Abstract
The human T cell lymphotropic virus type 2 (HTLV-2) is found mainly in Amerindians and in intravenous drug users (IDUs) from urban areas of the United States, Europe, and Latin America. Worldwide, HTLV-2a and HTLV-2b subtypes are the most prevalent. Phylogenetic analysis of HTLV-2 isolates from Brazil showed the HTLV-2a subtype, variant -2c, which spread from Indians to the general population and IDUs. The present study searched for the types of HTLV-2 that predominate among HIV-1-coinfected patients from southern and southeastern Brazil. Molecular characterization of the LTR, env, and tax regions of 38 isolates confirmed the HTLV-2c variant in 37 patients, and one HTLV-2b in a patient from Paraguay. Phylogenetic analysis of sequences showed different clades of HTLV-2 associated with risk factors and geographic region. These clades could represent different routes of virus transmission and/or little diverse evolutionary rates of virus. Taking into account the results obtained in the present study and the lack of the prototypic North American HTLV-2a strain and HTLV-2b subtypes commonly detected among HIV-coinfected individuals worldwide, we could speculate on the introduction of Brazilian HTLV-2 strains in such populations before the introduction of HIV.
Introduction
The human T cell lymphotropic virus type 2 (HTLV-2) is found mainly in American Indians and in intravenous drug users (IDUs) in urban areas of the United States, Europe, and Latin America.1–3 In Brazil, considered the country with the largest number of people infected by HTLVs,4 HTLV-2 is considered endemic among indigenous populations of the Amazon region and among Indians of the south region, and is present in IDUs of urban areas and in patients with HIV/AIDS.5–10
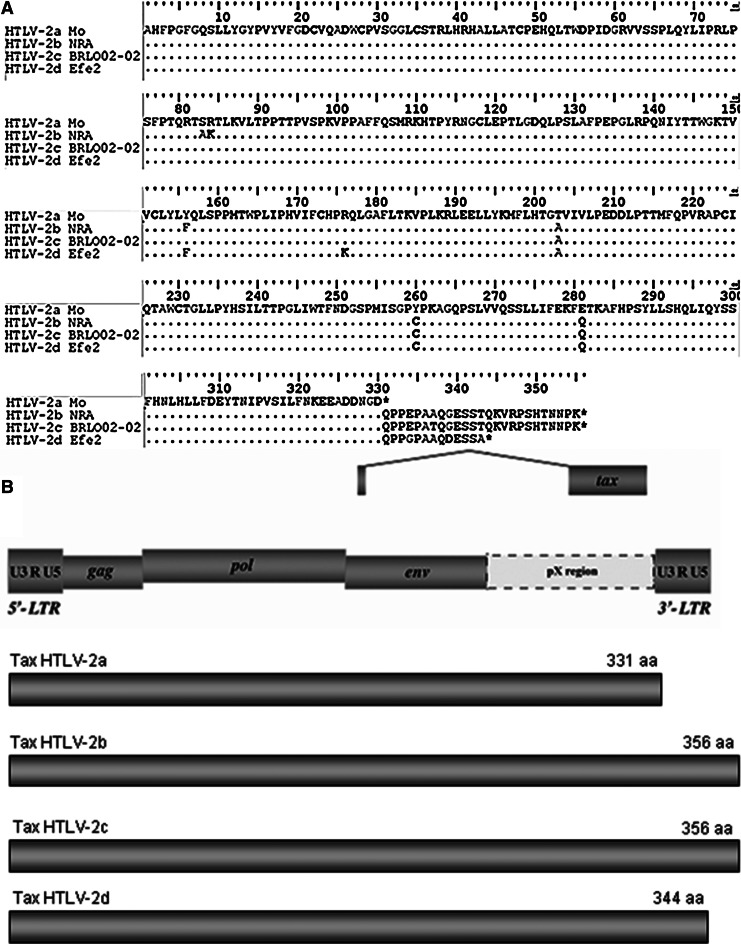
The HTLV-1/2 are low replicating viruses, thus have little genetic sequence variation.11,12 However, the env structural gene and the long terminal repeat (LTR) region containing the viral promoter are the most variable regions, thus are useful for virus subtyping.3 The LTR represents a noncoding region and consequently is not subject to the same evolutionary constraints as coding regions. The regulatory gene tax, which is engaged in viral transcription, diverges among isolates by nucleotide differences and size of the encoded Tax regulatory protein; thus this region is also valuable for molecular and epidemiological analysis.7,13,14 Still, the tax region allows us to discriminate the molecular variant of Brazilian strains named HTLV-2c, which encodes a long transactivating protein Tax because of the loss of the stop codon in the tax gene position of 2735 and the gain of 25 amino acids (Fig. 1).13 Interestingly, the Brazilian HTLV-2 strain has the tax region similar to the HTLV-2b subtype and the env and LTR genomic regions similar to the HTLV-2a subtype.7,13,15 The occurrence of the HTLV-2c variant in Brazil is intriguing and its origin is not completely clarified. It is thought that HTLV-2 reached the New World approximately 15,000 years ago by the human (Paleo Indians) migratory wave that crossed the Bering land bridge. Two different migratory routes paralleling the Andean Cordillera along the Pacific Coast and toward the Amazon region could have independently resulted in the emergence of the HTLV-2c strain, perhaps from a prototype 2a, or in the introduction of 2c exclusively into the Amazon region.7
FIG. 1.
Amino acid alignment of the tax region (A) and schematic Tax protein structure (B) among human T cell lymphotropic virus (HTLV)-2a (Mo prototype), HTLV-2b (NRA prototype), HTLV-2c (BRLO02-02), and HTLV-2d (Efe2) subtypes, highlighting the stop codon position in each sequence that results in an additional 25 amino acids at the C-terminal end of the Tax protein of HTLV-2b and HTLV-2c and 13 amino acids in HTLV-2d subtypes in relation to the HTLV-2a subtype. Note that the first amino acids are encoded in the env region not displayed in these sequence alignments.
Therefore, HTLV-2 molecular characterization worldwide disclosed four HTLV subtypes: HTLV-2a and HTLV-2b, the most prevalent among IDUs from urban areas of the Americas and Europe and in the indigenous population of the Americas, and with sporadic distribution in Asia and Africa; the HTLV-2c variant detected in the indigenous population of the Brazilian Amazon and in IDUs from urban populations in Brazil; and the HTLV-2d detected in pygmy tribes in Africa.13,16–21
In 2010, we started the characterization of HTLV-2 isolates that circulate in HIV-coinfected individuals from southern Brazil using nucleotide sequence and restriction fragment length polymorphism (RFLP) analysis of the LTR region, and confirmed that all HTLV-2 strains belonging to the 2a subtype. Moreover, we observed an association of molecular variants and sexual risk factor and injecting drug use.22 Now, in order to expand these findings, confirm the HTLV-2 subtype prevalent in HIV-coinfected individuals from the south and southeast regions, and provide information regarding the characterization and the origin of HTLV-2 in such populations, the present study was conducted.
Materials and Methods
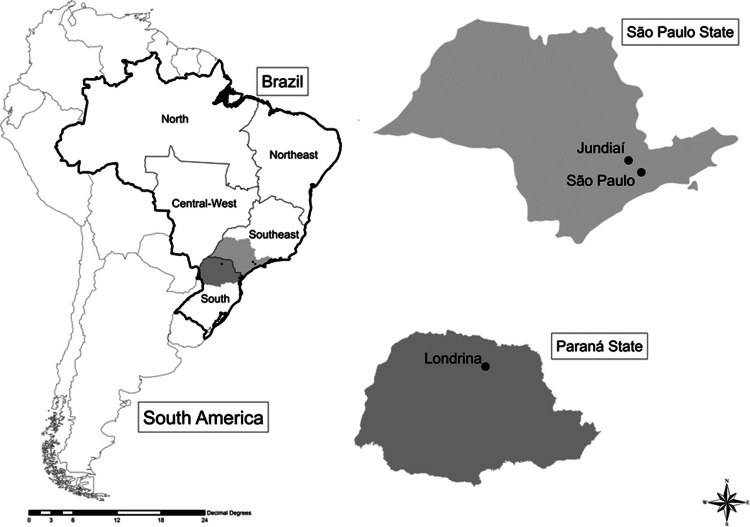
The samples analyzed consisted of blood samples obtained from 38 patients with HIV/AIDS attending the AIDS Reference Centers who were coinfected with HTLV-2: 28 from Londrina city and vicinities in Paraná (PR) state (BRLO, southern Brazil) of whom we have complete epidemiological data,9,22 and 10 from São Paulo and Jundiaí cities in São Paulo (SP) state (BRSP, southeastern Brazil) of whom we have only data concerning gender and age. The available data on such patients are described in Table 1 and the map showing the location and distances among the cities is presented in Fig. 2. Signed informed consent was obtained from patients, and the study was approved by the Ethics Committee of all participant institutions.
Table 1.
Epidemiological Data and Human T Cell Lymphotropic Virus Type 2 Sequences Accession Numbers Obtained from 38 HIV/Human T Cell Lymphotropic Virus Type 2-Coinfected Patients Attending AIDS Reference Centers in Paraná (South) and São Paulo (Southeast) States of Brazil
| Patient code | Gender/age (years) | Locality (city–state) | Risk factor | GenBank AN LTR | GenBank AN env | GenBank AN tax |
|---|---|---|---|---|---|---|
| BRLO02-02 | F/26 | Londrina–PR | IDU | HM770414 | JN887712 | |
| BRLO03-02 | F/39 | Londrina–PR | Sexual | HM770390 | JN887713 | |
| BRLO05-02 | M/56 | Londrina–PR | IDU | HM770391 | ||
| BRLO07-02 | M/41 | Londrina–PR | IDU | GU573730a | HM770392 | |
| BRLO09-02 | M/30 | Londrina–PR | IDU | GU573731a | HM770393 | JN887714 |
| BRLO10-02 | M/45 | Londrina–PR | Sexual | HM770394 | ||
| BRLO11-02 | M/35 | Londrina–PR | IDU | HM770395 | ||
| BRLO12-02 | F/33 | Londrina–PR | IDU | GU573732a | HM770396 | JN887715 |
| BRLO18-02 | F/29 | Londrina–PR | Sexual | GU573733a | HM770397 | |
| BRLO19-02 | M/34 | Londrina–PR | IDU | GU573734a | HM770398 | JN887716 |
| BRLO21-02 | F/30 | Londrina–PR | Sexual | GU573735a | HM770399 | JN887717 |
| BRLO22-02 | M/46 | Londrina–PR | Sexual | GU573736a | HM770400 | JN887718 |
| BRLO23-02 | F/39 | Londrina–PR | Sexual | GU573737a | HM770401 | |
| BRLO24-02 | M/37 | Londrina–PR | Sexual | GU573738a | HM770402 | |
| BRLO25-02 | F/33 | Londrina–PR | IDU | HM770415 | ||
| BRLO26-02 | M/37 | Londrina–PR | IDU | GU573739a | HM770403 | JN887719 |
| BRLO27-02 | F/31 | Londrina–PR | Sexual | GU573740a | HM770404 | JN887720 |
| BRLO28-02 | M/42 | Londrina–PR | Sex+IDU | GU573741a | HM770405 | JN887721 |
| BRLO29-02 | F/34 | Londrina–PR | Blood | GU573742a | HM770406 | |
| BRLO31-02 | M/29 | Londrina–PR | IDU | GU573743a | JN887722 | |
| BRLO32-02 | M/34 | Londrina–PR | IDU | HM770407 | JN887723 | |
| BRLO35-02 | M/34 | Londrina–PR | IDU | HM770408 | JN887724 | |
| BRLO37-02 | M/46 | Londrina–PR | Sex+IDU | JQ435902 | JQ435911 | JN887725 |
| BRLO38-02 | F/27 | Londrina–PR | Sexual | HM770409 | JN887726 | |
| BRLO39-02 | M/32 | Londrina–PR | IDU | HM770410 | JN887727 | |
| BRLO43-02 | M/34 | Londrina–PR | IDU | HM770411 | JN887728 | |
| BRLO45-02 | M/45 | Londrina–PR | Sexual | HM770412 | JN887729 | |
| BRLO49-02 | M/35 | Londrina–PR | Sex+IDU | HM770413 | JN887730 | |
| BRSP91-08 | M/47 | São Paulo–SP | unknown | JQ435903 | HM770416 | |
| BRSP111-08 | M/42 | São Paulo–SP | unknown | HM770417 | ||
| BRSP160-08 | M/46 | São Paulo–SP | unknown | JQ435904 | HM770418 | |
| BRSP171-08 | F/33 | Jundiaí–SP | unknown | JQ435905 | HM770423 | JN887731 |
| BRSP172-08 | M/46 | Jundiaí–SP | unknown | HM770424 | JN887732 | |
| BRSP239-08 | M/52 | São Paulo–SP | unknown | JQ435906 | HM770419 | JN887733 |
| BRSP319-08 | F/47 | São Paulo–SP | unknown | JQ435907 | HM770420 | JN887734 |
| BRSP348-08 | M/35 | Jundiaí–SP | unknown | JQ435908 | HM770425 | JN887735 |
| BRSP84-09 | M/36 | Jundiaí–SP | unknown | JQ435909 | HM770421 | |
| BRSP130-09 | M/29 | Jundiaí–SP | unknown | JQ435910 | HM770422 | JN887736 |
Sequences published by Magri et al., 2010.22
M, male; F, female; PR, Paraná state; SP, São Paulo state; IDU, intravenous drug users; AN, accession numbers; LTR, long terminal repeat.
FIG. 2.
Map of Brazil showing the location of the São Paulo state (highlighting São Paulo and Jundiaí cities, distance 37 miles) and the Paraná state (highlighting Londrina city, 330 miles from São Paulo city).
DNA samples were extracted from peripheral blood leukocytes. The established protocol for amplification and sequencing the LTR region was previously published,22 and for the env and tax regions were based on protocols previously described,13,23,24 in which one primer has undergone minor nucleotide modification and thermal cycling conditions were adjusted (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid). Still, in both protocols the polymerase chain reaction (PCR) and nested PCR were improved in stability using the GoTaq Colorless Master Mix (Promega Corporation, Madison, WI). In addition, to recover a few cases of HTLV-2-positive tax nested PCR that generated bad sequences, the sequencing was repeated with the primers described in Supplementary Table S2 and using three additional primers: Px107 Forward 5′ACC CCA TGT CAT ATT CTG CCA3′ (nt. 7713–7733), Px108 Reverse 5′AGC CTT TAC TTG GGA TTG TTT3′ (nt. 8265–8285), and Px104 Reverse 5′AAG TTC TTC TAA TCG TTT TAG3′ (nt. 7771–7791).13
Sequencing was performed using an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster, CA). All of the sequencing chromatograms were assembled and edited with Sequencher 4.7 software. Multiple alignments were performed using the Clustal W multiple-sequence alignment tool from BioEdit Sequence Alignment Editor, version 7.0.5.3, software with a reference set available in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank), in which the nucleotide and amino acid substitutions were searched. HTLV-2 Mo and HTLV-2 NRA were used as prototypic examples of HTLV-2a and HTLV-2b subtypes, respectively; the first was identified in a T cell line derived from an atypical variant of hairy cell leukemia from patient Mo and the second from a patient with CD8+ T cell lymphoproliferative disorder, both in the United States.25 HTLV-2 subtyping was screened with the NCBI Genotyping (www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi) tool website. Neighbor-joining (NJ) and maximum likelihood (ML) phylogenetic trees were constructed based on appropriate nucleotide substitution models determined by Modeltest v3.7 using PAUP v4b10 software (HKY+i+G model for the LTR and env regions and TrN+G model for the tax region), and showed similar topologies. Bootstrapping was performed with the stepwise addition algorithm for 1,000 replicates. The Efe2 (HTLV-2d) sequence was used as the outgroup. HTLV-1 was not used as the outgroup because it was previously indicated that the analysis could decrease the signal-to-noise value, resulting in serious topological errors.26 MEGA4 software was used to estimate nucleotide distances.
Results
Although all efforts were made to improve the amplification and sequencing of all HTLV-2 isolates, we were not able to sequence some genomic segments (Table 1). The protocol established for the LTR allowed us to obtain sequences of 458 bp from 23 patients (15 from the southern region, 14 previously published,22 and 8 from southeastern Brazil), sequences of 1,065 bp for env from 37 patients (27 from the southern region and 10 from southeastern Brazil), and sequences of 1,068 bp for tax from 25 patients (19 from the south and 6 from the southeast regions of Brazil). All GenBank accession numbers are presented in Table 1 and in the section Sequence Data.
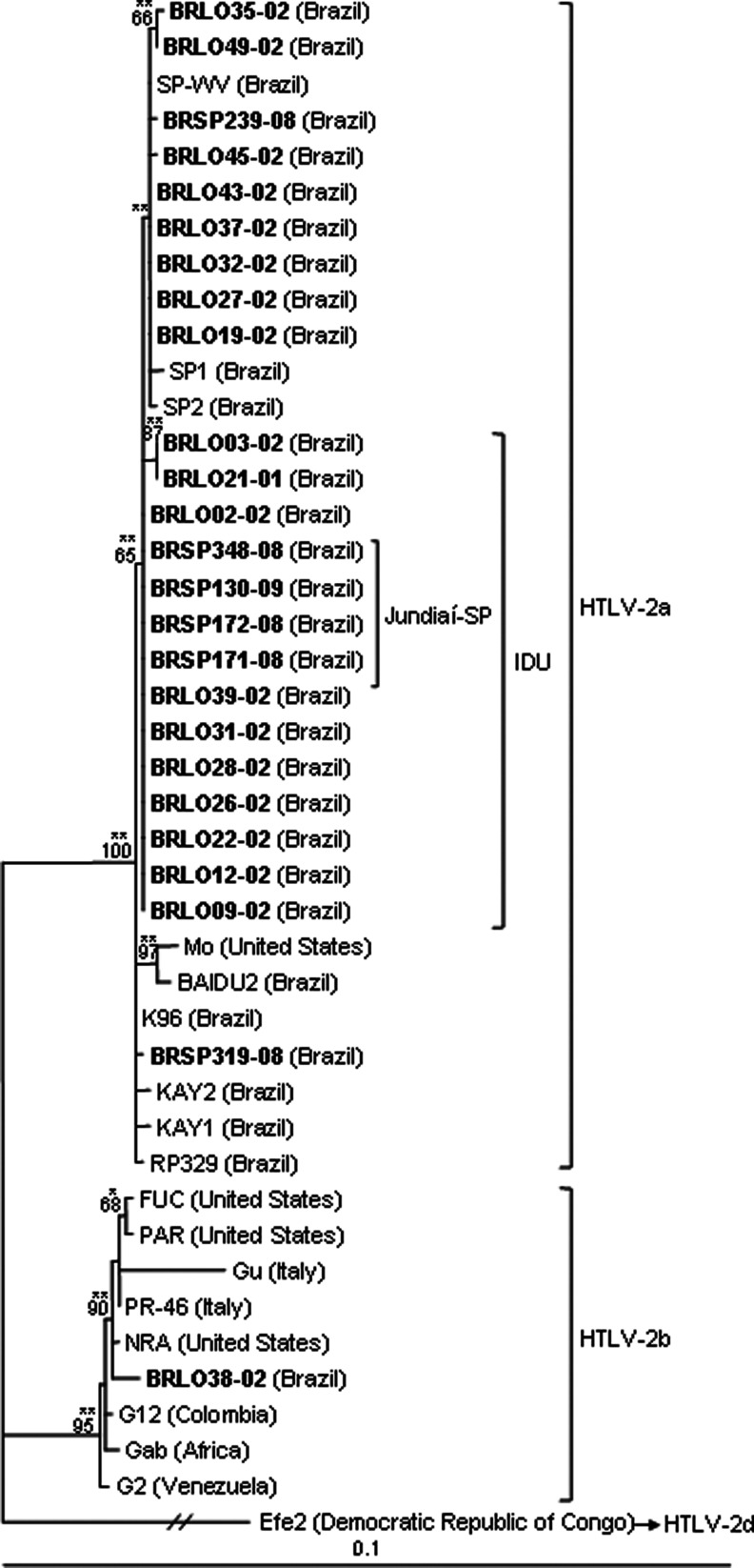
The sequences were analyzed by the NCBI Genotyping tool and by phylogeny and all belong to the HTLV-2a subtype (Figs. 3, 4, and 5), except one (BRLO38-02) that belongs to the HTLV-2b subtype (Figs. 4 and 5). The variant -2c was confirmed in all HTLV-2a isolates by tax sequence analysis using the Clustal W multiple-sequence alignment tool, which disclosed the loss of the stop codon in the tax gene position of 2735 and an additional 25 amino acids at the C-terminal end of the Tax protein (Fig. 1).
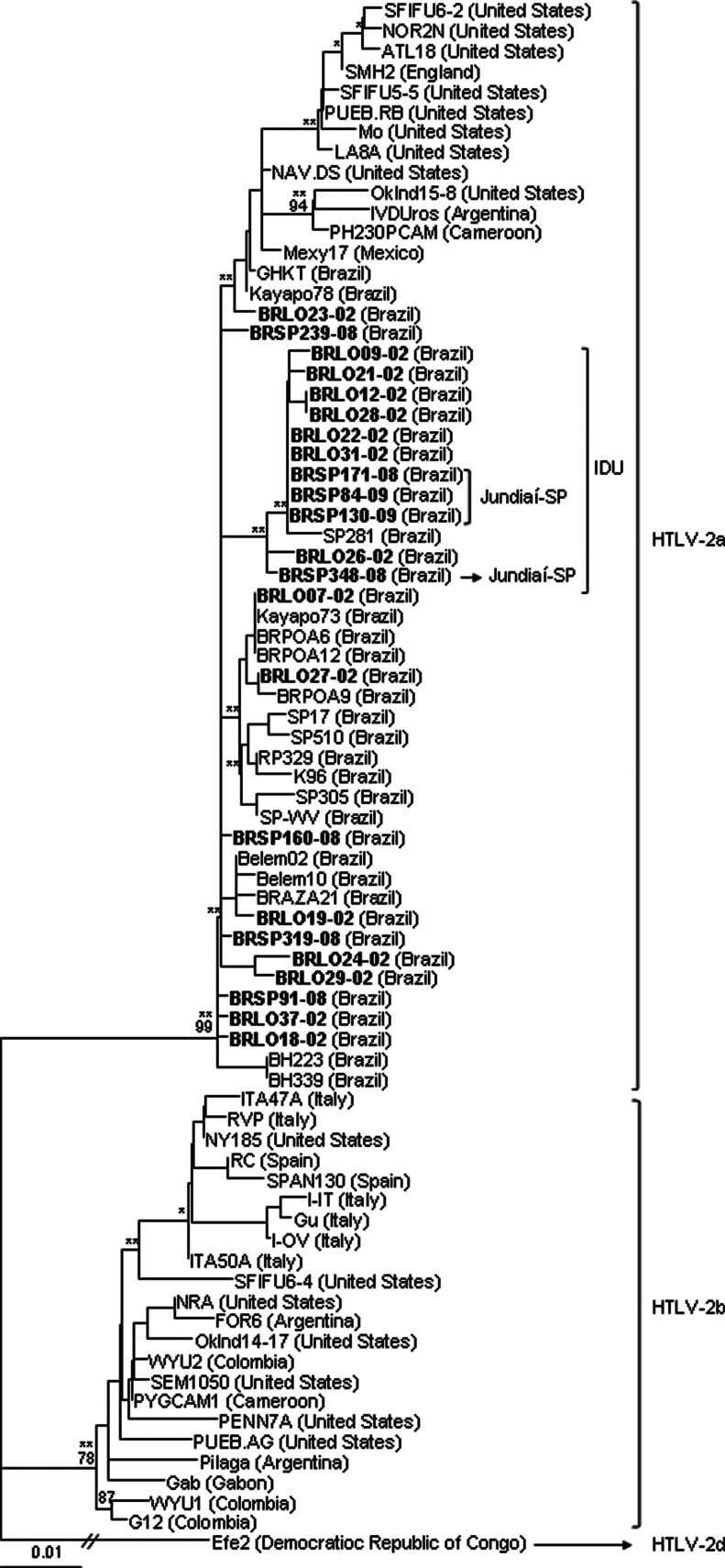
FIG. 3.
Dendrogram showing the phylogenetic relationship between 458 bp of the long terminal repeat (LTR) (nt. 181–638 in relation to the Mo prototype—AN M10060) region of the HTLV-2 strains, including sequences from the south and southeast regions of Brazil (GenBank AN GU573730–GU573743, and JQ435902–JQ435910) in bold. Bootstrap values above 65% and zero length using the likelihood ratio test with p<0.001 (**) and p≤0.05 (*) in key branches are depicted. The HTLV-2d Efe2 isolate was used as the outgroup.
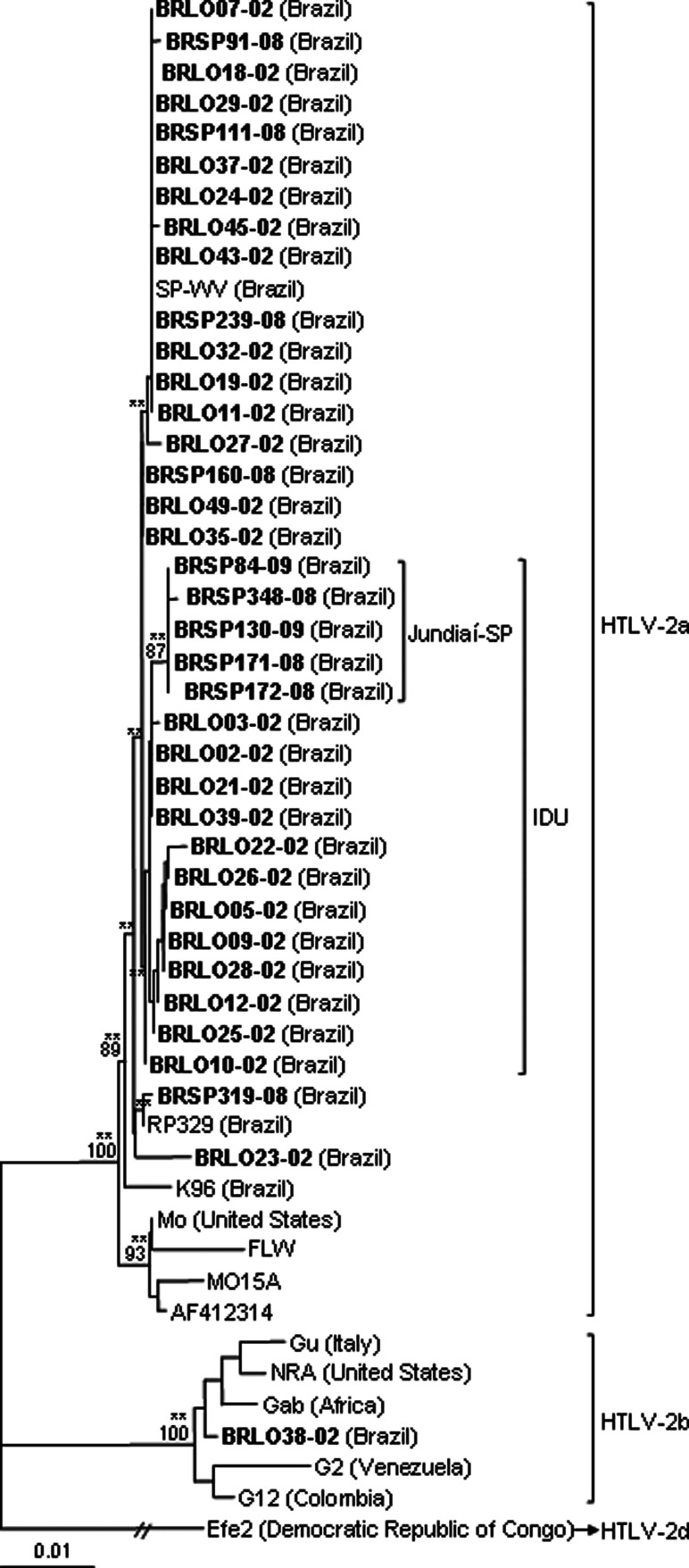
FIG. 4.
Dendrogram showing the phylogenetic relationship between 1,065 bp of the env (nt. 5573–6637 in relation to the Mo prototype—AN M10060) region of the HTLV-2 strains, including sequences from the south and southeast regions of Brazil (GenBank AN HM770390–HM770425 and JQ435911) in bold. Bootstrap values above 65% and zero length using the likelihood ratio test with p<0.001 (**) and p≤0.05 (*) in key branches are depicted. The HTLV-2d Efe2 isolate was used as the outgroup.
FIG. 5.
Dendrogram showing the phylogenetic relationship between 1,068 bp of the tax (nt. 7213–8280 in relation to the Mo prototype—AN M10060) region of the HTLV-2 strains, including sequences from the south and southeast regions of Brazil (GenBank AN JN887712–JN887736) in bold. Bootstrap values above 65% and zero length using the likelihood ratio test with p<0.001 (**) and p≤0.05 (*) in key branches are depicted. The HTLV-2d Efe2 isolate was used as the outgroup.
Phylogenetic sequence analysis of the env tree in relation to demographic and epidemiological data of patients disclosed an association between viral variants and type of exposure to the virus (IDU, p<0.001) and the place of origin of the patient (isolates from Jundiaí city in São Paulo State are present in a single clade, bootstrap=87%, p<0.001) (Fig. 4). But when the sequences were analyzed by the LTR and tax phylogenetic trees, it was observed that the sequences from Jundiaí clustered together on the IDU branch, not separately (LTR and tax: p<0.001) (Figs. 3 and 5, respectively).
Molecular analysis of the LTR, env, and tax sequences of HTLV-2, using MEGA4, showed high nucleotide similarity between the Brazilian sequences (99.3%, 99.6%, and 99.9%, respectively) and in relation to the HTLV-2a Mo prototype (97.8%, 99.2%, and 99.3%, respectively), in contrast to the low similarities detected in relation to the HTLV2-b NRA prototype (similarity of 95.2%, 95.7%, and 96.7%, respectively). When the sequences were stratified into two groups, one from Londrina and another from São Paulo, no differences were found. The similarities between the BRLO38-02 (HTLV-2b subtype) sequences and the NRA (HTLV-2b prototype) were 99.3% for the env and 99.5% for the tax region; unfortunately, the LTR region amplification did not function as required.
Regarding the presence of nucleotide substitutions in the LTR region of HTLV-2 in relation to the Mo prototype, in 100% of sequences G214A, T265C, C401T, and C551G substitutions were detected, while G316C, G317T, A448G, and T630C nucleotide substitutions were detected at frequencies of 96.3%, 96.3%, 92.6%, and 85.2%, respectively. The T315G, C320G, and G522A nucleotide substitutions were detected in 44.4% of sequences.
Concerning the presence of mutations in the env region, six nucleotide changes were very conserved among the isolates at positions T5726C, A5794G, T6109C, C6226T, C6379T, and A6580G with frequencies of 100%, 97.2%, 91.6%, 100%, 100%, and 100% relative to the Mo prototype, respectively. Among these nucleotide substitutions the only one that was a nonsynonymous mutation and thus generating an amino acid change was the T5726C. When the sequences were converted into amino acids the change S1909P was observed.
The sequences of the tax region of HTLV-2 were also analyzed for the presence of mutations in relation to the Mo prototype. The main nucleotide substitutions observed were A7819G, A7991G, A7798G, G8053C, and T8203A, with a frequency of 100%. Nucleotide substitutions C7611T, T7686C, and A7825G were detected at frequencies of 96%, 92%, and 36%, respectively. Amino acid changes of T2607A, Y2664C, and E2685Q and the loss of the stop codon 2735Q and the gain of 25 amino acids were detected in 100% of the sequences (Fig. 1). The tax sequence of the isolated BRLO38-02 was also aligned with the NRA prototype (HTLV-2b) and few nucleotide changes were detected (T7554C, T7611C, T7902C, T8031C, and G8142A), without amino acid change.
Discussion and Conclusions
The selected and optimized protocols in the present study proved useful to generate long fragments of env and tax regions of the HTLV-2 proviral genome and enabled robust phylogenetic analysis, although they still were not be able to amplify all the study samples. The sequencing of the tax region was nearly full: only lacking were the three initial nucleotides located at the beginning of the viral env gene that characterize the start codon of Tax (amino acid methionine), and by explicing encodes a protein of 356 amino acids in length (Fig. 1). The function of this long transactivator protein was poorly analyzed. There is only one study that compared the function of the Tax proteins of HTLV-1 and HTLV-2 subtypes a, b, c, and d, in CREB and NF-κB-mediated transactivation. Using the full-length HTLV-1 LTR and a 21-bp repeat reporter in 293T cells, no significant difference in Tax transactivation was detected, except for some HTLV-2a Tax isolates, including the Mo prototype. It was suggested that all HTLV-2 subtypes except the HTLV-2a subtype have a pathogenic potential equivalent to that of HTLV-1.27 Unfortunately this study was not designed to address this issue, but the Brazilian HTLV-2 isolates of the present study could help us to add information concerning this matter in the future.
The molecular characterization of the LTR, env, and tax region of HTLV-2 isolates from southern and southeastern Brazil corroborated the results obtained in a previous study of the LTR region22 and with data from the literature, in which Brazilian isolates clustered in the clade of the HTLV-2a subtype.13,15,21 The phylogenetic tree of the env region showed the presence of a group almost exclusively of IDUs, also found in the LTR and tax trees. These clades could represent a different route of virus transmission among IDUs or a little diverse evolutionary virus rate, as described in European IDUs.22,28 Furthermore, the clade in the env phylogenetic tree, which contains isolates from a specific geographic region (Jundiaí city 37 miles from São Paulo city), and the clades of the same sequences in the LTR and tax phylogenetic trees allow us to suggest that the risk factor for acquiring retroviruses in patients from Jundiaí was the use of injecting drugs. In addition, a single local monophyletic subcluster closed to IDUs suggests in situ dissemination of a local clade.
The HTLV-2b subtype found in one sample (BRLO38-02) of the present study (the env and tax regions) was associated with ethnic background; the patient was from Paraguay, a country endemic for HTLV-2b. Consistent with this finding, the HTLV-2b subtype has been found in the state of Rio Grande do Sul in Brazil,20 bordering countries where the HTLV-2b subtype prevails. We could speculate that with the commercial agreement of Mercosul, the increasing population flux across borders warrants monitoring this viral subtype in the country.
When the present sequences were compared with the HTLV-2 Mo prototype, a high nucleotide similarity of 99.2% in the env region was observed, and this result is in line with the results found in IDUs living in Salvador (99%), northeastern Brazil.19 Still, the amino acid change S1909P in 100% of the env sequences was also previously reported in Brazilian strains, and could represent a molecular signature of HTLV-2a (variant 2c) strains from Brazil.21,29
In relation to the tax sequences, the present results corroborated the previous reports that characterize the Brazilian HTLV-2c variant (often considered a molecular variant of -2a), highlighting the long Tax protein.7,13,15 In addition, high similarity in tax was observed among isolates from the present study (99.9%) and also in relation to the Mo prototype (99.3%). Therefore, it is necessary to emphasize that HTLV replication, unlike other retroviruses, is primarily through the clonal expansion of cells that are infected via mitosis and not so much by the use of the reverse transcriptase.30 During mitosis the cellular DNA polymerases are used and the new cells contain high fidelity copies of the original provirus.11 This fact could help to explain why HTLV has high genetic similarity, as opposed to HIV.31
Concerning the evolutionary rate of HTLV-2, Salemi and collaborators calibrated a molecular clock of the LTR region of HTLV-2 and estimated a fixation rate between 1.08×10−4 and 2.7×10−5 nucleotide substitutions per site per year for the 2a and 2b European IDU strains, and concluded that this rate is very low among RNA viruses.26 For example, HIV-1 (the env region) was measured up to 1.6×10−2 substitutions per site per year.32
High prevalences of HTLV-2 have been described in geographically isolated groups that should have different viral dynamics and evolution.11,26,33 Interestingly, phylogenetic analysis of LTR sequences from the Kayapo Indians of Brazil clustered the sequences in a phylogroup named A-II, in which sequences from one Mexican prostitute and two prostitutes from Ghana and Cameroon clustered together.34 This finding could suggest that HTLV-2a may have evolved from a common ancestor long before the HTLV-2a-infected ancestors of the Kayapo introduced this subtype into the Americas. Corroborating this hypothesis, Mauclère and collaborators proposed that HTLV-2 appeared in Africa, and then some of the strains left this continent during the period of human migration and became the ancestors of HTLV-2a/c or HTLV-2b. Additionally they also speculated that the HTLV-2b-bearing populations may have migrated through Asia and then separated into subgroups: some went to America and others returned to Africa. Lastly, the remaining virus became the HTLV-2d subtype.35 Also other studies agreed that HTLV-2 was originally brought from Asia into the Americas during the migration of the Asian populations over the Bering land bridge.33,36,37 These data and Mauclère's data accommodate the hypothesis of the entrance of HTLV-2 into the Americas by the Bering land bridge a long time ago.
In Europe it is believed that the -2a subtype was introduced to IDUs in Eastern Europe and the -2b in Western Europe in at least two separate periods.37 In Spain and Italy, for example, HTLV-2b remains the most prevalent subtype, although today new cases of this infection are decreasing.38,39 In Brazil, all data point to the HTLV-2c molecular variant as formerly present in Indians native tribes, with posterior dissemination to the urban population during its formation, through interethnic contact during the intense event of miscegenation, by sexual intercourse, and is maintained in Indians mostly by breast feeding.7,13,19,33,34
In conclusion, since we demonstrated the absence of the HTLV-2b and prototype North American HTLV-2a subtypes in the present study population, we suggest that these individuals had little interaction with individuals or blood products from other geographic areas, and also with individuals coinfected with HIV/HTLV-2 outside Brazil. In addition, we could speculate that there were different routes and origins of HIV and HTLV-2 in Brazil, probably through prior infection by HTLV-2 among IDUs and later on HIV. Because of the development and recent increase in the population movement in South American migration in the past years, surveillance of HTLV-2 infection is required and opportune in Brazil.
Sequence Data
The GenBank accession numbers of the 71 HTLV-2 new sequences obtained in our laboratory and included in the phylogenetic analysis are as follows: LTR: BRLO37-02 (JQ435902), BRSP91-08 (JQ435903), BRSP160-08 (JQ435904), BRSP171-08 (JQ435905), BRSP239-08 (JQ435906), BRSP319-08 (JQ435907), BRSP348-08 (JQ435908), BRSP84-09 (JQ435909), BRSP130-09 (JQ435910); env: BRLO02-02 (HM770414), BRLO03-02 (HM770390), BRLO05-02 (HM770391), BRLO07-02 (HM770392), BRLO09-02 (HM770393), BRLO10-02 (HM770394), BRLO11-02 (HM770395), BRLO12-02 (HM770396), BRLO18-02 (HM770397), BRLO19-02 (HM770398), BRLO21-02 (HM770399), BRLO22-02 (HM770400), BRLO23-02 (HM770401), BRLO24-02 (HM770402), BRLO25-02 (HM770415), BRLO26-02 (HM770403), BRLO27-02 (HM770404), BRLO28-02 (HM770405), BRLO29-02 (HM770406), BRLO32-02 (HM770407), BRLO35-02 (HM770408), BRLO37-02 (JQ435911), BRLO38-02 (HM770409), BRLO39-02 (HM770410), BRLO43-02 (HM770411), BRLO45-02 (HM770412), BRLO49-02 (HM770413), BRSP91-08 (HM770416), BRSP111-08 (HM770417), BRSP160-08 (HM770418), BRSP171-08 (HM770423), BRSP172-08 (HM770423), BRSP239-08 (HM770419), BRSP319-08 (HM770420), BRSP348-08 (HM770425), BRSP84-09 (HM770421), BRSP130-09 (HM770422); tax: BRLO02-02 (JN887712), BRLO03-02 (JN887713), BRLO09-02 (JN887714), BRLO12-02 (JN887715), BRLO19-02 (JN887716), BRLO21-02 (JN887717), BRLO22-02 (JN887718), BRLO26-02 (JN887719), BRLO27-02 (JN887720), BRLO28-02 (JN887721), BRLO31-02 (JN887722), BRLO32-02 (JN887723), BRLO35-02 (JN887724), BRLO37-02 (JN887725), BRLO38-02 (JN887726), BRLO39-02 (JN887727), BRLO43-02 (JN887728), BRLO45-02 (JN887729), BRLO49-02 (JN887730), BRSP171-08 (JN887731), BRSP172-08 (JN887732), BRSP239-08 (JN887733), BRSP319-08 (JN887734), BRSP348-08 (JN887735), BRSP130-09 (JN887736).
The GenBank accession numbers of the HTLV-2 reference sequences included in the phylogenetic study are as follows: LTR: BRLO7-02 (GU573730), BRLO9-02 (GU573731), BRLO12-02 (GU573732), BRLO18-02 (GU573733), BRLO19-02 (GU573734), BRLO21-02 (GU573735), BRLO22-02 (GU573736), BRLO23-02 (GU573737), BRLO24-02 (GU573738), BRLO26-02 (GU573739), BRLO27-02 (GU573740), BRLO28-02 (GU573741), BRLO29-02 (GU573742), BRLO31-02 (GU573743), SFIFU6 2 (U73022), NOR2N (U10258), ATL18 (U10252), SMH2 (Y09148), SFIFU5 5 (U73010), PUEBRB (U10262), Mo (M10060), LA8A (U10256), NAV.DS (U10257), Oklnd15 8 (U73015), IVDUros (AF054272), PH230PCAM (Z46838), Mexy17 (L42510), GHKT (L42507), RP329 (AF326583), K96 (AF326584), Kayapo78 (AF139388), SP-WV (AF139382), Kayapo73 (L42509), BRPOA6 (DQ028606), BRPOA12 (DQ028613), BRPOA9 (DQ028608), Belem10 (AF139393), Belem02 (AF139392), BH223 (AY509600), BH339 (AY509602), RC (L77235), SPAN130 (U10266), PortVs (AY622979), PortNn (AY622978), I-IT (Y09151), Gu (X89270), I-OV (Y09155), ITA47A (U10254), NY185 (U10259), RVP (L77244), ITA50A (U10255), SFIFU6-4 (U73018), BRPOA10 (DQ028611), FOR6 (AF054273), NRA (L20734), Oklnd14-17 (U73009), WYU2 (U12794), SEM1050 (U10263), PYGCAM1 (Z46888), PENN7A (U10260), PUEB AG (U10261), Pilaga (AF054271), Gab (Y13051), WYU1 (U12792), G12 (L11456), BRPOA11 (DQ028612), Efe2 (Y14365); env: Mo (M10060), MO15A (K02024), RP329 (AF326583), Gab (Y13051), Gu (X89270), FLW (S67545), AF412314 (AF412314), SP-WV (AF139382), G2 (AF074965), G12 (L11456), NRA (L20734), k96 (AF326584), Efe2 (Y14365); tax: Mo (M10060), K96 (AF326584), NRA (L20734), G12 (L11456), RP329 (AF326583), BAIDU2 (AF401496), SP-WV (AF139382), SP2 (U32872), SP1 (U32873), FUC (U32882), PAR (U32880), KAY1 (U32875), KAY2 (U32874), G2 (AF074965), Gab (Y13051), Gu (X89270), PR-46 (DQ022075), Efe2 (Y14365).
Supplementary Material
Acknowledgments
This study was supported by the Ministério da Ciência e Tecnologia/Conselho Nacional de Desenvolvimento Científico e Tecnológico (MCT/CNPq), Brazil (Universal Grant 481040/2007-2), a fellowship to A.C.A. (grant CNPq–PQ 303545/2012-7), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil (MSc fellowship to H.K.M. and PhD fellowship to M.C.M.), and Instituto Adolfo Lutz (grants 33/07 and 39/07). The authors are grateful to Michelle Cristina Araujo Picoli for map design.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Murphy EL. Mahieux R. De Thé G, et al. Molecular epidemiology of HTLV-II among United States blood donors and intravenous drug user: An age-cohort effect for HTLV-II RFLP type a. Virology. 1998;242:425–434. doi: 10.1006/viro.1997.9009. [DOI] [PubMed] [Google Scholar]
- 2.Zella D. Mori L. Ferrante P. Casoli C. Magnani G. Achilli G. Cattaneo E. Lori F. Bertazzoni U. HTLV-II infection in Italian drug users. Lancet. 1990;335:575–576. doi: 10.1016/0140-6736(90)92140-d. [DOI] [PubMed] [Google Scholar]
- 3.Taylor GP. The human T cell lymphotropic viruses. In: Zuckerman AJ, editor; Banatvala JE, editor; Pattison JR, editor; Griffiths PD, editor; Schoub BD, editor. Principles and Practice of Clinical Virology. 5th. John Wiley & Sons Ltd; Piscataway, NJ: 2004. pp. 759–777. [Google Scholar]
- 4.Catalan-Soares B. Carneiro-Proietti AB. Proietti FA Interdisciplinary HTLV Research Group. Heterogeneous geographic distribution of human T-cell lymphotropic viruses I and II (HTLV-I/II): Serological screening prevalence rates in blood donors from large urban areas in Brazil. Cadern Saúde Públ (Rio de J) 2005;21:926–931. doi: 10.1590/s0102-311x2005000300027. [DOI] [PubMed] [Google Scholar]
- 5.Gabbai AA. Bordini JO. Vieira-Filho JPB, et al. Selectivity of human T lymphotropic vírus type-1 (HTLV-1) and HTLV-2 infection among different populations in Brazil. Am J Trop Med Hyg. 1993;49:664–671. doi: 10.4269/ajtmh.1993.49.664. [DOI] [PubMed] [Google Scholar]
- 6.Etzel A. Shibata GY. Rozman M. Jorge MLSG. Damas CD. Segurado AAC. HTLV-1 and HTLV-2 infections in HIV infected individuals from Santos, Brazil: Seroprevalence and risk factors. J Acquir Immune Defic Syndr. 2001;26:185–190. doi: 10.1097/00042560-200102010-00015. [DOI] [PubMed] [Google Scholar]
- 7.Ishak R. Vallinoto ACR. Azevedo VN. Ishak MQ. Epidemiological aspects of retrovirus (HTLV) infection among Indian populations in the Amazon Region of Brazil. Cad Saúde púb (Rio de J) 2003;19:901–914. doi: 10.1590/s0102-311x2003000400013. [DOI] [PubMed] [Google Scholar]
- 8.Menna-Barreto M. Bender AL. Bonatto SL, et al. Human T cell lymphotropic virus type II in Guaraní Indians, Southern Brazil. Cad Saúde Púb (Rio de J) 2005;21:1947–1951. doi: 10.1590/s0102-311x2005000600045. [DOI] [PubMed] [Google Scholar]
- 9.Morimoto HK. Caterino-de-Araujo A. Morimoto AA, et al. Seroprevalence and risk factors for human T-cell lymphotropic virus type 1 and 2 infection in human immunodeficiency virus (HIV)-infected patients attending AIDS Referral Center Health Units in Londrina and other communities in Paraná, Brazil. AIDS Res Hum Retroviruses. 2005;21:256–262. doi: 10.1089/aid.2005.21.256. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira EH. Oliveira-Filho AB. Souza LA, et al. Human T-cell lymphotropic virus in patients infected with HIV-1: Molecular epidemiology and risk factors for transmission in Piauí, North-eastern Brazil. Curr HIV Res. 2012;10:700–707. doi: 10.2174/1570162x11209080700. [DOI] [PubMed] [Google Scholar]
- 11.Lemey P. Pybus OG. Van Dooren S. Vandamme AM. A Bayesian statistical analysis of human T-cell lymphotropic virus evolutionary rates. Infect Genet Evol. 2005;5:291–298. doi: 10.1016/j.meegid.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Bangham CRM. Osame M. Cellular immune response to HTLV-1. Oncogene. 2005;24:6035–6046. doi: 10.1038/sj.onc.1208970. [DOI] [PubMed] [Google Scholar]
- 13.Eiraku N. Novoa P. Ferreira MC, et al. Identification and characterization of new and distinct molecular subtype of human T-cell lymphotropic vírus type 2. J Virol. 1996;70:1481–1492. doi: 10.1128/jvi.70.3.1481-1492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feuer G. Green PL. Comparative biology of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2. Oncogene. 2005;24:5996–6004. doi: 10.1038/sj.onc.1208971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covas DM. Kashima S. Complete nucleotide sequences of the genomes of two Brazilian specimens of human T lymphotropic virus type 2 (HTLV-2) AIDS Res Hum Retroviruses. 2003;19:689–697. doi: 10.1089/088922203322280919. [DOI] [PubMed] [Google Scholar]
- 16.Ishak R. Harrington WJ. Azevedo VN, et al. Identification of human T-cell lymphotropic virus type IIa infection in the Kayapo, an indigenous population of Brazil. AIDS Res Hum Retroviruses. 1995;11:813–821. doi: 10.1089/aid.1995.11.813. [DOI] [PubMed] [Google Scholar]
- 17.Vandamme AM. Salemi M. Van Brussel M, et al. African origin of human T-lymphotropic virus type 2 (HTLV-2) supported by a potential new HTLV-2d subtype in Congolese Bambuti Efe Pygmies. J Virol. 1998;72:4327–4340. doi: 10.1128/jvi.72.5.4327-4340.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slattery JP. Franchini G. Gessain A. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 1999;9:525–540. [PubMed] [Google Scholar]
- 19.Alcantara LCJ. Shindo N. Van Doren S, et al. Brazilian HTLV type 2a strains from intravenous drug users (IDUs) appear to have originated from two sources: Brazilian Amerindians and European/North American IDUs. AIDS Res Hum Retroviruses. 2003;19:519–523. doi: 10.1089/088922203766774577. [DOI] [PubMed] [Google Scholar]
- 20.Renner JDP. Laurino JP. Menna-Barreto M. Schmitt VM. Molecular evidence of HTLV-II subtype B among an urban population living in South Brazil. AIDS Res Hum Retroviruses. 2006;22:301–306. doi: 10.1089/aid.2006.22.301. [DOI] [PubMed] [Google Scholar]
- 21.Novoa P. Penalva de Oliveira AC. Posada-Vergara MP. Duarte AJS. Casseb J. Molecular characterization of human T-cell lymphotropic virus type 2 (HTLV-II) from people living in urban areas of Sao Paulo city: Evidence of multiple subtypes circulation. J Med Virol. 2007;79:182–187. doi: 10.1002/jmv.20775. [DOI] [PubMed] [Google Scholar]
- 22.Magri MC. Morimoto HK. Brígido LFM. Rodrigues R. Caterino-de-Araujo A. Long terminal repeat sequence analysis of HTLV-2 molecular variants identified in Southern Brazil. AIDS Res Hum Retroviruses. 2010;26:1327–1331. doi: 10.1089/aid.2010.0121. [DOI] [PubMed] [Google Scholar]
- 23.Egan JF. O'Leary B. Lewis MJ, et al. High rate of human T lymphotropic virus type IIa infection in HIV type 1-infected intravenous drug abusers in Ireland. AIDS Res Hum Retroviruses. 1999;15:699–705. doi: 10.1089/088922299310782. [DOI] [PubMed] [Google Scholar]
- 24.Shindo N. Alcantara LC. Van Dooren S, et al. Human retroviruses (HIV and HTLV) in Brazilian Indians: Seroepidemiological study and molecular epidemiology of HTLV type 2 isolates. AIDS Res Hum Retroviruses. 2002;18:71–77. doi: 10.1089/088922202753394736. [DOI] [PubMed] [Google Scholar]
- 25.Lee H. Idler KB. Swanson P, et al. Complete nucleotide sequence of HTLV-II isolate NRA: Comparison of envelope sequence variation of HTLV_II isolates from U.S. blood donors and U.S. and Italian IV drug users. Virology. 1993;196:57–69. doi: 10.1006/viro.1993.1454. [DOI] [PubMed] [Google Scholar]
- 26.Salemi M. Vandamme AM. Gradozzi C, et al. Evolutionary rate and genetic heterogeneity of human T-cell lymphotropic virus type II (HTLV-II) using isolates from European injecting drug users. J Mol Evol. 1998;46:602–611. doi: 10.1007/pl00006340. [DOI] [PubMed] [Google Scholar]
- 27.Lewis MJ. Sheehy N. Salemi M. VanDamme AM. Hall WW. Comparison of CREB- and NK-B-mediated transactivation by human T lymphotropic virus type II (HTLV-II) and type I (HTLV-I) Tax proteins. Virology. 2002;295:182–189. doi: 10.1006/viro.2002.1357. [DOI] [PubMed] [Google Scholar]
- 28.Salemi M. Lewis M. Egan JF. Hall WW. Desmyter J. Vandamme A. Different population dynamics of human T cell lymphotropic virus type II in intravenous drug users compared with endemically infected tribes. Proc Natl Acad Sci USA. 1999;96:13253–13258. doi: 10.1073/pnas.96.23.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olah I. Fukumori LMI. Smid J, et al. Neither molecular diversity of the envelope, immunosuppression status, nor proviral load causes indeterminate HTLV Western blot profiles in samples from human T-cell lymphotropic virus type 2 (HTLV-2)-infected individuals. J Med Virol. 2010;82:837–842. doi: 10.1002/jmv.21718. [DOI] [PubMed] [Google Scholar]
- 30.Wattel E. Vartanian JP. Pannetier C. Wain-Hobson S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol. 1995;69:2863–2868. doi: 10.1128/jvi.69.5.2863-2868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemey P. Kosakovsky Pond SL. Drummond AJ, et al. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3(2):e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn BH. Shaw GM. Taylor ME, et al. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986;232:1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- 33.Biggar RJ. Taylor ME. Neel JV, et al. Genetic variants or human T lymphotropic virus type II in American Indian groups. Virology. 1996;216:165–173. doi: 10.1006/viro.1996.0043. [DOI] [PubMed] [Google Scholar]
- 34.Switzer WM. Black FL. Pieniazek D. Biggar RJ. Lal RB. Heneine W. Endemicity and phylogeny of the human T cell lymphotropic vírus type II subtype A from the Kayapo Indians of Brazil: Evidence for limited regional dissemination. AIDS Res Human Retroviruses. 1996;12:635–640. doi: 10.1089/aid.1996.12.635. [DOI] [PubMed] [Google Scholar]
- 35.Mauclère P. Afonso PV. Meertens L, et al. HTLV-2B strains, similar to those found in several Amerindian tribes, are endemic in central African Bakola pygmies. J Infect Dis. 2011;203:1316–1323. doi: 10.1093/infdis/jir031. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki Y. Gojobori T. The origin and evolution of the human T-cell lymphotropic virus type I and II. Virus Genes. 1998;16:69–84. doi: 10.1023/a:1007953826869. [DOI] [PubMed] [Google Scholar]
- 37.Salemi M. Vandamme AM. Desmyter J. Casoli C. Bertazzoni U. The origin and evolution of human T-cell lymphotropic virus type II (HTLV-II) and the relationship with its replication strategy. Gene. 1999;234:11–21. doi: 10.1016/s0378-1119(99)00169-9. [DOI] [PubMed] [Google Scholar]
- 38.Abad M. Dronda F. Dominguez E. Moreno S. Vallejo A. HTLV-2b among HIV type 1-coinfected injecting drug users in Spain. AIDS Res Hum Retroviruses. 2011;27:579–583. doi: 10.1089/AID.2010.0263. [DOI] [PubMed] [Google Scholar]
- 39.Salemi M. Cattaneo E. Casoli C. Bertazzoni U. Identification of IIa and IIb molecular subtypes of human T-cell lymphotropic virus type II among Italian injecting drug users. J Acquir Immune Defic Syndr. 1995;8:516–520. doi: 10.1097/00042560-199504120-00013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.