Abstract
Infections following repeated, low-dose (RLD), mucal S(H)IV exposures of macaques are used to model sexual HIV exposures for biomedical prevention testing. Different susceptibilities among animals can complicate study designs. In rhesus macaques, TRIM5 alleles Q, CypA, and TFP are resistance factors for infection with some S(H)IV strains, but not for SIVmac239 due to its capsid properties. SIVmac239-derived SHIVSF162P3 has been demonstrated to reproducibly infect mucosally in vaginal and rectal RLD models. To further test the suitability of SHIVSF162P3 for RLD models, we studied the influence of the TRIM5 genotype on susceptibility to rectal RLD infection and on plasma viremia by analyzing 43 male Indian rhesus macaques from control arms of completed studies. The median number of exposures required for infection was three (Q/Q, n=4) (TRIM5 alleles, number of macaques, respectively), four (Q/CypA, n=7), three (TFP/Q, n=15), three (TFP/TFP, n=15), and two (TFP/CypA, n=2); TRIM5CypA/CypA was not represented in our study. Median peak viremia (log10 viral copies/ml) in infected animals was 7.4 (Q/Q, n=4), 7.2 (Q/CypA, n=6), 7.3 (TFP/Q, n=13), 7.1 (TFP/TFP, n=15), and 6.5 (TFP/CypA; n=2). Neither susceptibility nor peak viremia was significantly different (log rank test, Kruskal–Wallis test, respectively). Rhesus macaques' susceptibility to RLD SHIVSF162P3 is independent of the TRIM5 TFP, CypA, and Q alleles, with the limitation that the power to detect any impact of CypA/CypA and TFP/CypA genotypes was nonexistent or low, due to absence or infrequency, respectively. The finding that TRIM5 alleles do not restrict mucosal infection or ensuing replication rates suggests that SHIVSF162P3 is indeed suitable for RLD experimentation.
The repeated-low dose (RLD) model of SIV or SHIV infection in nonhuman primates (NHPs), particularly macaques, has become a powerful tool to inform HIV biomedical intervention studies. There are many advantages to this approach as a model for HIV transmission, namely the similarity in inoculum and multiplicity in virus exposure before systemic infection to humans.1 The use of higher physiological doses has been thought to dilute the effects of microbicide or vaccine-induced immunity, thereby underestimating the potential of candidate biomedical preventions. In the RLD model, it is possible to measure susceptibility as the number of challenges required to infect with the assumption that each animal is vulnerable to infection with each challenge.2,3
One concern as it relates to NHP models is the variability in susceptibility to infection. In humans, major histocompatibility complex alleles play a role in susceptibility to HIV infection4 and they also impact the outcome of SIV infection in nonhuman primates.5,6 Another aspect of variability is the control of retroviral infection by antiviral restriction factors that are a part of innate immunity. The tripartite motif protein, TRIM5α, belongs to the tripartite motif-containing (TRIM) superfamily of proteins and has been identified as an important post-entry host restriction factor7 that inhibits retrovirus infection in a species-specific manner.8–10 The exact mechanism of TRIM5α restriction is not clear but it is known that TRIM5α proteins aggregate on the incoming retroviral capsid and decrease the stability of the capsid.9,11,12 In rhesus macaques, TRIM5 gene variants can be grouped into three allelic classes, TRIM5Q, TRIM5CypA, and TRIM5TFP, which yield six possible genotypes: TRIM5TFP/TFP, TRIM5TFP/Q, TRIM5TFP/CypA, TRIM5Q/Q, TRIM5Q/CypA, and TRIM5CypA/CypA. Cell culture assays and animal infections have been used to elucidate the role that allelic variation in the rhesus macaque TRIM5 gene has in susceptibility to SIV infection and viral replication,13–16 while other species such as cynomolgus macaques are also under investigation.17 The goal of this study was to define the influence of the TRIM5 genotype on susceptibility to SHIVSF162P3 infection in a RLD model.
We analyzed 43 male Indian rhesus macaques from control arms of studies completed at the CDC.18–21 They were purchased from diverse U.S. vendors, they were housed at the CDC according to the standards established in the Guide for the Care and Use of Laboratory Animals,22 and their use was approved by the Animal Care and Use Committee (IACUC) of the CDC. All were rectally challenged with 10 TCID50 of the same SHIVSF162P3 stock [obtained in 2001, National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (ARRRP)] for up to 14 or 16 exposures, once or twice a week. The virus stocks were not further propagated, which eliminated the potential variable of enriching a virus stock with capsid adaptations due to TRIM5 selective pressure during propagation in peripheral blood mononuclear cells (PBMCs) from individual macaques, another potential confounder in NHP challenge models. Plasma viremia was assessed by polymerase chain reaction (PCR) with a detection limit of 50 copies/ml as described previously.23 We determined the TRIM5 genotype by isolation of genomic DNA from PBMCs using the QIAamp DNA Blood Mini kit (QIAGEN, Valencia, CA) and direct bulk sequencing of a PCR fragment as previously described.13
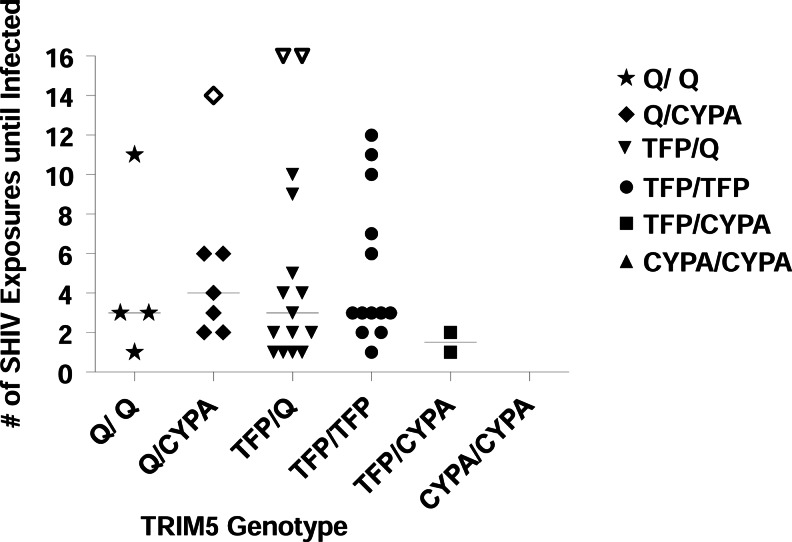
TRIM5 allelic frequencies in our cohort were as follows: 7% of the macaques were TRIM5Q/Q (n=4), 16% TRIM5Q/CypA (n=7), 35% TRIM5TFP/Q (n=15), 35% TRIM5TFP/TFP (n=15), and 5% TRIM5TFP/CypA (n=2); no macaque was TRIM5CypA/CypA. These percentages were similar to distributions recently reported from another major U.S. facility for nonhuman primate research.13,14 The median number of exposures required for infection was three (TRIM5Q/Q), four (TRIM5Q/CypA), three (TRIM5TFP/Q), three (TRIM5TFP/TFP), and two (TRIM5TFP/CypA) (Fig. 1). For statistical analysis, the three macaques that remained uninfected were assessed at the maximum number of challenges received. Differences in the number of exposures required to infect were tested using global log rank statistics. There was no statistically significant difference in the number of exposures needed for infection between the genotype groups (4 degrees of freedom, p=0.37). This suggests that the TRIM5 genotype has no marked effect on rhesus macaque susceptibility to SHIVSF162P3 or the number of challenges it takes to infect using SHIVSF162P3 in an RLD exposure model. Of note, the three uninfected animals in this study had Q/CYPA (n=1) and TFP/Q (n=2) alleles, and not TFP/CypA as reported for animals resistant to SIVsmE660 infection.14 Rather, the two macaques of the TFP/CypA phenotype were infected quickly, at exposures one and two, respectively. Moreover, animals requiring six or more exposures were found in each allele type except for TFP/CYPA (n=2).
FIG. 1.
TRIM5 genotypes versus susceptibility (number of SHIVSF162P3 challenges until Infection). The number of exposures until infection was compared in 43 rhesus macaques of indicated Trim5 genotypes. There was no correlation between the genotype and the number of exposures with SHIVSF162P3 that it took to infect (p=0.37, global log rank test). One week was subtracted for the eclipse phase when determining date of infection. The solid lines indicate the median number of SHIV exposures until infected. The open symbols represent the three uninfected animals in this study that had Q/CYPA (censored at 14 exposures) and TFP/Q (both were censored at 16 exposures) alleles.
The three uninfected macaques had Q/CYPA (n=1) and TFP/Q (n=2) alleles. The underlying resistance mechanisms remain unknown. MHC class I analysis for common Mamu alleles A*01, A*02, A*08, A*11, B*01, B*03, B*04, and B*17 revealed the presence of Mamu-A*08 in one resistant macaque, Mamu-B*01 in another, and none of these in the third macaque. Mamu-A*08 and Mamu-B*01 have not previously been associated with increased viral control.5,6 It is possible that uncharacterized Mamu alleles contributed to resistance. Many other host factors can also contribute to resistance to infection; e.g., we have previously shown that high systemic RANTES levels can render macaques resistant to SHIVSF162P3 infection in the RLD model,18 although the RANTES status of the three resistant macaques in this study was not available.
Previous work has provided evidence that TRIM5 alleles affect SIVsmE660 RLD rectal acquisition14 and penile acquisition.15 The alleles did not impact SIVmac23914 or SIVmac25124 RLD rectal acquisition, although an effect on SIVmac251 replication after intravenous acquisition was found.16,25 Contrary to SIVsmE660 challenges, we found no evidence that Q alleles conferred increased susceptibility or that TFP and CypA alleles conferred resistance to SHIVSF162P3 infection. In conclusion, SHIVSF162P3 is an addition to the list of viable strains for rhesus macaque RLD infections not affected by TRIM5 alleles. Finding a similarly suited virus strain may be the key to the development of RLD models in other species that have not been studied yet with regard to TRIM5 or other genetic restriction.
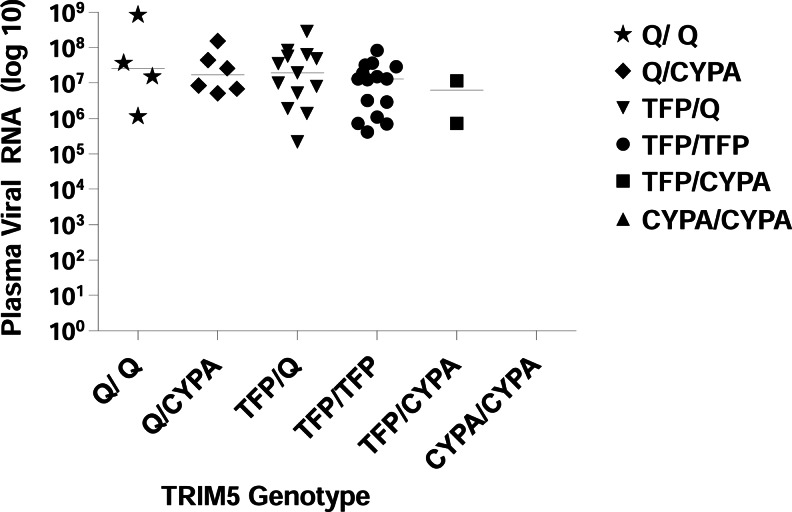
For SHIVSF162P3, peak viremia levels were also similar for macaques of different TRIM5 genotypes (Fig. 2). The Kruskal–Wallis rank sum statistic was used to test for genotype group differences in peak viral load. Following peak viremia, there were no significant differences in viral load or in viral load decay between host TRIM5 genotypes (4 degrees of freedom, p=0.45; these analyses were done using all data within 10 weeks of or after the peak viral load). A mixed-effects regression model was used to assess group differences in rate of decay after peak viral load and identified no differences (p>0.05). Last, to compare total viral burden by group, area under the curve (AUC) was determined and group comparisons were tested using Kruskal–Wallis tests, which again found no significant differences (p=0.20).
FIG. 2.
TRIM5 genotype versus peak plasma viremia with SHIVSF162P3. There was no association between the genotype and the amount of plasma viral RNA after infection with SHIVSF162P3 (p=0.45; Kruskal–Wallis test). The solid lines indicate median RNA copies/ml.
A recent study by Yeh et al. reports that susceptibility to repeated penile SIVsmE660 infection in rhesus macaques is highly dependent on the TRIM5 genotype.15 It is possible that penile or other transmission routes are more susceptible to the impact of TRIM5-mediated effects than during rectal transmission. Kirmaier et al. found that two restrictive alleles (TRIM5TFP/TFP and TRIM5TFP/CypA) led to lower SIVsmE543-3 replication than in homozygous TRIM5Q/Q macaques.13 Here, SHIVSF162P3 replication peaked at medians of 7.1, 6.5, and 7.4 log10 viral copies/ml for TFP/TFP, TFP/CypA, and Q/Q alleles, respectively, but these differences were not statistically significant. It is possible that a subtle TRIM5α impact on SHIVSF162P3 replication nevertheless exists, but could not be measured in our small sample macaque groups. Utilizing cell culture lines to measure infectivity and replication of SIVmac239 and SHIVSF162P3 for each expressed common allele would shed further light on what role TRIM5 may have in influencing susceptibility to infection.
In conclusion, our results demonstrate that rhesus macaques' susceptibility to RLD SHIVSF162P3 is independent of TRIM5 TFP, CypA, or Q alleles, with the caveat that the power to detect any impact of rare TRIM5CypA/CypA and TRIM5TFP/CypA genotypes was nonexistent or low, due to absence or infrequency, respectively. That TRIM5 alleles do not restrict mucosal infection or ensuing replication rates is perhaps not surprising because SHIVSF162P3 is a derivative of SIVmac239, which has previously been shown to infect cells and rhesus macaques irrespective of TRIM5 genotype.13,14 Our retrospective study served to provide an in vivo experimental demonstration that rhesus macaques can be reliably infected with rectal SHIVSF162P3 exposure irrespective of TRIM5 genotype. Although the result was expected due to similar studies with parent strain SIVmac23914 and due to in vitro infectivity experiments with SIVmac239,13 our findings further document in more animals that TRIM5 alleles do not confound macaque studies when SHIVSF162P3 is used at the reported dose, and using the reported stock preparation with all potential virus adaptations that can happen during virus propagation. This further documents the usefulness of this unique RLD model for preclinical HIV prevention trials.
Acknowledgments
This work was supported by the Centers for Disease Control and Prevention (CDC) and partially supported by Interagency Agreement Y1-AI-0681-02 between the CDC and the National Institutes of Health (NIH). This work was also partially supported by grants AI083118 and AI095092 from the National Institutes of Health (NIH). We would like to thank the veterinarians and animal technicians who performed the studies from which samples were obtained retrospectively. We would also like to thank past members of our research team for residual specimens that were used for this study. All authors critically reviewed this manuscript. E.N.K. thanks Gil Kersh for his support.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Author's contributions: K.B. and E.K. coordinated samples, analyzed data, and wrote the manuscript. J.M. and W.J. genotyped the samples. D.H. performed statistical analyses. D.A., G.G., W.H., and D.E. gave samples with infection and viral load data. R.M.H., J.M.N., W.H., and W.J. provided input in the study design and result interpretation. E.K. designed and led the study. All authors read, commented on, and approved the final manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Otten RA. Adams DR. Kim CN, et al. Multiple vaginal exposures to low doses of R5 simian-human immunodeficiency virus: Strategy to study HIV preclinical interventions in nonhuman primates. J Infect Dis. 2005;191(2):164–173. doi: 10.1086/426452. [DOI] [PubMed] [Google Scholar]
- 2.Regoes RR. Longini IM. Feinberg MB. Staprans SI. Preclinical assessment of HIV vaccines and microbicides by repeated low-dose virus challenges. PLoS Med. 2005;2(8):e249. doi: 10.1371/journal.pmed.0020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudgens MG. Gilbert PB. Assessing vaccine effects in repeated low-dose challenge experiments. Biometrics. 2009;65(4):1223–1232. doi: 10.1111/j.1541-0420.2009.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald KS. Fowke KR. Kimani J, et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J Infect Dis. 2000;181(5):1581–1589. doi: 10.1086/315472. [DOI] [PubMed] [Google Scholar]
- 5.Bontrop RE. Watkins DI. MHC polymorphism: AIDS susceptibility in non-human primates. Trends Immunol. 2005;26(4):227–233. doi: 10.1016/j.it.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Yant LJ. Friedrich TC. Johnson RC, et al. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80(10):5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8(1):55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stremlau M. Perron M. Lee M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman RM. Hall L. Kirmaier A, et al. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4(2):e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebastian S. Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Griffero F. Kar A. Perron M, et al. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J Virol. 2007;81(19):10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirmaier A. Wu F. Newman RM, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus, selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8(8) doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds MR. Sacha JB. Weiler AM, et al. The TRIM5{alpha} genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J Virol. 2011;85(18):9637–9640. doi: 10.1128/JVI.05074-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh WW. Rao SS. Lim SY, et al. The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J Virol. 2011;85(19):10389–10398. doi: 10.1128/JVI.00854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim SY. Rogers T. Chan T, et al. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 2010;6(1):e1000738. doi: 10.1371/journal.ppat.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietrich EA. Brennan G. Ferguson B. Wiseman RW. O'Connor D. Hu SL. Variable prevalence and functional diversity of the antiretroviral restriction factor TRIMCyp in Macaca fascicularis. J Virol. 2011;85(19):9956–9963. doi: 10.1128/JVI.00097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kersh EN. Luo W. Adams DR, et al. Repeated rectal SHIVSF162P3 exposures do not consistently induce sustained T cell responses prior to systemic infection in the repeat-low dose preclinical macaque model. AIDS Res Hum Retroviruses. 2009;25(9):905–917. doi: 10.1089/aid.2008.0287. [DOI] [PubMed] [Google Scholar]
- 19.Parikh UM. Dobard C. Sharma S, et al. Complete protection from repeated vaginal simian-human immunodeficiency virus exposures in macaques by a topical gel containing tenofovir alone or with emtricitabine. J Virol. 2009;83(20):10358–10365. doi: 10.1128/JVI.01073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kersh EN. Luo W. Adams DR, et al. No evidence of occult SHIV infection as demonstrated by CD8+ cell depletion after chemoprophylaxis-induced protection from mucosal infection in rhesus macaques. AIDS Res Hum Retroviruses. 2008;24(4):543–546. doi: 10.1089/aid.2007.0222. [DOI] [PubMed] [Google Scholar]
- 21.Aidoo M. Otten RA. Rodriguez V, et al. Absence of SHIV infection in gut and lymph node tissues in rhesus monkeys after repeated rectal challenges following HIV-1 DNA/MVA immunizations. Vaccine. 2007;25(35):6474–6481. doi: 10.1016/j.vaccine.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Council NR. 8th. The National Academies Press; Washington, D.C.: 2011. Guide for the Care and Use of Laboratory Animals. [PubMed] [Google Scholar]
- 23.Subbarao S. Otten RA. Ramos A, et al. Chemoprophylaxis with tenofovir disoproxil fumarate provided partial protection against infection with simian human immunodeficiency virus in macaques given multiple virus challenges. J Infect Dis. 2006;194(7):904–911. doi: 10.1086/507306. [DOI] [PubMed] [Google Scholar]
- 24.Fenizia C. Keele BF. Nichols D, et al. TRIM5alpha does not affect simian immunodeficiency virus SIV(mac251) replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure. J Virol. 2011;85(23):12399–12409. doi: 10.1128/JVI.05707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim SY. Chan T. Gelman RS, et al. Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 2010;6(6):e1000997. doi: 10.1371/journal.pgen.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]