Abstract
The Investigator DIPplex® kit (Qiagen) contain components for the simultaneous amplification and analysis of 30 biallelic autosomal INDELs and amelogenin. The objective of this study was to estimate the diversity of the 30 markers in Polish (N P = 122) and Taiwanese (N T = 126) population samples and to evaluate their usefulness in forensic genetics. All amplicon lengths were shorter than 160 base pairs. The DIPplex genotype distributions showed no significant deviation from Hardy–Weinberg rule expectations (Bonferroni corrected) except for DLH39 in the Taiwanese population. Among the Poles and the Taiwanese the mean observed heterozygosity values are 0.4385 and 0.4079, and the combined matching probability values are 7.98 × 10−14 and 1.22 × 10−11, respectively. The investigated marker set has been confirmed as a potential extension to standard short tandem repeat-based kits or a separate informative system for individual identification and kinship analysis. Eight INDELs have been selected as possible ancestry informative single-nucleotide polymorphisms for further analyses.
Keywords: INDEL polymorphism, Investigator DIPplex, Poland, Taiwan, Population genetics, Forensic efficiency
Introduction
INDELs (insertion–deletion) or DIPs (deletion–insertion polymorphisms) are short length diallelic polymorphisms, consisting of the presence or absence of short sequences (typically 1–50 bp). They are relatively common throughout the human genome representing 15–20 % of all polymorphisms [1] with the total number estimated at about 2 million [2]. Short amplicon size (50–150 bp), low mutation rate (<2 × 10−8), and capacity to multiplex (30–40 markers) and type using a single multiplexed PCR with fluorescently labeled primers followed by capillary electrophoresis (a current technology for human identification) [3–5] are the main advantages that make INDELs useful in forensic genetics applications including individual identification, kinship testing, population studies and ancient DNA analysis [6–8]. The Investigator DIPplex® kit (Qiagen) contain components for the simultaneous amplification and analysis of 30 biallelic autosomal INDELs and amelogenin. The INDELs are distributed over 19 autosomes at the minimum distance of 10 Mbp to routinely used STR and SNP markers. The allele length variations of the INDELs are between 4 and 22 bp, and all amplicons are shorter than 160 bp.
DNA extraction
Buccal swabs were anonymized and collected from unrelated volunteers along with information on the birthplace and ethnicity of the donor. Signed informed consents were obtained from all the participants and this study complied with the protocol approved by the Ethical Committee of Poznan University of Medical Sciences (Ref: 139/13). The population sample sizes were: Poles (N P = 122), and Taiwanese (N T = 126). The extraction of genomic DNA was carried out using QIAamp® DNA Mini Kit (Qiagen). The quantitation was performed using Quantifiler™ Human DNA Quantification Kit on a 7500 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s specifications. The samples were then normalized to 100 pg/μl and stored at −20 °C until amplification.
Amplification and genotyping
PCR conditions were applied according to the protocol recommended by the manufacturer of the Investigator DIPplex Kit (Qiagen) in PCR System 9700 (Applied Biosystems, USA) with a total reaction volume adjusted to 5 μl containing 1.8 μl nuclease-free water, 1.0 μl reaction mix A, 1.0 μl primer mix, 0.2 μl MultiTaq2 polymerase, and 100 pg DNA template. Control DNA XY5 was used to test performance of the DIPplex Kit. The amplification was performed with 30 PCR cycles. Electrophoresis and typing were performed in 3130 Genetic Analyzer (Applied Biosystems, USA) using a 36 cm capillary array and a denaturing polymer POP-4. BTO 550 (Qiagen) was used as the internal lane standard spanning fragments from 60 to 550 bps. Prior to the analysis, a five dye matrix standard (BT5) was established with the fluorescent labels dyes 6-FAM, BTG, BTY, BTR, and BTO under the Any5Dye virtual filter. Samples were injected for 10 s at 3 kV and electrophoresed for 1000 s at 15 kV at a run temperature of 60 °C. The data were collected using Data Collection v3.0 software. GeneMapper® ID-X v1.1.1 software was used for the INDELs classification.
Statistical analysis
Estimates for genetic diversity (allele frequencies, heterozygosity), conformance to expectations of the Hardy–Weinberg equilibrium (HWE) and for independence (Linkage Disequilibrium, LD) were obtained using GDA v1.0 software [9]. For multiple comparisons, the original significance levels achieved (P values) were transformed by the Bonferroni correction procedure [10], i.e. 30 markers per database yield an actual significance level of 0.0016667. Forensic informativeness was estimated by calculating discrimination power (DP), match probability (MP), polymorphic information content (PIC), typical paternity index (TPI), and power of paternity exclusion (PE) using Powerstats v1.2 spreadsheet (Promega) [11]. Comparison of allele frequency distributions was performed by means of a pairwise population comparison test (R × C contingency test; G. Carmody, Ottawa, Canada). AMOVA and population differentiation exact test were calculated with the Arlequin v.3.5 software [12].
Results and discussion
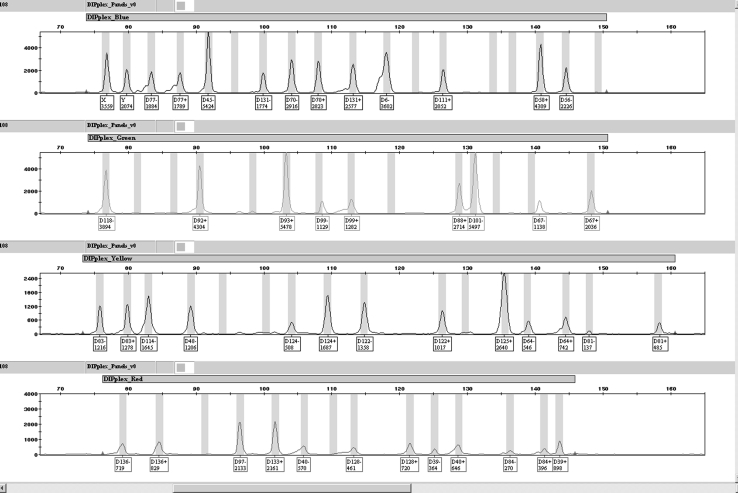
A representative DIPplex profile obtained from amplification of 100 pg DNA template is presented in Fig. 1. In the Polish population sample the INDELs frequency distributions showed no deviations from HWE (Bonferroni corrected, 0.0025 < P < 1.0000) evaluated by randomization procedure (10,000 cycles). Pairwise comparison using the exact test disequilibrium analysis with 16,000 permutation steps yielded departures from independence for 93 out of 435 pairs of INDELs under the analysis (0.0019 < P < 0.0480) (data not shown). The departures appeared statistically insignificant when the Bonferroni correction was used for the number of analysed loci. Observed heterozygosity for all the systems ranged 0.3525–0.5164, with an average of 0.4385, which is slightly lower than the values reported for Czech [6], German [13], Danish [14], Finnish [15], Central Spain, and the Basque Country populations [16]. In the Taiwanese population sample the INDELs frequency distributions showed no deviations from HWE (0.0032 < P < 1.0000) except for DLH39 (P = 0.0005). There were no statistically significant departures from independence between any pair-wise combination of INDELs (0.0018 < P < 0.0597) (data not shown). Observed heterozygosity for all the systems ranged 0.1270–0.6191, with an average of 0.4079, which corresponds to the values reported for Asian-Americans, and African-Americans [17]. The highest DP loci were HLD114 (DP = 0.660) for Poles and HLD118 (DP = 0.656) for Taiwanese. Based on data of the 30 INDELs the combined MP value among Poles amounts 7.98 × 10−14 which is more than two orders of magnitude lower than the value calculated for the Taiwanese population (1.22 × 10−11). Both parameters however, indicate a favourable value of a random match comparable with that of AmpFlSTR SGM kit [18, 19]. The combined values of PE are 0.9900 versus 0.9884, correspondingly (Table 1).
Fig. 1.
Representative DIPplex profile obtained from amplification of 100 pg DNA template
Table 1.
Population data and forensic efficiency parameters for 30 DIPplex INDELs in Polish (N P = 122) and Taiwanese (N T = 126) population samples
| HLD | Chromosomal location | GenBank SNP ID | Poles | Taiwanese | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -/DIP/ | P | Ho | He | PIC | MP | PE | TPI | -/DIP/ | P | Ho | He | PIC | MP | PE | TPI | ||||
| 1 | 77 | 7q31.1 | rs1611048 | 0.4221 | 0.0859 | 0.4098 | 0.4899 | 0.37 | 0.352 | 0.115 | 0.84 | 0.5079 | 0.5971 | 0.4762 | 0.5019 | 0.37 | 0.364 | 0.168 | 0.95 |
| 2 | 45 | 2q31.1 | rs2307959 | 0.4672 | 0.7044 | 0.4754 | 0.4999 | 0.37 | 0.366 | 0.167 | 0.95 | 0.3253 | 0.0417 | 0.5238 | 0.4408 | 0.34 | 0.449 | 0.209 | 1.05 |
| 3 | 131 | 7q36.2 | rs1611001 | 0.3811 | 0.6875 | 0.4508 | 0.4767 | 0.36 | 0.382 | 0.148 | 0.91 | 0.7063 | 0.8310 | 0.4286 | 0.4165 | 0.33 | 0.432 | 0.132 | 0.88 |
| 4 | 70 | 6q16.1 | rs2307652 | 0.5000 | 0.1113 | 0.4262 | 0.5021 | 0.38 | 0.346 | 0.131 | 0.87 | 0.3413 | 0.2355 | 0.3968 | 0.4514 | 0.35 | 0.390 | 0.112 | 0.83 |
| 5 | 6 | 16q13 | rs1610905 | 0.4509 | 0.0025 | 0.3607 | 0.4972 | 0.37 | 0.340 | 0.096 | 0.79 | 0.4762 | 0.4729 | 0.5397 | 0.5009 | 0.37 | 0.398 | 0.225 | 1.09 |
| 6 | 111 | 17p11.2 | rs1305047 | 0.4754 | 0.0181 | 0.3934 | 0.5008 | 0.37 | 0.340 | 0.110 | 0.82 | 0.8333 | 0.1096 | 0.2381 | 0.2789 | 0.24 | 0.569 | 0.041 | 0.66 |
| 7 | 58 | 5q14.1 | rs1610937 | 0.5738 | 1.0000 | 0.4918 | 0.4911 | 0.37 | 0.382 | 0.180 | 0.98 | 0.5635 | 0.5866 | 0.5238 | 0.4939 | 0.37 | 0.396 | 0.209 | 1.05 |
| 8 | 56 | 4q25 | rs2308292 | 0.3320 | 0.6944 | 0.4262 | 0.4481 | 0.35 | 0.401 | 0.125 | 0.86 | 0.3810 | 1.0000 | 0.4762 | 0.4735 | 0.36 | 0.392 | 0.168 | 0.95 |
| 9 | 118 | 20p11.1 | rs16438 | 0.5902 | 0.0975 | 0.4098 | 0.4857 | 0.37 | 0.359 | 0.125 | 0.86 | 0.0953 | 0.0139 | 0.1270 | 0.1730 | 0.16 | 0.725 | 0.013 | 0.57 |
| 10 | 92 | 11q22.2 | rs17174476 | 0.5820 | 0.1844 | 0.4262 | 0.4886 | 0.37 | 0.360 | 0.131 | 0.87 | 0.5317 | 0.1494 | 0.4286 | 0.5000 | 0.37 | 0.349 | 0.132 | 0.88 |
| 11 | 93 | 12q22 | rs2307570 | 0.4631 | 1.0000 | 0.5000 | 0.4993 | 0.37 | 0.378 | 0.188 | 1.00 | 0.4206 | 0.4664 | 0.5238 | 0.4893 | 0.37 | 0.400 | 0.209 | 1.05 |
| 12 | 99 | 14q23.1 | rs2308163 | 0.4057 | 1.0000 | 0.4836 | 0.4842 | 0.37 | 0.385 | 0.174 | 0.97 | 0.1587 | 0.0032 | 0.1905 | 0.2681 | 0.23 | 0.597 | 0.027 | 0.62 |
| 13 | 88 | 9q22.32 | rs8190570 | 0.5697 | 0.3678 | 0.4508 | 0.4923 | 0.37 | 0.364 | 0.148 | 0.91 | 0.4524 | 0.0301 | 0.3968 | 0.4974 | 0.37 | 0.344 | 0.112 | 0.83 |
| 14 | 101 | 15q26.1 | rs2307433 | 0.4180 | 0.5856 | 0.4590 | 0.4886 | 0.37 | 0.370 | 0.154 | 0.92 | 0.5397 | 0.2804 | 0.4444 | 0.4978 | 0.37 | 0.360 | 0.143 | 0.90 |
| 15 | 67 | 5q33.2 | rs1305056 | 0.3934 | 0.0450 | 0.3852 | 0.4775 | 0.36 | 0.361 | 0.110 | 0.82 | 0.3413 | 0.6911 | 0.4286 | 0.4514 | 0.35 | 0.397 | 0.132 | 0.88 |
| 16 | 83 | 8p22 | rs2308072 | 0.5287 | 0.8519 | 0.5164 | 0.5004 | 0.37 | 0.385 | 0.202 | 1.03 | 0.5794 | 0.1457 | 0.5556 | 0.4893 | 0.37 | 0.420 | 0.241 | 1.13 |
| 17 | 114 | 17p13.3 | rs2307581 | 0.6885 | 0.3900 | 0.3934 | 0.4307 | 0.34 | 0.410 | 0.110 | 0.82 | 0.7143 | 1.0000 | 0.4127 | 0.4098 | 0.32 | 0.435 | 0.122 | 0.85 |
| 18 | 48 | 2q11.2 | rs28369942 | 0.4385 | 0.1988 | 0.4344 | 0.4945 | 0.37 | 0.356 | 0.136 | 0.88 | 0.6667 | 1.0000 | 0.4444 | 0.4462 | 0.35 | 0.407 | 0.143 | 0.90 |
| 19 | 124 | 22q12.3 | rs6481 | 0.3606 | 0.4328 | 0.4262 | 0.4631 | 0.35 | 0.385 | 0.131 | 0.87 | 0.4641 | 0.2831 | 0.5556 | 0.5015 | 0.37 | 0.408 | 0.241 | 1.13 |
| 20 | 122 | 21q22.11 | rs8178524 | 0.5451 | 0.7106 | 0.5164 | 0.4980 | 0.37 | 0.388 | 0.202 | 1.03 | 0.8333 | 0.7486 | 0.2698 | 0.2789 | 0.24 | 0.562 | 0.052 | 0.68 |
| 21 | 125 | 22q11.23 | rs16388 | 0.5779 | 0.8525 | 0.4754 | 0.4886 | 0.37 | 0.379 | 0.174 | 0.97 | 0.5238 | 0.5990 | 0.4762 | 0.5009 | 0.37 | 0.365 | 0.168 | 0.95 |
| 22 | 64 | 5q12.3 | rs1610935 | 0.4631 | 0.1375 | 0.4262 | 0.4987 | 0.37 | 0.347 | 0.125 | 0.86 | 0.1429 | 1.0000 | 0.2540 | 0.2459 | 0.21 | 0.598 | 0.046 | 0.67 |
| 23 | 81 | 7q21.3 | rs17879936 | 0.5574 | 0.4697 | 0.4590 | 0.4954 | 0.37 | 0.364 | 0.154 | 0.92 | 0.2778 | 0.0069 | 0.3016 | 0.4028 | 0.32 | 0.434 | 0.064 | 0.72 |
| 24 | 136 | 22q13.1 | rs16363 | 0.5328 | 1.0000 | 0.4918 | 0.4999 | 0.37 | 0.373 | 0.180 | 0.98 | 0.5635 | 0.0062 | 0.6191 | 0.4939 | 0.37 | 0.464 | 0.314 | 1.31 |
| 25 | 133 | 3p22.1 | rs2067235 | 0.4877 | 0.1463 | 0.4344 | 0.5018 | 0.37 | 0.349 | 0.136 | 0.88 | 0.6429 | 1.0000 | 0.4603 | 0.4610 | 0.35 | 0.398 | 0.155 | 0.93 |
| 26 | 97 | 13q12.3 | rs17238892 | 0.5041 | 0.0728 | 0.4180 | 0.5020 | 0.37 | 0.364 | 0.148 | 0.91 | 0.6111 | 0.7100 | 0.4603 | 0.4772 | 0.36 | 0.382 | 0.155 | 0.93 |
| 27 | 40 | 1p32.3 | rs2307956 | 0.4959 | 0.2106 | 0.4426 | 0.5021 | 0.37 | 0.353 | 0.145 | 0.90 | 0.3571 | 0.1250 | 0.3968 | 0.4610 | 0.35 | 0.388 | 0.116 | 0.84 |
| 28 | 128 | 1q31.3 | rs2307924 | 0.5451 | 0.0175 | 0.3852 | 0.4980 | 0.37 | 0.341 | 0.107 | 0.82 | 0.6508 | 0.0775 | 0.3810 | 0.4563 | 0.35 | 0.380 | 0.106 | 0.82 |
| 29 | 39 | 1p22.1 | rs17878444 | 0.5820 | 0.0059 | 0.3525 | 0.4842 | 0.37 | 0.347 | 0.083 | 0.76 | 0.8254 | 0.0005 | 0.1905 | 0.2894 | 0.25 | 0.576 | 0.027 | 0.62 |
| 30 | 84 | 8q24.12 | rs3081400 | 0.4631 | 0.1550 | 0.4344 | 0.5012 | 0.37 | 0.364 | 0.148 | 0.91 | 0.2698 | 0.0390 | 0.3175 | 0.3956 | 0.32 | 0.440 | 0.071 | 0.73 |
-/DIP/ frequency of deletion allele, P probability value for HWE, H o observed heterozygosity, H e expected heterozygosity, PIC polymorphic information content, MP match probability, PE power of exclusion, TPI typical paternity index
A pairwise testing for heterogeneity using the χ2-test was applied to compare allelic distributions. Minor or no significant differences were found between the Polish sample and Czech [6], Danish [14], Finnish [15], and American-Caucasian [17] data sets. Correspondingly, the comparison between the Taiwanese sample and Asian-Americans [17] yielded no significant differences (0.032 < P < 1.000). On the other hand, among differences revealed between the Poles and the Taiwanese at 14 INDELs (P < 0.05), these at HLD131, HLD111, HLD118, HLD99, HLD48, HLD122, HLD64, HLD81, HLD39, and HLD84 remained significant after the critical value was corrected for multiple testing (Table 2). It is noteworthy that the same loci significantly accounted for diversity between Caucasian and Asian samples, based on North American datasets published elsewhere [17].
Table 2.
P values of population differentiation tested by an exact test and population specific F ST indices per polymorphic locus (absolute values)
| HLD | P value | F ST Poles | F ST Taiwanese | Average F ST |
|---|---|---|---|---|
| 77 | 0.202 | 0.0108 | 0.0107 | 0.0107 |
| 45 | 0.043 | 0.0371 | 0.0376 | 0.0373 |
| 131 | 0.000 | 0.1893 | 0.1897 | 0.1895 |
| 70 | 0.022 | 0.0463 | 0.0468 | 0.0466 |
| 6 | 0.771 | −0.0027 | −0.0028 | −0.0028 |
| 111 | 0.000 | 0.2450 | 0.2468 | 0.2459 |
| 58 | 0.887 | 0.0331 | 0.0331 | 0.0331 |
| 56 | 0.460 | 0.0013 | 0.0011 | 0.0012 |
| 118 | 0.000 | 0.4260 | 0.4282 | 0.4271 |
| 92 | 0.477 | 0.0011 | 0.0010 | 0.0011 |
| 93 | 0.569 | −0.0004 | −0.0003 | −0.0004 |
| 99 | 0.000 | 0.1361 | 0.1381 | 0.1371 |
| 88 | 0.120 | 0.0232 | 0.0232 | 0.0232 |
| 101 | 0.089 | 0.0253 | 0.0252 | 0.0253 |
| 67 | 0.463 | 0.0017 | 0.0019 | 0.0018 |
| 83 | 0.477 | 0.0011 | 0.0012 | 0.0012 |
| 114 | 0.758 | −0.0026 | −0.0024 | −0.0025 |
| 48 | 0.001 | 0.0962 | 0.0966 | 0.0964 |
| 124 | 0.151 | 0.0440 | 0.0437 | 0.0439 |
| 122 | 0.000 | 0.1732 | 0.1751 | 0.1741 |
| 125 | 0.394 | 0.0019 | 0.0018 | 0.0019 |
| 64 | 0.000 | 0.2132 | 0.2154 | 0.2143 |
| 81 | 0.000 | 0.1452 | 0.1459 | 0.1455 |
| 136 | 0.670 | −0.0022 | −0.0021 | −0.0021 |
| 133 | 0.032 | 0.0438 | 0.0441 | 0.0440 |
| 97 | 0.118 | 0.0189 | 0.0191 | 0.0190 |
| 40 | 0.046 | 0.0346 | 0.0349 | 0.0347 |
| 128 | 0.113 | 0.0189 | 0.0192 | 0.0190 |
| 39 | 0.000 | 0.1287 | 0.1305 | 0.1296 |
| 84 | 0.000 | 0.0733 | 0.0742 | 0.0737 |
Italicised significant differentiation test P values, after Bonferroni correction
Wright’s F ST was analysed to measure population substructure effects [20]. AMOVA results revealed that most of the molecular variation was due to variation within the analysed populations (92.54 %) rather than among them, with average fixation index values of 0.0743 and 0.0749 (Poles and Taiwanese, respectively). Our findings correspond to those presented by other authors who used AMOVA to compare the allelic frequencies for each DIPplex locus in populations of Europe, Africa, Asia and North America [16, 17]. Moreover, in our analysis individual INDELs displayed noticeable disparities in fixation index spanning from −0.0004 to −0.0003 (HLD93) to 0.4260 and 0.4282 (HLD118) for Poles and Taiwanese, respectively (Table 2). The individual mutation rate of a locus is one of the factors that may explain the observed discrepancy [21]. However, when compared with mutation rates of 10−3–10−5 for STRs [22, 23], SNPs have essentially mutation rates estimated at as low as 10−8 [24]. From the point of view of forensic genetics, markers with high heterozygosity and very low F ST are potentially advantageous due to relatively high discrimination efficiency irrespective of population of origin [24, 25]. High heterozygosity enhances the polymorphism information at each SNP and low F ST diminishes the chance of interpopulation effects. Some SNPs are reported to have remarkably little variation in allele frequency around the world [26]. On the other hand, ancestry informative single-nucleotide polymorphisms (AISNPs) are required to show low heterozygosity and high allele frequency divergence between different ancestral or geographically distant populations (F ST values). These genetic markers are especially useful in establishing the high probability of an individual’s biogeographical ancestry [27, 28]. We have selected eight INDELs (HLD131, HLD111, HLD118, HLD99, HLD122, HLD64, HLD81, HLD39) with F ST higher than 0.1 between Poles and Taiwanese as potential AISNPs for further analyses. Other sets of population data are needed to verify the robustness of these loci.
References
- 1.Weber JL, David D, Heil J, Fan Y, Zhao C, Marth G. Human diallelic insertion/deletion polymorphisms. Am J Hum Genet. 2002;71:854–862. doi: 10.1086/342727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullaney JM, Mills RE, Pittard WS, Devine SE. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet. 2010;19:R131–R136. doi: 10.1093/hmg/ddq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li C, Zhao S, Zhang S, Li L, Liu Y, Chen J, et al. Genetic polymorphism of 29 highly informative InDel markers for forensic use in the Chinese Han population. Forensic Sci Int Genet. 2011;5:e27–e30. doi: 10.1016/j.fsigen.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Pimenta JR, Pena SDJ. Efficient human paternity testing with a panel of 40 short insertion–deletion polymorphisms. Genet Mol Res. 2010;9:601–607. doi: 10.4238/vol9-1gmr838. [DOI] [PubMed] [Google Scholar]
- 5.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zidkova A, Horinek A, Kebrdlova V, Korabecna M. Application of the new insertion–deletion polymorphism kit for forensic identification and parentage testing on the Czech population. Int J Leg Med. 2013;127:7–10. doi: 10.1007/s00414-011-0649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amorim A, Pereira L. Pros and cons in the use of SNPs in forensic kinship investigation: a comparative analysis with STRs. Forensic Sci Int. 2005;150:17–21. doi: 10.1016/j.forsciint.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Hollard C, Mendisco F, Keyser C, Crubézy E, Ludes B. First application of the Investigator DIPplex indels typing kit for the analysis of ancient DNA samples. Forensic Sci Int Genet Suppl Series. 2011;3:e393–e394. doi: 10.1016/j.fsigss.2011.09.058. [DOI] [Google Scholar]
- 9.Lewis PO, Zaykin D (2011) Genetic data analysis: computer program for the analysis of allelic data. Version 1.0 (d16c). Free program distributed by the authors over the internet from http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php. Accessed 11 Aug 2012
- 10.Weir BS (1990) Multiple tests. In: Genetic data analysis. Vol. 109. Sinauer Associates Inc., Sunderland, p 109–110
- 11.Tereba A (1999) Tools for analysis of population statistics. PowerStats Program Version 1.2. Promega Corporation. Free program distributed by Promega Corporation over the internet from http://www.promega.com/geneticidtools/powerstats. Accessed 05 Aug 2012
- 12.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 13.Qiagen (2010) Supplementary material: population data for analysis of results from the Investigator DIPplex kit Qiagen
- 14.Friis SL, Børsting C, Rockenbauer E, Poulsen L, Fredslund SF, Tomas C, et al. Typing of 30 insertion/deletions in Danes using the first commercial indel kit - Mentype® DIPplex. Forensic Sci Int Genet. 2012;6:e72–e74. doi: 10.1016/j.fsigen.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Neuvonen AM, Palo JU, Hedman M, Sajantila A. Discrimination power of Investigator DIPplex loci in Finnish and Somali populations. Forensic Sci Int Genet. 2012;6:e99–e102. doi: 10.1016/j.fsigen.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Martín P, García O, Heinrichs B, Yurrebaso I, Aguirre A, Alonso A. Population genetic data of 30 autosomal indels in Central Spain and the Basque Country populations. Forensic Sci Int Genet. 2013;7(2):e27–30. doi: 10.1016/j.fsigen.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 17.LaRue BL, Ge J, King JL, Budowle B. A validation study of the Qiagen Investigator DIPplex® kit; an INDEL-based assay for human identification. Int J Legal Med. 2012;126:533–540. doi: 10.1007/s00414-012-0667-9. [DOI] [PubMed] [Google Scholar]
- 18.Tucholska-Lenart A, Wujec J, Samborski J, Jakubowska E. Allele frequencies for 10 STR loci in a population from central Poland. Forensic Sci Int. 2002;129:131–133. doi: 10.1016/S0379-0738(02)00201-3. [DOI] [PubMed] [Google Scholar]
- 19.Wang CW, Chen DP, Chen CY, Lu SC, Sun CF. STR data for the AmpFlSTR SGM plus and profiler loci from Taiwan. Forensic Sci Int. 2003;138:119–122. doi: 10.1016/j.forsciint.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Weir B, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- 21.Wolanska-Nowak P. On the subtle southern Polish population subdivision. Problems of Forensic Sciences. 2007;71:265–279. [Google Scholar]
- 22.Huang QY, Xu FH, Shen H, Deng HY, Liu YJ, Liu YZ, et al. Mutation patterns at dinucleotide microsatellite loci in humans. Am J Hum Genet. 2002;70:625–634. doi: 10.1086/338997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupuy BM, Stenersen M, Egeland T, Olaisen B. Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum Mutat. 2004;23:117–124. doi: 10.1002/humu.10294. [DOI] [PubMed] [Google Scholar]
- 24.Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Candidate SNPs for a universal individual identification panel. Hum Genet. 2007;121:305–317. doi: 10.1007/s00439-007-0342-2. [DOI] [PubMed] [Google Scholar]
- 25.Budowle B, van Daal A. Forensically relevant SNP classes. Biotechniques. 2008;44:603–610. doi: 10.2144/000112806. [DOI] [PubMed] [Google Scholar]
- 26.Kidd KK, Pakstis AJ, Speed WC, Grigorenko EL, Kajuna SLB, Nganyirwa J, et al. Developing a SNP panel for forensic identification of individuals. Forensic Sci Int. 2006;164:20–32. doi: 10.1016/j.forsciint.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Frudakis T, Venkateswarlu K, Thomas MJ, Gaskin Z, Ginjupalli S, Dunturi S, et al. A classifier for the SNP-based inference of ancestry. J Forensic Sci. 2003;48:771–782. [PubMed] [Google Scholar]
- 28.Phillips C, Salas A, Sanchez JJ, Fondevila M, Gomez-Tato A, Álvarez-Dios J, et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet. 2007;1:273–280. doi: 10.1016/j.fsigen.2007.06.008. [DOI] [PubMed] [Google Scholar]