ART duration was 22 (14–36) months. Median fasting triglycerides were 1.60 (1.13–1.75) and 1.04 (0.83–1.45) mmol/liter among L3e1 (n=22) and other subhaplogroups, respectively (p=0.003). Subhaplogroup L3e1 [adjusted OR (aOR) 3.16 (95% CI: 1.11–8.96); p=0.03] and exposure to lopinavir/r [aOR 2.98 (95% CI: 1.02–8.96); p=0.05] were independently associated with hypertriglyceridemia, after adjusting for age, sex, and ART duration. There were no significant associations between mtDNA haplogroups and cholesterol, dysglycemia, hyperlactatemia, or lipoatrophy, or DSP. Subhaplogroup L3e1 and lopinavir/r exposure were independently associated with hypertriglyceridemia in black South Africans on ART. This is the first report to link an African mtDNA variant with hypertriglyceridemia. If replicated, these findings may provide new insights into host factors affecting metabolic complications.
Introduction
Exposure to antiretroviral therapy (ART) has been associated with metabolic adverse effects such as dyslipidemia, insulin resistance, dysglycemia, central fat accumulation, peripheral fat loss (lipoatrophy), and peripheral neuropathy.1,2 Many of these adverse effects are thought to be related to nucleoside reverse transcriptase inhibitor (NRTI)-related mitochondrial toxicity, at least in part due to inhibition of mitochondrial DNA (mtDNA) gamma polymerase and subsequent mitochondrial dysfunction.2 Other antiretroviral classes have also been associated with mitochondrial dysfunction.3,4
Mitochondrial DNA is distinct from nuclear DNA and codes for 13 polypeptides essential for oxidative phosphorylation.5 Mitochondrial DNA exhibits abundant genetic variation across the 16.6 kb mitochondrial genome. Human mtDNA sequences have diverged over approximately the last 150,000 years due to natural selection and human migration, resulting in distinct patterns of single nucleotide polymorphisms (SNPs), called haplogroups.6 Evidence for associations between mtDNA haplogroups and outcomes in HIV-infected participants has been reported for CD4 count recovery,7 AIDS progression,8 and ART-related complications.9,10,11,12,13,14,15 Studies in HIV-uninfected populations (primarily European and Asian) have reported associations between mtDNA and metabolic complications including dyslipidemia and diabetes.16,17,18,19,20
The majority of HIV-infected persons worldwide reside in sub-Saharan Africa,21 but this region has been underrepresented in genetic studies. Studies in African-Americans showed associations between African mtDNA subhaplogroup L1c and peripheral neuropathy,15 and between haplogroup L2 and CD4 T cell recovery.7 To date, no published studies have investigated associations between African mtDNA haplogroups and important metabolic complications including lipoatrophy, dysglycemia, or dyslipidemia. In this study, we report the prevalence of mtDNA haplogroups in a South African population enrolled in an ART program, and explore associations between African mtDNA and ART-associated complications including metabolic complications and distal sensory polyneuropathy (DSP). Because ART-associated adverse effects in HIV-infected persons, and metabolic diseases in general, are believed to result in part from mitochondrial dysfunction, we hypothesized that mtDNA variation would be associated with susceptibility to ART-associated lipoatrophy, dysglycemia, hypertriglyceridemia, and/or distal peripheral neuropathy in this South African population.
Materials and Methods
Study design and participants
This analysis was conducted by retrospectively analyzing data and specimens that had been collected during a prospective, cross-sectional study of the prevalence of metabolic and neuropathy complications of ART.22,23 Participants were ambulatory HIV-infected African black adults who presented for a routine follow-up visit at public-sector antiretroviral clinics in Cape Town. Participants were recruited by convenience sampling between February 2007 and September 2008. They were eligible if they were on ART therapy for at least 6 months, with no renal or hepatic disease, no active opportunistic infections, or no known history of diabetes or dyslipidemia. Participants on lipid or glucose-lowering therapy were also excluded. The University of Cape Town research ethics committee approved the primary study and all participants who signed informed consent for the genetic study and had clinical and laboratory evaluations and/or whole body dual-energy X-ray absorptiometry (DEXA) were included in this analysis. The Vanderbilt University Institutional Review Board approved the use of de-identified DNA and clinical data for these analyses.
Clinical and laboratory evaluations
Participants fasted overnight and underwent an oral glucose tolerance test (OGTT). Impaired fasting glucose (IFG), impaired glucose tolerance (IGT), and diabetes were defined according to the American Diabetes Association criteria.24 Fasting triglyceride concentrations were determined at the start of the OGTT and were measured at the National Health Laboratory Services. Hypertriglyceridemia was defined according to NCEP III criteria.25 Point-of-care measurement of lactate was done before glucose loading using the Accutrend lactate meter (Roche, Basel, Switzerland). Hyperlactatemia was defined as a lactate concentration ≥2.5 mmol/liter.26 For the present study, DSP was defined as the presence of at least two of the following signs: reduced or absent reflexes and impaired vibratory or pinprick sensation with or without neuropathic symptoms (pain, paresthesia, or numbness). A DSP diagnosis was considered only if signs and symptoms were bilateral, of symmetrical onset, and present for at least 2 weeks. Limb fat was measured by DEXA scan and percentage of limb fat was used to determine lipoatrophy, which was defined as percentage limb fat below the median stratified by sex. We reviewed medical records to determine duration on antiretroviral therapy, current CD4+ lymphocyte counts, and viral load within 3 months of the study visit, where available.
Mitochondrial DNA isolation and sequencing
Chromosomal and mtDNA were isolated from buffy coats using Maxwell16® DNA Purification Kits (Promega Corporation, Madison, WI). Full mitochondrial DNA sequencing was performed using the GeneChip Human Resequencing Array v2.0 (Affymetrix Inc., Santa Clara, CA). The full sequence data were used to assign haplogroups and subhaplogroups based on PhyloTree.27
Statistical analysis
Kruskal–Wallis and Wilcoxon rank-sum tests were used to compare medians between haplogroups. Linear regression models were used to compare continuous outcomes between subhaplogroups. For analysis of binary outcomes, a case-control design was used. The Pearson Chi-squared test was used to compare proportions of subjects with dichotomous metabolic outcomes. Logistic regression models were fitted to assess the association between subhaplogroups and dysglycemia, DSP, hyperlactatemia, hypertriglyceridemia, and lipoatrophy. Age, sex, lopinavir exposure, and duration on ART were included in all multivariate regression models. Age and sex were included a priori as potential confounders. Lopinavir was included as the potential confounder as it had been associated with metabolic complications. The duration on ART was included in all multivariate models as it differed between the major haplogroups. Subhaplogroups with fewer than 10 participants were excluded from analyses. Analyses were performed using Stata release 11 (StataCorp LP, College Station TX). All tests were two-sided, and a p value<0.05 was considered significant. As this was an exploratory study, we did not formally correct for multiple comparisons.
Results
Participant characteristics and mitochondrial haplogroups
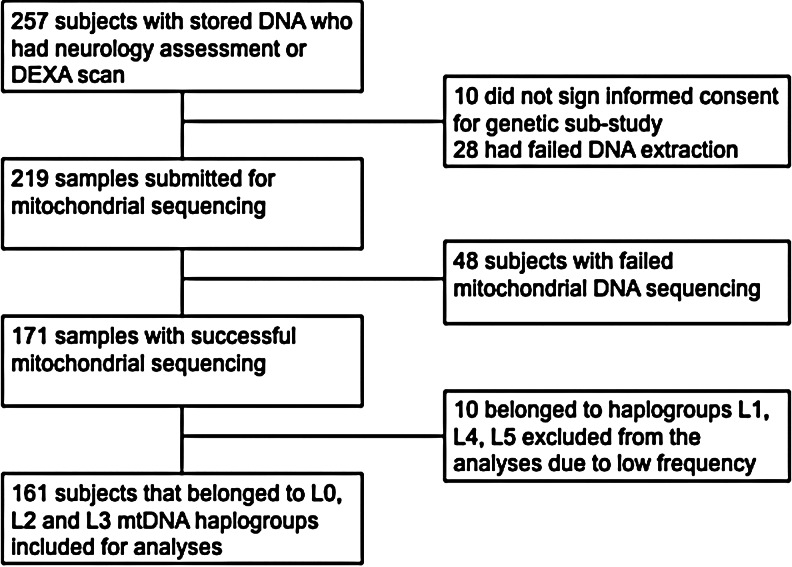
A total of 219 participants provided DNA for analysis, and mitochondrial sequencing was successful in 171 (78%). The primary reason for lack of sequencing data was insufficient or poor quality DNA. Figure 1 shows the flow chart of participants in this study. We restricted our analyses to the 161 participants who belonged to the L0 (n=78), L2 (30), and L3 (53) major haplogroups (Table 1) because the numbers in the other haplogroups were small [L4 (1), L1 (3), and L5 (6)]. Most participants (78%) were female, which reflects the population treated in the referring clinics. Current ART regimens contained efavirenz, nevirapine, and lopinavir/ritonavir in 33%, 25%, and 42%, respectively. Consistent with South African HIV treatment guidelines at the time, most participants on a first-line regimen were treated with stavudine/lamivudine (66%) or zidovudine/lamivudine (34%) combinations as the NRTI backbone, while most of those on second-line regimens were treated with zidovudine/didanosine (58%) or zidovudine/lamivudine (24%) combinations. Duration on ART differed across haplogroups (Kruskal–Wallis p=0.02; Table 1), with the L0 major haplogroup having significantly shorter ART duration (p=0.01), and the L3 major haplogroup having longer ART duration compared to the others (p=0.01). However, the duration on lopinavir/ritonavir did not differ among the major haplogroups (Kruskal–Wallis p=0.88; Table 1). The L2 major haplogroup had fewer females than other haplogroups (p=0.04). Otherwise, baseline characteristics did not differ significantly by major mtDNA haplogroup.
FIG. 1.
Flow chart of the study population.
Table 1.
Study Population Characteristics Including Metabolic and Neuropathic Data, by Major Haplogroups
| |
Study population submitted for sequencing |
Major haplogroups analyzed |
|||
|---|---|---|---|---|---|
| Variable | All subjects N=219 | Subjects L0, L2, L3 N=161 | L0 N=78 | L2 N=30 | L3 N=53 |
| Age (years) | 36 (31–42) | 36 (32–42) | 35 (31–40) | 36 (31–41) | 37 (32–44) |
| Female n/N (%)a | 179/219 (82) | 125/161 (78) | 63/78 (80) | 19/30 (63) | 43/53 (81) |
| BMI (kg/m2) | 26 (23–31) | 25 (23–28) | 25 (23–29) | 24 (21–28) | 27 (24–34) |
| N=216 | N=158 | N=77 | N=30 | N=51 | |
| CD4 count at start of treatment (cells/mm3) | 97 (48–151) | 97 (54–153) | 93 (40–155) | 79 (33–130) | 113 (75–179) |
| N=199 | N=146 | N=69 | N=28 | N=49 | |
| CD4 count at study enrollment (cells/mm3) | 385 (247–520) | 382 (247–518) | 346 (238–510) | 349 (201–518) | 447 (288–524) |
| N=217 | N=160 | N=77 | N=30 | N=53 | |
| Viral load (VL) log10 | |||||
| VL before ART (copies/ml) | 4.64 (3.68–5.26) | 4.97 (4.49–5.45) | 4.98 (4.56–5.49) | 4.98 (4.55–5.38) | 4.85 (4.48–5.33) |
| N=100 | N=75 | N=30 | N=14 | N=31 | |
| VL<400 at study enrollment n/N (%) | 142/163 (87) | 109/127 (86) | 50/58 (86) | 23/27 (85) | 36/42 (85) |
| Blood pressure (mm Hg) | 109/73 (101/66–119/80) | 107/73 (101/57–119/81) | 108/71 (101/63–119/77) | 109/75 (100/64–120/79) | 107/73 (102/67–120/81) |
| Waist:hip ratio | 0.88 (0.84–0.94) | 0.88 (0.83–0.94) | 0.86 (0.82–0.94) | 0.88 (0.84–0.93) | 0.89 (0.85–0.94) |
| NRTI backbone n/N (%) | |||||
| Stavudine/lamivudine | 97/218 (45) | 69/160 (43) | 41/77 (53) | 8/30 (27) | 20/53 (38) |
| Zidovudine/lamivudine | 66/218 (30) | 46/160 (28) | 19/77 (25) | 10/30 (33) | 17/53 (32) |
| Zidovudine/didanosine | 46/218 (21) | 38/160 (23) | 12/77 (16) | 12/30 (40) | 14/53 (26) |
| Other | 9/218 (4) | 7/160 (5) | 5/77 (7) | 0/30 (0) | 2/53 (4) |
| NNRTI/PI n/N (%) | |||||
| Efavirenz | 82/219 (37) | 54/161 (33) | 27/78 (35) | 6/30 (20) | 21/53 (40) |
| Nevirapine | 57/219 (26) | 40/161 (25) | 24/78 (30) | 9/30 (30) | 7/53 (13) |
| Lopinavir/ritonavir | 80/219 (37) | 67/161 (42) | 27/78 (35) | 15/30 (50) | 25/53 (47) |
| Duration (months) | |||||
| Stavudine | 15 (9–23) | 15 (10–22) | 14 (9–19) | 14 (10–21) | 17 (12–30) |
| N=187 | N=137 | N=70 | N=25 | N=42 | |
| Zidovudine | 16 (10–26) | 16 (10–27) | 16 (11–27) | 16 (7–27) | 16 (10–26) |
| N=114 | N=88 | N=35 | N=22 | N=31 | |
| Lopinavir/ritonavir | 18 (10–26) | 18.5 (10–26) | 18 (12–30) | 18 (7–27) | 20 (10.5–26) |
| N=76 | N=66 | N=27 | N=15 | N=24 | |
| Any ARTb | 23 (14–33) | 22 (14–36) | 19 (11–30) | 22 (15–33) | 30 (17–41) |
| N=214 | N=157 | N=76 | N=30 | N=51 | |
| Lipids (mmol/liter) | N=217 | N=159 | N=78 | N=30 | N=51 |
| Total cholesterol | 4.34 (3.75–5.25) | 4.33 (3.79–5.28) | 4.34 (3.86–5.15) | 4.23 (3.95–4.72) | 4.45 (3.66–5.49) |
| LDL-C | 2.70 (2.20–3.22) | 2.71 (2.20–3.20) | 2.71 (2.20–3.08) | 2.75 (2.21–3.10) | 2.80 (2.19–3.49) |
| HDL-C | 1.03 (0.82–1.30) | 1.03 (0.81–1.29) | 1.07 (0.88–1.27) | 0.96 (0.80–1.29) | 1.01 (0.75–1.32) |
| Triglycerides | 1.11 (0.85–1.49) | 1.11 (0.85–1.59) | 1.07 (0.87–1.41) | 0.99 (0.78–1.62) | 1.23 (0.87–1.81) |
| Glucose (mmol/liter) | |||||
| Fasting | 5.3 (4.8–5.6) | 5.1 (4.8–5.6) | 5.0 (4.7–5.4) | 5.3 (4.9–5.6) | 5.1 (4.8–5.6) |
| N=217 | N=159 | N=78 | N=30 | N=51 | |
| 2 h | 6.1 (5.2–7.1) | 6.0 (5.2–7.3) | 5.8 (5.05–6.95) | 6.15 (5.70–7.80) | 6.2 (5.3–7.5) |
| N=215 | N=157 | N=76 | N=30 | N=51 | |
| Lactate (mmol/liter) | 1.9 (1.3–2.5) | 1.9 (1.3–2.5) | 1.9 (1.3–2.5) | 1.9 (1.7–2.5) | 1.8 (1.3–2.8) |
| N=207 | N=155 | N=75 | N=29 | N=51 | |
| Percentage limb fat (%)c | 17 (12–21) | 17 (12–21) | 18 (14–21) | 13 (9–20) | 17 (13–21) |
| N=172 | N=131 | N=62 | N=28 | N=41 | |
| DSP n/N (%) | 149/219 (68) | 106/161 (65) | 54/78 (69) | 21/30 (70) | 31/53 (58) |
Chi-square test shows that the L2 major haplogroup had fewer females than other haplogroups (p=0.04).
Kruskal–Wallis test showed that ART duration differed across the haplogroups (p=0.02). Wilcoxon rank sum test showed that the L0 major haplogroup had significantly shorter ART duration than others (p=0.01) and the L3 major haplogroup had significantly longer ART duration than the others (p=0.01).
Wilcoxon rank sum test showed that the L2 major haplogroup had significantly lower percentage limb fat than other groups. Other baseline characteristics did not significantly differ by major mtDNA haplogroup.
Data are reported in median (IQR) unless specified. When N is not specified, there was no missing data. BMI, body mass index; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; ART, antiretroviral therapy; PI, protease inhibitor; DSP, distal sensory polyneuropathy.
Metabolic complications and mitochondrial haplogroups
Median [interquartile range (IQR)] lipid values in this cohort were total cholesterol 4.33 (3.79–5.28) mmol/liter, LDL 2.71 (2.20–3.20) mmol/liter, HDL 1.03 (0.81–1.29) mmol/liter, and triglycerides 1.11 (0.85–1.59) mmol/liter. Simple regression analyses showed no statistically significant associations between the metabolic parameters and the three haplogroups (p>0.05 for each analysis, Table 2).
Table 2.
Univariate Analyses of Metabolic Complications and Major Mitochondrial Haplogroups
| |
Major haplogroups |
|||||
|---|---|---|---|---|---|---|
| |
L0 |
L2 |
L3 |
|||
| Variable | Beta coeff (95% CI) | p | Beta coeff (95% CI) | p | Beta coeff (95% CI) | p |
| Total cholesterol | −0.01 (−0.47–0.27) | 0.59 | −0.12 (−0.51–0.26) | 0.53 | 0.20 (−0.25–0.65) | 0.38 |
| Triglycerides | −0.10 (−0.34–0.13) | 0.39 | −0.11 (−0.35–0.13) | 0.35 | 0.20 (−0.06–0.46) | 0.13 |
| Fasting glucose | −0.18 (−0.63–0.27) | 0.42 | −0.06 (−0.44–0.30) | 0.73 | 0.26 (−0.40–0.91) | 0.44 |
| 2 h glucose | −0.69 (−1.60–0.22) | 0.14 | 0.32 (−0.62–1.27) | 0.50 | 0.56 (−0.70–1.81) | 0.38 |
| Lactate | −0.07 (−0.38–0.24) | 0.65 | −0.01 (−0.35–0.33) | 0.95 | 0.09 (−0.28–0.46) | 0.64 |
Fasting glucose and lipid data were available from 159 participants. For 2 h glucose and lactate, data were available only from 157 and 155 participants, respectively. All variables were measured in mmol/liter. p values are comparing variables for each haplogroup with the other two haplogroups combined.
Metabolic complications were common in this cohort. Hypercholesterolemia was present in 31%, hypertriglyceridemia in 18%, dysglycemia in 34%, and hyperlactatemia in 28%. Table 3 shows unadjusted and adjusted odds ratios for metabolic complications. In these analyses, there was a statistically significant association between the L3 haplogroup and hypertriglyceridemia [unadjusted odds ratio (OR) 2.58 (95% CI 1.14–5.84), p=0.02; Table 3]. This association remained significant after adjusting for age, sex, current lopinavir-based ART, and overall ART duration [adjusted odds ratio (aOR) 2.44(95% CI 1.03–5.77), p=0.04]. As expected, lopinavir/r use was also independently associated with hypertriglyceridemia [aOR 3.55 (95% CI 1.21–10.39), p=0.02].
Table 3.
Univariate and Multivariate Analyses of Metabolic Complications and Major Haplogroups
| |
Major haplogroups |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
L0 |
L2 |
L3 |
|||||||||
| |
Univariate |
Multivariate |
Univariate |
Multivariate |
Univariate |
Multivariate |
||||||
| Variable | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p |
| Hypercholesterolemia | 0.88 (0.45–1.74) | 0.72 | 0.95 (0.48–1.91) | 0.90 | 0.63 (0.25–1.59) | 0.33 | 0.59 (0.23–1.47) | 0.26 | 1.5 (0.76–3.14) | 0.23 | 1.51 (0.74–3.07) | 0.23 |
| Hypertriglyceridemia | 0.44 (0.19–1.04) | 0.06 | 0.56 (0.22–1.40) | 0.21 | 0.83 (0.29–2.40) | 0.73 | 0.63 (0.21–1.94) | 0.43 | 2.58 (1.14–5.83) | 0.02 | 2.44 (1.03–5.77) | 0.04 |
| Dysglycemia | 0.77 (0.40–1.49) | 0.44 | 0.82 (0.41–1.63) | 0.57 | 1.62 (0.72–3.66) | 0.24 | 1.58 (0.72–3.48) | 0.26 | 0.95 (0.47–1.92) | 0.88 | 0.89 (0.42–1.87) | 0.76 |
| Hyperlactatemia | 0.96 (0.47–1.94) | 0.92 | 0.93 (0.45–1.96) | 0.85 | 0.95 (0.39–2.35) | 0.92 | 0.90 (0.35–2.30) | 0.83 | 1.08 (0.51–2.26) | 0.84 | 1.16 (0.54–2.49) | 0.96 |
Adjusted for age, sex, lopinavir treatment, and treatment duration. p values are comparing variables for each haplogroup with the other two haplogroups combined.
Every mitochondrial haplogroup is further divided into subhaplogroups.27 A weak association with a haplogroup might be driven by a stronger association with a subhaplogroup. Because of the availability of full mtDNA sequence data in this study, we were able to explore whether a subhaplogroup within L3 was associated with hypertriglyceridemia. The frequencies of the L3 subhaplogroups in the study population were L3e1 (23), L3e2 (16), L3d1 (4), L3d3 (3), L3e4 (3), L3f1 (2), L3b1 (1), and L3e3 (1). Based on the infrequency of non-L3e1 and non-L3e2 subhaplogroups, we chose to analyze subhaplogroups L3e1 and L3e2 separately, and to combine the remaining participants as “other L3 subhaplogroups.”
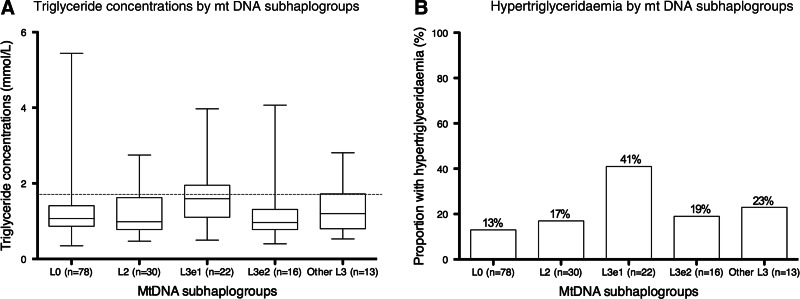
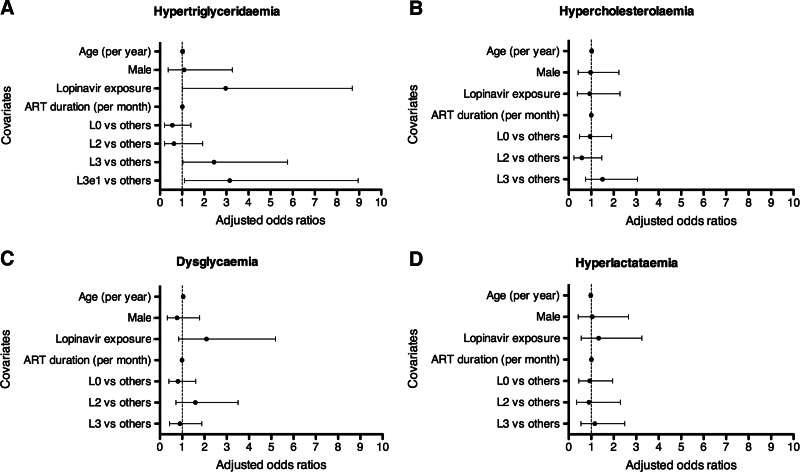
Figure 2 shows the triglyceride concentrations (Fig. 2A) and hypertriglyceridemia (Fig. 2B) in the L3e1 subhaplogroup, other L3 subhaplogroups, and the haplogroups L0 and L2. The triglyceride concentrations differed significantly between the mtDNA subhaplogroups (Kruskal–Wallis p=0.047). The L3e1 subhaplogroup had significantly higher triglyceride concentrations compared to other subhaplogroups combined (Wilcoxon p=0.003; Fig. 2A). Linear regression showed that mean difference in triglyceride concentrations between the L3e1 subhaplogroup and other subhaplogroups combined was 0.40 mmol/liter [(95% CI 0.06–0.75 mmol/liter), p=0.02]. After adjusting for age, sex, lopinavir/ritonavir-based ART, and ART duration; the association was no longer statistically significant [mean difference 0.35 (95% CI −0.01–0.72) mmol/liter, p=0.06]. The prevalence of hypertriglyceridemia in the L0 and L2 haplogroups and L3 subhaplogroups was L0 (13%) and L2 (17%), L3e1 (41%), L3e2 (19%), and other L3 subhaplogroups (23%). When comparing L3e1 to other subhaplogroups combined, the prevalence of hypertriglyceridemia was 41% and 15%, respectively (p=0.004; Fig. 2B). By logistic regression, there was a statistically significant association between the L3e1 subhaplogroup and hypertriglyceridemia [unadjusted OR 3.82 (95% CI 1.45–10.10), p=0.007]. When adjusted for age, sex, current lopinavir treatment, and ART duration, there remained a significant association with the L3e1 subhaplogroup [aOR 3.16 (95% CI 1.11–8.96), p=0.03]. Lopinavir treatment was also independently associated with hypertriglyceridemia [aOR 2.97 (95% CI 1.02–8.69), p=0.05]. Figure 3 summarizes various multivariate models for the metabolic complications.
FIG. 2.
Fasting triglyceride concentrations and hypertriglyceridemia by mitochondrial DNA (mtDNA) subhaplogroups. (A) Box plots of triglyceride concentrations in the L3e1 subhaplogroup, other L3 subhaplogroups, as well at the other haplogroups L0 and L2. Boxes represents medians and interquartile ranges; whiskers are the range. Triglycerides differed by mtDNA haplogroups (Kruskal–Wallis p=0.047), with L3e1 having significantly higher concentrations than other haplogroups (Wilcoxon p=0.003). The gray dashed line indicates the cutoff value of 1.7 mmol/liter used to categorize hypertriglyceridemia.25 (B) The proportion of participants with hypertriglyceridemia by mtDNA subhaplogroups, with 41% in the L3e1 subhaplogroup and 15% in other subhaplogroups combined (Chi square test p=0.004).
FIG. 3.
Associations between mitochondrial DNA subhaplogroups and metabolic complications in HIV-infected South African adults on antiretroviral therapy (ART), shown as adjusted odds ratios (aORs). aORs were obtained from separate models: L0, L2, L3, and L3e1. The covariates age, sex, lopinavir/ritonavir (LPV/r) exposure, and ART duration displayed in (A) are from the L3e1 model. (B–D) The same covariates are from the L0 model. After adjusting for age, sex, LPV/r exposure, and ART duration, L3 [aOR 2.44 (95% CI: 1.03–5.77) and p=0.04], L3e1 [aOR 3.16 (95% CI: 1.11–8.96); p=0.03] were independently associated with hypertriglyceridemia. LPV/r exposure [aOR 2.97 (95% CI: 1.02–8.69); p=0.047] was also independently associated with hypertriglyceridemia and this was consistent in all subhaplogroup models.
There were no other significant associations between haplogroups and metabolic complications other than hypertriglyceridemia. In multivariate analyses for each haplogroup, increasing age was an independent predictor of dysglycemia after adjusting for mtDNA haplogroup, sex, current lopinavir/r treatment, and duration on ART (p for age in each model<0.05; Fig. 3C).
Peripheral lipoatrophy and mitochondrial haplogroups
Overall median (IQR) percentage limb fat by DEXA was 17 (12–21)% (Table 1). Percentage limb fat differed by sex, with males having a median (IQR) of 8 (6–10)% and females having a median (IQR) of 18 (15–22)%. Initial unadjusted analyses demonstrated significantly lower percentage limb fat in the L2 haplogroup compared with the other haplogroups (p=0.04). However, the L2 haplogroup also had a lower percentage of females than the other haplogroups (p=0.04). In view of these findings, peripheral lipoatrophy was defined as percentage limb fat below the median in sex-stratified populations (male n=30; female n=101). Logistic regression analyses stratified by sex showed no associations between L2 haplogroup and peripheral lipoatrophy by this definition [males: unadjusted OR 1.33 (95% CI 0.30–5.91), p=0.71; females: unadjusted OR 1.17 (95% CI 0.41–3.36), p=0.71]. Adjusting for age and ART regimen also did not show significant associations [males: unadjusted OR 1.17 (95% CI 0.24–5.72), p=0.85; females: unadjusted OR 1.28 (95% CI 0.46–3.61), p=0.64].
Distal sensory polyneuropathy (DSP) and mitochondrial haplogroups
The overall prevalence of DSP was 66% (Table 1). There were no statistically significant associations between DSP and mitochondrial haplogroups, either when treated as a composite variable or when assessing symptomatic and asymptomatic DSP separately (all p>0.1; data not shown).
Discussion
We explored associations between African mtDNA haplogroups and ART-related complications in a cohort of HIV-infected South African adults. We found that the L3e1 subhaplogroup was significantly associated with hypertriglyceridemia independent of lopinavir/ritonavir exposure. This is the first report to link an African mtDNA variant to hypertriglyceridemia. We found no associations between mtDNA haplogroups and other metabolic complications, including hypercholesterolemia, dysglycemia, lipoatrophy, or DSP.
Mitochondrial DNA haplogroups have been mostly studied to understand human evolution and migration.28 However, recent studies have investigated the association of mtDNA haplogroups with human diseases and longevity.7–15 We described the frequencies of the mitochondrial haplogroups in our South African cohort recruited from two Cape Town clinics, the majority of which belonged to the L0 haplogroup. There are limited data regarding the distribution of the mitochondrial haplogroups in the South African population. However, the distribution of mitochondrial haplogroups we found was similar to a recently published South African study conducted in 71 children.29 None of the participants analyzed belonged to the European mitochondrial haplogroups.
The association between L3e1 subhaplogroup and hypertriglyceridemia was independent of LPV/r use, which is a well-documented cause of elevated triglycerides.30 In a recent study of 174 non-Hispanic white HIV-infected clinical trial participants in the United States, the European mtDNA haplogroup I was associated with a decrease in triglycerides over 96 weeks of ART when compared with non-I mtDNA haplogroups.10 To the best of our knowledge, the association between African mtDNA haplogroups and hypertriglyceridemia has not previously been studied.
An association between an mtDNA polymorphism and plasma triglycerides was previously reported in a Canadian cohort.31 Variation at mtDNA position 16517 was associated with significantly higher fasting plasma triglyceride concentrations in this population. These data support the hypothesis that mitochondrial DNA variation may influence fatty acid and lipid metabolism. Fatty acids derived from plasma triglycerides are precursors of acyl CoA, which is used in the mitochondrial ß oxidation cycle of fatty acid metabolism. Genetic variation affecting mitochondrial function might also affect the utilization of fatty acid in ß oxidation.31 Differences in cellular utilization may then alter the demand of fatty acids from triglycerides, which may influence plasma concentrations.31 However, the exact mechanism is still unclear.
We found no association between mitochondrial DNA haplogroups and hypercholesterolemia. In the study referenced above, European mtDNA haplogroup I was associated with higher baseline (pre-ART) non-HDL cholesterol and a significant decrease in non-HDL cholesterol after 96 weeks of ART.10 In a study of 83 elite athletes and 61 patients with sedentary lifestyles, mtDNA clade HV was significantly associated with higher LDL cholesterol in the elite athletes, but not in those with a sedentary lifestyle.19 The lack of an association between mtDNA haplogroups and hypercholesterolemia observed in our cohort may be due to several factors. First, the studies discussed above investigated only European haplogroups,10 not African haplogroups. Second, nuclear genetic factors known to influence cholesterol (e.g., LPL, APOC3, and APOA2) may have a stronger influence on cholesterol than mitochondrial genetic variation, or mediate relationships between mitochondrial variation and phenotype. Third, data on lifestyle factors known to influence plasma cholesterol were not available in our cohort.
We observed no statistically significant associations between African mtDNA haplogroups and fasting dysglycemia. Multivariate analyses showed that older age was an independent predictor of dysglycemia, independent of haplogroup, sex, lopinavir treatment, or duration on ART. To our knowledge, this is the first study to investigate an association between this outcome and mtDNA haplogroups in an African cohort. Several inborn mitochondrial diseases include diabetes,32 and common mitochondrial DNA mutations have also been associated with type 2 diabetes mellitus in Asians and Europeans.33,34 Studies investigating mitochondrial haplogroups and type 2 diabetes have shown conflicting results. Although the prevalence of dysglycemia was high (33%) in our cohort, only 8 of 159 participants had diabetes and therefore diabetes was not included as an outcome in this analysis. Hyperlactatemia is a recognized consequence of mitochondrial dysfunction associated with d-drug use.35 We found no association between African mtDNA haplogroups and hyperlactatemia, which is similar to the finding from another South African study.36
There are inconsistent data regarding the influence of specific European mtDNA haplogroups on lipoatrophy.11,12 The AIDS Clinical Trials Group (ACTG) cohort found that the mtDNA haplogroup J tended to be protective against lipoatrophy.11 The Multicenter AIDS Cohort Study (MACS), conducted in 410 U.S. men (100% white), found that mtDNA haplogroup H was significantly associated with clinically defined lipoatrophy in the arms and legs, and that mtDNA haplogroup T tended to be protective.12 In contrast, a recent Italian study showed that mtDNA haplogroup T tended to increase the risk of lipoatrophy when compared to mtDNA haplogroup H.37 These studies are limited by diverse populations and heterogeneous outcome ascertainment. It is hypothesized that variations in mtDNA haplogroups may influence lipoatrophy at the adipocyte level, affecting cellular energy production efficiency, reactive oxygen species generation, and/or levels of apoptosis, all of which may be exacerbated by use of NRTIs. However, the exact mechanism remains unclear. The lack of an association between African mtDNA haplogroups and lipoatrophy could have been influenced by the way lipoatrophy was defined in our cohort. Due to significant differences between men and women, lipoatrophy was defined as percentage limb fat below the median in the sex-stratified populations. We also compared groups in the lowest and highest tertiles and quartiles and found no significant associations (data not shown). Notably, only a subset had DEXA scans, therefore we had limited power to detect differences. Larger prospective studies are needed to determine if an association between lipoatrophy and African mtDNA haplogroups exists.
Finally, we found no association between African mtDNA haplogroups and DSP. A study conducted in non-Hispanic black participants from ACTG study 384 reported an association between the African mtDNA L1c and increased susceptibility to peripheral neuropathy during NRTI treatment.15 As discussed above, the L1 haplogroup was found in only three participants in our cohort and they were excluded from our analyses, so we were unable to replicate this reported association. Here, DSP was frequent and no association was observed with a particular haplogroup. We were also unable to adjust for the effect of d-drugs, an important cause of DSP, as >90% of participants had been exposed to stavudine and/or didanosine. All participants were examined by either of two trained clinicians, and DSP was more rigorously defined compared with many other reports. The prevalence of DSP in our cohort was high (66%), compared with that reported in the ACTG study (33%).15
Our study had several limitations. The design was cross-sectional, with participants of different HIV disease stages and different ART durations. We excluded ART-naive participants and participants with current opportunistic infections, known hepatic or renal disease, or with known dyslipidemia (or taking lipid-lowering therapy) or dysglycemia (or taking antidiabetic drugs). Viral load data were missing in the majority of participants, and were therefore not included in the analysis as a covariate. Our sample size was relatively small and we did not formally adjust for multiple comparisons. However, when considering the five mtDNA subhaplogroups we analyzed, only the unadjusted analyses between subhaplogroup L3e1 and triglycerides concentrations (Wilcoxon p=0.003), or hypertriglyceridemia (p=0.007) remained significant when our level of significance was corrected to p<0.01.
In conclusion, we observed a novel association between African mtDNA subhaplogroup L3e1 and hypertriglyceridemia. Functional studies are needed to unravel the mechanism by which this subhaplogroup may increase triglycerides. We did not find associations between African mtDNA haplogroups and the other ART-related complications studied, but we may have been underpowered to identify smaller associations. Larger studies in sub-Saharan Africa, which has the highest HIV burden in the world but few pharmacogenomic studies, are needed to confirm the association we found between African mtDNA subhaplogroup L3e1 and hypertriglyceridemia, and to explore associations between mtDNA haplogroups and other HIV- and ART-related complications.
Acknowledgments
This work was supported in part by NIH grants AI-077505 and AI-054999 (D.W.H.); K23AI073141 and P30AI 060354 (C.W.W.); CIDRI and Discovery foundation awards (P.Z.S.); and Department of Health of South Africa and the World Diabetes Foundation (N.S.L., J.D.). Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1 TR000445. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
These data were presented at the 19th Conference on Retroviruses and Opportunistic Infections, Seattle, Washington, March 2012 [Abstract #605].
Author Disclosure Statement
D.W.H. has been principal investigator on research grants to Vanderbilt University from Boehringer-Ingelheim, Bristol-Gilead Sciences, and Merck. T.H. has been principal investigator on research grants to Vanderbilt University from Merck.
References
- 1.Kohler JJ. Lewis W. A brief overview of mitochondrial toxicity from NRTIs. Environ Mol Mutagen. 2007;48:166–172. doi: 10.1002/em.20223. [DOI] [PubMed] [Google Scholar]
- 2.Falutz J. HIV infection, body composition changes and related metabolic complications: Contributing factors and evolving management strategies. Curr Opin Clin Nutr Metab Care. 2011;14:255–260. doi: 10.1097/MCO.0b013e3283457a8f. [DOI] [PubMed] [Google Scholar]
- 3.Karamchand L. Dawood H. Chuturgoon AA. Lymphocyte mitochondrial depolarization and apoptosis in HIV-1-infected HAART patients. J Acquir Immune Defic Syndr. 2008;48:381–388. doi: 10.1097/QAI.0b013e3181799662. [DOI] [PubMed] [Google Scholar]
- 4.Viengchareun S. Caron M. Auclair M, et al. Mitochondrial toxicity of indinavir, stavudine and zidovudine involves multiple cellular targets in white and brown adipocytes. Antivir Ther. 2007;12:919–929. [PubMed] [Google Scholar]
- 5.Wallace DC. Brown MD. Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 6.Saxena R. de Bakker PIW. Singer K. Mootha V. Burtt N. Hirshchorn M, et al. Comprehensive association testing of common mitochondrial DNA variation. Am J Hum Genet. 2006;76:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grady B. Samuels DC. Robbins GK. Selph B. Canter JA. Pollard RB, et al. Mitochondrial genomics and CD4 T-cell count recovery after antiretroviral therapy initiation in AIDS Clinical Trials Group Study 384. J Acquir Immune Defic Syndr. 2011;58:363–370. doi: 10.1097/QAI.0b013e31822c688b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrickson SL. Hutcheson HB. Ruiz-Pesini E. Poole JC. Lautenberger J. Sezgin E, et al. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Micheloud D. Berenguer J. Guzmán-Fulgencio M. Campos Y. Garcia-Álvarez M. Catalán P, et al. European mitochondrial DNA haplogroups and metabolic disorders in HIV/HCV coinfection. J Acquir Immune Defic Syndr. 2011;58:371–378. doi: 10.1097/QAI.0b013e31822d2629. [DOI] [PubMed] [Google Scholar]
- 10.Hulgan T. Haubrich R. Riddler SA. Tebas P. Ritchie MD. McComsey GA. Haas DW. Canter JA. European mitochondrial DNA haplogroups and metabolic changes during antiretroviral therapy in AIDS Clinical Trials Group Study A5142. AIDS. 2011;25:37–47. doi: 10.1097/QAD.0b013e32833f9d02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulgan T. Tebas P. Canter JA. Mulligan K. Haas DW. Dubé M. Grinspoon S, et al. Hemochromatosis gene polymorphisms, mitochondrial haplogroups, and peripheral lipoatrophy during antiretroviral therapy. JID. 2008;197:858–866. doi: 10.1086/528697. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson SL. Kingsley LA. Ruiz-Pesini E. Poole JC. Jacobson LP. Palella FJ, et al. Mitochondrial DNA haplogroups influence lipoatrophy after highly active antiretroviral therapy. JAIDS. 2009;51:111–116. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulgan T. Haas DW. Haines JL. Ritchie MD. Robbins GK. Shafer RW, et al. Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: An adult AIDS clinical trials group study. AIDS. 2005;19:1341–1349. doi: 10.1097/01.aids.0000180786.02930.a1. [DOI] [PubMed] [Google Scholar]
- 14.Canter JA. Haas DW. Kallianpur AR. Ritchie MD. Robbins GK. Shafer W, et al. The mitochondrial pharmacogenomics of haplogroup T: MTND2*LHON4917G and antiretroviral therapy-associated peripheral neuropathy. Pharmacogenom J. 2008;8:71–77. doi: 10.1038/sj.tpj.6500470. [DOI] [PubMed] [Google Scholar]
- 15.Canter JA. Robbins GK. Selph D. Clifford DB. Kallianpur AR. Shafer R, et al. for the ACTG 384, New Work Concept Sheet 273 Study Teams. African mitochondrial DNA subhaplogroups and peripheral neuropathy during antiretroviral therapy. JID. 2010;201:1703–1707. doi: 10.1086/652419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokaze A. Ishikawa M. Matsunaga N. Yoshida M. Sekine Y. Teruya K, et al. Association of the mitochondrial DNA 5178 A/C polymorphism with serum lipid levels in the Japanese population. Hum Genet. 2001;109:521–525. doi: 10.1007/s004390100602. [DOI] [PubMed] [Google Scholar]
- 17.Lal S. Madhavan M. Heng CK. The association of mitochondrial DNA 5178 C>A polymorphism with plasma lipid levels among three ethnic groups. Ann Hum Genet. 2005;69:639–644. doi: 10.1111/j.1529-8817.2005.00192.x. [DOI] [PubMed] [Google Scholar]
- 18.Park KS. Chan JC. Chuang LM. Suzuki S. Araki E. Nanjo K, et al. A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologia. 2008;51:602–608. doi: 10.1007/s00125-008-0933-z. [DOI] [PubMed] [Google Scholar]
- 19.Dahmani Y. Marcuello A. Diez-Sanchez C. Ruiz-Pesini E. Montoya J. Lopez-Perez J. Association of human mitochondrial DNA variants with plasma LDL levels. Mitochondrion. 2008;8:247–253. doi: 10.1016/j.mito.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Feder J. Ovadia O. Blech I. Cohen J. Wainstein J. Harman-Boehm I, et al. Parental diabetes status reveals association of mitochondrial DNA haplogroup J1 with type 2 diabetes. BMC Genet. 2009;10 doi: 10.1186/1471-2350-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UNAIDS report 2010. http://www.unaids.org/documents/20101123_FS_SSA_em_en.pdf. Aug 1, 2011. http://www.unaids.org/documents/20101123_FS_SSA_em_en.pdf
- 22.Dave JA. Lambert EV. Badri M. West S. Maartens G. Levitt NS. Effect of nonnucleoside reverse transcriptase inhibitor-based antiretroviral therapy on dysglycaemia and insulin sensitivity in South African HIV-infected patients. J Acquir Immune Defic Syndr. 2011;57:284–289. doi: 10.1097/QAI.0b013e318221863f. [DOI] [PubMed] [Google Scholar]
- 23.Maritz J. Benatar M. Dave J. Harrison TB. Badri M. Levitt NS. Heckmann JM. HIV neuropathy in South Africans: Frequency, characteristics, and risk factors. Muscle Nerve. 2010;41:599–606. doi: 10.1002/mus.21535. [DOI] [PubMed] [Google Scholar]
- 24.American Diabetes Association: Diagnosis and classification of diabetes mellitus. Diabetes Care. 2008;31:S55–S60. doi: 10.2337/dc08-S055. [DOI] [PubMed] [Google Scholar]
- 25.Expert Panel on Detection, Evaluation, Treatment of High Blood Cholesterol in Adults. Executive summary of the third report of the of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) JAMA. 2001;285:2489–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 26.McComsey GA. Yau L. Asymptomatic hyperlactataemia: Predictive value, natural history and correlates. Antiviral Ther. 2004;9:205–221. [PubMed] [Google Scholar]
- 27.van Oven M. Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. http://www.phylotree.org. Hum Mutat. 2009;30(2):E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 28.Gonder MK. Mortensen HM. Reed FA. de Sousa A. Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 29.van der Walt EM. Smuts I. Taylor RW. Elson JL. Turnbull DM. Louw R. van der Westhuizen FH. Characterization of mtDNA variation in a cohort of South African paediatric patients with mitochondrial disease. Eur J Hum Genet. 2012;20:650–656. doi: 10.1038/ejhg.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montes ML. Pulido F. Barros C. Condes E. Rubio R. Cepeda C, et al. Lipid disorders in antiretroviral-naïve patients with lopinavir-ritonavir based HAART: Frequency, characterization and risk factors. JAC. 2005;55:800–804. doi: 10.1093/jac/dki063. [DOI] [PubMed] [Google Scholar]
- 31.Hegele RA. Zinman B. Hanley JG. Harris S. Connelly PW. A common mtDNA polymorphism associated with variation in plasma triglyceride concentration. Am J Hum Genet. 1997;60:1552–1555. doi: 10.1016/S0002-9297(07)64252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maechler P. Wollheim CB. Mitochondrial function in normal and diabetics ß-cells. Nature. 2001;414:807–812. doi: 10.1038/414807a. [DOI] [PubMed] [Google Scholar]
- 33.Park KS. Chan JC. Chuang L-M. Suzuki S. Araki E. Nanjo K, et al. A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologica. 2008;58:602–608. doi: 10.1007/s00125-008-0933-z. [DOI] [PubMed] [Google Scholar]
- 34.Poulton J. Luan J. Macaulay V. Hennings S. Mitchell J. Wareham J. Type 2 diabetes is associated with a common mitochondrial variant: Evidence from a population-based case-control study. Hum Mol Genet. 2002;11:1581–1583. doi: 10.1093/hmg/11.13.1581. [DOI] [PubMed] [Google Scholar]
- 35.Hocqueloux L. Alberti C. Feugeas J-P. Lafaurie M. Lukasiewicz E. Bagnard G, et al. Prevalence, risk factors and outcome of hyperlactataemia in HIV-infected patients. HIV Med. 2003;4:18–23. doi: 10.1046/j.1468-1293.2003.00132.x. [DOI] [PubMed] [Google Scholar]
- 36.Arenas-Pinto A. Weller I. Ekong R. Grant A. Karstaedt A. Reiss P, et al. Common inherited mitochondrial DNA mutations and nucleoside reverse transcriptase inhibitor-induced severe hyperlactataemia in HIV-infected adults: An exploratory study. Antivir Ther. 2012;17:275–282. doi: 10.3851/IMP1947. [DOI] [PubMed] [Google Scholar]
- 37.De Luca A. Nasi M. Di Giambenedetto S. Cozzi-Lepri A. Pinti M. Marzochetti A, et al. Mitochondrial DNA haplogroups and incidence of lipodystrophy in HIV infected patients on long-term antiretroviral therapy. J Acquir Immune Defic Syndr. 2012;59:113–120. doi: 10.1097/QAI.0b013e31823daff3.. [DOI] [PubMed] [Google Scholar]